Abstract
Delivery of proteins and organelles to the vacuole by autophagy and the cytoplasm to vacuole targeting (Cvt) pathway involves novel rearrangements of membrane resulting in the formation of vesicles that fuse with the vacuole. The mechanism of vesicle formation and the origin of the membrane are complex issues still to be resolved. Atg18 and Atg21 are proteins essential to vesicle formation and together with Ygr223c form a novel family of phosphoinositide binding proteins that are associated with the vacuole and perivacuolar structures. Their localization requires the activity of Vps34, suggesting that phosphatidylinositol(3)phosphate may be essential for their function. The activity of Atg18 is vital for all forms of autophagy, whereas Atg21 is required for the Cvt pathway but not for nitrogen starvation-induced autophagy. The loss of Atg21 results in the absence of Atg8 from the pre-autophagosomal structure (PAS), which may be ascribed to a reduced rate of conjugation of Atg8 to phosphatidylethanolamine. A similar defect in localization of a second ubiquitin-like conjugate, Atg12-Atg5, suggests that Atg21 may be involved in the recruitment of membrane to the PAS.
INTRODUCTION
The eukaroytic cell is a complex, living unit that depends on the spatial confinement of a variety of specialized tasks to specific compartments or organelles. Various strategies have been developed by the cell to accommodate the efficient and specific sorting of the proteins that define the function of each of these compartments, and most of these involve the recognition of a signal within the protein to be targeted by a receptor or an adaptor. One such protein sorting event in yeast is the cytoplasm to vacuole targeting (Cvt) pathway (reviewed in Strømhaug and Klionsky, 2004). This biosynthetic pathway is used for the delivery of the precursor form of the resident hydrolase aminopeptidase I (prApe1) to the vacuole. Precursor Ape1 is synthesized on free ribosomes in the cytosol and rapidly forms dodecamers that aggregate into a large complex (the Ape1 complex). The sorting signal in prApe1 is then recognized by the receptor/adaptor protein Atg19 to form a Cvt complex. Atg19 further interacts with Atg11, which is a large coiled-coil protein that functions in part to recruit prApe1 to the pre-autophagosomal structure (PAS); the PAS is thought to be a nucleating or organizing site for formation of a double membrane vesicle that sequesters the Cvt complex and subsequently fuses with the vacuole to deliver the cargo into the lumen.
Several rounds of mutagenesis as well as the screening of diploid and haploid yeast deletion libraries have resulted in the identification of >20 genes required for the formation of the Cvt vesicle (reviewed in Klionsky et al., 2003; Strømhaug and Klionsky, 2004). The majority of these genes also are required for the formation of autophagosomes. Autophagosomes are also double membrane vesicles, but they are considerably larger than Cvt vesicles and contain cytoplasm destined for degradation within the vacuole. Autophagy and the Cvt pathway therefore serve opposite purposes but share mostly the same machinery for vesicle formation. Autophagy is furthermore a ubiquitous pathway present in all eukaryotic cells, whereas the Cvt pathway seems to have evolved specifically in Saccharomyces cerevisiae for the sorting and transport of aminopeptidase I. However, due to the genetic and mechanistic overlap with autophagy, any discovery regarding the molecular mechanism of the Cvt pathway is pertinent to all eukaryotic cells.
The study of the Cvt pathway and autophagy in yeast has started to reveal the molecular basis for a novel vesicle formation process involving several unique components. One of these components is a protein complex that contains the phosphatidylinositol (PtdIns) 3-kinase activity of Vps34 (Herman and Emr, 1990). Vps34 bound to Vps15 is required for a number of vesicular transport events in the cell, and a complex also containing Vps30/Atg6 and Atg14 is essential for the formation of Cvt vesicles and autophagosomes (Kihara et al., 2001). The activity of the PtdIns 3-kinase complex is required for the recruitment of several Atg proteins to the PAS, including Atg8.
Atg8 has been proposed to play a role in a membrane expansion phase after nucleation of the autophagosome (Abeliovich et al., 2000; reviewed in Noda and Ohsumi, 2004). Atg8 is a small protein containing a three-dimensional structure resembling ubiquitin (Paz et al., 1999). The Atg8 protein undergoes a series of posttranslational modifications resulting in conjugation to phosphatidylethanolamine (PE), thereby anchoring Atg8 to membranes. Stability of the conjugate in vivo depends on the presence of a protein conjugate of Atg12 and Atg5. The presence of the Atg12-Atg5 conjugate is critical for vesicle formation; however, some autophagosomes of reduced size may form upon starvation in the absence of Atg8 (Abeliovich et al., 2000).
In the current work, we have characterized the Atg21 protein as being required for efficient localization and lipidation of Atg8. Furthermore, Atg8 cannot be detected at the PAS by fluorescent microscopy in the atg21 mutant. Atg21 belongs to a novel family of phosphoinositide binding proteins localized to the vacuole and vacuole-associated structures. Although another member of this family, Atg18, is required for both the Cvt pathway and autophagy (Barth et al., 2001; Guan et al., 2001), Atg21 is required for the Cvt pathway, but not for autophagy induced by nitrogen starvation.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions
The yeast strains used in this study are listed in Table 1. Synthetic minimal (SMD), nitrogen starvation (SD-N), oleic acid (YTO), synthetic glycerol (SGd), and rich (YPD) media were as described previously (Hutchins et al., 1999). S. cerevisiae strains were generally grown at 30°C to early mid-log phase. Temperature-sensitive strains were grown at 26°C and shifted to 38°C for the appropriate time.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | Robinson et al. (1988) |
