Graphical abstract
Keywords: Schistosoma, Reproduciton, Stem cell, Vitellaria
Highlights
-
•
Transcriptional profiling identifies candidate factors associated with the schistosome vitellarium.
-
•
In situ hybridization confirms many new markers of this tissue.
-
•
New cell type-specific markers for various stages of vitellocyte development are reported.
Abstract
Schistosomes cause significant morbidity and mortality in millions of the world’s poorest people. While parasite egg-induced inflammation is the primary driver of host pathology, relatively little is known at the molecular level about the organ systems that participate in schistosome egg production (i.e., testes, ovaries and vitellaria). Here we use transcriptional profiling and in situ hybridization to characterise the vitellarium of Schistosoma mansoni. We uncovered several previously uncharacterised vitellaria-specific factors and defined molecular markers for various stages in the vitellocyte differentiation process. These data provide the framework for future in-depth molecular studies exploring the biology of this important parasite organ.
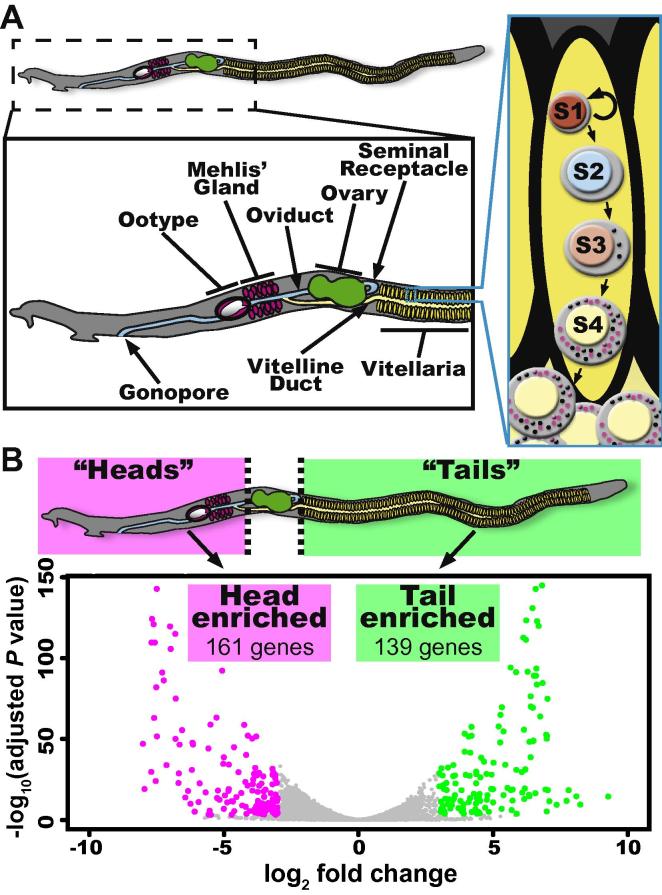
A schistosome worm pair can lay an egg every 1–5 min in vivo (Basch, 1991). Similar to all neoophoran flatworms (i.e., all parasitic flatworms and a subset of derived free-living worms (Martin-Duran and Egger, 2012, Collins and Newmark, 2013, Laumer et al., 2015), schistosomes produce ectolecithal (i.e., yolk on the outside) eggs consisting of an oocyte surrounded by specialised “yolk” cells known as vitellocytes (Gönnert, 1955, Basch, 1991, Shinn, 1993, Collins et al., 2011). Vitellocytes provide both nutrition for the developing zygote and constituents essential for the construction of the polyphenolic egg shell (Basch, 1991, Shinn, 1993). Vitellocytes are produced by a specialised organ, the vitellarium, that occupies the posterior two-thirds of the female schistosome body (Fig. 1A). This tissue is composed of a network of thousands of follicles (vitellaria) in which specialised stem cells, called S1 cells, differentiate to ultimately produce mature vitellocytes (S4 cells) (Fig. 1A) (Erasmus, 1975). These mature vitellocytes are fed anteriorly though the vitelline duct and are joined with fertilised oocytes in the ootype where the mature egg is formed.
Fig. 1.
