Abstract
Rab 11 GTPase is an important regulator of endocytic membrane traffic. Recently, we and others have identified a novel family of Rab11 binding proteins, known as Rab11-family interacting proteins (FIPs). One of the family members, Rab coupling protein (RCP), was identified as a protein binding to both Rab4 and Rab11 GTPases. RCP was therefore suggested to serve a dual function as Rab4 and Rab11 binding protein. In this study, we characterized the cellular functions of RCP and mapped its interactions with Rab4 and Rab11. Our data show that RCP interacts only weakly with Rab4 in vitro and does not play the role of coupling Rab11 and Rab4 in vivo. Furthermore, our data indicate that the RCP–Rab11 complex regulates the sorting of transferrin receptors from the degradative to the recycling pathway. We therefore propose that RCP functions primarily as a Rab11 binding protein that regulates protein sorting in tubular endosomes.
INTRODUCTION
Eukaryotic cells internalize cell surface proteins via a process known as endocytosis. Endocytosed proteins are first delivered to the multifunctional organelles often referred to as early or sorting endosomes (EEs). From there, proteins are either recycled back to the plasma membrane or transported to late endosomes and lysosomes for degradation. Sorting and recycling of endocytosed proteins are required for proper cellular function and growth, yet we still understand little about the mechanisms involved. At least some of the sorting seems to occur in the EEs via generation of EE tubular extensions that give rise to recycling endosomes (REs) and ensure the delivery of proteins back to the plasma membrane. In addition to RE-dependent recycling, protein also can be recycled directly from EEs to the plasma membrane via the “fast” recycling pathway (Mellman, 1996; Robinson et al., 1996).
Members of the Rab GTPase family have emerged as important regulators of vesicular traffic, governing specific membrane trafficking steps (Gonzalez and Scheller, 1999; Prekeris, 2003). At least six Rab proteins (Rabs 4, 5, 7, 9, 11, and 15) regulate the traffic and sorting of endocytosed material between endosomes, lysosomes, and the plasma membrane (van der Sluijs et al., 1992; Ullrich et al., 1996; Chavrier and Goud, 1999). Rab11 in particular plays an essential role in protein recycling (Ullrich et al., 1996; Prekeris, 2003) and also has been implicated in regulating several other membrane transport pathways, including phagocytosis, polarized epithelial transport, and the delivery of the insulin-dependent glucose transporter to the plasma membrane (Cox et al., 2000; Kessler et al., 2000; Wang et al., 2000; Wilcke et al., 2000).
Despite considerable efforts, the mechanism of Rab11 action remains to be fully understood. Cycling between GTP- and GDP-bound forms of Rab proteins regulates the recruitment of various effector proteins to cellular membranes, thereby affecting the targeting and fusion of transport vesicles. Because the ability of GTP-Rabs to interact with several different effector molecules could be the basis for the specific functions of Rab proteins in a variety of cellular processes, much effort has been invested in identifying effector proteins for Rab11. We recently identified Rip11, a novel Rab11-interacting protein (Prekeris et al., 2000), and subsequently five more related proteins have been found (Hales et al., 2001; Prekeris et al., 2001; Lindsay et al., 2002; Hickson et al., 2003), forming the Rab11-family interacting proteins (FIPs). Based on sequence homology, FIPs can be divided into three classes (Meyers and Prekeris, 2002): class I FIPs (Rip11, RCP, and FIP2/nRip11) contain a C2 domain at the N-terminal end of the protein; class II FIPs (FIP3/Eferin and FIP4) contain two EF-hands and a proline-rich region; and class III has only one member, FIP1, with no homology to known protein domains. The common feature of all FIP proteins is the presence of a highly conserved, 20-amino acid motif at the C terminus of the protein, known as the Rab11/25 binding domain (RBD) (Prekeris et al., 2001). The RBD seems to be in a predominantly α-helical conformation, allowing the highly conserved hydrophobic residues to form a hydrophobic Rab11 binding patch (Meyers and Prekeris, 2002).
Although FIPs were identified as Rab11 binding proteins, their role in regulating membrane traffic remains to be determined. Given that most mammalian cells express several FIP proteins, one possible role could be the formation of mutually exclusive complexes with Rab11, thereby directing the recruitment of Rab11 to different membrane trafficking pathways (Meyers and Prekeris, 2002). Indeed, Rip11 can bind to membranes in the absence of Rab11, although Rab11 binding is required for its appropriate localization (Meyers and Prekeris, 2002). Furthermore, because several FIP-interacting proteins have been identified (Shin et al., 1999; Lapierre et al., 2001; Cullis et al., 2002; Lindsay et al., 2002; Hickson et al., 2003), FIPs may serve as adaptor proteins regulating the recruitment of additional Rab11 effector proteins.
Recent data suggest that FIPs may play a key role in regulating endocytic membrane transport. RCP, a class I FIP, was proposed to regulate protein recycling from the EE and RE to the plasma membrane (Lindsay et al., 2002). What makes RCP especially interesting is that it was originally identified as a protein binding to both Rab11 and Rab4 GTPases, hence its name Rab coupling protein, and was suggested to regulate protein traffic from EEs to REs (Lindsay et al., 2002). Similarly, Rabenosin-5 was recently identified as a divalent Rab effector for Rab5 and Rab4 (de Renzis et al., 2002). Because Rab4, Rab5, and Rab11 regulate sequential steps along the endocytic recycling pathway, it is tempting to speculate that RCP regulates protein recycling and sorting by connecting Rab4 and Rab11 domains on EEs and REs (de Renzis et al., 2002).
The aim of this work was to map RCP interactions with Rab4 and Rab11and to analyze the role of RCP in endocytic membrane traffic. Our data suggest that RCP does not play a role as a Rab11- and Rab4-coupling protein, but it complexes with Rab11 to regulate transferrin receptor (TfR) sorting into the recycling pathway instead of the degradative pathway. Thus, we propose that in addition to its other functions, when bound to RCP, Rab11 is also involved in sorting of proteins for recycling to the plasma membrane.
MATERIALS AND METHODS
Reagents and Plasmids
Cell culture reagents were obtained from Invitrogen (Carlsbad, CA) unless otherwise specified. Most chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Mouse monoclonal anti-myc antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Transferrin conjugated to Texas Red (Tf-TxR) was purchased from Molecular Probes (Eugene, OR). Fluorescein isothiocyanate-labeled anti-rabbit IgG and Texas Red-labeled anti-mouse IgG antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Mouse monoclonal anti-EEA1 antibody was purchased from BD Biosciences (San Diego, CA). Mouse monoclonal anti-transferrin receptor antibodies were obtained from Zymed Laboratories (South San Francisco, CA). The polyclonal rabbit anti-Rip11 antibody was described previously (Prekeris et al., 2000). The polyclonal rabbit anti-RCP antibody was prepared by immunization with recombinant RCP, expressed, and purified from Escherichia coli as a glutathione S-transferase (GST)-fusion protein. cDNAs encoding RCP and Rip11 were cloned in pEGFP-N1 or pEYFP-N1. Polyclonal rabbit anti-GST was prepared by immunization with recombinant GST. Mouse anti-transferrin receptor-phycoerythrin (TfR-PE) was purchased from BD Biosciences. Mouse anti-LDLR monoclonal antibody (15CA) was obtained from Oncogene (San Diego, CA). Mouse anti-human EGFR antibody (clone 13A9) (Fendly et al., 1990) was conjugated to Alexa488 by using a protein labeling kit according to manufacturer's instructions (Molecular Probes).
Cell Culture, Immunofluorescence Microscopy, and Fluorescence Energy Transfer (FRET) Analysis
HeLa cells were cultured as described previously (Meyers and Prekeris, 2002). For immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.4% saponin, and nonspecific sites were blocked with phosphate-buffered saline containing 0.2% bovine serum albumin, 0.4% saponin, and 1% fetal bovine serum. After incubation with specific antibodies, samples were extensively washed and mounted in VectaShield (Vector Laboratories, Burlingame, CA). Cells were imaged with an inverted Axiovert 200M deconvolution microscope (Carl Zeiss, Thornwood, NY). Image processing was done using Intelligent Imaging Innovations three-dimensional rendering and exploration software. For quantitation of colocalization of transiently transfected HeLa cells, only cells expressing low amounts of protein were analyzed, as defined by the fluorescence of the green fluorescent protein (GFP)-tagged protein being similar (no more than 2-fold higher) to the fluorescence obtained from the antibodies against the endogenous protein with which it was costained. All images were digitally deconvolved before analysis. The background fluorescence was then subtracted from all images before quantitation.