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | ResGen |
| TN124 | MATα leu2-3,112 trp1 ura3-52 pho8::pho8Δ60 pho13::LEU2 | Noda et al. (1995) |
| TN125 | MATα leu2-3,112 trp1 ura3-52 pho8::pho8Δ60 pho13::TRP1 | Noda et al. (1995) |
| AHY001 | SEY6210; atg11Δ::HIS3 | Kim et al. (2001b) |
| atg11Δ | BY4742; atg11Δ::Kan | ResGen |
| atg18Δ | BY4742; atg18Δ::Kan | ResGen |
| atg21Δ | BY4742; atg21Δ::Kan | ResGen |
| EMY119 | SEY6210; fab1-2 | Yamamoto et al. (1995) |
| JGY3 | SEY6210; atg18Δ::HIS5 S.p. | Guan et al. (2001) |
| JGY20 | TN125; atg18Δ::LEU2 | This study |
| JKY007 | SEY6210; atg9Δ::HIS3 | Noda et al. (2000) |
| pep4Δ | BY4742; pep4Δ::Kan | ResGen |
| PJ69-4A | MATα leu2-3,112 trp1-Δ901 ura3-52 his3-Δ200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
| PSY2 | BY4742; atg21Δ::Kan pep4Δ::URA3 | This study |
| PSY5 | SEY6210; atg21Δ::Kan | This study |
| PSY6 | SEY6210; atg21Δ::HIS3 S.k. | This study |
| PSY7 | SEY6210; atg21Δ::TRP1 | This study |
| PSY62 | SEY6210; ATG18-GFP::HIS3 S.k. | This study |
| PSY63 | SEY6210; ATG21-GFP::HIS3 S.k. | This study |
| PSY66 | SEY6210; ATG5-HA::HIS3 S.k. | This study |
| PSY100 | SEY6210; YGR223c-GFP::HIS3 S.k. | This study |
| PSY107 | TN124; atg21Δ::TRP1 | This study |
| PSY128 | SEY6210; atg4Δ::LEU2 atg21Δ::TRP1 | This study |
| PSY139 | SEY6210; atg8Δ::LEU2 atg21Δ::TRP1 | This study |
| PSY146 | SEY6210; fab1-2tsf ATG18-GFP::HIS3 S.k. | This study |
| PSY155 | SEY6210; ATG5-HA::HIS3 S.k. atg21Δ::TRP1 | This study |
| PSY167 | SEY6210; atg21Δ::Kan atg18Δ::LEU2 | This study |
| PSY190 | SEY6210; atg11Δ::HIS3 atg21Δ::Kan | This study |
| PSY191 | SEY6210; atg9Δ::HIS3 atg21Δ::Kan | This study |
| PSY226 | SEY6210 atg8Δ::LEU2 atg21Δ::HIS3 GFP-ATG8::URA3 | This study |
| PSY227 | SEY6210; ATG1-GFP::HIS3 S.k. atg21Δ::TRP1 RFP-APE1::LEU2 | This study |
| PSY228 | SEY6210; ATG14-GFP::HIS3 S.k. atg21Δ::TRP1 RFP-APE1::LEU2 | This study |
| PSY285 | SEY6210; atg8Δ::LEU2 ATG5-GFP::HIS3 S.k. RFP-APE1::URA3 | This study |
| PSY286 | SEY6210; atg8Δ::LEU2 ATG5-GFP::HIS3 S.k. atg21Δ::TRP1 RFP-APE1::URA3 | This study |
| PSY290 | SEY6210; fab1-2tsf ATG21-GFP::HIS3 S.k. | This study |
| PSY291 | SEY6210; vps34Δ::TRP1 VPS34tsf-416 ATG18-GFP::HIS3 S.k. | This study |
| PSY292 | SEY6210; vps34Δ::TRP1 VPS34tsf-416 ATG21-GFP::HIS3 S.k. | This study |
| PSY293 | SEY6210; vps34Δ::TRP1 VPS34tsf-416 YGR223c-GFP::HIS3 S.k. | This study |
| PSY303 | PJ69-4A; atg21Δ::TRP1 | This study |
| vps34tsf | SEY6210; vps34Δ::TRP1 pvps34tsf::URA3 | Stack et al. (1995) |
| WPHYD2 | SEY6210; atg4Δ::LEU2 | Kim et al. (2001a) |
| WPHYD7 | SEY6210; atg8Δ::LEU2 | Kim et al. (2001a) |
Reagents and Antisera/Antibodies
Oxalyticase was from Enzogenetics (Corvallis, OR). Other reagents were obtained from sources described previously (Guan et al., 2001; Kim et al., 2002). The pFA6a knockout and tagging vectors containing TRP1, HIS3, or KanMX markers were generous gifts from Dr. Mark Longtine (Oklahoma State University, Stillwater, OK) (Longtine et al., 1998). Cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) tagging vectors were from the Yeast Resource Center (University of Washington, Seattle, WA). Antisera against Ape1 (Klionsky et al., 1992) and Fox3 (Hutchins et al., 1999) have been described previously. Antibody against Pho8 was from Molecular Probes (Eugene, OR), anti-green fluorescent protein (GFP) was from Covance Research Products (Berkeley, CA), and the anti-hemagglutinin (HA) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Screening the Gene-Deletion Libraries
Diploid and MATα haploid gene-deletion libraries were obtained from ResGen/Invitrogen (Huntsville, AL). The mutants provided from the company were inoculated on YPD plates and incubated at 30°C for 12–24 h. To screen for cvt mutants, the cells on YPD plates were collected and subjected to immunoblot analysis by using anti-Ape1 antiserum.
Disruption, Epitope Tagging, Gene Cloning, Mutagenesis, and Protein Purification
The chromosomal tagging and deletions were performed by a polymerase chain reaction (PCR)-based procedure (Longtine et al., 1998). Putative knockout strains were checked by Western blotting, by PCR, or both.
ATG18 and ATG21 without endogenous promoters were cloned by PCR by using genomic DNA as template. The genes were put into pCuGFP(416) (Kim et al., 2001a) or pMAL-c2 (New England Biolabs, Beverly, MA) resulting in plasmids pPS123 (pCuGFPAtg21(416)), pPS122 (pMBPAtg21), and pPS175 (pMBPAtg18). The mRFP1 plasmid (Campbell et al., 2002) was a gift from Dr. Roger Y. Tsien (University of California, San Diego, San Diego, CA) and was cloned into a BglII site in front of the start codon of APE1 after PCR-mediated incorporation of flanking BamHI sites in pRS305 (pPS128), pRS306 (pPS129), and pRS414 (pPS130). The pPS128 and pPS129 plasmids were integrated into the genome after cutting with AflII and AvrII, respectively. The Atg8-GFP, GFP-AUT7ΔR, AUT7GFP(416), and pCuGFP-AUT7(416) plasmids (Kim et al., 2001a), and the pPS97 (pCuYFPAtg11(426)) and pCVT19CFP(414) (Kim et al., 2002) plasmids have all been described. Plasmid PS132 (pGFP-Atg8(303)) was made by moving pGFP-Atg8 from pRS316 GFP-AUT7 (Suzuki et al., 2001) into pRS303.
To generate yeast two-hybrid plasmids, DNA fragments encoding ATG3 and ATG7 were generated by PCR and cloned as a BamHI-SalI fragment into both pGAD-C1 and pGBDU-C1 vectors (James et al., 1996), creating pGAD-ATG3, pGAD-ATG7, pGBDU-ATG3, and pGBDU-ATG7, respectively. The plasmid pGAD-ATG8 (pPS140) was made by moving ATG8 from pCuGFP-AUT7(416) to pGAD-C2. Plasmid pGBD-Aut7 was described previously (Shintani et al., 2002). These plasmids and the vectors without inserts were cotransformed in the appropriate combinations into both the PJ69-4A and PJ69-4A atg21Δ (PSY303) strains. Interactions were monitored by the ability of the transformed strains to grow on plates lacking histidine.
