Abstract
We have a limited understanding of the site specificity of multisubunit lysine acetyltransferase (KAT) complexes for histone-based substrates, especially in regards to the different complexes formed during nucleosome assembly. Histone complexes could be a major factor in determining the acetylation specificity of KATs. In the present study, we utilized a label-free quantitative mass spectrometry-based method to determine the site specificity of acetylation catalyzed by Piccolo NuA4 on (H3/H4)2 tetramer, tetramer bound DNA (tetrasome), and nucleosome core particle (NCP). Our results show that Piccolo NuA4 can acetylate multiple lysine residues on these three histone complexes, of which NCP is the most favorable, (H3/H4)2 tetramer is the second, and tetrasome is the least favorable substrate for Piccolo NuA4 acetylation. Although Piccolo NuA4 preferentially acetylates histone H4 (H4K12), the site specificity of the enzyme is altered with different histone complex substrates. Our results show that before nucleosome assembly is complete, H3K14 specificity is almost equal to that of H4K12 and DNA-histone interactions suppress the acetylation ability of Piccolo NuA4. These data suggest that the H2A/H2B dimer could play a critical role in the increase of acetylation specificity of Piccolo NuA4 for NCP. This demonstrates that histone complex formation can alter the acetylation preference of Piccolo NuA4. Such findings provide valuable insight into regulating Piccolo NuA4 specificity by modulating chromatin dynamics, and in turn manipulating gene expression.
Keywords: histone acetylation, histone complex, Piccolo NuA4, KAT kinetics, mass spectrometry
INTRODUCTION
Covalent modifications on histones are believed to be one of the major mechanisms for the regulation of compaction of chromatin in eukaryotic cells [1]. For example, it has been observed that hyperacetylation of histone tail domains in euchromatin is linked to gene activation, whereas hypoacetylation is mostly found in heterochromatin and related to gene repression [1–3]. Histone acetylation has been shown to mitigate the DNA-histone interactions by charge neutralization and is associated with gene transcriptional activation, DNA replication, and DNA damage repair [4–8]. To maintain homeostasis, histones can be acetylated and deacetylated by two groups of enzymes: lysine acetyltransferases (KATs) and lysine deacetylases (KDACs), respectively. KATs can acetylate multiple lysine residues on histones [9–11], and different acetylated sites can result in different downstream biological results. For example, H3K56 acetylation is linked to DNA damage repair [8], while H3K14 acetylation has been associated with gene transcriptional activation [12].
Several known KATs belonging to the MYST family are not only involved in gene regulations but also related to major human disease progression. For example, Tip60 (Tat-interactive protein, 60 kDa; the first discovered human MYST protein) promotes HIV replication [13], and is also linked to metastasis and malignancy of many cancers [14–16]. In addition to Tip60, MOZ (monocytic leukemia zinc finger protein), MORF (MOZ-related factor), and HBO1 (histone acetyltransferase bound to ORC1) have been identified as human KATs in the MYST family, and importantly, these KATs play important roles in oncogenesis [17,18]. Although these KATs are grouped together due to their high sequence similarities, a wide range of regulatory functions of individual KATs has been reported, while the roles of others still remain to be determined [11,18]. One of the possible explanations for this difference in regulatory activity could be that individual KATs possess unique acetylation specificity, which could result in diverse regulatory effects.
In the present study, we focused on the acetylation specificity of Piccolo NuA4, because: 1) The catalytic subunit, Esa1 (one MYST protein), and the NuA4 complex are both essential for cell viability of Saccharomyces cerevisiae [5,19]. Esa1 is highly similar to human Tip60 and is linked to the function of DNA damage repair and cell cycle progression [19–22]. Both the catalytic subunit and HAT complex are structurally and functionally conserved from yeast to humans [23,24]. 2) Piccolo NuA4 demonstrates the ability for H4 acetylation in in vitro and in vivo assays [22,25], while the NuA4 complex, composed of multiple polypeptide subunits, is also able to catalyze histone H2A and H4 N-terminal acetylation [22]. Piccolo NuA4, an Esa1-Epl1-Yng2 trimeric complex, can be purified and separated from the larger NuA4 complex from yeast extracts [22]. The Epl1 subunit is a homolog of the Drosophila Enhancer of Polycomb, which is a suppressor of position-effect variegation and an enhancer of Polycomb mutations [26,27], while Yng2 is a homolog of the ING1 tumor suppressor, involved in DNA damage response, cell proliferation, and apoptosis [28–31]. However, the functions of the subunits of the full NuA4 complex are not fully understood.
Previous studies have shown that Esa1 itself can only acetylate histones associated with no DNA, while the Piccolo NuA4 complex prefers to catalyze nucleosome [20,22]. Similarly, different substrate preference and different acetylation specificity for the lysine residues has been addressed in the studies between Gcn5 (a catalytic subunit) and ADA or SAGA complex (a multisubunit complex incorporating Gcn5) [32–35]. It has been shown that other subunits in the KAT complex can directly and/or indirectly facilitate the recognition of nucleosomes as a substrate, increase the efficiency of acetylation, and alter the specificity of a KAT [36]. Moreover, it has been reported that NuA4 may work along with other protein complexes [23,37–39]. Thus, we chose Piccolo NuA4 as a KAT to perform a comprehensive study of its site-specific acetylation patterns. Importantly, Piccolo NuA4 has a strong substrate preference to chromatin over free histones and is able to catalyze nontargeted acetylation on both histone H2A and H4 [22].
Not only can the protein-protein interactions within an enzyme complex significantly change its acetylation patterns, but also the composition of the substrate can influence acetylation rates, resulting in different site-specific specificity and selectivity. In our previous studies, we observed that p300, CBP, as well as the Rtt109-Vps75 complex have altered lysine acetylation specificity for histone H3 alone compared to tetramer (H3/H4)2 [40,41]. Although most KATs have broad acetylation spectra on histones, acetylation on nonnucleosomal histones versus nucleosomes could represent different biological effects. For example, acetylation on nascent histones is most related to histone deposition [32], whereas nucleosomal acetylation can be linked to transcriptional activation [9]. We hypothesized that a KAT (such as Piccolo NuA4) should demonstrate a preference for a specific histone complex. At present we lack a comparison of site-specific kinetics for modification of histones in the presence and absence of DNA, such as (H3/H4)2 tetramer, tetrasome, and nucleosome core particle (NCP). We also lack quantitative analysis of modification of individual histone residues. To address these deficiencies, we have performed a series of kinetic assays, analyzed by a mass spectrometry (MS)-based method, to determine the specificity of Piccolo NuA4 for individual lysine residues. After examining the activity of this enzyme on tetramer, tetrasome, and nucleosome, we find that the altered site specificity of Piccolo NuA4 is due to the additional protein-protein and/or protein-DNA interactions. The results of this study on Piccolo NuA4 provides valuable, detailed insight into the factors regulating KAT specificity in chromatin dynamics.
EXPERIMENTAL PROCEDURES
Reagents
All Chemicals and solvents were purchased from Sigma-Aldrich or Fisher meeting the highest commercial grade or LC/MS grade. Ultrapure water was generated from a Millipore Direct-Q 5 ultrapure water system.