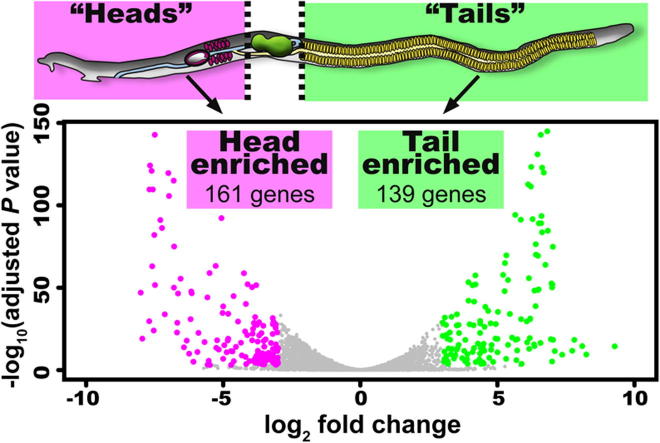
Identification of genes associated with the schistosome vitellarium. (A) Cartoon of the schistosome reproductive system and stages of vitellocyte development (inset boxed in cyan). Within the vitellaria, S1 cells proliferate and differentiate to ultimately generate S4 vitellocytes that are rich in lipid (magenta circles) and vitelline (black circles) droplets. These mature vitellocytes pass to the vitelline duct and travel anteriorly to the ootype. (B) Cartoon depicts the strategy to identify vitellaria associated transcripts. To identify genes associated with the vitellaria, RNA from amputated female heads and tails were subjected to RNAseq. The Volcano Plot depicts the 161 head-enriched (magenta dots) and 139 tail-enriched (green dots) transcripts. Genes not reaching our fold-change and significance thresholds are depicted as grey dots.
In both a biological and therapeutic sense, the vitellarium is an intriguing organ. From a biological perspective, vitellaria and ectolecithal eggs are evolutionarily-derived features absent from basal flatworms, where the oocytes are the exclusive source of yolk (Martin-Duran and Egger, 2012). How did the production of ectolecithal eggs evolve? What advantage does ectolecithal egg production afford these worms? What is the developmental relationship between the vitellaria and the female germ line? From a therapeutic viewpoint, the vitellarium represents an attractive therapeutic target since egg production promotes both disease transmission and host pathology (Pearce and MacDonald, 2002). Moreover, since vitellaria are a flatworm-specific innovation, it is possible that parasite-specific therapeutics targeting this organ could be developed. Unfortunately, no systematic molecular characterisation of the schistosome vitellarium has been reported to date. Here we describe our initial efforts to characterise this organ at a molecular level.
Recently, important tools to study the schistosome vitellarium, including tissue isolation (Hahnel et al., 2013) and Fluorescence Activated Cell Sorting (Lu et al., 2015), have been reported. While potentially powerful, these approaches are technically demanding, requiring a large number of parasites to generate sufficient material for standard transcriptional profiling. Therefore, we took advantage of the fact that the posterior of the female schistosome is almost entirely vitellaria (Fig. 1A). Since tissues such as the intestine, tegument and protonephridia are evenly distributed in the worm, we reasoned that comparing the transcriptional profiles of “heads” versus “tails” would allow us to identify transcripts enriched in the vitellarium. To perform these experiments, we separated adult male and female Schistosoma mansoni (recovered from mice 7–8 weeks p.i.) as previously described (Collins et al., 2013). Using a sharpened tungsten needle, we amputated females both anterior to and posterior to the ovary (Fig. 1B). We then prepared RNA from ∼100 anterior or posterior worm fragments using Trizol (Invitrogen, Carlsbad, CA, USA) and DNase treatment (DNA-free RNA Kit, Zymo Research, Irvine, CA, USA). Three independent rounds of RNA extraction were performed and samples were submitted to the University of Texas Southwestern Medical Center Genomics Core Facility, USA, for RNAseq library preparation (TruSeq Stranded RNA LT Kit, Illumina, San Diego, CA, USA) and sequencing (Illumina HiSeq2500, San Diego, CA, USA). Reads were mapped to the schistosome genome (Berriman et al., 2009, Protasio et al., 2012) with STAR (v 2.5.0b) (Dobin et al., 2013). The genome sequence and gff3 files were downloaded from Wormbase Parasite (v4) (Howe et al., 2016). Differential gene expression analysis was performed using DESeq v1.22 (Anders and Huber, 2010) (Supplementary Table S1).
