To the Editor: Colistin is an old-generation antimicrobial agent; however, because it is one of the few agents that remain effective against multidrug-resistant gram-negative bacteria (e.g., carbapenem-resistant Pseudomonas aeruginosa and Enterobacteriaceae), its clinical usefulness is being increasingly recognized (1). Previous reports have described the mechanisms of colistin resistance (2) as being chromosomally mediated and not associated with horizontal gene transfer. However, from 2011 through 2014, a plasmid-encoded colistin-resistance gene, mcr-1, was identified in colistin-resistant Escherichia coli isolated in China, particularly from animals. Specifically, mcr-1–positive isolates were found in 21% of healthy swine at slaughter, 15% of marketed pork and chicken meat, and 1% of hospitalized human patients (3). A study of E. coli isolated from healthy cattle, swine, and chickens in Japan during 2000–2014 found only 2 (0.02%) of 9,308 isolates positive for mcr-1 (4). We report the rates at which mcr-1 was detected in our stored collection of E. coli isolates from diseased swine (swine with diarrhea or edema disease), hereafter referred to as swine-pathogenic E. coli.
We recently analyzed swine-pathogenic E. coli strains isolated from diseased swine throughout Japan during 1991–2014 (5). We analyzed all swine disease-associated E. coli strains isolated from the 23 Livestock Hygiene Service Centers in Japan (including prefectures that covered 75% of total swine production in Japan in 2014) and sent to the National Institute of Animal Health for diagnostic purposes during 1991–2014. Among the 967 strains examined, 684 (71%) belonged to E. coli serogroup O139, O149, O116, or OSB9.
In the study reported here, we investigated these 684 strains for susceptibility to colistin and for mcr-1 carriage. The strains from the 4 predominant serogroups (Technical Appendix Table) can be considered representative of swine-pathogenic E. coli strains isolated from farm animals, but not food products, in Japan. MICs were determined by using the agar dilution method according to the recommendations of the Clinical and Laboratory Standards Institute (6). The presence of mcr-1 was detected by PCR (3).
Among the 684 strains examined, colistin MICs exhibited a bimodal distribution of 0.25–128 μg/mL and peaked at 0.5 and 16 μg/mL (Technical Appendix Figure). According to the European Committee on Antimicrobial Susceptibility Testing criterion (7), in which isolates with an MIC of >4 μg/mL are considered colistin resistant, 309 (45%) of the 684 strains were classified as colistin resistant. The gene mcr-1 was detected in 90 (13%) strains, and the MICs for these mcr-1–positive strains ranged from 8 to 128 μg/mL (Technical Appendix Figure). Among the 309 colistin-resistant strains, mcr-1–positive and mcr-1–negative isolates had the same 50% and 90% MICs, 16 and 32 μg/mL, respectively. These results indicate that a high proportion of swine-pathogenic E. coli in Japan are resistant to colistin, that mcr-1 has already been widely disseminated among these strains, and that the level of colistin resistance mediated by mcr-1 is similar to that mediated by mcr-1–independent mechanisms.
In 2004, colistin-resistant E. coli already represented 77% of the isolates, and the positivity rates varied from year to year (26%–82%) (Figure). First detection of mcr-1–positive strains was in 2007, and the proportion of mcr-1 positivity has risen, especially since 2009 (Figure). During 2013–2014, approximately half of the strains isolated were mcr-1 positive (Figure), and most colistin-resistant strains isolated during these 2 years carried mcr-1 (85% and 62% in 2013 and 2014, respectively). Of note, the rates of mcr-1–positive strains among the 4 serogroups isolated from 2010 through 2014 did not differ significantly (χ2 test): 22 (20%) of 110 in O139, 38 (38%) of 100 in O149, 19 (26%) of 73 in O116, and 6 (32%) of 19 in OSB9. This finding suggests that the sharp rise in the proportion of mcr-1–positive strains has been driven by plasmid-mediated horizontal gene transfer, not by the expansion of a specific clone.
Figure.