Mutagenesis of pPS123 (pCuGFPAtg21(416)) to generate pPS125 containing the Arg343Gly Arg344Gly double mutation was performed using QuikChange (Stratagene, La Jolla, CA) and verified by sequencing.
MBP, MBP-Atg18, MBP-Atg21, and MBP-Ygr223c were purified from BL-21 Codon Plus cells (Stratagene) by using maltose-coupled Sepharose from New England Biolabs.
Immunoblot and Lipid Overlay Analysis, Pulse/Chase Labeling, and Immunoprecipitation
Immunoblot analysis was carried out essentially as described previously (Hutchins et al., 1999; Reggiori et al., 2003). For kinetic analysis of Atg8 lipidation, yeast cells were grown to A600 = 1.0 and labeled with 20 μCi of Tran[35S] label for 5 min, followed by washing and chasing in SD-N. Samples were removed at the indicated time points and subjected to immunoprecipitation as described previously using an antiserum against Atg8 (Klionsky et al., 1992). Separation of Atg8-PE from Atg8 was obtained by adding 6 M urea to 12.5% SDS polyacrylamide gels (Kirisako et al., 2000).
For the in vitro lipid binding assay, PIP Strips (Echelon, Salt Lake City, UT) were blocked in 2% fat-free bovine serum albumin (BSA) for 30 min, probed with 100 ng/ml protein in 2% fat-free BSA for 1 h, followed by an antibody against maltose binding protein (MBP) (New England Biolabs) (1:3000 in 2% fat-free BSA) (1 h) and secondary antibody conjugated to horseradish peroxidase (1 h).
Analyses of the Cvt Pathway and Autophagy
Peroxisome degradation rates, and the protease-protection and alkaline phosphatase assays have been described previously (Noda et al., 1995; Hutchins et al., 1999; Guan et al., 2001).
Microscopy
All strains used for microscopy were grown in YPD or SMD to early mid-log phase. For the starvation time course, cells were grown in YPD to early log phase before being shifted to SD-N medium. FM 4-64 staining and microscopy analyses were performed as described previously (Kim et al., 2001b, 2002).
RESULTS
Screen for Mutants Defective in Autophagy-related Pathways
The commercial availability of a yeast deletion library has resulted in the functional description of a variety of previously uncharacterized genes. To find new genes required for the Cvt pathway and autophagy, we screened the deletion library for strains defective in maturation of the cargo protein precursor aminopeptidase I (prApe1). Precursor Ape1 normally undergoes proteolytic removal of its N-terminal propeptide upon vacuolar delivery (Klionsky et al., 1992); this processing event results in a molecular mass shift that can easily be detected by Western blot. Several novel genes were identified that were required for prApe1 maturation, and one of these, ATG21, had some similarity to ATG18 as well as another gene, YGR223c. We have previously characterized ATG18 as a gene required for the formation of Cvt vesicles and autophagosomes (Guan et al., 2001). During the course of these studies, ATG21 was published as MAI1 and shown to be required for prApe1 transport (Barth et al., 2002). Recently, a unified nomenclature was adopted for yeast AuTophaGy-related genes (Klionsky et al., 2003). Accordingly, we hereafter refer to the gene as ATG21 and to the protein as Atg21.
Atg21 Is Required for the Formation of Cvt Vesicles
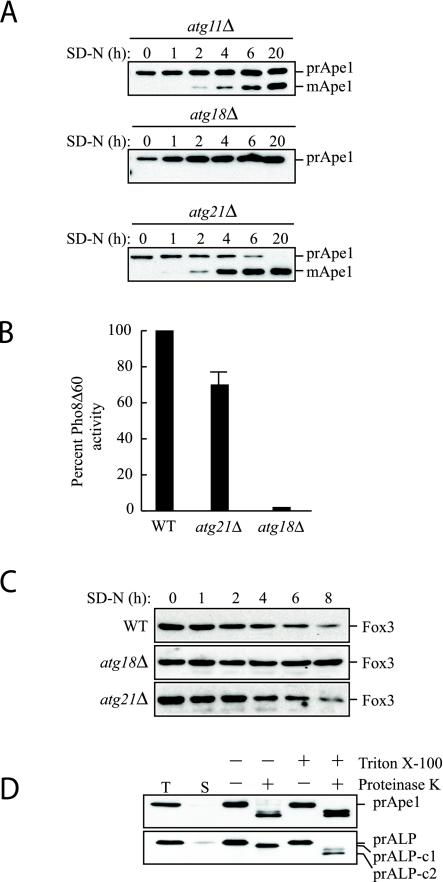
Precursor Ape1 transits to the vacuole by two pathways: the Cvt pathway is used under vegetative conditions, and autophagy is required for delivery during starvation. We verified the results from the initial screen for new mutants by examining cells from log phase cultures. As expected, the atg21Δ mutant was defective for the maturation of prApe1 in rich media (Figure 1A, zero-hour time point). However, prApe1 was matured upon nitrogen starvation (Figure 1A). As controls, we examined the atg11Δ and atg18Δ strains. Atg11 is required for the Cvt pathway but is not essential for autophagy; atg11Δ cells are able to mature ∼50% of the accumulated prApe1 when cells are shifted to starvation conditions to induce autophagy (Figure 1A; Kim et al., 2001b). In contrast, Atg18 is required for both the Cvt pathway and autophagy (Guan et al., 2001). The atg18Δ mutant was unable to mature prApe1 even in starvation conditions, in agreement with its being defective for both pathways. The ygr223cΔ strain was not defective in the Cvt pathway, and an atg21Δ ygr223cΔ double mutant was phenotypically identical to an atg21Δ single mutant (our unpublished data). These data suggest that Atg21 is defective for the Cvt pathway but is not required for vacuolar import of prApe1 by the autophagy pathway.
Figure 1.
Atg21 is required for the Cvt pathway. (A) atg21Δ cells were grown in YPD and shifted to SD-N to induce autophagy. The atg21Δ cells mature prApe1 under starvation conditions more rapidly than atg11Δ, whereas no maturation is seen in atg18Δ cells. (B) Pho8Δ60, a marker for nonspecific autophagy, is delivered to the vacuole of atg21Δ cells in nitrogen starvation conditions. WT (TN124), atg18Δ (JGY20), and atg21Δ (PSY107) cells were grown as described in A. Cells were shifted to SD-N, samples collected, and protein extracts assayed for alkaline phosphatase activity. The activity in the wild-type sample was set to 100% and the other activities normalized relative to wild-type. The atg21Δ results represent the mean and S.E. of three experiments, whereas the atg18Δ results are based on duplicate samples from the same experiment. (C) atg21Δ and ygr223cΔ cells are not deficient in peroxisome degradation. Cells were shifted from oleic acid-containing medium to SD-N, and pexophagy was monitored by Western blot by using an antibody against peroxisomal thiolase (Fox3). The result for the ygr223cΔ strain was essentially the same as that shown for atg21Δ. (D) Atg21 is required for Cvt vesicle formation. atg21Δ pep4Δ (PSY2) cells were converted to spheroplasts and subjected to a protease protection assay performed as described in MATERIALS AND METHODS.