Our RNAseq analysis identified 161 head-enriched (log2 fold change ⩽ −3, P < 0.001) and 139 tail-enriched (log2 fold change ⩾ 3, P < 0.001) transcripts (Fig. 1B, Supplementary Table S2). Our tail-enriched dataset included many of the known vitellaria-specific factors including two tyrosinases (Smp_050270; Smp_013540) (Fitzpatrick et al., 2007), a superoxide dismutase (Smp_095980) (Cogswell et al., 2011), female specific protein 800 (Smp_000290) (Reis et al., 1989), p48 (Smp_014610) (Chen et al., 1992), and p14 (Smp_131110) (Bobek et al., 1988) (Supplementary Table S3), confirming the utility of our approach. Additionally, the tail-enriched data set included nine of the 11 genes in the S. mansoni genome annotated as containing a “Trematode Egg Shell” domain (Pfam ID: PF08034) (Supplementary Table S3). These proteins and others are thought to be the major structural components of the egg shell in schistosomes and other trematodes (Ebersberger et al., 2005). Given that the vitellarium is a flatworm-specific organ, it is not surprising that a large fraction of the genes in this dataset are annotated as hypothetical proteins.
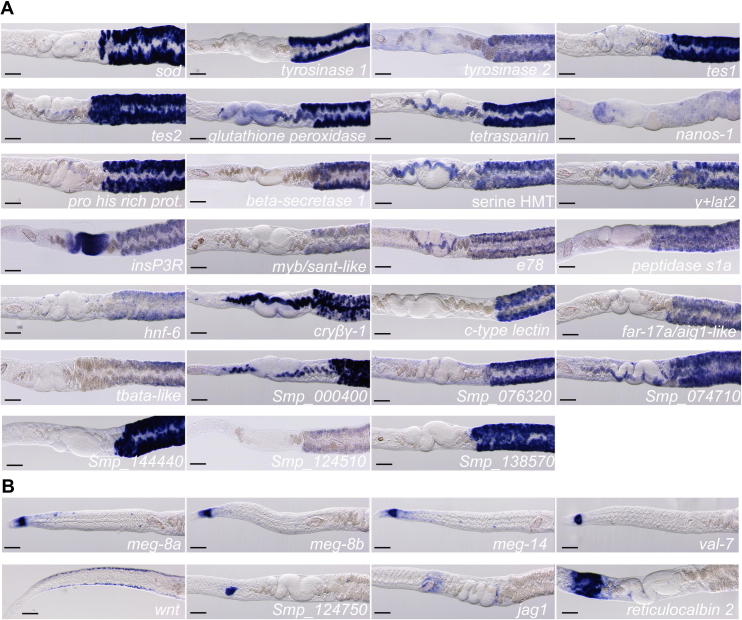
To further examine our RNAseq dataset, we cloned cDNA fragments for 54 genes (42 tail-enriched and 12 head-enriched; Supplementary Table S4) as described previously (Collins et al., 2010). Some of the genes selected were previously shown to be expressed in the vitellaria (e.g., tyrosinase 1); the remainder were selected essentially at random. Using these cloned cDNAs, we generated antisense riboprobes and performed a whole-mount in situ hybridization (WISH) screen of female worms (Collins et al., 2013). Of the 42 tail-enriched genes screened, we found mRNAs for 32 genes that were either exclusive to or highly enriched in the vitellaria (Fig. 2A) (Supplementary Table S4). The remaining 10 tail-enriched transcripts were either not detected, expressed at low levels with unclear expression patterns, or ubiquitously expressed (Supplementary Table S4). Nevertheless, these data confirm the utility of our approach to identify vitellaria-expressed transcripts. In addition to confirming the expression of known vitellaria-specific factors (e.g., superoxide dismutase and both tyrosinases), we identified several novel markers for the schistosome vitellarium (Fig. 2A) (Supplementary Table S4). These included genes predicted to encode enzymes (e.g., a serine hydroxymethyltransferase, a peptidase and a glutathione peroxidase), an amino acid transporter, eggshell proteins (tes1 and tes2), a homologue of the Drosophila nuclear hormone receptor Ecdysone-Induced protein 78c (e78), and several hypothetical proteins. Nanos proteins are RNA-binding proteins that play essential roles in germ line development throughout metazoa (Seydoux and Braun, 2006). In free-living flatworms, nanos expression is nearly exclusive to the presumptive stem cells in the male and female germ line (Wang et al., 2010, Wang et al., 2007). Consistent with observations in free-living flatworms, we observed nanos-1 expression in the anterior portion of the ovary where the female germ line stem cells, or oogonia, are thought to exist (Fig. 2A) (Nollen et al., 1976). Interestingly, we also detected expression of nanos-1 mRNA in small patches of cells scattered throughout the vitellaria (Fig. 2A).