All colocalization data are the means of five randomly chosen cells. Colocalization analysis was done using Intelligent Imaging Innovations three-dimensional rendering and exploration software. The background fluorescence was determined by randomly sampling an area devoid of cells. The averaged background was then subtracted from the images before colocalization analysis.
For FRET analysis, cells were cotransfected with proteins tagged with yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP). Cells were imaged and corrected FRET (cFRET) was calculated using Intelligent Imaging Innovations three-dimensional rendering and exploration software as described previously with the equation cFRET = FRET - 0.4 × CFP - 0.037 × YFP (Sorkin et al., 2000). Normalized FRET (NFRET) was calculated using the equation NFRET = cFRET/CFP. Only cells expressing similar amounts of CFP- and YFP-tagged proteins (YFP/CFP = 0.5–2.0) were included in the FRET analysis.
Transferrin Recycling Assays
For imaging analysis of transferrin recycling, HeLa cells were plated on collagen-coated coverslips and grown to 60% confluence. Cells were washed with phosphate-buffered saline and incubated for 1 h at 4°C in serum-free, HEPES-buffered, DMEM with 20 μg/ml Tf-TxR. Cells were then washed extensively and returned to 37°C in serum-supplemented DMEM containing 0.2 mg/ml unlabeled transferrin. At each time point, cells were fixed with 4% paraformaldehyde and imaged using an inverted Axovert 200M deconvolution microscope (Carl Zeiss).
Overexpression and Fluorescence-activated Cell Sorting (FACS) Assays
For each DNA construct to be analyzed one 80–90% confluent T-75 cm2 flask of HeLa cells was transiently transfected using LipofectAMINE 2000 (Invitrogen). The cells were incubated with the transfection mixture (60 μg of DNA and 120 μl of LipofectAMINE) for 3 h, and the media were replaced. The next day the cells were trypsinized and resuspended first in complete media, and then in serum-free media and used for FACS analysis.
For uptake assays, the cells were incubated with 20 μg/ml Tf-Alexa 647 (Molecular Probes) or with 2 μg/ml anti-TfR-PE at 4°C for 30 min. Cells were then incubated at 37°C for various time intervals in the continuous presence of either 20 μg/ml Tf-Alexa 647 or with 2 μg/ml anti-TfR-PE (preincubation at 4°C was required to generate a sufficient signal for detection by flow cytometer). Cells were then washed and the experiment was completed by pelleting and resuspending the cells in 3% paraformaldehyde. Cell-associated Tf-Alexa647 was determined by FACS analysis.
For recycling assays, cells were incubated with Tf-Alexa 647 for 30 min at 4°C followed by internalization for 20 min at 37°C in continuous presence of Tf-Alexa647. Cells were then washed and incubated in complete media supplemented with 50 μg/ml unlabeled Tf for various times before fixation. Cell-associated Tf-Alexa647 was determined by FACS analysis.
FACS analysis was performed using a Cytomics FC 500 flow cytometer (Beckman Coulter, Fullerton, CA) equipped with 488- and 647-nm lasers, gating for 10,000 transfected (GFP-positive) cells, and the amount of internalized transferrin (Tf) was determined (Supplemental Figure 3).
RNA-Interference (RNAi) Analysis
For each small interfering RNA (siRNA) knock-down experiment, HeLa cells were transiently transfected with HPP grade siRNA (QIAGEN, Valencia, CA) based on human RCP or Rip11 sequences (for RCP 5′-CGCCTCTTTCCCAGTCCATGT-3′, for Rip11 5′-GAGCTGAGTGCTCAGGCTAAA-3′). The cells were incubated overnight with transfection mixture (12 μg of siRNA and 5 μl of LipofectAMINE 2000 per well of the plate), and then trypsinized and transferred to T-25-cm2 flasks. Control transfections received LipofectAMINE 2000 without siRNA. Cells were then incubated for 74 h, and Tf uptake and recycling were measured using FACS analysis (see description above). The extent of knock-down was estimated by Western blotting with anti-Rip11 and anti-RCP antibodies.
To determine the cell surface levels of TfR and enhanced epidermal growth factor receptor (EGFR), RNAi-treated cells were trypsinized, and incubated with either 2 μg/ml anti-TfR-PE or anti-EGFR-Alexa488 antibodies. To determine the total TfR or EGFR levels, cells were fixed and permeablized with 0.4% saponin before incubation with anti-TfR-PE or anti-EGFR-Alexa488 antibodies. In both cases, cells were then washed and analyzed by FACS (10,000 cells/sample).
Expression and Purification of Proteins
GST-gene fusion constructs were prepared by cloning Rab11a, RCP, Rip11, Rab4a, and Rab4b fragments into pGEX-KG (Amersham Biosciences, Piscataway, NJ) and transforming them into BL-21 Codon Plus E. coli (Stratagene). GST fusion proteins were expressed and purified as described previously (Meyers and Prekeris, 2002). Protein concentrations were determined using the Bradford assay.
In Vitro Binding Assays
In vitro binding assays were performed using 50 μl of packed glutathione beads coated with 5 μg of GST-fusion protein in a final volume of 0.5 ml and varying amounts of soluble proteins in 50 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, 0.1% bovine serum albumin, 1 mM phenylmethylsulfonyl fluoride. Guanosine 5′-3-O-(thio)triphosphate or guanine 5′-O-(2-thio)diphosphate was added where indicated. Reactions were incubated while rotating at 4°C for 1 h, pelleted at 2000 × g for 3 min, and washed three times in 1 ml of reaction buffer. Bound proteins were eluted with 1% SDS and separated on SDS-PAGE and either stained with Coomassie Blue or immunoblotted. For immunoblotting, after incubation with primary antibody, the blots were incubated with secondary antibodies against either anti-rabbit IgG or anti-mouse IgG conjugated to Cy5 fluorophore. Blots were imaged using Typhoon multipurpose scanner and quantified using ImageQuant software.
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a VP-ITC calorimeter from OriginLab (Northampton, MA). Rab4A, 4B, and 11A proteins were purified as described above, except after GST column chromatography the bound GST-Rab beads were processed through a series of nucleotide [guanosine 5′-(β,γ-imido)triphosphate (GppNHp) or GDP] exchange reactions in the presence of EDTA to purify the protein in either the GppNHp-bound or the GDP-bound form, as described previously for the Rab5 GTPase (Christoforidis and Zerial, 2000). The thrombin cleaved Rab proteins were used for ITC studies. Rab (8 μM) was loaded in the sample cell at 25°C (in phosphate-buffered saline [PBS] containing 5 mM MgCl2 and 0.5 mM GppNHp or GDP) in a volume of 1.426 ml, and titrated with GST-RCP (75 μM) in same buffer with each 5-μl injection to a total of 42 injections at 4-min intervals with stirring at 300 rpm to allow the baseline to stabilize. The data were fitted using the one set of sites model to calculate the binding constant (K) using Origin software, provided by OrginLab.
GST-RCP Recruitment Assays
HeLa cells were grown on collagen-coated coverslips to 60% confluence, washed with ice-cold reaction buffer (134 mM potassium acetate, 20 mM HEPES, pH 7.4, 2.5 mM magnesium acetate) and permeabilized with 10 μM digitonin for 15 min. Cells were then washed and overlaid with 500 μl of reaction buffer containing 0.5 mM ATP, 0.5 mM GTP, 25 μg/ml bovine serum albumin, 1 mg/ml rat brain cytosol, ATP regeneration system (80 mM creatine phosphate and 1 U/ml creatine kinase), and 5 μg/ml GST-tagged purified protein. Cells were then incubated at room temperature for 30 min., washed, fixed with 4% paraformaldehyde, and processed for immunofluorescence analysis. GST-tagged proteins were visualized using anti-GST antibodies.