Piccolo NuA4 preparation
Recombinant Piccolo NuA4 was coexpressed in E. coli and purified as described previously [42,43]. Briefly, hexahistidine tagged Epl1(51-380), Yng2(1-218) and Esa1(1-445) were coexpressed using the pST44 polycistronic expression vector [44]. To validate that the truncated Epl1 used in this study has no effects on histone acetylation, we carried out the MS-based analysis to measure individual lysine acetylation levels from whole cell histone extracts of wild-type, epl1(1-380), and epl1(1-485) yeast strains [22]. We found that when compared to wild type, the truncated Epl1 has no significant impact on H4 tail acetylation in vivo (Supplemental Figure 1). The Piccolo NuA4 complex was purified from the soluble E. coli extract using Talon metal affinity chromatography, followed by SourceQ anion-exchange, SourceS cation-exchange and SourceISO hydrophobic interaction chromatography.
DNA, (H3/H4)2 tetramer, tetrasome, and nucleosome preparation
Recombinant Xenopus histones H3 and H4 were purified by the Protein Purification Core at Colorado State University. The preparation of 207 bp Widom 601 DNA and histone (H3/H4)2 tetramer refolding were done using previously reported methods [45–47], and the concentrations of DNA and histone were determined by UV absorbance and calculated from the extinction coefficients [48,49]. The assembly of tetrasome was carried out following the published methods [50,51]. Nucleosome core particles (NCP) reconstituted from recombinant Xenopus core histones and 147 bp Widom 601 DNA positioning sequence were prepared as described previously [46], including anion-exchange purification.
KAT kinetic assays of Piccolo NuA4
A time course study was carried out to characterize the site-specific details of Piccolo NuA4 acetylation on two different histone complexes ((H3/H4)2 and NCP) beyond steady state. The experimental conditions were 5 µM (H3/H4)2 or NCP, 300 µM acetyl-CoA, and 75 nM Piccolo NuA4 in 100 mM ammonium bicarbonate and 50 mM HEPES buffer (pH 7.2) at 37°C. Steady-state (enzyme concentrations much less than substrate concentrations) assays for (H3/H4)2 tetramer, tetrasome, and NCP were performed under identical buffer conditions (100 mM ammonium bicarbonate and 50 mM HEPES buffer (pH 7.2)) at 37°C. The assays contained 0.02 to 0.8 µM Piccolo NuA4 (varied with substrate concentration) and 0.15 – 20 µM of (H3/H4)2, 0.15 – 15 µM of tetrasome, or 0.15 – 15 µM of NCP with saturating acetyl-CoA (300 µM). Sample preparation, including reaction quench, post-reaction propionylation, and tryptic digestion, were performed as in our previous published procedures [52,53].
DNA titration
To investigate if the amount of DNA is critical for affecting histone acetylation, we measured the apparent catalytic constant (kcat(app)) of Piccolo NuA4 by titrating different concentrations of DNA and maintaining saturated (H3/H4)2 and acetyl-CoA (15 and 300 µM, respectively) under steady-state conditions (0.8 µM Piccolo NuA4). Before the assay starts, the mixtures of (H3/H4)2 and different concentrations of DNA were pre-incubated in the aformentioned experimental buffer at 37°C for 30 min. All other sample preparations were the same as the above descriptions.
Sample analysis via UPLC-MS/MS
A Waters Acquity H-class UPLC coupled with a Thermo TSQ Quantum Access triple quadrupole mass spectrometer was used to quantify the acetylated lysines on H3 and H4 peptides. The UPLC and MS/MS settings, solvent gradient, and detailed mass transitions were reported in our previously published work [40,52,53] and Table 1. Retention time and specific mass transitions were both used to identify individual acetylated and/or propionylated peptides. The areas under individual resolved peaks were integrated using Xcalibur software (version 2.1, Thermo). Relative quantitative analysis was used to determine the amount of acetylation on individual lysines [52,53], and the time course kinetics of Piccolo NuA4-mediated acetylation for individual lysines can be plotted. We also noted that the ionization efficiency of H4 tail peptide (K5-R17) is about 10-fold less than that of H3 K9-R17 peptide. Thus, the samples of low concentration titrations (< 3 µM) have to be concentrated for the detection of histone acetylation.
Table 1.
MS detection parameters of tryptic peptides from histone H2A and H2B.
| Modification on lysinesa |
Peptide sequence | Precursor ion (m/z) |
Product ions (m/z) |
Collision energy (eV) |
|---|---|---|---|---|
| H2A K5ac- K9ac |
4GKaQGGKaTR11 | 458.254 | 503.294, 560.315, 688.374 |
17 |
| H2A K5ac- K9un |
4GKaQGGKpTR11 | 465.262 | 517.309, 574.331, 702.389 |
17 |
| H2A K5un- K9ac |
4GKpQGGKaTR11 | 465.264 | 503.294, 560.315, 688.374 |
17 |
| H2A K5un- K9un |
4GKpQGGKpTR11 | 472.270 | 517.309, 574.331, 702.389 |
17 |
| H2A K13ac- K15ac |
12AKaAKaTR17 | 379.730 | 446.272, 517.309 |
15 |
| H2A K13ac- K15un |
12AKaAKpTR17 | 386.737 | 460.288, 531.325 |
15 |
| H2A K13un- K15ac |
12AKpAKaTR17 | 386.739 | 446.272, 517.309 |
15 |
| H2A K13un- K15un |
12AKpAKpTR17 | 393.745 | 460.288, 531.325 |
15 |
| H2A K36ac | 36KaGNYAER42 | 440.220 | 538.262, 652.305, 709.326 |
16 |
| H2A K36un | 36KpGNYAER42 | 447.227 | 538.262, 652.305, 709.326 |
16 |
| H2A K74ac- K75ac |
72DNKaKaTR77 | 423.227 | 446.272, 616.378, 730.421 |
16 |
| H2A K74ac- K75un |
72DNKaKpTR77 | 430.235 | 460.288, 630.393, 744.436 |
16 |
| H2A K74un- K75ac |
72DNKpKaTR77 | 430.237 | 446.272, 630.393, 744.436 |
16 |
| H2A K74un- K75un |
72DNKpKpTR77 | 437.243 | 460.288, 644.409, 758.452 |
16 |
| H2B K85ac | 80LAHYNKaR86 | 472.259 | 322.187, 622.331, 759.390 |
17 |
| H2B K85un | 80LAHYNKpR86 | 479.267 | 322.187, 636.346, 773.405 |
17 |
Acetylation and no acetylation on lysine are indicated as ac and un, respectively.
Data analysis
All the steady-state data were fit with Prism (version 5.0d). The initial rates of acetylation on individual lysines were linearly regressed from the increase of acetylation as a function of time prior to a total 10% acetylation. For individual lysines, the apparent steady-state parameters kcat(app) and Km(app) were determined by fitting the Michaelis-Menten equation, where the substrate concentration is the concentration of individual histones (H3 or H4), and the enzyme concentration is the concentration of Piccolo NuA4. Note that the concentrations of histones (x-axis of Michaelis-Menten curves) are represented as the monomer concentrations of individual histones; for example, the titrated histone H4 concentration is two-fold the titrated concentration of (H3/H4)2 or NCP. To analyze the case of multiple, competitive sites with comparable acetylation rates, a one-site model is suitable to determine the specificity of each site, because the value of apparent specificity constant (kcat(app)/Km(app)) is conserved for individual sites [52]. Selectivity is typically measured by the ratio of kcat(app)/Km(app) for two residues of interest; the ratio of kcat(app) is also a viable alternative (eq. 1). This is due to the fact that the ratio of kcat(app)s for residues is equal to the ratio of kcat(app)/Km(app)s for the residues (eq. 2). In eq. 1, kx and Kx are the kcat and Km, respectively, for a specific site out of the four sites; and K1, K2, K3, and K4 are the Km for site 1–4, respectively. In the system with the competitive sites, the ratio of kcat(app)(s) can be used to determine the site preference of an enzyme (eq. 2). A two-tailed t test was applied to analyze if the steady-state kinetic parameters from two groups were significantly different.