The maturation of prApe1 in atg21Δ cells in medium lacking nitrogen suggested that autophagy may be functional in this mutant. Monitoring prApe1 maturation alone, however, is not an adequate measure of autophagy; prApe1 can be delivered to the vacuole in cells that are compromised for autophagic activity (Abeliovich et al., 2000). To examine the autophagic capacity of atg21Δ cells, we relied on an assay that measures nonspecific autophagy. In this assay, a truncated form of the vacuolar alkaline phosphatase precursor Pho8Δ60 is expressed in the cytosol (Noda et al., 1995). To be proteolytically activated, Pho8Δ60 needs to be transported to the vacuole by autophagy. Under nitrogen starvation conditions that induce autophagy, Pho8Δ60 activity in atg21Δ cells was similar, although slightly reduced, relative to that seen in the wild-type control (Figure 1B). In contrast, the atg18Δ strain showed an almost complete block in autophagy as measured by Pho8Δ60 activity. This result suggested that Atg21 was needed for a type of specific autophagy, import of prApe1, but not for nonspecific autophagy. Similarly, only a minor reduction in the autophagy rate was found for ygr223cΔ cells (our unpublished data).
We extended this analysis by examining a second type of specific autophagy, the degradation of peroxisomes, or pexophagy. Yeast cells were grown in oleic acid to induce peroxisome proliferation and then shifted to medium lacking nitrogen. We have previously demonstrated that these conditions induce pexophagy that is dependent upon the Atg proteins (Hutchins et al., 1999). Pexophagy was monitored by following degradation of the peroxisomal enzyme thiolase Fox3. Wild-type cells showed normal degradation of Fox3 after the shift to SD-N medium (Figure 1C). The atg18Δ mutant was completely blocked for Fox3 degradation, indicating that Atg18 is needed for pexophagy, in agreement with our previous results (Guan et al., 2001). The atg21Δ cells displayed a rate of peroxisome degradation that was similar to that seen with the wild-type strain. Similarly, the ygr223cΔ strain was not defective in pexophagy (our unpublished data). Atgt21 therefore does not fall into the same category as Atg11, or the Atg20 and Atg24 mutants, which are required for the Cvt pathway and pexophagy but not for nonselective autophagy (Nice et al., 2002).
To gain insight into the stage of the Cvt pathway that was blocked in the atg21Δ strain, we carried out a protease protection assay (Guan et al., 2001). In this assay, atg21Δ cells lacking the vacuolar protease Pep4 were osmotically lysed and treated with exogenously added proteinase K (Figure 1D). Precursor Ape1 was cleaved by proteinase K even in the absence of detergent, showing that prApe1 was not protected within a membranous compartment in atg21Δ cells. Vacuolar alkaline phosphatase was used as a control to show that vacuoles, and presumably Cvt vesicles, were not lysed upon cell disruption; only the cytosolic tail of Pho8 was cleaved in the absence of detergent, whereas the lumenally oriented propeptide also was processed when the vacuolar membrane was disrupted with detergent. These results suggest that the osmotic lysis did not disrupt intracellular compartments and that the protease sensitivity seen in the atg21Δ cells was due to a defect in Cvt vesicle formation.
Atg21 Is Associated with the Vacuole and Prevacuolar Structures
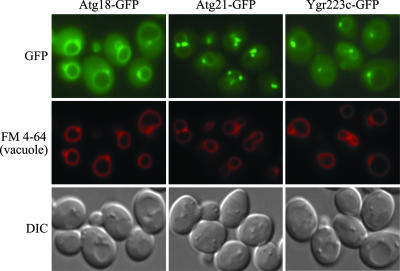
Most of the proteins involved in the Cvt and autophagy pathways associate at least transiently with the PAS. One of the exceptions is seen with Atg18; we have previously shown that Atg18 is predominantly associated with the vacuolar membrane (Guan et al., 2001). To gain further insight into the function of Atg21, we examined the localization of the protein in vivo. Similar to Atg18, a vacuolar association also was found for Atg21 that had been chromosomally tagged with GFP (Figure 2). However, Atg21-GFP also was associated with a number of punctate structures that were stained with the dye FM 4-64 that was taken up by endocytosis. These FM 4-64–stained structures also were associated with the vacuole. These results are in agreement with those of Barth et al. (2002) based on immunofluorescence microscopy. The majority of Atg21-GFP seemed to be associated with the punctate structures, whereas only a small fraction of Atg18-GFP showed this localization. Ygr223c-GFP also showed some vacuolar and punctate localization, with an expression level that was generally lower than Atg18-GFP and Atg21-GFP (Figure 2).
Figure 2.
Atg21 is associated with the vacuole and prevacuolar structures. Wild-type (SEY6210) cells were chromosomally tagged with GFP at the ATG18 (PSY62), ATG21 (PSY63), and YGR223c (PSY100) loci, respectively, and stained with FM 4-64 to label the vacuoles. Cells were grown to early mid-log phase in YPD before viewing. DIC, differential interference contrast.
As we have shown that Atg21 is required for the formation of Cvt vesicles (Figure 1D), it seemed plausible to assume that one or more of the punctate structures seen with Atg21-GFP corresponded to the PAS, which is thought to be involved in vesicle nucleation/formation. We examined localization of Atg21 relative to the PAS by monitoring cells expressing Atg21-GFP from the chromosomal locus and prApe1 fused to the red fluorescent protein as a marker for the pre-autophagosomal structure. The two fluorescent constructs did not display colocalization (our unpublished data). Although the lack of colocalization of Atg21 with prApe1 indicated that the visible fraction of Atg21 is not associated with the PAS, we cannot exclude that a minor amount of Atg21 also is active at this site; recently, we have shown that Atg18 interacts with Atg2 at the PAS, even though the majority of Atg18 does not seem to localize to the pre-autophagosomal structure (Reggiori et al., 2004).