Fig. 2.
Whole mount in situ hybridization for head- and tail-enriched schistosome transcripts. Expression of (A) tail-enriched or (B) head-enriched transcripts. Gene names are listed below each image. Genes without clear homologues of known function or recognisable domains are listed with their Smp number. Details of genes examined are provided in Supplementary Table S4. Over 75% of tail-enriched transcripts examined by whole mount in situ hybridization were expressed in the vitellaria, whereas the head-enriched transcripts were detected in the oesophageal glands, the ootype, Mehlis’ Gland, and in cells within the parenchyma. Images were captured using a Zeiss AxioZoom.V16 equipped with a transmitted light base and a Zeiss AxioCam 105 Color camera. Anterior is to the left. Scale bars = 100 μm.
Furthermore, we examined the expression of genes from our head-enriched dataset by whole mount in situ hybridization. Consistent with a previous study that compared gene expression in the head versus tail of male S. mansoni (Wilson et al., 2015), we identified several Venom Allergen proteins (VALs) and Mini Exon Genes (MEGs) that were expressed in the worm’s oesophageal gland (Fig. 2B) (Supplementary Table S4). We also observed an mRNA encoding a Wnt protein that was expressed along the lateral margins of female parasites (Fig. 2B). This expression started just behind the head of the worm and extended posteriorly approximately halfway down the length of the worm (Fig. 2B). Intriguingly, this Wnt protein appears to be orthologous to the protein encoded by the planarian wntP-2 gene. This planarian gene is expressed in a gradient along the planarian posterior and plays a role in re-establishing polarity during regeneration (Petersen and Reddien, 2009). Thus, this schistosome Wnt protein could play a role in establishing and maintaining axial polarity during parasite growth and homeostatic tissue renewal. Our analysis also identified new markers for two components of the female reproductive tract: Mehlis’ gland and the ootype. The function of Mehlis’ gland is unclear but it has been hypothesised to perform a variety of functions including providing lubrication for the reproductive tract, activating sperm and providing constituents important for egg shell formation (Smyth and Halton, 1983). The ootype sits just anterior to Mehlis’ gland and is responsible for eggshell formation (Fig. 1A). We observed cells in the proximity of Mehlis’ gland strongly expressing a homologue of the Ca2+ binding protein Reticulocalbin 2 and cells in the ootype expressing a schistosome-specific hypothetical protein (Smp_124750) (Fig. 2B). We also observed expression of a Jagged homologue (jag1) in the region of Mehlis’ gland (Fig. 2B). Jagged proteins are cell surface ligands that activate Notch signalling in adjacent cells (Kopan and Ilagan, 2009). With the expression of this Jagged molecule in Mehlis’ gland, it is possible that the Notch pathway becomes activated in cells (i.e., vitellocytes, oocytes or sperm) as they transit near Mehlis’ gland on their way through the oviduct. Such activation could be critical to initiating processes such as eggshell formation (in the case of vitellocytes or oocytes) or fertilisation (in the case of sperm). Examination of other Notch signalling components could provide support for this hypothesis.
Upon closer examination we noted that many tail-enriched genes were expressed in distinct cell types within the vitellaria. For instance, the expression of some genes was restricted to the vitellaria (e.g., tyrosinase 1), whereas others were expressed both in the vitellaria and vitelline duct (Smp_000400) (Fig. 2A). These two groups of expression could be divided even further, since some genes appeared to be expressed in subsets of cells within the vitellaria (e.g., cryβγ-1 and nanos-1). Since the cells within the vitelline duct represent the mature vitellocytes (S4 cells), we reasoned that genes whose expression is highly enriched in the vitellaria and not the vitelline duct might represent factors expressed at earlier stages of vitellogenesis. To explore this idea further, we conducted fluorescence in situ hybridization (FISH) experiments with Fluorescein- and Digoxigenin-labelled riboprobes as described previously (Collins et al., 2013). Detection of transcripts was performed using peroxidase-conjugated antibodies and sequential rounds of Tyramide Signal Amplification (TSA); residual peroxidase activity between rounds of TSA was quenched with 100 mM Sodium Azide in TNT (0.1 M Tris pH 7.5, 0.15 M NaCl and 0.1% Tween-20) for 45 min (King and Newmark, 2013). Following TSA detection, animals were incubated for 1 h in 10 mM CuSO4 and 50 mM NH4CH3CO2 pH 5.0 to quench autofluorescence within the vitellaria (King and Newmark, 2013).