| eq. 1 |
| eq. 2 |
RESULTS
Residues acetylated by Piccolo NuA4 on (H3/H4)2 and NCP beyond steady state
Before we could measure the specificity of Piccolo NuA4, we needed to determine which residues can be acetylated, including those residues acetylated beyond the conditions of steady-state. We expanded the coverage to include sites of histone H2A and H2B, because Piccolo NuA4 is known to display modest acetylation activity on H2A [24,25,54,55]. The detailed mass transitions for tryptic digestion of H2A and H2B are summarized in Table 1. For this initial investigation, we incubated 75 nM Piccolo NuA4 with 5 µM (H3/H4)2 or NCP for 25 min with 5 different time point sampling. As previously described [52,53], these samples were then propionylated and digested with trypsin prior to MS analysis.
Our multiplexed MS analysis is used to analyze peptides and quantitate the fraction of acetylation on individual lysine residues. Using this MS-based technique we can quantitate the individual lysine acetylation on core histones. Thus, these data allow us to quantitate each of the multiple lysines, on multiple histones, acetylated by Piccolo NuA4 at different time points (Fig. 1). Although we can measure acetylation at 29 different lysine residues, we found only 6 lysine residues on NCP to be acetylated by Piccolo NuA4 under the time frame of our assays.
Figure 1.
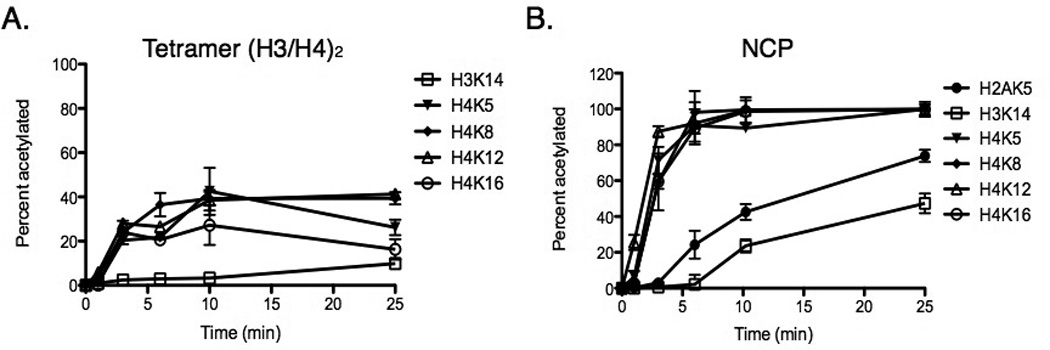
Site-specific time course kinetics of Piccolo NuA4 acetylation for (A) tetramer (H3/H4)2 and (B) NCP. 75 nM Piccolo NuA4, 300 µM acetyl-CoA, and either 5 µM tetramer (H3/H4)2 or 5 µM NCP were supplied in 100 mM ammonium bicarbonate and 50 mM HEPES buffer at 37°C and pH 7.2. The error bar indicates the standard error of three sets of assays.
Even under identical conditions, it is clear that (H3/H4)2 tetramer and NCP have different acetylation profiles. The highest acetylation levels on individual lysines of (H3/H4)2 are only about 40% (i.e., H4K8 and H4K12) after a 25-min incubation, but H4K5, H4K8, H4K12, and H4K16 on NCP reach almost 100% acetylation at 25 min. This implies that different histone complexes can influence Piccolo NuA4 acetylation, and NCP is a favored substrate compared to tetramer. Similar to past studies, we found that the H4 tail (i.e., H4K5, H4K8, H4K12, and H4K16) was preferentially acetylated by Piccolo NuA4, while H2AK5 and H3K14 were modestly acetylated.
Determining the specificity of Piccolo NuA4 for (H3/H4)2 tetramer
Studying the specificity of different KATs traditionally has been hindered by the difficulty in quantifying acetylation on multiple histone residues simultaneously [11,56]. Although most KATs are able to acetylate multiple lysine residues on histone proteins, this does not mean that they lack specificity. We used multiplexed MS analysis to simultaneously quantitate the acetylation of multiple lysines on histones. This type of multiplexed analysis provides a major advantage over conventional labeling and/or biochemical methods (e.g., Western blotting). For example, while the use of remote radioactive labeling or fluorescence labeling can be utilized in high throughput techniques, the lack of site-specific quantification is a significant drawback. On the other hand, site-specific antibody methods might show high sensitivity on detection of lysine-specific acetylation, but also due to high specificity and high selectivity of antibodies, the sample processing is arduous, especially for multiple sample/site analyses from KAT kinetic studies. In addition, antibody quality and epitope occlusion are the main concerns for antibody quantitative methods. The MS-based technique has the advantages of being able to quantitate multiple site-specific acetylation events from one sample injection. Moreover, the label-free and high throughput nature of this technique can efficiently analyze our numerous samples from KAT kinetic assays and may potentially provide the seamless transitions from in vitro to in vivo sample analysis. Thus, utilizing this MS-based method, our laboratory has demonstrated that Gcn5, p300 and CBP have their own site preference on histone H3 and H4, and that the acetylation specificity can be altered by different complexes of histones [40,52]. Enzymes such as p300 and CBP, once thought to be completely promiscuous, have been shown to act with a high level of specificity [40]. Aside from these studies, there is limited information in regards to the effect of different histone complexes on the catalytic reactivity of particular KATs, and even fewer studies provide quantitative information on the specificity of acetylation at individual lysines. In the present study, we performed steady-state kinetics using our multiplexed MS analysis to determine the specificity of Piccolo NuA4 for its primary acetylation sites. The merits of coupling enzyme kinetic assay and MS analysis are that we can quantify the specificity of individual sites and easily compare the selectivity between sites as well as histone complexes.
While our goal was to understand the impact of larger complexes, such as the nucleosome or tetrasome, on Piccolo NuA4 acetylation, we first needed to characterize the acetylation of (H3/H4)2 tetramer alone. Thus, we conducted a steady-state assay using (H3/H4)2 tetramer as a substrate, under conditions of saturating acetyl-CoA. We found that under steady-state conditions, Piccolo NuA4 acetylated 4 lysine residues (H3K14, H4K5, H4K8, and H4K12) (Fig. 2A), while other sites were not significantly acetylated prior to a total of 10% acetylation occurring (the upper limit for the steady-state assumption to hold true). We next looked at the difference in specificity for each residue acetylated by Piccolo NuA4: the order of specificity for (H3/H4)2 was H3K14 ≅ H4K12 > H4K5 > H4K8 (Table 2; Fig. 2B). Specificity is defined by kcat/Km, and the difference in kcat/Km between two residues is referred to as selectivity, but there is another way to calculate selectivity in this assay. Because all of the competing residues are contained on a single substrate (or substrate complex), the kcat(app) parameter becomes a viable alternative for calculating selectivity (eq. 2). In addition, using kcat(app) increases the accuracy and sensitivity for the comparisons between individual sites, as we have previously discussed [41]. We found that the site selectivity estimated by the ratio of kcat(app) matched the results based on the specificity constants of individual sites. While we were able to detect acetylation of H4K16 at long time points using Piccolo NuA4 (Fig. 1), lack of H4K16 acetylation under steady-state conditions (Fig. 2) indicates that H4K16 was the least favorable site of acetylation on the H4 tail, and H4K16 acetylation may occur after either H4K5 or H4K12 acetylation. In addition, the acetylation specificity between H3K14 and H4K12 was not significantly different, and H3K14 and H4K12 on (H3/H4)2 were the most favorable sites of Piccolo NuA4 acetylation. The specificity for H4K5 and H4K8 was ~2-fold and ~3-fold, respectively, less than the specificity for H3K14.