Atg18, Atg21, and Ygr223c Bind to Phosphoinositides
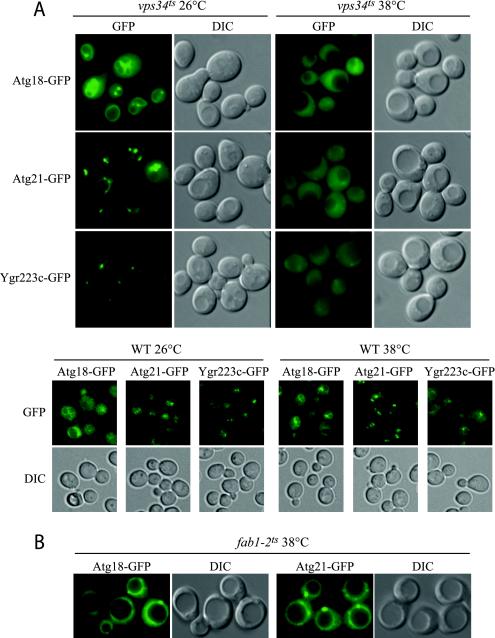
The majority of Atg proteins seem to function in vesicle formation. Because Atg21 does not seem to localize to the PAS, a possible explanation of its localization could be that the protein functions before the formation of the Cvt vesicle. Along these lines, we have not been able to detect interaction of Atg21 with other Cvt pathway and autophagy-specific proteins by affinity isolation or by two-hybrid analyses (our unpublished data). Similarly, the localization of Atg21-GFP did not change in any of the atg null mutant strains that we examined (our unpublished data). We therefore investigated whether the distribution of Atg21-GFP and its homologues changed in mutants known to affect vacuolar function. In two of the deletion mutants examined, vps34Δ and vps15Δ, all three proteins were found exclusively in the cytosol (our unpublished data). Because these mutants exhibit severe growth defects, we repeated the experiment by using a temperature-sensitive form of Vps34 (Figure 3A). Atg18-, Atg21- and Ygr223c-GFP showed normal distribution at the permissive temperature, but they were essentially all lost from the vacuole and associated structures in the vps34ts strain within minutes of shifting to the nonpermissive temperature. A change in localization of these proteins was not seen when wild-type cells were shifted from 26 to 38°C (Figure 3A), indicating that the loss of localization seen in the vps34ts strain was due to inactivation of the Vps34 protein.
Figure 3.
Atg18, Atg21, and Ygr223c localization is dependent on PtdIns 3-kinase activity. Strains PSY291, PSY292, PSY293, PSY62, PSY63, PSY100, PSY146, and PSY290 expressing Atg18-GFP, Atg21-GFP, or Ygr223c-GFP from the respective chromosomal loci in the vps34ts and wild-type SEY6210 (A) or fab1-2ts background (B) were grown at 26°C to early mid-log phase in SMD and shifted to 38°C for 10 (A) or 40 min (B). Cells were viewed by fluorescence microscopy as described in the legend to Figure 2. Loss of PtdIns 3-kinase but not PtdIns(3)P 5-kinase activity at the nonpermissive temperature resulted in loss of localization of Atg18 and Atg21. DIC, differential interference contrast.
Vps34 is the sole PtdIns 3-kinase in yeast, but PtdIns triphosphate (PtdIns(3)P) can further be converted into Pt-dIns(3,5)-bisphosphate (PtdIns(3,5)P2) by Fab1. The loss of localization in cells lacking Vps34 therefore could reflect the absence of either phosphoinositide. To distinguish between these possibilities, we examined localization in a fab1ts mutant strain. Chromosomally expressed Atg18-GFP and Atg21-GFP seemed to be present at the vacuole at the nonpermissive temperature in the fab1ts mutant (Figure 3B). However, Atg18-GFP in particular seemed to show less vacuolar association after >30 min at the nonpermissive temperature (Figure 3B). This result can possibly be ascribed to the severe swelling of the vacuole occurring as a result of the loss of PtdIns(3,5)P2 (Gary et al., 1998). Binding of Atg18-GFP and GFP-Atg21 to the vacuole in fab1Δ cells could easily be seen using high expression plasmids (our unpublished data).
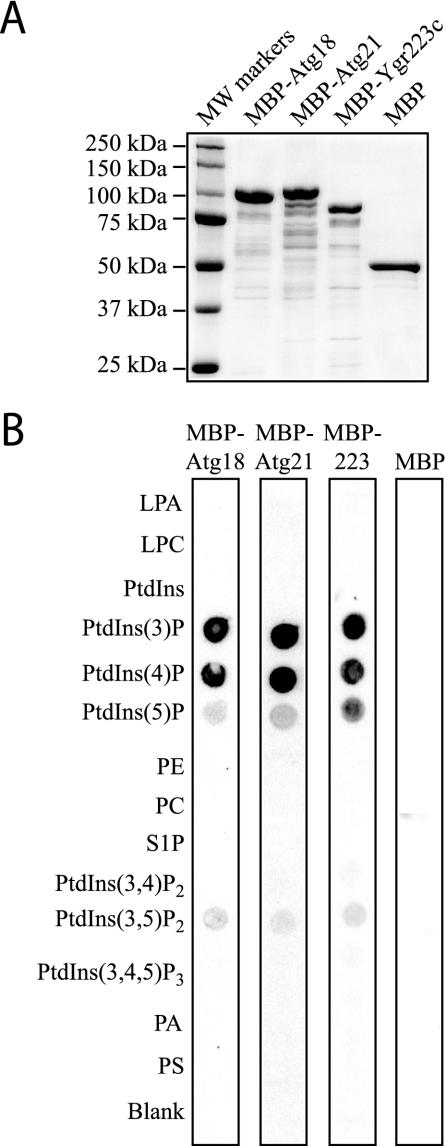
To explore the binding of Atg21 to phosphoinositides in greater detail, we purified full-length Atg18, Atg21, and Ygr223c fused to the MBP expressed in E. coli (Figure 4A). As expected, the Atg18, Atg21, and Ygr223c fusion proteins, but not MBP itself, bound to PtdIns(3)P that had been immobilized on nitrocellulose in vitro, but they also bound to PtdIns 4-phosphate with apparently equal affinity (Figure 4B). The proteins also had some affinity for PtdIns 5-phosphate and PtdIns(3,5)P2. All three proteins therefore bind to phosphoinositides, even though they do not contain any known phosphoinositide binding domains such as the FYVE, PX, or PH domains. This finding is in close agreement with a recent study showing that WIPI49, a mammalian homologue of Atg18, binds phosphoinositides, with an apparent preference for PtdIns(3)P and a reduced but significant binding of PtdIns 5-phosphate and PtdIns(3,5)P2 (Jeffries et al., 2004).
Figure 4.
Atg21 binds to phosphoinositides in vitro. (A) MBP, MBP-Atg18, MBP-Atg21, and MBP-Ygr223c were purified from E. coli as described in MATERIALS AND METHODS. Protein (1 μg) was loaded on a gel, and the gel was stained with Coomassie Brilliant Blue G. (B) PIP Strips (Echelon) containing lipids immobilized on nitrocellulose were incubated with 100 ng/ml MBP, MBP-Atg18, MBP-Atg21, or MBP-Ygr223c as described in MATERIALS AND METHODS.