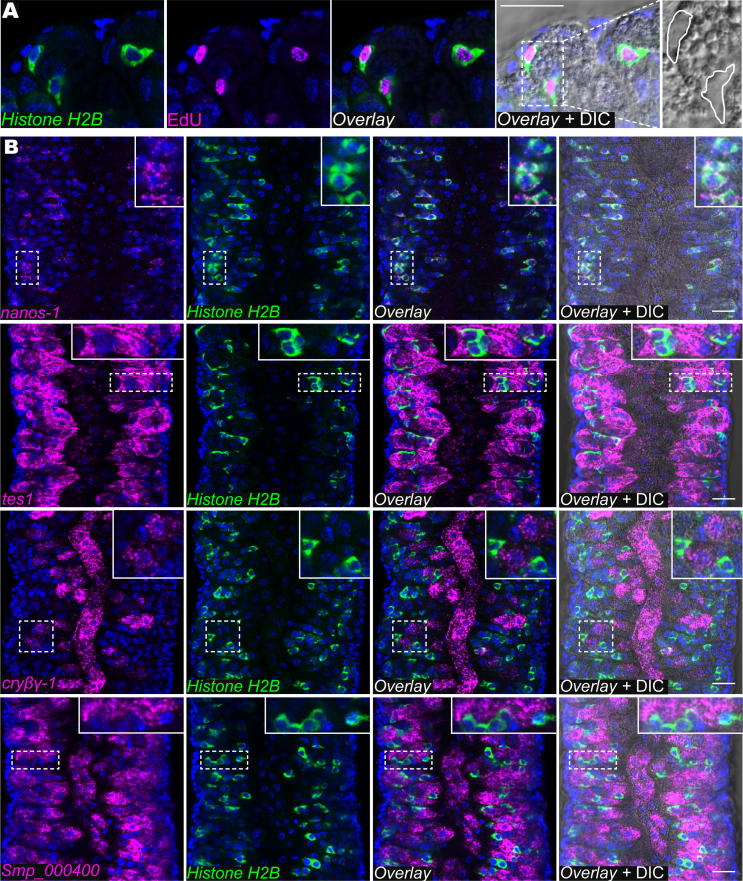
We performed FISH with riboprobes for nanos-1, tes1, cryβγ-1 or Smp_000400 with the proliferative cell marker Histone H2B. Since Histone H2B expression is restricted to proliferative cells in the schistosome soma and germ line (Collins et al., 2013), we reasoned that it would be expressed in the proliferative S1 stem cells and perhaps in their early differentiation progeny within the vitellaria. Consistent with this hypothesis, Histone H2B expression was limited to small cells that incorporated the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) and did not possess the cytoplasmic granularity characteristic of more differentiated cells in the vitellarium (Fig. 3A and B). By double FISH, we found that nearly all Histone H2B+ cells were also positive for nanos-1, indicating that nanos-1 is a marker for the schistosome S1 cells (Fig. 3B). Given the highly conserved role of Nanos proteins in germ cell development, this observation provides molecular evidence that the vitellocyte lineage may be evolutionarily derived from the germ line. In contrast to nanos-1, we did not observe co-expression of tes1, cryβγ-1, or Smp_000400 with Histone H2B, suggesting that these genes are not expressed in the S1 cells (Fig. 3B). Consistent with this idea, these genes were all expressed in cells possessing a highly granular cytoplasm (Fig. 3B). Similar to our WISH results, these genes all had distinct patterns of expression: tes1 was highly expressed in the majority of cells within the vitellaria and not highly expressed in the cells within the vitelline duct; cryβγ-1 was highly expressed in cells within the vitelline duct and a subset of cells in the vitellaria located proximal to the vitelline duct; and Smp_000400 appeared to be broadly expressed in both the vitellaria and cells within the vitelline duct. Based on these patterns of expression we suggest a model for vitelline cell development in which nanos-1+/Histone H2B+ S1 stem cells differentiate to tes1+/Smp_000400+ cells before becoming mature S4 vitellocytes that express high levels of both cryβγ-1 and Smp_000400. Whether the tes1+/Smp_000400+ cells represent the S2–S3 cells defined in the classic literature using electron microscopy (Fig. 1A) (Erasmus, 1975) cannot be determined at this time. Future studies utilising these markers in concert with EdU pulse-chase approaches (Collins et al., 2013) will provide unambiguous support for our proposed model of vitellocyte differentiation.