RESULTS
Rab11, but Not Rab4, Is an RCP Binding Protein In Vivo
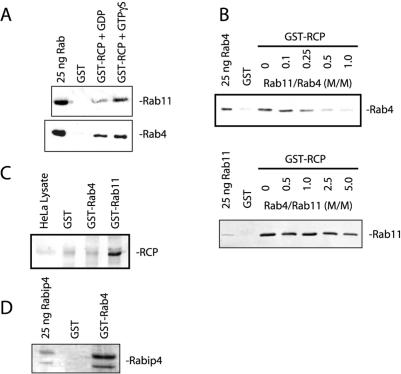
RCP was originally identified as a Rab4 binding protein in a yeast two-hybrid screen (Lindsay et al., 2002). Interestingly, sequence analysis revealed that RCP also contains the RBD domain that mediates interactions with Rab11 (Prekeris et al., 2001; Meyers and Prekeris, 2002). To test whether RCP interacts with both Rab4 and Rab11, we incubated GST or GST-RCP on glutathione beads with 1 μM of recombinant soluble Rab4A or Rab11A. As shown in Figure 1A, RCP interacted with Rab11A in a GTP-dependent manner (5.3-fold enhancement over GDP), whereas it bounds similarly to both GDP- and GTP-bound forms of Rab4A (1.2-fold enhancement over GDP). To determine whether RCP can simultaneously interact with both Rab GTPases, we incubated GST-RCP beads with recombinant Rab4A and increasing concentrations of Rab11A. As shown in the Figure 1B (top), Rab11A completely inhibited the binding of Rab4A to RCP, suggesting that these Rabs use the same or overlapping sites for interactions with RCP. These data suggest that Rab4A and Rab11A likely compete for binding to RCP in vivo. Interestingly, Rab11 seems to bind better to RCP, because it completely inhibited Rab4–RCP interaction at a 1:1 ratio (Rab11:Rab4) (Figure 1B). Consistent with this, Rab4 did not have any inhibitory effect on Rab11–RCP interactions until present at fivefold excess (Figure 1B, bottom).
Figure 1.
Rab11 and Rab4 bind to the same or overlapping site on RCP. (A) To characterize RCP interactions with Rab11 and Rab4, glutathione beads coated with either GST or GST-RCP were incubated with recombinant purified Rab11A and/or Rab4A in the presence of either GTPγS or GDP. Beads were then washed and bound proteins eluted with 1% SDS and analyzed by SDS-PAGE and immunoblotting with either anti-Rab11 or anti-Rab4 antibodies. (B) Glutathione beads coated with either GST alone or GST-RCP were incubated with 1 μM purified Rab4 (top) or Rab11 (bottom). To test whether Rab11 and Rab4 compete for RCP binding increasing concentrations of Rab11 (top) or Rab4 (bottom) were added to the binding assays. Beads were then washed and bound protein eluted with 1% SDS. The amounts of bound Rab4 (top) or Rab11 (bottom) were determined by immunoblotting. (C) HeLa cell Triton X-100 lysates were incubated with Affi-Beads coated with GST, GST-Rab4, or GST-Rab11. Beads were then washed, and the bound protein were eluted with 1% SDS and analyzed by immunoblotting. (D) Glutathione beads coated with either GST alone or GST-Rab4 were incubated with recombinant Rabip4. Beads were washed and bound protein eluted with 1% SDS. Eluates were then run on SDS-PAGE and stained with Coomassie stain.
Competition data suggest that Rab4 may interact with RCP at much lower affinity than Rab11 in vitro. Because Rab11 and Rab4 bind to the same (or at least overlapping) site, it is possible that in vivo Rab11 rather than Rab4 is the predominant RCP interacting protein. Consistent with this, when HeLa cell lysate was used as the source of full-length endogenous RCP (Figure 1C), it only bound to GST-Rab11A, not GST-Rab4A. In contrast, GST-Rab4A (Figure 1D) but not GST-Rab11A (our unpublished data) bound to Rabip4, a known Rab4 binding protein (Fouraux et al., 2004).
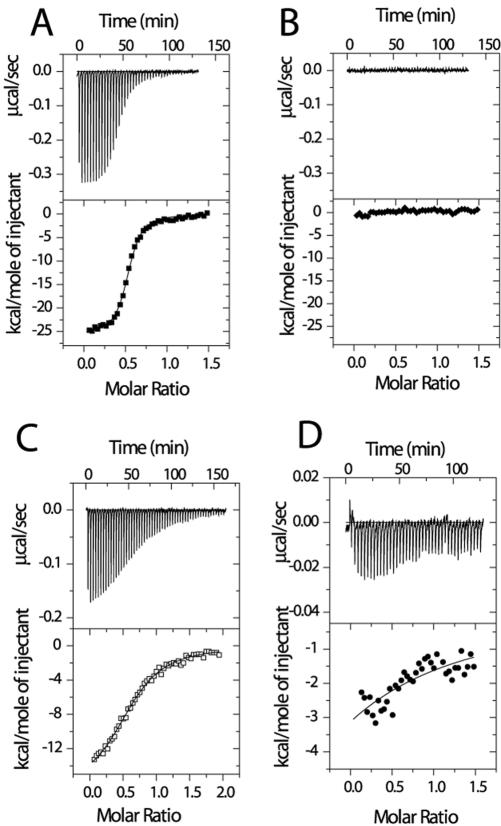
The property of Rab4A binding to RCP raises the question as to whether this interaction is physiologically relevant. We therefore performed quantitative ITC analysis of RCP–Rab11A and RCP–Rab4B complexes. For this analysis, we used a truncated form of RCP (aa 479–659) (GST-RCP-F1), because it can be expressed and purified from E. coli in the amounts needed for ITC and was originally identified as being sufficient for interaction with both Rab4A and Rab11A (Lindsay et al., 2002). As shown in Figure 2A, the titration of Rab11A with increasing amounts of GST-RCP-F1 in the presence of GppNHp resulted in a gradual decrease in the exothermic heat of binding with each successive injection until saturation was achieved. This was specific, because no exothermic heat response was generated with buffer alone or with other Rab GTPases (Figure 2B; our unpublished data), indicating that the ITC-based assay can be used to measure affinities between Rabs and their effector proteins. Similarly, titrating Rab4B produced very little exothermic heat response, even less than that seen with the GDP-bound form of Rab11A (Figure 2, C and D), suggesting that Rab4B binding to RCP is of very low affinity. The calculated affinity of RCP-F1 for the GppNHp-bound form of Rab11A (105 nM) was 200 times higher than that for GppNHp-bound Rab4b (20800 nM) and 11 times higher than the GDP-bound form of Rab11A (1180 nM). Similar results were observed for the interaction of RCP-F1 with Rab4A and Rab11A by using a GST bead pull-down assay (our unpublished data).
Figure 2.
RCP binds to Rab11 with much higher affinity than to Rab4. Raw data (top) from ITC analysis was obtained after 42 successive 5-μl injections of 75 μM GST-RCP into 8 μM Rab11 or Rab4 in PBS containing 5 mM MgCl2 and 0.5 mM GppNHp or GDP. Nonlinear least-squares fit of the heat released as a function of the added ligand also is shown (bottom). GST-RCP titration with buffer in the presence of GppNHp (B); with the GppNHp-bound form of Rab11A (A); with the GDP-bound form of Rab11A (C); and with the GppNHp-bound form of Rab4B (D). Note the smaller scale in D.