To gain further information about the domain(s) required for phosphoinositide binding, we mapped regions of the protein that displayed lipid-binding characteristics. Along these lines, we constructed a series of deletions within Atg21 and Atg18 and examined the ability of the mutated proteins to bind immobilized lipids. We found that deletions of either the extreme N termini of the proteins or a central region resulted in loss of lipid binding (our unpublished data). In contrast, deletions of the C termini of Atg21 or Atg18 seemed to eliminate the ability of the proteins to complement the prApe1 transport defect of the respective null mutants, but they did not affect phosphoinositide binding in vitro (our unpublished data), suggesting the possible presence of a protein-binding site. The central domain of Atg18, Atg21, and Ygr223c that we found to be required for phosphoinositide binding includes one of three WD-40 repeats. Alignment of the Atg21, Atg18, and Ygr223c protein sequences revealed a conserved Phe-Arg-Arg-Gly sequence within this central region. Mutation of two conserved arginine residues in the mammalian Atg18 homologue WIPI49 eliminated binding of the protein to PtdIns(3)P (Jeffries et al., 2004). We explored the nature of this lipid-binding region in Atg21 in greater detail by mutating the two arginine residues to lysine. The R343K R344K double mutation in Atg21 resulted in a protein with no activity (i.e., inability to import prApe1) and no association with the vacuole or punctate structures (Figure 5). However, the mutant protein still bound to the various phosphoinositides in vitro with similar affinities as wild-type Atg21 (our unpublished data). Identical results were obtained when the arginines were mutated into glutamines or alanines (our unpublished data). Thus, an apparently novel lipid-binding motif in combination with a putative protein binding motif may be responsible for functional binding of Atg21 to biological membranes in vivo.
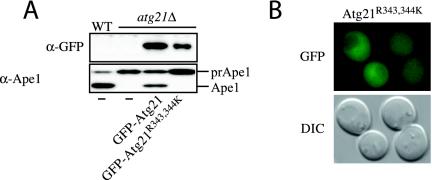
Figure 5.
Arginine343 and arginine344 are required for function of Atg21. (A) atg21Δ cells (PSY7) expressing GFP-Atg21 (pPS123) but not GFP-Atg21R343,344K (pPS125) mature prApe1. (B) GFP-Atg21R343,344K does not localize to the vacuole and punctate structures. DIC, differential interference contrast.
Atg21 Is Required for Efficient Localization of Atg8 to the PAS
As mentioned above, no Cvt pathway- or autophagy-specific proteins were found to be required for the localization of Atg21, and the Atg18, Atg21, and Ygr223c proteins were not dependent on each other for localization (our unpublished data). We have previously shown that Atg18 is required for the punctate localization of Atg2 (Guan et al., 2001). Accordingly, we next investigated whether Atg21 was required for association of proteins with the PAS. To minimize artifacts, these studies were either performed with chromosomally tagged proteins that were all fully functional (Atg1-GFP, Atg2-GFP, Atg14-GFP, and Atg9-YFP), with integrated plasmids expressing functional fluorescent-tagged proteins behind the corresponding gene's native promoter (GFP-Atg8 and RFP-prApe1), or with plasmids expressing near endogenous levels of protein (YFP-Atg11 and CFP-Atg19). We have previously shown that Atg11 recruits prApe1 to the PAS by binding to the prApe1 receptor Atg19 (Shintani et al., 2002), and we found here that the colocalization of Atg11 with Atg19 does not require Atg21 (Figure 6A). Using RFP-prApe1 as a marker for the PAS, we furthermore found that Atg1-GFP, Atg14-GFP, Atg9-YFP, and Atg2-GFP all were present at the PAS in atg21Δ cells (Figure 6B; our unpublished data). The punctate localization of Atg2-GFP has been shown to require Atg1, Atg9, and Atg14 and is thereby a control that the activities of these proteins are also normal. All four proteins showed increased punctate localization in atg21 cells compared with wild type, which can probably be ascribed to accumulation of prApe1 at the PAS. The only protein not seen associated with the PAS was GFP-Atg8, which displayed a diffuse cytosolic staining (Figure 6B).
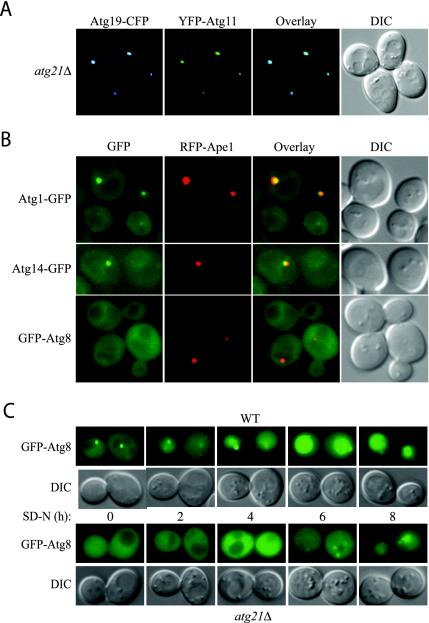
Figure 6.
Localization of Atg proteins to the PAS in atg21Δ cells. (A) Atg19-CFP and Atg11-YFP colocalize in atg21Δ cells. PSY6 (atg21Δ) cells were transformed with plasmids expressing Atg19-CFP (pCVT19CFP(414)) and YFP-Atg11 (pPS97), grown in SMD to mid-log phase and analyzed by fluorescent microscopy. (B) GFP-Atg8 is not localized to the PAS in atg21Δ cells. atg21Δ cells expressing Atg1-GFP (PSY227) or Atg14-GFP (PSY228) from the chromosomal loci, and also expressing RFP-prApe1 from the endogenous promoter on an integrated plasmid, or atg21Δ atg8Δ cells expressing GFP-Atg8 (PSY226) behind the endogenous promoter from an integrated plasmid, and also expressing RFP-prApe1 from the endogenous promoter on a centromeric plasmid, were grown in YPD to mid-log phase before viewing. Localization of Atg2-GFP was essentially the same as that shown for Atg1-GFP. (C) Transport of GFP-Atg8 to the vacuole is reduced in atg21Δ cells. Wild-type (WT, SEY6210) and atg21Δ cells (PSY7) were transformed with a plasmid expressing GFP-Atg8 from the CUP1-promoter (pCuGFP-AUT7(416)), grown to mid-log phase in SMD before the addition of 1 mM phenylmethylsulfonyl fluoride and starved for nitrogen for up to 8 h as indicated. DIC, differential interference contrast.