Fig. 3.
Fluorescence in situ hybridization identifies markers for unique cell populations within the schistosome vitellaria. (A) 5-ethynyl-2′-deoxyuridine labelling and FISH for Histone H2B. Parasites were treated for 4 h with the thymidine analogue 5-ethynyl-2′-deoxyuridine, fixed and processed for FISH as previously described (Collins et al., 2013). 5-ethynyl-2′-deoxyuridine incorporation following a 4 h pulse appeared to be exclusive for Histone H2B+ cells in the vitellaria, indicating that these cells are the proliferative cells of the vitellaria and are likely to represent S1 cells. Differential Interference Contrast imaging (left) showed that these cells lack the granularity characteristic of differentiated vitelline cells, consistent with the idea that these cells are undifferentiated S1 cells. (B) Double fluorescence in situ hybridization for Histone H2B with nanos-1, tes1, cryβγ-1, or Smp_000400. nanos-1 expression was restricted to Histone H2B+ cells indicating that nanos-1 is likely a marker for S1 cells. tes1, cryβγ-1 and Smp_000400 are not expressed in Histone H2B+ S1 cells; rather, their expression was observed in distinct sets of more differentiated vitelline cells. tes1 expression was highest in cells within the vitellaria and lower in the most differentiated vitelline cells within the vitelline duct. cryβγ-1 was expressed at high levels in cells within the vitelline duct and in cells adjacent to the duct. Smp_000400 appeared to be ubiquitously expressed in all cells of the vitelline lineage with the exception of the S1 cells. Thus, we speculate that cells expressing high levels of tes1 and Smp_000400 are likely to represent an intermediate step in the differentiation process of mature vitellocytes that express cryβγ-1 and Smp_000400. Insets show magnified views of boxed regions. In all images, nuclei are in blue. Images were acquired using an Zeiss LSM700 laser scanning confocal microscope with either a Plan-Apochromat 63×/1.4 Oil DIC or EC Plan-Neofluar 40×/1.30 Oil DIC objective lens. Scale bars = 20 μm.
Here we provide an initial characterisation of the schistosome vitellarium. In sum, we validated several novel factors expressed in this organ system and identified new cell type-specific markers for S1 cells and their differentiation progeny. Combination of the techniques outlined here with the emerging approaches to isolate distinct vitelline cell populations (Lu et al., 2015), should allow for a higher resolution description of the developmental events culminating in vitellocyte production. Furthermore, since vitellaria are a flatworm-specific evolutionary innovation, we suspect these data will provide a starting point for the molecular characterisation of this organ in other flatworms. Indeed, identification of molecules essential for vitellocyte production in more experimentally tractable flatworm models (e.g., planarians) could provide a path towards understanding the biology of this fascinating organ while simultaneously identifying therapeutic targets against schistosomes and other parasitic flatworms.
Acknowledgments
We thank Julie Collins, George Wendt and Anushka Wikramarante for comments on the manuscript. This work was supported by National Institutes of Health (NIH), USA, R01AI121037 (JJC) and the Wellcome Trust, United Kingdom, grant number 107475/Z/15/Z. Mice and B. glabrata snails were provided by the National Institutes for Allergy and Infectious Disease Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD, USA) through NIH-NIAID Contract HHSN272201000005I for distribution through BEI Resources. In adherence to the Animal Welfare Act and the Public Health Service Policy on Humane Care and Use of Laboratory Animals, all experiments with and care of vertebrate animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of University of Texas Southwestern Medical Center, USA (protocol approval number APN 2014-0072). The RNAseq dataset is available at NCBI under the accession number GSE78967.