Figure 2.
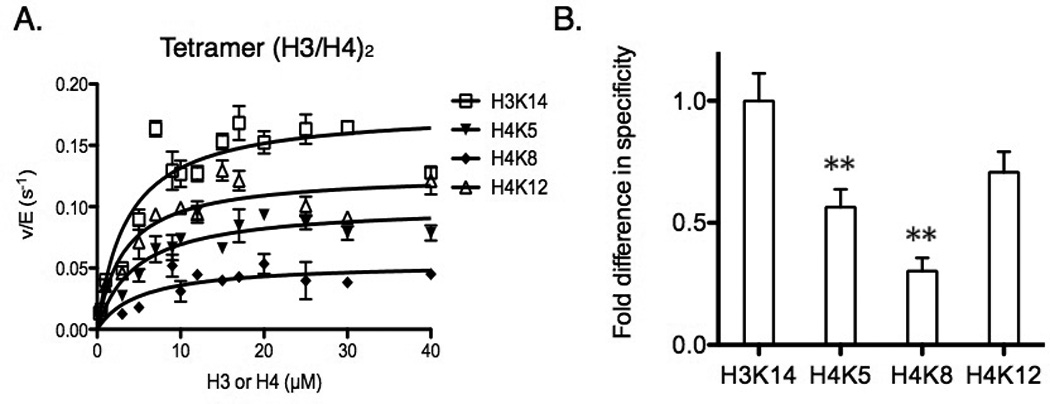
Specificity comparison between lysine residues in (H3/H4)2. (A) Determination of steady-state parameters for individual lysines (H3K14, H4K5, H4K8, and H4K12) of (H3/H4)2 acetylation catalyzed by Piccolo NuA4 in the presence of saturating acetyl-CoA. The error bar represents the standard error of v/E. The apparent kinetic parameters are summarized in Table 2. (B) The fold difference in specificity is determined by the ratio of kcat(app), and each site is normalized to the highest kcat(app) of H3K14. ** p < 0.01 when compared to H3K14. The specificity for H3K14 and H4K12 are not significantly different. The error bar represents the standard error of specificity measurements.
Table 2.
Kinetic parameters from titration of different histone complexes (mean ± standard error).
| Substrate Titrated |
Residue Acetylated |
kcat(app) (× 10−2s−1) |
Km(app) (µM) |
kcat/Km (× 10−2µM−1s−1) |
|---|---|---|---|---|
| H3/H4 | H3K14 | 17.9 ± 1.4 | 3.6 ± 1.2 | 4.9 ± 1.6 |
| H3/H4 | H4K5 | 10.1 ± 1.0 | 4.6 ± 1.8 | 2.2 ± 0.9 |
| H3/H4 | H4K8 | 5.4 ± 0.9 | 5.1 ± 3.1 | 1.1 ± 0.7 |
| H3/H4 | H4K12 | 12.7 ± 1.1 | 3.2 ± 1.2 | 3.9 ± 1.5 |
| NCP | H4K5 | 29.1 ± 2.3 | 1.4 ± 0.6 | 20.3 ± 7.8 |
| NCP | H4K12 | 49.6 ± 4.1 | 1.8 ± 0.7 | 27.0 ± 10.3 |
| Tetrasome | H3K14 | 0.93 ± 0.04 | 1.1 ± 0.3 | 0.85 ± 0.21 |
| Tetrasome | H4K5 | 0.56 ± 0.06 | 1.6 ± 0.9 | 0.35 ± 0.21 |
| Tetrasome | H4K8 | 0.34 ± 0.03 | 1.3 ± 0.7 | 0.27 ± 0.15 |
| Tetrasome | H4K12 | 1.06 ± 0.11 | 1.5 ± 0.8 | 0.72 ± 0.42 |
Alterations of the specificity of Piccolo NuA4 for NCP
Next we wanted to examine if the NCP substrate (2 H2A/H2B, 2 H3/H4, and 147 bp Widom 601 DNA) alters the site specificity of Piccolo NuA4. Thus, we performed the same steady-state assay as described above, but instead used NCP as the substrate. Under steady-state conditions, we found the primary acetylation sites of Piccolo NuA4 were reduced from four to two (for tetramer vs. nucleosome substrates). On the nucleosome, only H4K5 and H4K12 were found to be significantly acetylated prior to reaching a total of 10% acetylation (Fig. 3A), while H4K8 and H3K14 were not significantly acetylated. In addition, based on the kinetic parameters fit from Fig. 3A, the specificity of H4K12 was about 2-fold higher than that of H4K5 (Table 2; Fig. 3B), and the preference of Piccolo NuA4 acetylation between these two primary sites remained the same (H4K12 > H4K5) as on the tetramer. Although fewer primary sites were detected, the larger kcat(app) and the smaller Km(app) of the residues on NCP suggested that, compared to the secondary sites, the primary acetylation sites on NCP are more predominant than on tetramer. However, fewer acetylated sites detected from these steady-state assays of NCP did not imply that acetylation of Piccolo NuA4 for NCP was only limited to these two residues: beyond the initial 10% acetylation, we have demonstrated that Piccolo NuA4 can acetylate H4K5, H4K8, H4K12, H4K16, H3K14, and H2AK5 on NCP (Fig. 1). Those kinetic parameters, accompanied with the results of Fig. 1, indicate that NCP is a preferred substrate for Piccolo NuA4 acetylation. While our observation that acetylation is stimulated on NCP is consistent with the published reports [22,25,42], our findings build on these results by providing a detailed, site-specific analysis of lysine acetylation.
Figure 3.
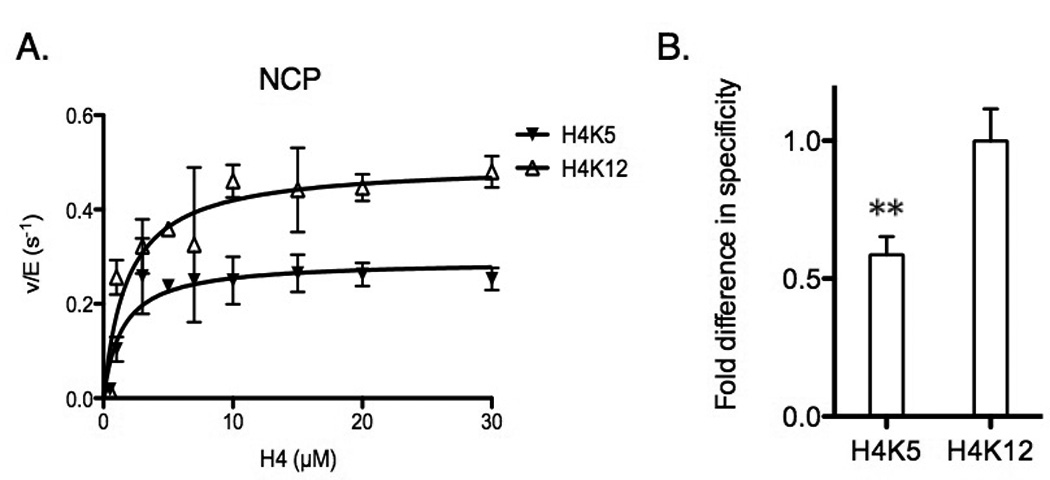
Specificity comparison between lysine residues in NCP. (A) Determination of steady-state parameters for individual lysines (H4K5 and H4K12) of NCP acetylation catalyzed by Piccolo NuA4 in the presence of saturating acetyl-CoA. The error bar represents the standard error of v/E. The apparent kinetic parameters are summarized in Table 2. (B) The fold difference in specificity is determined by the ratio of kcat(app), and each site is normalized to the highest kcat(app) of H4K12. ** p < 0.01 when compared to H4K12. The error bar represents the standard error of specificity measurements.