The absence of GFP-Atg8 from the PAS in cells lacking Atg21 was surprising. Atg8 plays a role in the Cvt pathway and is required for efficient nonselective autophagy, yet we found that atg21Δ cells displayed essentially normal uptake of Pho8Δ60 (Figure 1B). Atg8 is transported to the vacuole on the inside of the autophagosome and accumulates in the vacuole in protease-deficient mutants. We used this phenotype to further examine the effect of the atg21 mutation on Atg8 localization. Wild-type and atg21Δ cells expressing GFP-Atg8 were treated with the protease inhibitor phenylmethylsulfonyl fluoride under autophagy-inducing conditions to allow for the accumulation of autophagic bodies (Figure 6C). In wild-type cells, GFP-Atg8 can be seen to accumulate in the vacuole after 2 h, with strong staining of the vacuole after 4 h. In atg21Δ cells, there was almost no vacuolar accumulation of GFP-Atg8 even after 4 h; these cells generally had to be starved for 8 h to detect even a low level of GFP-Atg8 in the vacuole. This result showed that the itinerary of Atg8 was not completely blocked in atg21Δ cells, but rather that a lower level of Atg8 may be directed to the site of autophagosome formation. This reduced amount of Atg8 was apparently still sufficient for a near normal rate of autophagy.
Atg21 Is Required for Efficient Lipidation of Atg8
Atg8 is a small protein that is conjugated to PE (Kirisako et al., 2000), and we extended our analysis by examining lipidation of Atg8. Atg8-PE can be separated from Atg8 by carrying out SDS-PAGE in the presence of urea (Kirisako et al., 2000). Exponentially growing cells in rich media contain only minor amounts of lipidated Atg8, and there was only a very minor decrease in atg21Δ cells compared with the wild type (Figure 7A). In comparison, atg18Δ and pep4Δ cells displayed a slight increase in the lipidated form of Atg8. After starvation, however, atg21Δ cells accumulated only unlipidated Atg8. In contrast, atg18Δ and pep4Δ cells accumulated Atg8-PE; accumulation was in the vacuole in the pep4Δ mutant. Wild-type cells do not accumulate Atg8-PE because the protein is degraded after vacuolar delivery. To verify that Atg8-PE was not degraded in the vacuole in atg21Δ cells, we examined the protein in an atg21Δ pep4Δ double mutant strain. There was only a small increase in the level of Atg8-PE in the atg21Δ pep4Δ strain, indicating that vacuolar degradation was not the primary reason for the decreased signal seen in the atg21Δ cells. This result confirmed the reduced transport of GFP-Atg8 to the vacuole by autophagy seen in atg21Δ cells (Figure 6C).
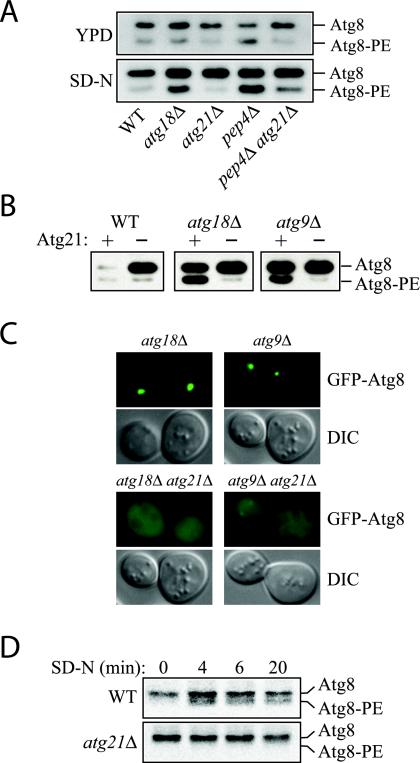
Figure 7.
Lipidation of Atg8 is reduced in atg21Δ cells. (A) Atg8 lipidation in rich medium versus nitrogen starvation medium. Cells were grown in YPD to early mid-log phase and then starved for 3 h in SD-N. Atg8-PE was separated from Atg8 by 12% SDS page gels containing 6% urea (Kirisako et al., 2000). All strains are of BY4742 background. (B) Deletion of ATG21 antagonizes the accumulation of Atg8-PE in autophagy mutants. WT (SEY6210), atg21Δ (PSY7), atg18Δ (JGY3), atg18Δ atg21Δ (PSY167), atg9Δ (JKY007), and atg9Δ atg21Δ (PSY191) cells were starved for 4 h in SD-N, and Atg8-PE was analyzed by Western blot. (C) Deletion of ATG21 antagonizes the accumulation of GFP-Atg8 at the PAS in autophagy mutants. Strains used in B containing a plasmid expressing GFP-Atg8 from the CUP1 promoter were grown to mid-log phase in SMD and then starved for 4 h in SD-N. (D) atg21Δ cells have a reduced rate of Atg8 lipidation. Cells of BY4742 background were labeled and Atg8 immunoprecipitated as described in MATERIALS AND METHODS and analyzed by 12% SDS-urea gels.
Atg8-PE also was seen to accumulate in other strains defective in the Cvt pathway and autophagy including atg9Δ (Figure 7B). The Atg8-PE that accumulated in the atg18Δ or atg9Δ strains after cells were starved for 3 h was abolished by removal of Atg21 (Figure 7B). Consistent with the lipidation data, atg18Δ atg21Δ and atg9Δ atg21Δ cells expressing GFP-Atg8 did not show the punctate structures seen in atg18Δ and atg9Δ single mutants (Figure 7C).
The decreased amount of lipidated Atg8 in cells lacking Atg21 could either be due to a decreased rate of lipidation or due to decreased stability of the Atg8-PE conjugate (Suzuki et al., 2001). To test this, we carried out a pulse-chase analysis of Atg8 lipidation (Figure 7D). Cells were labeled during a 5-min pulse and then subjected to a nonradioactive chase. Lipidated Atg8 occurred after a 4-min chase in starvation medium in control cells. The pool of lipidated Atg8 did not change significantly during a 20-min chase, suggesting that there was not a rapid turnover of lipidated Atg8 under these conditions. Cells lacking Atg21 did not display any lipidated Atg8 during the course of the experiment, showing that the lipidation rate itself was considerably slowed down in the absence of Atg21.
One possible explanation for the reduced lipidation of Atg8 seen in the atg21Δ strain would be a defect in the enzymes that carry out the posttranslational modifications of Atg8 (reviewed in Ohsumi 2001). Atg8 is initially synthesized with a C-terminal arginine that is removed by the Atg4 protease. The E1-like protein Atg7 through interaction with the newly exposed C-terminal glycine then activates Atg8. Finally, Atg8 is transferred to the conjugating enzyme Atg3, which covalently attaches it to PE. To examine whether the initial processing step was defective, we analyzed the ability of atg21Δ cells to carry out the C-terminal cleavage step, by using a chimeric protein with GFP fused to the C terminus of Atg8 to monitor the cleavage event. In wild-type cells, Atg8-GFP is cleaved by the Atg4 protease, resulting in the removal of the GFP moiety (Figure 8A; Kim et al., 2001a). The atg21Δ strain displayed efficient cleavage of Atg8-GFP in both rich and starvation conditions, whereas an atg4Δ mutant was completely blocked in the removal of GFP, suggesting that the initial Atg4-dependent processing step was unaffected by the absence of Atg21. Analysis of prApe1 processing in these cells allowed verification that the strains were otherwise displaying the expected Cvt- and autophagy-defective phenotypes.