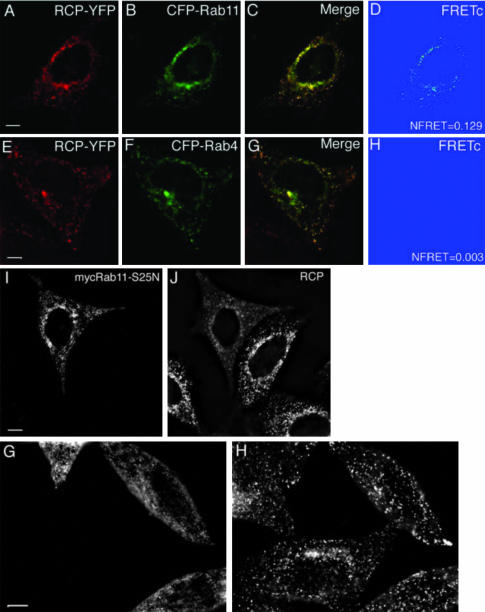
Although our in vitro data suggest that RCP binds to Rab11A with at least 200-fold higher affinity, it remained possible that in vivo RCP could be recruited to endosomes that are highly enriched in Rab4, thus compensating for its relatively low affinity. To test this possibility, we measured FRET between RCP-YFP/CFP-Rab11A and RCP-YFP/CFP-Rab4. To achieve “FRETing” between CFP-Rab11A and RCP, we had to attach the YFP tag to the C terminus of the protein. Because RBD also is localized at the C terminus of FIP proteins, this raised the possibility that the YFP tag may interfere with RCP binding to Rab11 or Rab4. However, the staining of HeLa cells expressing RCP-YFP with anti-Rab11 antibodies showed that RCP-YFP was still capable of appropriate targeting in the cell, because it localizes to a punctuate endocytic structures containing endogenous Rab11 (Supplemental Figure 1). Furthermore, the localization of RCP-YFP was similar to the localization of endogenous RCP, which also colocalized with Rab11 (Supplemental Figure 2). That is in agreement with earlier published data that attachment of a GST-tag to the C terminus of the RBD domain has no influence on its binding to Rab11 in vitro (Prekeris et al., 2001). As shown in Figure 3, A–D, we could detect FRET between RCP-YFP and CFP-Rab11A (NFRET = 0.129), suggesting that RCP and Rab11 form a complex in vivo. The FRET was specific to Rab11A, because CFP-Rab5 did not elicit any FRET with RCP-YFP (NFRET = 0.002). Despite colocalization, we could not detect any FRET between RCP-YFP and CFP-Rab4, suggesting that although RCP and Rab4 may be present on the same organelles, they are not likely to form complex in vivo (Figure 3, E–H).
Figure 3.
RCP interacts with Rab11 but not Rab4 in vivo. HeLa cells were transfected with ether RCP-YFP/CFP-Rab11A (A–D) or RCP-YFP/CFP-Rab4A (E–H), fixed and imaged. cFRET images were generated as described in MATERIALS AND METHODS. NFRET was calculated using the equation NFRET = cFRET/CFP and is an average from at least five randomly picked cells. The yellow signal in C and G represents the overlap between CFP and YFP. In I–H, HeLa cells were transfected with either myc-tagged Rab11-S25N (I and J) or myc-tagged Rab4-I121N (G and H) and costained with anti-myc (I and G) and anti-RCP (J and H) antibodies. Bars, 2 μm.
The FRET studies suggested that RCP and Rab4 may not interact in vivo. However, the lack of FRET between two proteins does not always mean lack of binding. Because subcellular localization of Rab effector proteins usually is dependent on their interactions with Rab GTPases, we tested the effect of Rab4 and Rab11 dominant negative mutants on RCP localization. To this end, we transected cells with either myc-tagged dominant negative Rab11 (S25N) or Rab4 (N121I) constructs. Consistent with our data, only Rab11-S25N resulted in partial redistribution of RCP from endosomes to the cytosol (Figure 3, I and J), whereas Rab4-N121I did not (Figure 3, G and H). Together, these data indicate that RCP interacts with Rab11, but not Rab4, in vivo.
The RCP–Rab11 Complex Is Localized to Recycling Endosomes
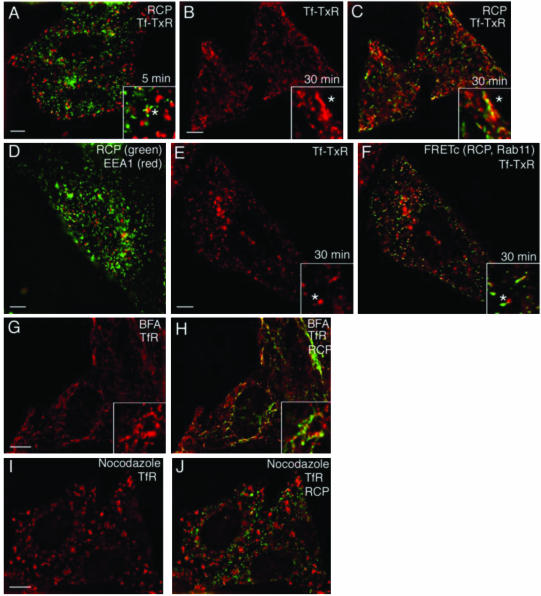
Subcellular localization often yields clues as to a protein's cellular function. To determine the localization of RCP, we bound Tf-TxR to HeLa cells at 4°C for 1 h, then washed and incubated them at 37°C with unlabeled Tf for either 5 or 30 min. These conditions are well known to label predominantly the EEs after 5 min and REs after 30 min. As shown in Figure 4 A, only a very small fraction of RCP colocalized with Tf-TxR after the 5-min incubation (11.5 ± 0.2% colocalization). Consistent with this, there was also very little colocalization between RCP and EEA1, a marker of EE (Figure 4D). Interestingly, although there was very little colocalization with the vesicular EEs, RCP staining could be seen in the adjacent tubules that seemed to be tubular extensions of the EE (Figure 4 A, asterisk in inset). After the 30-min incubation, RCP exhibited much better colocalization with Tf-TxR (47.6 ± 3.6% colocalization) (Figure 4, B and C), suggesting that RCP is present in REs but not EEs. To determine whether the RCP that is localized to RE is complexed with Rab11, we coimaged FRET between RCP–YFP and CFP–Rab11 with TfR and found substantial overlap (Figure 4, E and F). In addition, RCP–YFP/CFP–Rab11 complexes also could be detected on tubular extensions of vesicular endosomes, presumably EEs (Figure 4, E and F, asterisk in inset), supporting a possible role of the RCP–Rab11 complex in the regulation of protein recycling.
Figure 4.
Subcellular localization of RCP. Tf-TxR (A–C, red) was bound to HeLa cells for 1 h at 4°C and then incubated at 37°C for 5 min (A) or 30 min (B and C). Cells were then fixed and stained with anti-RCP antibodies (A and C, green). In D, fixed HeLa cells were costained with anti-RCP (green) and anti-EEA1 (red) antibodies. In E and F, HeLa cells transfected with RCP-YFP and CFP-Rab11A were incubated with Tf-TxR for 1 h at 4°C and then incubated at 37°C for 30 min. Colocalization between Tf-TxR (E and F, red) and RCP–YFP/CFP–Rab11 complex was then determined using FRET (F, green). In G–J, HeLa cells were treated with either nocodazole (I and J) or brefeldin A (G and H), fixed, and stained with anti-RCP (H and J, green) or anti-TfR (G–J, red) antibodies. The yellow signal in C, F, H, and J, represents overlap. Bars, 2 μm.
The dynamics of endocytic proteins in the presence of brefeldin A (BFA) and nocodazole can reveal the features of their native localization and trafficking patterns. BFA causes tubulation of the trans-Golgi network (TGN) and recycling endosomes (Lippincott-Schwartz et al., 1991; Robinson and Kreis, 1992), eventually collapsing them into a continuous network around the microtubule-organizing center (Lippincott-Schwartz et al., 1991). BFA treatment resulted in tubulation of RCP in a pattern reminiscent of the tubulation of RE (Figure 4, G and H) (Lippincott-Schwartz et al., 1991). Consistent with this, the RCP-containing tubules partially colocalized with TfR (49.8 ± 6.7% colocalization) but far less with EEA1 (10.2 ± 0.4% colocalization; our unpublished data). The microtubule-depolymerizing agent nocodazole blocks protein exit from EEs (Yamashiro et al., 1984), causing the accumulation of TfR in large peripheral EEs. However, RCP does not colocalize (only 7.1 ± 0.6% overlap) with these TfR-containing endosomes (Figure 4, I and J), being instead in small puncta dispersed through the entire cytoplasm (Figure 4H). Because the stability of RE also depends on the microtubular cytoskeleton, it is possible that these puncta represent fragmented REs.