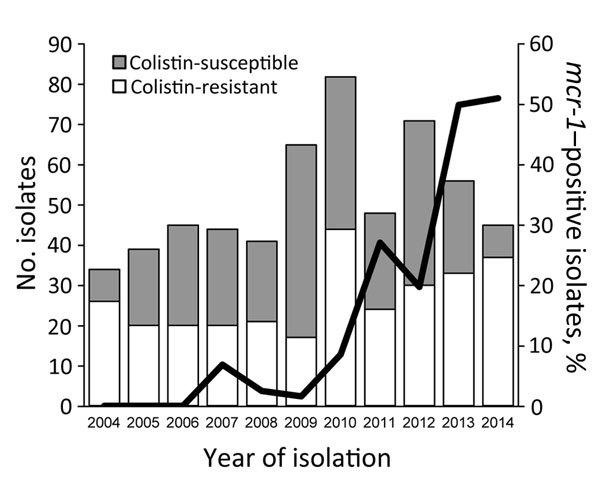
Changes in the numbers of colistin-susceptible and colistin-resistant Escherichia coli isolated from swine with diarrhea or edema disease, Japan, 2004–2014. The line shows the changes in proportion of mcr-1–positive isolates among the total isolates for each year.
In Japan, rates of isolation of colistin-resistant and mcr-1–positive E. coli strains from healthy animals are low, 1.00% and 0.02% of 9,308 strains examined, respectively (4). These low rates may be the result of the prudent use of colistin in Japan. During 2000–2007 in Japan, colistin use in swine did not increase significantly (8). However, our data show that mcr-1 has recently been disseminated among swine-pathogenic E. coli in Japan, which might be associated with the use of colistin to treat disease in swine. Although mcr-1–positive bacteria have not yet been isolated from humans in Japan (4), the sharp increase in swine-pathogenic E. coli in animal strains implies a risk for transmission of mcr-1 from these strains to human-pathogenic bacteria, a serious concern for human medicine. More active surveillance of mcr-1–positive colistin-resistant bacteria in human and animal environments is needed.
Information about isolates used in study of mcr-1–positive colistin-resistant pathogenic Escherichia coli in swine, Japan, 2007–2014, and MICs of colistin for E. coli isolated from diseased swine in Japan 1991–2014.
Acknowledgments
We thank Yuna Hikoda for technical assistance and all of the prefectural Livestock Hygiene Service Centers for providing E. coli isolates and strain information.
This work was supported by a Grant-in-Aid for Scientific Research (category C), KAKENHI grant no. 15K08484 to M.K. from the Japan Society for the Promotion of Science, and a grant from the Research Program on Emerging and Re-emerging Infectious Diseases (no. 15fk0108021h0002) from the Japan Agency for Medical Research and Development.
Footnotes
Suggested citation for this article: Kusumoto M, Ogura Y, Gotoh Y, Iwata T, Hayashi T, Akiba M. Colistin-resistant mcr-1–positive pathogenic Escherichia coli in swine, Japan, 2007−2014 [letter]. Emerg Infect Dis. 2016 Jul [date cited]. http://dx.doi.org/10.3201/eid2207.160234
References
- 1.Ruppé E, Woerther PL, Barbier F. Mechanisms of antimicrobial resistance in gram-negative bacilli. Ann Intensive Care. 2015;5:61. 10.1186/s13613-015-0061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 4.Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis. 2016;16:284–5. 10.1016/S1473-3099(16)00008-6 [DOI] [PubMed] [Google Scholar]
- 5.Kusumoto M, Hikoda Y, Fujii Y, Murata M, Miyoshi H, Ogura Y, et al. Emergence of a multidrug-resistant Shiga toxin-producing enterotoxigenic Escherichia coli lineage in diseased swine in Japan. J Clin Microbiol. 2016;54:1074–81. 10.1128/JCM.03141-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 24th informational supplement. Document M100–S24. Wayne (PA): The Institute; 2014. [Google Scholar]
- 7.European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 60 [cited 2016 Feb 6]. http://www.eucast.org/clinical_breakpoints/
- 8.Koike R, Asai T, Ozawa M, Ishikawa H. The use of therapeutic antimicrobials for food-producing animals [in Japanese]. Ann Rep Natl Vet Assay Lab. 2009;45:30–3. Summary in English [cited 2016 Apr 6]. http://www.maff.go.jp/nval/kouhou/nenpo/no45/pdf/no45-040-043-p030-033-gaku-gi-3.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information about isolates used in study of mcr-1–positive colistin-resistant pathogenic Escherichia coli in swine, Japan, 2007–2014, and MICs of colistin for E. coli isolated from diseased swine in Japan 1991–2014.