DNA in the absence of NCP suppresses H3/H4 acetylation by Piccolo NuA4
After demonstrating the preference of Piccolo NuA4 for NCP, we examined whether the presence of the DNA is key to increasing in the specificity of Piccolo NuA4 for NCP. During in vitro nucleosome assembly, the tetrasome is one of the intermediate states between the (H3/H4)2 tetramer and the nucleosome. To our knowledge, there has been no acetylation kinetic studies conducted regarding this intermediate, and therefore no acetylation specificity has been reported on tetrasome. In the nucleus, there should be few randomly bound histones, due to chaperone-mediated assembly and energy demands for importing free histones. In vitro, tetrasomes can be formed via a salt gradient similar to nucleosomes, but mixing H3/H4 and DNA does not result in classical tetrasomes [50,51]. The two main possibilities are that DNA could either stimulate or inhibit Piccolo NuA4 acetylation. It is also possible that DNA-H3/H4 complex and tetrasome formation could have different effects on Piccolo NuA4.
For these experiments, various quantities of DNA were added to (H3/H4)2 stock solutions (mixed at low salt assay buffer), and the individual titrations were pre-incubated at 37°C for 30 min. Then we measured kcat(app) under conditions of saturating (H3/H4)2 and acetyl-CoA (15 and 300 µM, respectively) with variable concentrations of DNA (207 bp Widom 601 DNA) ranging from 4.5–30 µM. Intriguingly, we found that when at least 4.5 µM of DNA was present in the solution, the acetylation rate of Piccolo NuA4 was significantly suppressed. Additionally, as the amount of DNA was increased, the acetylation rate and the specificity for individual sites remains approximately the same (Fig. 4). These results indicated that DNA may not change the site preference of Piccolo NuA4 acetylation, but the interactions between DNA and (H3/H4)2 could ablate acetylation catalyzed by Piccolo NuA4. There is no significant change in acetylation detected when increasing DNA-to-(H3/H4)2 ratios from 0.3 to 2, suggesting that more than one (H3/H4)2 can interact with DNA when mixed at low salt. We also examined the effect of free DNA on NCP by adding free DNA to (5 µM) NCP (1:1 DNA:NCP), and found that the primary sites catalyzed by Piccolo NuA4 remained the same (H4K5 and H4K12). In addition, the acetylation rates for 5 µM NCP were 0.215 ± 0.024 s−1 of H4K5 and 0.337 ± 0.049 s−1 of H4K12, which are not significantly different from the NCP assays without free DNA (0.238 ± 0.013 s−1 of H4K5 and 0.350 ± 0.010 s−1 of H4K12 (from Fig. 3A)). These data suggest that DNA is acting to suppress the acetylation of H3/H4 in the absence of the NCP. To determine if the DNA dependent inhibition of H3/H4 acetylation was due to incorrect complex formation, we used salt gradients to form tetrasomes (2:1 H3/H4 to DNA). We observed no difference between the random histone DNA interaction and preformed tetrasome.
Figure 4.
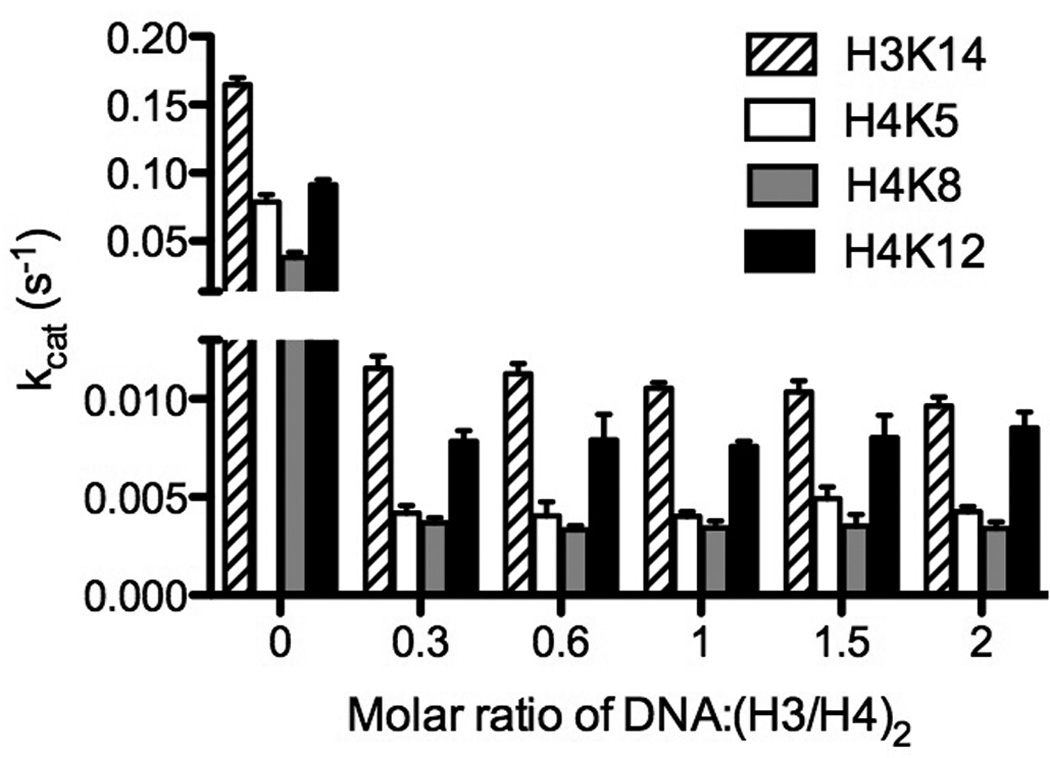
Reduction of histone acetylation by Piccolo NuA4 by adding DNA in the presence of saturating acetyl-CoA and (H3/H4)2. The error bar represents the standard error of kcat(app).
To compare our specificity results, we next used tetrasome as a substrate and carried out a steady-state assay to determine the residue specificity for Piccolo NuA4 on tetrasome. Similar to the results of (H3/H4)2 tetramer, Piccolo NuA4 targeted the same four acetylated lysine residues (H3K14, H4K5, H4K8, and H4K12) under steady-state conditions (Fig. 5A). We found the individual site specificity of Piccolo NuA4 for tetrasome to be H3K14 ≅ H4K12 > H4K5 > H4K8, and even the differences (ratios) between those four sites were similar to (H3/H4)2 tetramer (Table 2; Fig. 5B). However, compared with the kinetic parameters from tetramer, there was a much smaller kcat(app) and comparable Km(app) for tetrasome, indicating that the specificity of Piccolo NuA4 is lowest on tetrasome (Fig. 6). By directly comparing specificity constants, it is clear that NCP was the most preferential substrate for acetylation mediated by Piccolo NuA4, with H4K12 being the most favorable residue for acetylation on this substrate. (H3/H4)2 tetramer was the second most preferred substrate, and tetrasome was the least. On both tetramer and tetrasome, H3K14 and H4K12 were the most preferred sites of Piccolo NuA4-mediated acetylation.