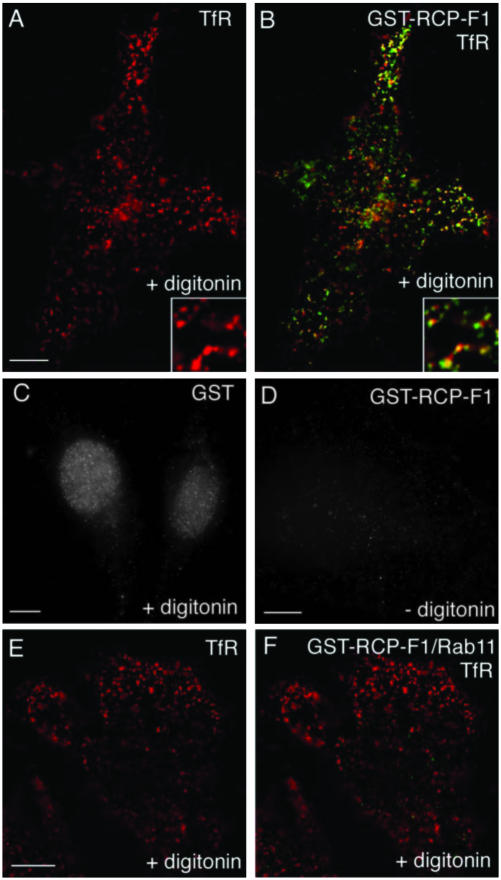
The immunofluorescence data suggest that RCP is recruited to REs, most likely via its interactions with Rab11. To test this, we used digitonin-permeabilized HeLa cell recruitment assays. To differentiate between endogenous and recombinant RCP, we used the GST-RCP-F1 fusion protein, which binds to Rab11 with ∼100 nM affinity (Figure 2). The GST-RCP-F1 protein was indeed recruited to endosomes containing TfR (62.2 ± 1.9% colocalization) (Figure 5, A and B). In contrast, EEA1-positive EEs did not recruit GST-RCP-F1 (8.2 ± 0.4% colocalization; our unpublished data). The endosomal binding was mediated by RCP-F1, because GST alone did not bind to endocytic membranes (Figure 5C) and was dependent on membrane permeabilization, because no signal was obtained in the absence of digitonin (Figure 5D). To test whether GST-RCP-F1 recruitment was dependent on binding to Rab11, recombinant soluble Rab11A or Rab4A (both lacking their geranylgeranylation motifs) were added to permeabilized cells along with GST-RCP-F1. As shown in Figure 5, E and F, the presence of soluble recombinant Rab11A inhibited GST-RCP-F1 recruitment to TfR-containing endosomes, whereas soluble Rab4A did not have any effect of GST-RCP-F1 recruitment (our unpublished data).
Figure 5.
RCP recruitment to endosomes is dependent on binding to Rab11. HeLa cells were incubated in the presence (A–C, E–F) or absence (D) of digitonin. Cells were then washed and incubated with either GST (C) or GST-RCP-F1 (A and B, D–F) in the presence (E and F) or absence (A–D) of soluble recombinant Rab11. Cells were fixed and stained with anti-GST (B and F, green; and C and D, gray) and anti-TfR (A and B and E and F, red) antibodies. Yellow in B and F represents the degree of overlap. Bars, 3 μm.
Overexpression of GFP-RCP-F1 Inhibits TfR Recycling
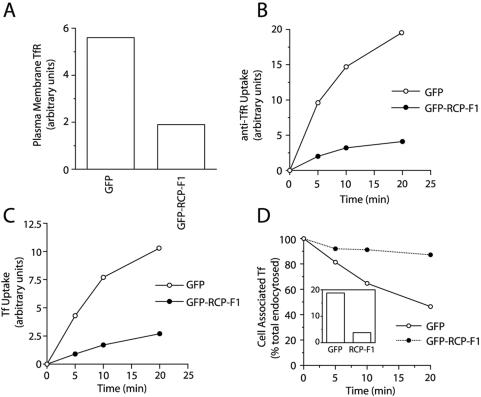
Our data suggest that the RCP–Rab11 complex is localized to recycling endosomes, where it may regulate protein recycling. To test this, we used FACS analysis to measure intracellular trafficking of TfR in wild-type or GFP-RCP-F1–expressing HeLa cells (for details, see MATERIALS AND METHODS and Supplemental Figure 3). We have shown that the RCP-F1 truncation mutant binds Rab11 in vitro and in vivo (Figures 1, 2, and 5), thus it should act as a dominant negative mutant by competing with endogenous RCP for binding to Rab11. To this end, we measured the effect of GFP-RCP-F1 overexpression on the levels of plasma membrane TfR as well as TfR endocytosis. To measure the levels of plasma membrane TfR, we incubated HeLa cells at 4°C with PE-conjugated anti-TfR antibodies (anti-TfR-PE). As shown in Figure 6A, the overexpression of GFP-RCP-F1 significantly decreased the amount of TfR present at the plasma membrane. To measure the rate of TfR endocytosis, we incubated HeLa cells at 37°C in the continuous presence of anti-TfR-PE. Consistent with the possible role of RCP in TfR traffic, overexpression of GFP-RCP-F1 inhibited anti-TfR-PE uptake (Figure 6B). Similar results also were obtained when we used Alexa647-conjugated transferrin (Tf-Alexa647) (Figure 6C).
Figure 6.
Overexpression of RCP-F1 inhibits Tf recycling and uptake. (A and B) HeLa cells expressing either GFP or GFP-RCP-F1 were incubated at 4°C for 30 min with 2 μg/ml anti-TfR-PE antibody. The amount of plasma membrane bound anti-TfR-PE antibody after this incubation was quantitated by FACS and is shown in A. Cells were then incubated at 37°C for varying amounts of time in the continuous presence of 2 μg/ml anti-TfR-PE antibody. The amount of internalized anti-TfR-PE antibody was quantitated by FACS analysis and is shown in B. The data shown are the means of at least three independent experiments. To better compare the rates of uptake, the plasma membrane-bound anti-TfR-PE after 4°C incubation was subtracted from the data in A. (C) HeLa cells expressing either GFP or GFP-RCP-F1 were incubated at 4°C for 30 min with 20 μg/ml Tf-Alexa647. Cells were then incubated at 37°C for varying amounts of time in the continuous presence of 20 μg/ml Tf-Alexa647. The amount of internalized Tf-Alexa647 was quantitated by FACS analysis. The data shown are the means of at least three independent experiments. (D) HeLa cells expressing either GFP or GFP-RCP-F1 were incubated for 30 min with Tf-Alexa647 at 4°C. Cells were then incubated for an additional 20 min at 37°C in the presence of Tf-Alexa647. The amounts of internalized Tf-Alexa647 after incubation at 37°C are shown in the inset. Cells were then washed and incubated at 37°C for varying amounts of time in the presence of unlabeled Tf. The Tf-Alexa647 remaining in the cells was quantitated using FACS analysis and expressed as a percentage of total endocytosed (at time 0) Tf-Alexa647.
The reduction of plasma membrane TfR as well as inhibition of anti-TfR-PE and Tf-Alexa647 uptake could be the result of impaired recycling by trapping the TfR in endosomes. Indeed, FACS analysis of Tf-Alexa647 recycling showed that overexpression of GFP-RCP-F1 inhibited the release of cell-associated Tf-Alexa647 (Figure 6D).
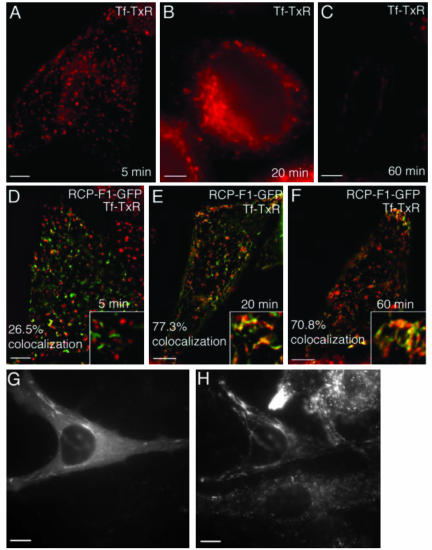
To confirm our FACS analysis of TfR recycling, we bound untransfected or GFP-RCP-F1–expressing HeLa cells with Tf-TxR for1hat4°C and then incubated in media containing unlabeled Tf for 5, 20, and 60 min before fixation and imaging. After 5 min in both untransfected (Figure 7A) and GFP-RCP-F1–expressing cells (Figure 7D) Tf-TxR was localized predominantly in EEs. These EEs were largely devoid of GFP-RCP-F1 (26.5% colocalization). After 20-min “chase,” a large portion of the Tf-TxR could be observed in tubular endosomes containing GFP-RCP-F1 (77.3% colocalization). Interestingly, the majority of Tf-TxR in untransfected HeLa cells was in perinuclear recycling endosomes (Figure 7B), whereas in the cells overexpressing GFP-RCP-F1, it was found in tubular organelles that accumulated in the periphery of the cell (Figure 7E). These organelles are likely recycling endosomes, because it was previously reported that overexpression of the RCP-F1 fragment causes tubulation of this compartment (Lindsay et al., 2002). Sixty-minute chase resulted in almost complete loss of Tf-TxR fluorescence in nontransfected HeLa cells (Figure 7C), whereas cells expressing GFP-RCP-F1 retained Tf-TxR in a pattern similar to that after a 20-min chase, confirming that RCP-F1 overexpression resulted in accumulation of Tf-TxR in tubular endosomes (Figure 7F). Staining of GFP-RCP-F1–expressing cells with anti-TfR antibody revealed that TfR also was predominantly accumulated in endocytic tubules (Figure 6, G and H). Similar GFP-RCP-F1–induced tubulation also was observed for low-density lipoprotein receptor, suggesting that RCP may regulate the bulk recycling of plasma membrane receptor proteins (Supplemental Figure 3, G and H).