Footnotes
Note: RNAseq data was submitted to NCBI under accession number GSE78967.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijpara.2016.03.004.
Appendix A. Supplementary data
Gene expression data for all schistosome genes.
Gene expression data for schistosome head-enriched transcripts (adj. P value < 0.001, log 2 Fold Change > −3).
Gene expression data for schistosome tail-enriched transcripts (adj. P value < 0.001, log 2 Fold Change < 3).
Details of schistosome genes examined in this study.
References
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch P.F. Oxford University Press; New York: 1991. Schistosomes: Development, Reproduction, and Host Relations. [Google Scholar]
- Berriman M., Haas B.J., LoVerde P.T., Wilson R.A., Dillon G.P., Cerqueira G.C., Mashiyama S.T., Al-Lazikani B., Andrade L.F., Ashton P.D., Aslett M.A., Bartholomeu D.C., Blandin G., Caffrey C.R., Coghlan A., Coulson R., Day T.A., Delcher A., DeMarco R., Djikeng A., Eyre T., Gamble J.A., Ghedin E., Gu Y., Hertz-Fowler C., Hirai H., Hirai Y., Houston R., Ivens A., Johnston D.A., Lacerda D., Macedo C.D., McVeigh P., Ning Z., Oliveira G., Overington J.P., Parkhill J., Pertea M., Pierce R.J., Protasio A.V., Quail M.A., Rajandream M.A., Rogers J., Sajid M., Salzberg S.L., Stanke M., Tivey A.R., White O., Williams D.L., Wortman J., Wu W., Zamanian M., Zerlotini A., Fraser-Liggett C.M., Barrell B.G., El-Sayed N.M. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobek L.A., Rekosh D.M., LoVerde P.T. Small gene family encoding an eggshell (chorion) protein of the human parasite Schistosoma mansoni. Mol. Cell. Biol. 1988;8:3008–3016. doi: 10.1128/mcb.8.8.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., Rekosh D.M., LoVerde P.T. Schistosoma mansoni p48 eggshell protein gene: characterization, developmentally regulated expression and comparison to the p14 eggshell protein gene. Mol. Biochem. Parasitol. 1992;52:39–52. doi: 10.1016/0166-6851(92)90034-h. [DOI] [PubMed] [Google Scholar]
- Cogswell A.A., Collins J.J., III, Newmark P.A., Williams D.L. Whole mount in situ hybridization methodology for Schistosoma mansoni. Mol. Biochem. Parasitol. 2011;178:46–50. doi: 10.1016/j.molbiopara.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.J., III, Hou X., Romanova E.V., Lambrus B.G., Miller C.M., Saberi A., Sweedler J.V., Newmark P.A. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.J., III, King R.S., Cogswell A., Williams D.L., Newmark P.A. An atlas for Schistosoma mansoni organs and life-cycle stages using cell type-specific markers and confocal microscopy. PLoS Negl. Trop. Dis. 2011;5:e1009. doi: 10.1371/journal.pntd.0001009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.J., III, Newmark P.A. It’s no fluke: the planarian as a model for understanding schistosomes. PLoS Pathog. 2013;9:e1003396. doi: 10.1371/journal.ppat.1003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.J., III, Wang B., Lambrus B.G., Tharp M.E., Iyer H., Newmark P.A. Adult somatic stem cells in the human parasite Schistosoma mansoni. Nature. 2013;494:476–479. doi: 10.1038/nature11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I., Knobloch J., Kunz W. Cracks in the shell–zooming in on eggshell formation in the human parasite Schistosoma mansoni. Dev. Genes. Evol. 2005;215:261–267. doi: 10.1007/s00427-005-0467-z. [DOI] [PubMed] [Google Scholar]
- Erasmus D.A. Schistosoma mansoni: development of the vitelline cell, its role in drug sequestration, and changes induced by Astiban. Exp. Parasitol. 1975;38:240–256. doi: 10.1016/0014-4894(75)90027-2. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick J.M., Hirai Y., Hirai H., Hoffmann K.F. Schistosome egg production is dependent upon the activities of two developmentally regulated tyrosinases. FASEB J. 2007;21:823–835. doi: 10.1096/fj.06-7314com. [DOI] [PubMed] [Google Scholar]
- Gönnert R. Schistosomiasis studies. II. Oogenesis of Schistosoma mansoni and the development of the eggs in the host organism. Z. Tropenmed. Parasitol. 1955;6:33–52. [PubMed] [Google Scholar]