Figure 5.
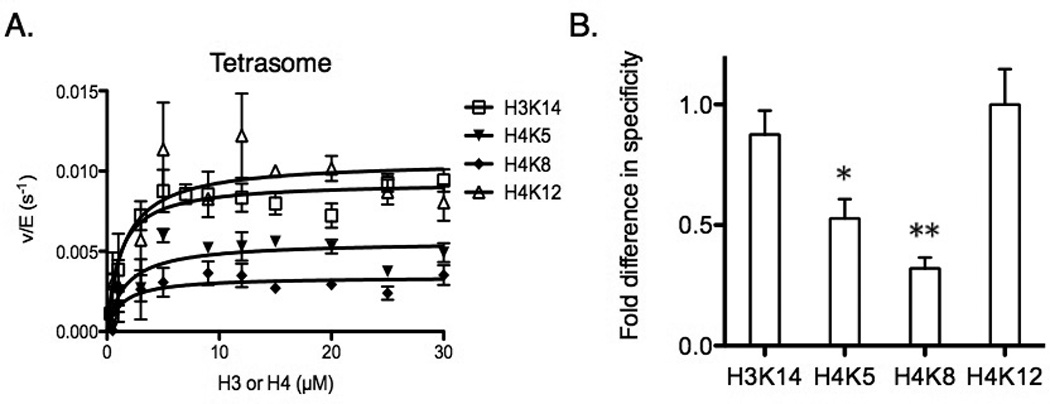
Specificity comparison between lysine residues in tetrasome. (A) Determination of steady-state parameters for individual lysines (H3K14, H4K5, H4K8, and H4K12) of tetrasome acetylation catalyzed by Piccolo NuA4 in the presence of saturating acetyl-CoA. The error bar represents the standard error of v/E. The apparent kinetic parameters are summarized in Table 2. (B) The fold difference in specificity is determined by the ratio of kcat(app), and each site is normalized to the highest kcat(app) of H4K12. * p < 0.05, and ** p < 0.01 when compared to H4K12. The specificity for H3K14 and H4K12 has no significant differences in tetrasome. The error bar represents the standard error of specificity measurements.
Figure 6.
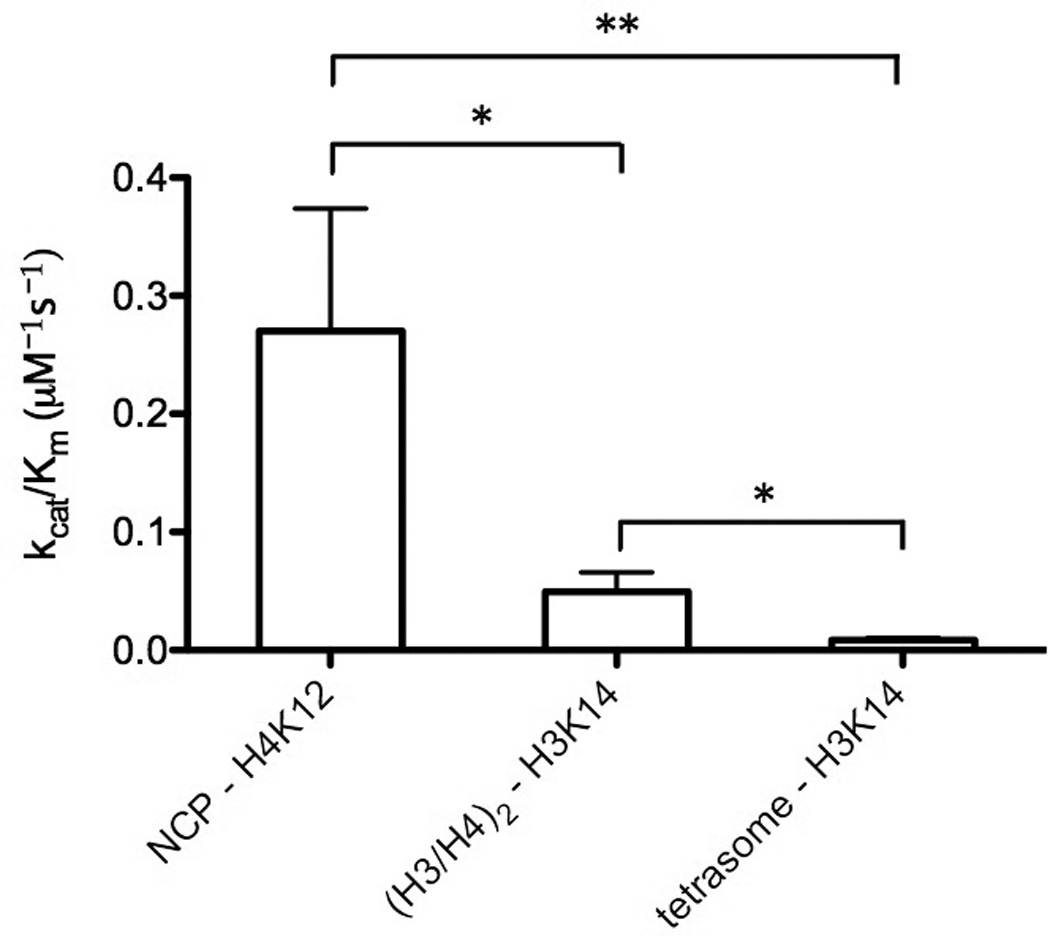
Comparison of specificity of Piccolo NuA4 among different histone complexes. By comparing the highest specificity constant from the individual site of different histone complexes, NCP is the most preferential substrate and tetrasome is the least. * p < 0.05, and ** p < 0.01. The error bar represents the standard error of specificity constant.
Truncated Piccolo NuA4 complex shares the histone acetylation specificity of the wild type in vivo
To confirm that the truncated version of Piccolo NuA4 complex used in this study was not influencing the specificity of acetylation, we looked at truncations of the same complex in vivo. These two truncated Piccolo NuA4 complexes (epl1(1-380) and epl1(1-485)) have previously been shown to have activities that are comparable to wild-type [22,42]. However, little work had been done to quantitatively measure that there was no change in histone acetylation across multiple sites. We therefore utilized our quantitative MS-based assay to determine whether the mutant Piccolo NuA4 had any influence on histone acetylation in vivo. We observed no change in H4 tail acetylation between wild-type and mutants (Supplemental Figure 1). Our data together with previously published results [22,42,57,58] suggest that when the enhancer of polycomb A (EPcA) domain remains intact (i.e., Epl1(51-380)), the Piccolo NuA4 complexes possess the same histone acetylation specificity.