Figure 7.
Overexpression of GFP-RCP-F1 inhibits TfR recycling from endosomes to the plasma membrane. HeLa cells were mock transfected (A–C) or transfected with GFP-RCP-F1 (D–F). Cells were incubated with Tf-TxR for 1 h at 4°C; washed; incubated at 37°C for 5 min (A and D), 20 min (B and E), and 60 min (C and F); and fixed. The localization and fluorescence intensities of Tf-TxR and GFP-RCP-F1 were then imaged. The colocalization between Tf-TxR and RCP-F1-GFP are the means of five randomly picked cells. In G and H, HeLa cells were transfected with GFP-RCP-F1 (G) and stained with anti-TfR antibodies (H).
RCP Regulates TfR Sorting to the Recycling Pathway Instead of the Degradative Pathway
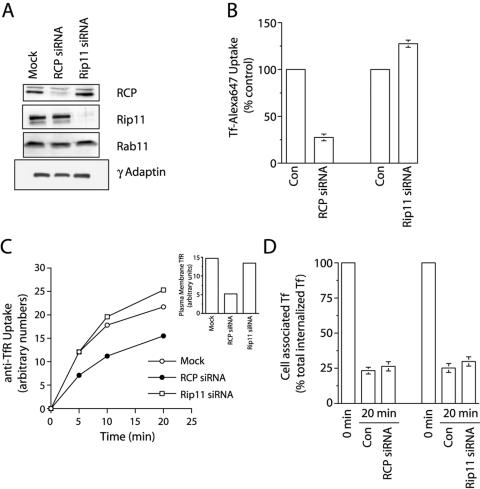
Overexpression of the dominant negative mutant of RCP, RCP-F1, inhibited Tf recycling from REs to plasma membranes. However, it is likely that the effect of RCP-F1 is due to sequestration of Rab11 from other FIP proteins, rather than direct inhibition of endogenous RCP. Indeed, we have previously reported that overexpression of RBD domain can sequester Rab11 (Meyers and Prekeris, 2002). Furthermore, overexpression of the equivalent dominant negative truncation mutant of Rip11 (aa 490–652) also inhibited Tf uptake and recycling in FACS assays (our unpublished data). Thus, it is likely that overexpression of the RBD from all FIPs inhibits Rab11 and does not represent the RCP function. To test more specifically whether RCP regulates Tf recycling, we knocked-down RCP and Rip11 by using siRNAs based on the human RCP and Rip11 sequences. As shown in Figure 8A, siRNA treatment knocked-down RCP and Rip11 levels by >90%, as assessed by Western blotting. The knock-down was specific, because RCP and Rip11 siRNAs had no effect on each other, nor on Rab11 or γ-adaptin (Figure 8A). To test the effect of RCP and Rip11 knock-down on TfR recycling, RCP siRNA or Rip11 siRNA-transfected HeLa cells were incubated for 30 min at 4°C and then for varying time intervals at 37°C in the presence of Tf-Alexa647 and analyzed by FACS. Consistent with the role of RCP in regulating TfR traffic, RCP but not Rip11 knock-down significantly inhibited anti-Tf-PE antibody uptake (Figure 8B). Similar results were also obtained with anti-TfR-PE (Figure 8C).
Figure 8.
Effect of RCP knock-down on TfR endocytosis, lysosomal degradation and recycling. (A) HeLa cells were either mock transfected or transfected with Rip11 or RCP siRNAs, and Triton X-100 extracts of them were analyzed by Western blotting with anti-RCP, anti-Rip11, anti-Rab11, and anti-γ adaptin antibodies to determine the extent and specificity of the knock-down. (B) HeLa cells were either mock transfected (control) or transfected with Rip11 or RCP siRNAs. After 72 h, cells were incubated for 30 min at 4°C with 20 μg/ml Tf-Alexa647. Cells were then incubated for additional 30 min at 37°C in the presence of Tf-Alexa647. Cells were then washed, fixed in 3% paraformaldehyde, and amounts of internalized Tf-Alexa647 were quantitated by FACS. The data presented are means ± SE of three independent experiments. (C) HeLa cells were either mock transfected or transfected with Rip11 or RCP siRNAs. After 72 h, cells were incubated for 30 min at 4°C with 2 μg/ml anti-TfR-PE antibody. The amount of plasma membrane bound anti-TfR-PE antibody is shown in the inset. Cells were then moved to at 37°C and incubated for varying amounts of time in the presence of 20 μg/ml Tf-Alexa647. Cells were then washed, fixed in 3% paraformaldehyde, and the time course of anti-TfR-PE antibody internalization was quantitated by FACS analysis. (D) HeLa cells were either mock transfected (control) or transfected with Rip11 or RCP siRNAs. After 72 h, cells were incubated for 30 min with Tf-Alexa647 at 4°C followed by additional 20-min incubation at 37°C. Cells were then washed and incubated at 37°C for 20 min in the presence of unlabeled Tf. The amounts of cell associated Tf-Alexa647 was quantitated by FACS. The data presented are means ± SE of three independent experiments.
To test whether the decrease in anti-TfR-PE uptake is due to the inhibition of TfR recycling, we measured the release of cell-associated Tf-Alexa647 from HeLa cells treated with RCP or Rip11 siRNA. Surprisingly, RCP knock-down had no effect on Tf-Alexa647 recycling (Figure 8D). Because RCP knock-down had no effect on Tf-Alexa647 recycling, we tested whether RCP siRNA treatment caused changes in the levels of the TfR. As shown in Figure 9A, inset, RCP knock-down resulted in a dramatic decrease in the total levels of TfR.
Figure 9.
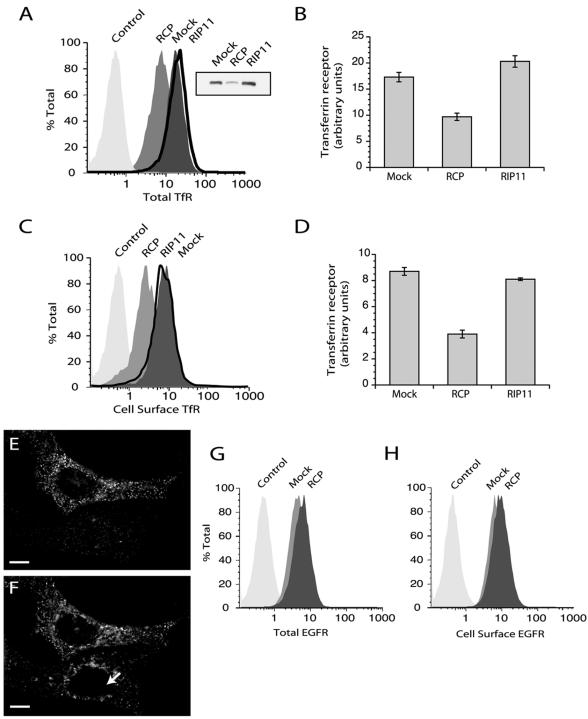
RCP knock-down increases lysosomal degradation of TfR but not EGFR. (A and C) HeLa cells were either mock transfected or transfected with RCP or Rip11 siRNAs. After 74-h incubation, cells were either permeabilized with saponin to measure total TfR (A) or nonpermeabilized to measure cell surface TfR (C), stained with anti-TfR-PE antibodies, and quantitated by FACS analysis or Western blotting (A, inset). (B and D) Quantitation of data from A and C. The presented data are means ± SD of three independent experiments. (E and F) Cells were transfected with RCP siRNA, fixed, and stained with anti-RCP (E) and anti-TfR (F) antibodies. Arrow points to a HeLa cell with down-regulated RCP. Bars, 2 μM. (G and H) HeLa cells were either mock transfected or transfected with RCP siRNAs. After 74-h incubation, cells were either permeabilized to measure total EGFR (G) or nonpermeabilized to measure cell surface EGFR (H). The levels of EGFR were quantitated by FACS analysis.