- Hahnel S., Lu Z., Wilson R.A., Grevelding C.G., Quack T. Whole-organ isolation approach as a basis for tissue-specific analyses in Schistosoma mansoni. PLoS Negl. Trop. Dis. 2013;7:e2336. doi: 10.1371/journal.pntd.0002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C., Harris T.W., Kishore R., Lee R., Lomax J., Li Y., Muller H.M., Nakamura C., Nuin P., Paulini M., Raciti D., Schindelman G., Stanley E., Tuli M.A., Van Auken K., Wang D., Wang X., Williams G., Wright A., Yook K., Berriman M., Kersey P., Schedl T., Stein L., Sternberg P.W. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44:D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R.S., Newmark P.A. In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev. Biol. 2013;13:8. doi: 10.1186/1471-213X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M.X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumer C.E., Hejnol A., Giribet G. Nuclear genomic signals of the ’microturbellarian’ roots of platyhelminth evolutionary innovation. eLife. 2015;4 doi: 10.7554/eLife.05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Quack T., Hahnel S., Gelmedin V., Pouokam E., Diener M., Hardt M., Michel G., Baal N., Hackstein H., Grevelding C.G. Isolation, enrichment and primary characterisation of vitelline cells from Schistosoma mansoni obtained by the organ isolation method. Int. J. Parasitol. 2015;45:663–672. doi: 10.1016/j.ijpara.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Martin-Duran J.M., Egger B. Developmental diversity in free-living flatworms. Evodevo. 2012;3:7. doi: 10.1186/2041-9139-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen P.M., Floyd R.D., Kolzow R.G., Deter D.L. The timing of reproductive cell development and movement in Schistosoma mansoni, S. japonicum, and S. haematobium, using techniques of autoradiography and transplantation. J. Parasitol. 1976;62:227–231. [PubMed] [Google Scholar]
- Pearce E.J., MacDonald A.S. The immunobiology of schistosomiasis. Nature Rev. Immunol. 2002;2:499–511. doi: 10.1038/nri843. [DOI] [PubMed] [Google Scholar]
- Petersen C.P., Reddien P.W. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protasio A.V., Tsai I.J., Babbage A., Nichol S., Hunt M., Aslett M.A., De Silva N., Velarde G.S., Anderson T.J., Clark R.C., Davidson C., Dillon G.P., Holroyd N.E., LoVerde P.T., Lloyd C., McQuillan J., Oliveira G., Otto T.D., Parker-Manuel S.J., Quail M.A., Wilson R.A., Zerlotini A., Dunne D.W., Berriman M. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 2012;6:e1455. doi: 10.1371/journal.pntd.0001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis M.G., Kuhns J., Blanton R., Davis A.H. Localization and pattern of expression of a female specific mRNA in Schistosoma mansoni. Mol. Biochem. Parasitol. 1989;32:113–119. doi: 10.1016/0166-6851(89)90062-5. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Braun R.E. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shinn G.L. Formation of egg capules by flatworms (Phylum Platyhelminthes) Trans. Am. Microsc. Soc. 1993;112:18–34. [Google Scholar]
- Smyth J.D., Halton D.W. second ed. Cambridge University Press; Cambridge: 1983. The Physiology of Trematodes. [Google Scholar]
- Wang Y., Stary J.M., Wilhelm J.E., Newmark P.A. A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes Dev. 2010;24:2081–2092. doi: 10.1101/gad.1951010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zayas R.M., Guo T., Newmark P.A. Nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.A., Li X.H., MacDonald S., Neves L.X., Vitoriano-Souza J., Leite L.C., Farias L.P., James S., Ashton P.D., DeMarco R., Castro Borges W. The schistosome esophagus is a ’hotspot’ for microexon and lysosomal hydrolase gene expression: implications for blood processing. PLoS Negl. Trop. Dis. 2015;9:e0004272. doi: 10.1371/journal.pntd.0004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression data for all schistosome genes.
Gene expression data for schistosome head-enriched transcripts (adj. P value < 0.001, log 2 Fold Change > −3).
Gene expression data for schistosome tail-enriched transcripts (adj. P value < 0.001, log 2 Fold Change < 3).
Details of schistosome genes examined in this study.