DISCUSSION
We have detected that on NCP, Piccolo NuA4 can predominately acetylate four H4 lysine residues (Lys5, Lys8, Lys12 and Lys16), and moderately acetylate H2A on one lysine residue (Lys5) and one lysine residue on H3 (Lys14). No other acetylated residues were detected on H4 even after these 4 lysine residues of H4 were >90% acetylated (i.e., after a 10 min incubation in the NCP assay, Fig. 1). Although it has been demonstrated that Piccolo NuA4 can acetylate H2A, H3, and H4 on NCP, and that the acetylation stoichiometry at reaction completion is 4:1:1 [25], the order of acetylation was thought to be random on nucleosomal H4 [55]. Our data suggests that order may not be random: we found that while H4K16 is acetylated by Piccolo NuA4, this appears to be a secondary site of acetylation, or occurs only after acetylation of other sites. This suggests that the acetylation on H4K5 and/or H4K12 is required for H4K16 acetylation; a similar result is also observed from the acetylation of H4K8 on NCP (Fig. 1). These data indicate that Piccolo NuA4 specifically acetylates the H4 tail domain (Lys5, Lys8, Lys12, and Lys16) on NCP. In addition to H2A, H3, and H4, we expanded our assay to include sites of H2B. While our assay currently only detects one site of H2B, H2BK85, (due to the length of peptides created by tryptic digestion) we found no significant acetylation at this site, consistent with other reports [25,55].
In order to understand the contribution of NCP on Piccolo NuA4 specificity we needed to look at the acetylation of other histone complexes. We looked at the specificity for either histone H3 or H4 on tetramer and tetrasome. On these two histone complexes, H4 had ~2-fold higher specificity than H3 (Table 2). However, the specificity for individual H4 sites was either similar to or even lower than the specificity for H3K14, which is the only site on H3 that is significantly acetylated during the time course assay of Piccolo NuA4 (Fig. 1, 2B, and 5B). H4 on NCP is the only primary target for Piccolo NuA4 acetylation, because no other lysines on histone H2A, H2B, and H3 of NCP demonstrated significant acetylation prior to a total 10% acetylation occuring. When only the individual histones are considered, H4 is always the primary histone acetylated by Piccolo NuA4, regardless of the histone complex. We note that when nucleosome assembly is complete, the prevalence of H3K14 acetylation by Piccolo NuA4 is suppressed. These results highlight that, without this type of site-specific analysis, we could have a biased interpretation of histone preference while also losing the detailed examination of individual lysines.
Comparing site-specific MS-based data to radioactive filter-binding assays or coupled assays (e.g., pyruvate dehydrogenase convention of NAD to NADH) [25,59] is not possible, as they only interpret the kinetics regarding the total acetylation amount. However, either radioactive filter-binding assays or coupled assays do provide some perspectives on the specificity of the Piccolo NuA4 complex. For example, Berndsen et al. have reported that Piccolo NuA4 has 3–35 fold higher specificity for free histone H4 compared to the other free histones (H2A, H2B, and H3), and the specificity for NCP is almost 20-fold higher than for free histone H4 [25]. In the present study, we investigate not only the Piccolo NuA4 selectivity between sites or individual histones within three specific histone complexes, but also the Piccolo NuA4 specificity for histones during the stages of nucleosome assembly. In order to model the specificity alterations for lysine acetylation during the nucleosome assembly in the nucleus, we chose the starting material of (H3/H4)2 tetramer instead of free histones. In addition, we considered NCP and the intermediate substrate, tetrasome or (H3/H4)2 bound DNA, which lies between free (H3/H4)2 tetramer and NCP. Then, we conducted site-specific kinetic analysis of Piccolo NuA4 for primary acetylation sites and demonstrated that, regardless of the histone complex, H4K12 is the preferential site of Piccolo NuA4. We do, however, note that on both tetramer and tetrasome, H3K14 and H4K12 had high specificities, and ratio differences (selectivity) between the four-lysine residues (H3K14, H4K5, H4K8, and H4K12) remained about the same from the tetramer to tetrasome state (Fig. 2B and 5B). Thus, it suggests that from the stage of tetramer to tetrasome, DNA wrapping causes the suppression of acetylation but has no significant alteration on selectivity between primary sites.
In NCP, specificity for H4K12 remained higher than any other site. H4K5ac specificity was about half of H4K12, no matter what the histone complex is. However, the preference of H3K14 and H4K8 acetylation by Piccolo NuA4 seems to be lost due to the completion of nucleosome assembly (from tetrasome to NCP). By comparing the highest specificity constants of individual sites measured from these three histone complexes, we demonstrate the order of preference: NCP > (H3/H4)2 tetramer > tetrasome (Table 2 and Fig. 6). Furthermore, the value of specificity for NCP increases ~6-fold from the tetrameric complex, and tetramer is ~6-fold higher than tetrasome. Therefore, the composition of the histone complex is a key factor in altering Piccolo NuA4 acetylation specificity. Although contacts on histone-fold domain of H4 are crucial to more efficient histone acetylation by Piccolo NuA4 on NCP [25], it has also been observed that the Epl1 EPcA domain can cross-link to the N-terminal tail of histone H2A [36,58]. Remarkably, we did not observe progressively increasing acetylation specificity with the progress of nucleosome assembly. Because tetrasome shows a decrease in specificity from tetramer, and DNA interactions in the absence of NCP result in suppression of H3/H4 acetylation, this suggests: 1) that H2A/H2B plays an important role in facilitating Piccolo NuA4 recognition of H4 on NCP, and 2) DNA-histone interactions are not a factor contributing to more NCP recognition by Piccolo NuA4.
The increase in Piccolo NuA4 specificity for the NCP as compared to free H3/H4 is different than other KATs that have been studied [33,60]. The study of the former observation claims that histone-histone complex and/or DNA-histone complex causes more structural complexity and thus should be more sterically hindering to interactions with KATs; for example, Rtt109 [60]. Conversely, Piccolo NuA4 can more efficiently acetylate histones in the nucleosome complex, because, in fact, Esa1 on Piccolo NuA4 can recognize the histones folded in the nucleosome and overcome the steric hindrance [25], and Epl1 and Yng2 also play critical roles in assisting nucleosome acetylation [42]. Similarly, the human counterpart of the KAT complex, the Tip60 complex, can acetylate either free or nucleosomeal histones, while Tip60, its catalytic subunit, is only able to acetylate free histones [11,24]. Thus, steric hindrance of histone-histone complexes or DNA-histone complexes cannot be universally applied to explain how a histone complex alters the KAT residue specificity. And histone assembly could be key to understanding changes in KAT specificity (e.g., the role of H2A/H2B dimer in Piccolo NuA4 acetylation for NCP). Additionally, lysine residues (H4K5, H4K8, H4K12, H4K16, H3K14, and H2AK5) acetylated by Piccolo NuA4 are all involved in gene transcriptional activation [12,54,61–65]. Together, these suggest that Piccolo NuA4, acting as a KAT in the nuclei, is well link to transcriptional activation instead of nascent histone deposition (i.e., Rtt109 in cytoplasm). Thus, we propose that due to various functions of KATs, each KAT and/or its subunit is able to select its targeted substrate via its preference for specific histone complexes.
In summary, based on the steady-state assays, we can create a detailed picture of site-specific Piccolo NuA4-mediated acetylation on histones. These data provide viable information on site-specific acetylation, and the specificity for primary acetylation sites. These data also provide detailed information regarding how nucleosome assembly plays a role in Piccolo NuA4-mediated acetylation. Such knowledge could be valuable for modulating Piccolo NuA4 activity during changes in chromatin dynamics, and as a tool for further regulating gene expression.
Supplementary Material
Acknowledgments
We acknowledge the financially supporting from the Canadian Institutes of Health Research (MOP-64289) to J. C and the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM102503 to A. J. A. and R01GM088236 to S. T.