To confirm the immunoblotting data, we stained mock-, RCP-, or Rip11 siRNA-treated cells with anti-TfR antibodies in the presence or absence of saponin to determine the effects on total and plasma membrane-associated amounts of TfR. Cell staining was quantitated using FACS analysis. As shown in Figure 9, A and B, RCP knock-down resulted in a significant decrease in total TfR levels, which was confirmed by immunofluorescence microscopy (Figure 9, E and F, arrow points to cell transfected with RCP siRNA). A similar decrease also was seen in TfR levels at the plasma membrane (Figure 9, C and D). The ratio between total and cell surface TfR in RCP siRNA transfected cells was not different to the ratio in mock-transfected cells, confirming that the rates of TfR endocytosis and exocytosis are not affected by RCP knock-down. It is possible that the loss of RCP leads to the missorting of the TfR, so it is transported to lysosomes for degradation. Indeed, treatment of HeLa cells with either bafilomycin (an inhibitor of late endosome acidification) or leupeptin and pepstatin (inhibitors of lysosomal proteases) resulted in increased levels of cellular TfR (Supplemental Figure 4B). Thus, RCP seems to play a role in sorting of TfR away from the lysosomal degradation pathway into recycling pathway.
EGFR is known to be endocytosed upon ligand binding and sorted in EEs for either degradation in lysosomes or recycling back to the plasma membrane. Although EGFR endocytosis seems to use a different molecular machinery than TfR, it remains unclear whether sorting and recycling of EGFR uses the same transport vesicles/tubules (Marks et al., 1996). To test this, we used FACS analysis to determine whether RCP knock-down has an effect on the levels of cellular EGFR. In contrast to TfR, RCP knock-down did not cause a significant decrease in EGFR levels (Figure 9, G and H), suggesting that EGFR and TfR are perhaps not recycled in the same vesicles.
DISCUSSION
A key step in understanding endocytic traffic is the characterization of interactions between Rab GTPases and their effector proteins. Emerging data suggest that Rab11 may play a role in multiple membrane transport steps by forming mutually exclusive complexes with various effector proteins (Meyers and Prekeris, 2002). In this study, we demonstrate that RCP, a member of FIPs, plays a key role in regulating sorting TfR in recycling endosomes.
RCP was originally identified in a yeast two-hybrid screen as a putative dual Rab4 and Rab11 interacting protein (Lindsay et al., 2002). However, our in vitro binding experiments clearly demonstrate that RCP cannot act as a Rab11 and Rab4 coupling protein, because these two Rabs compete with each other for binding to RCP. Surprisingly, all our experiments indicated that RCP binds to Rab4 with very low affinity compared with Rab11, suggesting that Rab4 may not interact with RCP in vivo. The discrepancy with yeast two-hybrid data (Lindsay et al., 2002) is likely explained by the sensitivity of two-hybrid system, allowing detection of the low-affinity RCP–Rab4 interaction. Although the low affinity interaction with Rab4 could be compensated by a higher cellular concentration of Rab4 than Rab11, this seems not to be the case in HeLa cells because both Rab GTPases are expressed in comparable amounts (Rab11, 270 nM; Rab4, 120 nM). Interestingly, we estimate cellular concentration of RCP in HeLa cells to be 530 nM, sufficient for the recruitment to the membranes by Rab11-GTP (binding affinity 105 nM), but not Rab4-GTP (binding affinity 20,800 nM). Consistent with this, FRET experiments only detected RCP interactions with Rab11 but not Rab4 in vivo. Furthermore, overexpression of Rab4 dominant negative mutant does not have any effect on RCP distribution. Thus, our data suggest that RCP primarily acts by binding to Rab11, although we cannot fully discount the possibility that some of RCP interactions with Rab4 may occur in vivo.
If RCP does not serve as a Rab11 and Rab4 coupling protein, what is its role in endocytic membrane traffic? The most surprising finding in this study is that RCP siRNA dramatically altered the cellular levels of the TfR while having no effect on TfR transport from REs to the plasma membrane. This is an unexpected finding, because traditionally Rab11 has been implicated in protein transport from REs to plasma membrane rather than sorting. Indeed, overexpression of the RBD domain, an inhibitor of the Rab11 GTPase, does dramatically inhibit TfR delivery to the plasma membrane by trapping it in RE. Thus, Rab11 probably regulates multiple steps of protein recycling. It is tempting to speculate that differential interaction with various FIP proteins may regulate different endocytic traffic steps. Our studies indicate that the Rab11–RCP complex may be involved in sorting of proteins from degradative to recycling pathway.
Because RCP does not colocalize with vacuolar EEs, how can it regulate the sorting of proteins from there? One possibility is that RCP is recruited to the tubular EE extensions that mediate protein traffic to RE. Indeed, we occasionally observed RCP-containing tubules in proximity to vacuolar EEs, although we could not determine whether these tubules were actually connected to vacuolar EEs due to the limitations of light microscopy. The formation of EE tubular extensions is thought to mediate membrane protein (including TfR) sorting away from internal soluble EE proteins via high surface-to-volume ratio. Thus, the RCP–Rab11 complex may play a role in initiation and extension of these tubules. The inhibition of EE tubule formation would result in accumulation of TfR in EEs and eventual degradation in lysosomes. Another possibility is that the RCP–Rab11 complex regulates protein sorting at RE. Emerging evidence suggests that at the RE, specific proteins are loaded into distinct carrier vesicles, a process that could be governed by different Rab11–FIP complexes. A defect in TfR sorting into such carrier vesicles could lead to the transport of TfR to the lysosomes via the AP-3 coat-dependent transport pathway, although the existence of lysosomal sorting from RE is still controversial (Dell'Angelica et al., 1998, 1999; Peden et al., 2004). It should be noted that this RCP-dependent recycling pathway is not the only means of TfR transport to plasma membrane, because RCP only partially colocalizes with TfR and its knock-down did not affect TfR recycling. At least part of TfR is recycled directly from the EE to the plasma membrane via a fast recycling pathway (Mellman, 1996; Robinson et al., 1996). It is also likely that TfR can enter several different types of carrier vesicles at RE. Thus, blocking the RCP–Rab11 recycling pathway may only cause partial TfR missorting to lysosomes. Nevertheless, because the rate of TfR recycling is much higher then the half-life of the TfR molecule, a small increase in lysosomal missorting at every round of recycling could lead to a significant increase in TfR degradation (Weissman et al., 1986).
Interestingly, knock-down of Rip11 by using siRNA stimulated Tf uptake. We have previously shown that Rip11 does not colocalize with TfR in epithelial cells (Prekeris et al., 2000). Because Rip11 also has a limited colocalization with TfR in HeLa cells (our unpublished data), it is not likely that Rip11 plays a direct role in TfR endocytic traffic. One possible explanation for the increased uptake is that Rip11 and RCP may compete for binding to Rab11. Indeed, we previously showed that these and other FIPs form mutually exclusive complexes with Rab11 (Meyers and Prekeris, 2002). Because small Rab GTPases, including Rab11, usually are not very abundant, it is likely that various FIPs would compete for Rab11 binding. Consistent with this, overexpression of the RBD domain of various FIPs has been shown to sequester Rab11 and have strong inhibitory effect on a variety of Rab11-dependent membrane trafficking steps (Meyers and Prekeris, 2002). Thus, it is possible that knock-down of Rip11, an abundant protein in HeLa cells, would liberate Rab11, allowing it to interact with RCP and regulate TfR sorting.
In summary, our data suggest that RCP is a Rab11 effector protein that regulates TfR sorting. RCP is likely to be recruited either to the tubular extensions of the EE or directly to the RE though interactions with Rab11. Formation of the RCP–Rab11 complex then regulates the sorting of TfR away from the degradative lysosomal pathway, directing it instead to the plasma membrane. Because the Rip11–Rab11 protein complex does not directly affect TfR transport from REs to the plasma membrane, it is tempting to speculate that recruitment of specific Rab11–FIP complexes to the EE or RE may play a role in the endocytic sorting of different cargo molecules. However, many questions remain to be answered. Although the RCP–Rab11 interaction is probably partly regulated by competition with other FIP proteins that regulate sorting and traffic of distinct cargo proteins, other regulatory mechanisms remain to be determined. Further characterization of the roles and regulation of different Rab11–FIP complexes will shed light on the mechanisms governing endocytic protein sorting and recycling.