REFERENCES
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein M, Hecht A, Fisher-Adams G, Wan J, Mann R, Strahl-Bolsinger S, Laroche T, Gasser S. The regulation of euchromatin and heterochromatin by histones in yeast. J. Cell Sci. 1995;1995:29–36. doi: 10.1242/jcs.1995.supplement_19.4. [DOI] [PubMed] [Google Scholar]
- 3.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 5.Carrozza MJ, Utley RT, Workman JL, Côté J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 6.Bickmore WA, van der Maarel SM. Perturbations of chromatin structure in human genetic disease: recent advances. Hum. Mol. Genet. 2003;12:R207–R213. doi: 10.1093/hmg/ddg260. [DOI] [PubMed] [Google Scholar]
- 7.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 9.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 10.Lau OD, Courtney AD, Vassilev A, Marzilli LA, Cotter RJ, Nakatani Y, Cole PA. p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate. J. Biol. Chem. 2000;275:21953–21959. doi: 10.1074/jbc.M003219200. [DOI] [PubMed] [Google Scholar]
- 11.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mann RK, Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. The EMBO journal. 1992;11:3297. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 14.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 15.Sakuraba K, Yokomizo K, Shirahata A, Goto T, Saito M, Ishibashi K, Kigawa G, Nemoto H, Hibi K. TIP60 as a potential marker for the malignancy of gastric cancer. Anticancer Res. 2011;31:77–79. [PubMed] [Google Scholar]
- 16.Chen G, Cheng Y, Tang Y, Martinka M, Li G. Role of Tip60 in human melanoma cell migration, metastasis, and patient survival. Journal of Investigative Dermatology. 2012;132:2632–2641. doi: 10.1038/jid.2012.193. [DOI] [PubMed] [Google Scholar]
- 17.Borrow J, Stanton VP, Andresen JM, Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dubé I, Frischauf AM. The translocation t (8; 16)(p11; p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB–binding protein. Nat. Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 18.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 19.Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Côté J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM - related cofactor Tra1p. The EMBO journal. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyon Y, Côté J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Doyon Y, Selleck W, Lane WS, Tan S, Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berndsen CE, Selleck W, McBryant SJ, Hansen JC, Tan S, Denu JM. Nucleosome recognition by the Piccolo NuA4 histone acetyltransferase complex. Biochemistry. 2007;46:2091–2099. doi: 10.1021/bi602366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinclair DA, Clegg NJ, Antonchuk J, Milne TA, Stankunas K, Ruse C, Grigliatti TA, Kassis JA, Brock HW. Enhancer of Polycomb is a suppressor of position-effect variegation in Drosophila melanogaster. Genetics. 1998;148:211–220. doi: 10.1093/genetics/148.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stankunas K, Berger J, Ruse C, Sinclair D, Randazzo F, Brock HW. The enhancer of polycomb gene of Drosophila encodes a chromatin protein conserved in yeast and mammals. Development. 1998;125:4055–4066. doi: 10.1242/dev.125.20.4055. [DOI] [PubMed] [Google Scholar]
- 28.Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV. The candidate tumour suppressor p33ING1cooperates with p53 in cell growth control. Nature. 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 29.Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33ING1 are associated with histone acetyltransferase activities. Mol. Cell. Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nourani A, Doyon Y, Utley RT, Allard S, Lane WS, Côté J. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 2001;21:7629–7640. doi: 10.1128/MCB.21.22.7629-7640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng X, Hara Y, Riabowol K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002;12:532–538. doi: 10.1016/s0962-8924(02)02391-7. [DOI] [PubMed] [Google Scholar]
- 32.Kuo M-H, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 33.Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 34.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 35.Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 36.Lalonde M-E, Cheng X, Côté J. Histone target selection within chromatin: an exemplary case of teamwork. Genes Dev. 2014;28:1029–1041. doi: 10.1101/gad.236331.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 38.Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 2009;29:6473–6487. doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, Côté J. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A. Z by the SWR1 complex. J. Biol. Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henry RA, Kuo Y-M, Andrews AJ. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry. 2013;52:5746–5759. doi: 10.1021/bi400684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo YM, Henry RA, Huang L, Chen X, Stargell LA, Andrews AJ. Utilizing targeted mass spectrometry to demonstrate Asf1-dependent increases in residue specificity for Rtt109-Vps75 mediated histone acetylation. PloS one. 2015;10:e0118516. doi: 10.1371/journal.pone.0118516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selleck W, Fortin I, Sermwittayawong D, Côté J, Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 2005;25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrios A, Selleck W, Hnatkovich B, Kramer R, Sermwittayawong D, Tan S. Expression and purification of recombinant yeast Ada2/Ada3/Gcn5 and Piccolo NuA4 histone acetyltransferase complexes. Methods. 2007;41:271–277. doi: 10.1016/j.ymeth.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expression Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Lowary P, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 46.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 47.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2003;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 48.Gill SC, Von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 49.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 50.Donham DC, Scorgie JK, Churchill ME. The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4–DNA complexes. Nucleic Acids Res. 2011;39:5449–5458. doi: 10.1093/nar/gkr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scorgie JK, Donham DC, III, Churchill ME. Analysis of histone chaperone antisilencing function 1 interactions. Methods Enzymol. 2012;512:223–241. doi: 10.1016/B978-0-12-391940-3.00010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuo Y-M, Andrews AJ. Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PloS one. 2013;8:e54896. doi: 10.1371/journal.pone.0054896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuo Y-M, Henry RA, Andrews AJ. A quantitative multiplexed mass spectrometry assay for studying the kinetic of residue-specific histone acetylation. Methods. 2014;70:127–133. doi: 10.1016/j.ymeth.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohba R, Steger DJ, Brownell JE, Mizzen CA, Cook RG, Côté J, Workman JL, Allis CD. A novel H2A/H4 nucleosomal histone acetyltransferase in Tetrahymena thermophila. Mol. Cell. Biol. 1999;19:2061–2068. doi: 10.1128/mcb.19.3.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnold KM, Lee S, Denu JM. Processing mechanism and substrate selectivity of the core NuA4 histone acetyltransferase complex. Biochemistry. 2011;50:727–737. doi: 10.1021/bi101355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinert BT, Iesmantavicius V, Moustafa T, Schölz C, Wagner SA, Magnes C, Zechner R, Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 2014;10 doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chittuluru JR, Chaban Y, Monnet-Saksouk J, Carrozza MJ, Sapountzi V, Selleck W, Huang J, Utley RT, Cramet M, Allard S. Structure and nucleosome interaction of the yeast NuA4 and Piccolo–NuA4 histone acetyltransferase complexes. Nat. Struct. Mol. Biol. 2011;18:1196–1203. doi: 10.1038/nsmb.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang J, Tan S. Piccolo NuA4-catalyzed acetylation of nucleosomal histones: Critical roles of an Esa1 Tudor/chromo barrel loop and an Epl1 enhancer of polycomb A (EPcA) basic region. Mol. Cell. Biol. 2013;33:159–169. doi: 10.1128/MCB.01131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim Y, Tanner KG, Denu JM. A continuous, nonradioactive assay for histone acetyltransferases. Anal. Biochem. 2000;280:308–314. doi: 10.1006/abio.2000.4546. [DOI] [PubMed] [Google Scholar]
- 60.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang LH, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikeda K, Steger DJ, Eberharter A, Workman JL. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 63.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 64.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 65.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