Acknowledgments
We thank Gayle Wilson and Drs. Kathryn Howell and Suzie Scales for critical reading of the manuscript. We also are grateful to Drs. Alexander Sorkin for providing CFP-Rab11a, CFP-Rab4, and CFP-Rab5 constructs; Mary McCaffrey for human RCP cDNA; and Dr. Peter van der Sluijs for providing GST-Rabip4 construct. This work was supported in part by the grants from American Diabetes Association J02JF37 (to R.P.) and Howard Hughes Medical Institute Junior Faculty Award (to R.P.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-12-0918. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-12-0918.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Callaghan, J., Simonsen, A., Gaullier, J.M., Toh, B.H., and Stenmark, H. (1999). The endosome fusion regulator early-endosomal autoantigen 1 (EEA1) is a dimer. Biochem. J. 338, 539-543. [PMC free article] [PubMed] [Google Scholar]
- Chavrier, P., and Goud, B. (1999). The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 11, 466-475. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., Zerial, M. (2000). Purification and identification of novel Rab effectors using affinity chromatography. Methods 20, 403-410. [DOI] [PubMed] [Google Scholar]
- Cox, D., Lee, D.J., Dale, B.M., Calafat, J., and Greenberg, S. (2000). A Rab11-containing rapidly recycling compartment in macrophages that promotes phagocytosis. Proc. Natl. Acad. Sci. USA 97, 680-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis, D.N., Philip, B., Baleja, J.D., and Feig, L.A. (2002). Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J. Biol. Chem. 277, 49158-49166. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., Klumperman, J., Stoorvogel, W., and J.S. Bonifacino. (1998). Association of the AP-3 adaptor complex with clathrin. Science 280, 431-434. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., Shotelersuk, V., Aguilar, R.C., Gahl, W.A., and J.S. Bonifacino. (1999). Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A-subunit of the AP-3 adaptor. Mol. Cell. 3, 11-21. [DOI] [PubMed] [Google Scholar]
- de Renzis, S., Sonnichsen, B., and Zerial, M. (2002). Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat. Cell Biol. 4, 124-133. [DOI] [PubMed] [Google Scholar]
- Fendly, B.M., Winget, M., Hudziak, R.M., Lipari, M.T., Napier, M.A., and Ullrich, A. (1990). Characterization of murine monoclonal antibodies reactive to either the human epidermal growth factor receptor of HER2/neu gene product. Cancer Res. 50, 1550-1558. [PubMed] [Google Scholar]
- Fouraux, M.S., Deneka, M., Ivan, V., van der Heijden, A., Raymackers, J., van Suylekom, D., van Venrooij, W.J., van der Sluijs, P., and Pruijn, G.J. (2004). Rabip4 is an effector of Rab5 and Rab4 and regulates transport through early endosomes. Mol. Biol. Cell 15, 611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, L., Jr., and Scheller, R.H. (1999). Regulation of membrane trafficking: structural insights from a Rab/effector complex. Cell 96, 755-758. [DOI] [PubMed] [Google Scholar]
- Hales, C.M., Griner, R., Hobdy-Henderson, K.C., Dorn, M.C., Hardy, D., Kumar, R., Navarre, J., Chan, E.K., Lapierre, L.A., and Goldenring, J.R. (2001). Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276, 39067-39075. [DOI] [PubMed] [Google Scholar]
- Hickson, G.R.X., Matheson, J., Riggs, B., Maier, V.H., Fielding, A.B., Prekeris, R., Sullivan, W., Barr, F.A., and Gould, G.W. (2003). Arfophilins are dual Arf/Rab11 binding proteins that regulate recycling endosome distribution and are related to Drosophila nuclear fallout. Mol. Biol. Cell 14, 2908-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, A., Tomas, E., Immler, D., Meyer, H.E., Zorzano, A., and Eckel, J. (2000). Rab11 is associated with GLUT4-containing vesicles and redistributes in response to insulin. Diabetologia 43, 1518-1527. [DOI] [PubMed] [Google Scholar]
- Lapierre, L.A., Kumar, R., Hales, C.M., Navarre, J., Bhartur, S.G., Burnette, J.O., Provance, D.W. Jr., Mercer, J.A., Bahler, M., and Goldenring, J.R. (2001). Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 12, 1843-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, A.J., Hendrick, A.G., Cantalupo, G., Senic-Matuglia, F., Goud, B., Bucci, C., and McCaffrey, M.W. (2002). Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 277, 12190-12199. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Yuan, L., Tipper, C., Amherdt, M., Orci, L., and Klausner, R.D. (1991). Brefeldin A's effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 67, 601-616. [DOI] [PubMed] [Google Scholar]
- Marks, M.S., Woodruff, L., Ohno, H., and Bonifacino, J.S. (1996). Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J. Cell Biol. 135, 341-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman, I. (1996). Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12, 575-625. [DOI] [PubMed] [Google Scholar]
- Meyers, J.M., and Prekeris, R. (2002). Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J. Biol. Chem. 277, 49003-49010. [DOI] [PubMed] [Google Scholar]
- Peden, A.A., Oorschot, V., Hesser, B.A., Austin, C.D., Scheller, R.H., and J. Klumperman. (2004). Localisation of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 164, 1065-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris, R. (2003). Rabs, Rips, FIPs, and endocytic membrane traffic. Sci. World J. 3, 870-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris, R., Davies, J.M., and Scheller, R.H. (2001). Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 276, 38966-38970. [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Klumperman, J., Chen, Y.A., and Scheller, R.H. (1998). Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 143, 957-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris, R., Klumperman, J., and Scheller, R.H. (2000). A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell 6, 1437-1448. [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Yang, B., Oorschot, V., Klumperman, J., and Scheller, R.H. (1999). Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol. Biol. Cell 10, 3891-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.S., and Kreis, T.E. (1992). Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: effects of brefeldin A and G protein activators. Cell. 69, 129-138. [DOI] [PubMed] [Google Scholar]
- Robinson, M.S., Watts, C., and Zerial, M. (1996). Membrane dynamics in endocytosis. Cell 84, 13-21. [DOI] [PubMed] [Google Scholar]
- Shin, O.H., Ross, A.H., Mihai, I., and Exton, J.H. (1999). Identification of arfophilin, a target protein for GTP-bound class II ADP-ribosylation factors. J. Biol. Chem. 274, 36609-36615. [DOI] [PubMed] [Google Scholar]
- Sorkin, A., McClure, M., Huang, F., and Carter, R. (2000). Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr. Biol. 10, 1395-1398. [DOI] [PubMed] [Google Scholar]
- Ullrich, O., Reinsch, S., Urbe, S., Zerial, M., and Parton, R.G. (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135, 913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs, P., Hull, M., Webster, P., Male, P., Goud, B., and Mellman, I. (1992). The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70, 729-740. [DOI] [PubMed] [Google Scholar]
- Wallace, D.M., Lindsay, A.J., Hendrick, A.G., and McCaffrey, M.W. (2002). The novel Rab11-FIP/Rip/RCP family of proteins displays extensive homo- and hetero-interacting abilities. Biochem. Biophys. Res. Commun. 292, 909-915. [DOI] [PubMed] [Google Scholar]
- Wang, X., Kumar, R., Navarre, J., Casanova, J.E., and Goldenring, J.R. (2000). Regulation of vesicle trafficking in Madin-Darby canine kidney cells by Rab11a and Rab25. J. Biol. Chem. 275, 29138-29146. [DOI] [PubMed] [Google Scholar]
- Weissman, A.M., Klausner, R.D., Rao, K., and Harford, J.B. (1986). Exposure of K562 Cells to anti-receptor monoclonal antibody OKT9 results in rapid redistribution and enhanced degradation of transferrin receptor. J. Cell Biol. 102, 951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcke, M., Johannes, L., Galli, T., Mayau, V., Goud, B., and Salamero, J. (2000). Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 151, 1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro, D.J., Tycko, B., Fluss, S.R., and Maxfield, F.R. (1984). Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell 37, 789-800. [DOI] [PubMed] [Google Scholar]