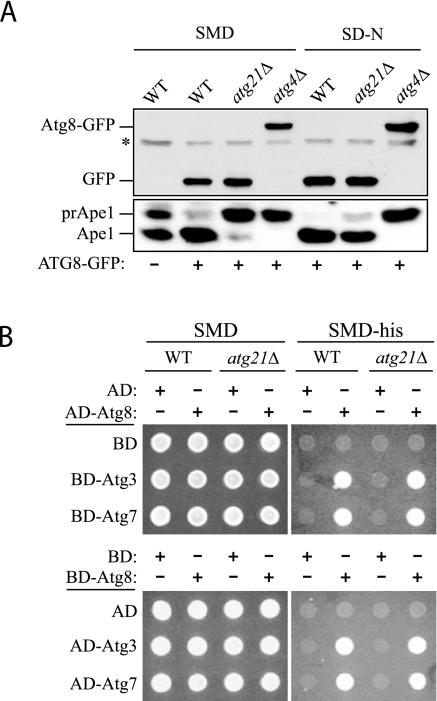
Figure 8.
The conjugation system responsible for posttranslational modification of Atg8 is functional in the atg21Δ mutant. (A) Atg4 is able to recognize Atg8 and carry out proteolytic cleavage at the C terminus. A plasmid encoding Atg8-GFP was transformed into the wild-type (SEY6210), atg4Δ (WPHYD2) or atg21Δ (PSY5) strain. Cells were grown in SMD to early log phase (A600 = 0.6) and then either retained in SMD or nitrogen starved in SD-N medium for 3 h. Protein extracts were analyzed by Western blot by using antiserum to Ape1 or anti-GFP monoclonal antibodies. The asterisk denotes a contaminating band that cross-reacts with the anti-GFP antibody. (B) Atg7 E1-like and Atg3 E2 enzymes are able to activate and conjugate Atg8. A yeast two-hybrid analysis was carried out as described in MATERIALS AND METHODS.
We next examined the ability of the Atg7 E1-like enzyme to activate, and the Atg3 E2 conjugating enzyme to interact with Atg8, through a yeast two-hybrid analysis (Figure 8B). ATG8 was fused to the sequence encoding the GAL4 activating or DNA binding domain and cotransformed into wild-type or atg21Δ cells with plasmids encoding ATG3 or ATG7 fused to the corresponding GAL4 domain. The empty vectors were used as controls. Transformants were selected and then analyzed for growth on plates lacking histidine. Growth on the minus histidine plates indicated that Atg8 was able to interact with Atg3 and Atg7 in the presence or absence of Atg21 (Figure 8B). The level of growth was similar to that seen with an interaction between Atg19 and Atg11, two proteins that have previously been shown to interact by the yeast two-hybrid system (our unpublished observations). In contrast, none of the strains containing an empty vector were able to grow in the absence of histidine, indicating that the constructs expressing Atg3, Atg7, or Atg8 were not able to induce expression from the GAL promoter independently. These results suggest that the activating and conjugating enzymes are able to interact with and modify Atg8 in the absence of Atg21. Finally, we examined the ability of GFP-Atg8ΔR to interact with Atg7 and Atg3 through microscopy analysis. The removal of the ultimate arginine residue bypasses the need for the initial Atg4-dependent processing step. In atg4Δ cells, GFP-Atg8ΔR can be seen to accumulate on the vacuole. Atg8-PE that is present on the external surface of the Cvt vesicle or autophagosome is normally removed after a second cleavage event by Atg4; in the absence of this cleavage, Atg8-PE accumulates on the vacuole surface after vesicle fusion with this organelle. We observed that GFP-Atg8ΔR accumulated on the vacuole surface in atg4Δ atg21Δ cells (our unpublished data). This localization confirms that Atg8 was competent for activation and modification (addition of PE) by Atg7 and Atg3, respectively, in the absence of Atg21.
Atg21 Is Required for Efficient Localization of Atg5 to the PAS
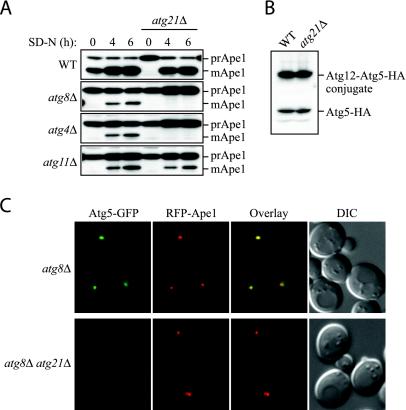
The results in Figure 7 showed that Atg21 was essential for the increased amount of Atg8-PE formed upon starvation but that Atg21 was not absolutely required for lipidation; cells lacking Atg21 still had a low level of Atg8-PE (Figure 7, A and B). The decreased formation of Atg8-PE in cells lacking Atg21 could be an indirect effect resulting from the loss of Atg21 activity rather than an indication that Atg21 is directly involved in the lipidation reaction itself; an indirect effect is also in agreement with the finding that posttranslational modification of Atg8 can occur normally in atg21Δ cells (Figure 8). To test between these alternatives, we made double deletion strains with atg8Δ, atg4Δ, and atg11Δ. As single mutants, these cells are all blocked in prApe1 transport in rich medium, but they are able to transport some prApe1 to the vacuole upon starvation (Figure 9A). If the only effect of removing Atg21 is inactivating Atg8, an additional deletion of Atg21 from atg8Δ cells should not cause a greater defect in prApe1 transport to the vacuole upon starvation. The atg8Δ atg21Δ and atg4Δ atg21Δ double mutants, however, did not mature prApe1 upon starvation, whereas an atg11Δ atg21Δ double mutant still transported prApe1 to the vacuole under these conditions. This latter result is in agreement with the near normal rate of starvation-induced autophagy seen in the atg11Δ atg21Δ double mutant (our unpublished data). Overall, these results suggest that Atg21 plays a role in prApe1 import that extends beyond its function in allowing efficient lipidation and/or stability of Atg8.
Figure 9.
Atg21 is required for Atg5-GFP accumulation at the PAS. (A) Cells lacking both Atg8 and Atg21 do not mature prApe1 upon starvation. WT (SEY6210), single mutants (WPHYD7, WPHYD2, AHY001, and PSY7) and double mutants (PSY139, PSY128, and PSY190) were grown in YPD to mid-log phase and starved for 4 and 6 h before being analyzed by Western blot by using an antibody against Ape1. (B) Atg21 is not required for conjugation of Atg12 to Atg5. WT (PSY66) and atg21Δ (PSY155) cells expressing Atg5-HA from the chromosomal locus were grown to early mid-log phase in YPD and subjected to Western blot by using an antibody against HA. (C) Atg21 is required for Atg5-GFP accumulation at the PAS. atg8Δ (PSY285) and atg8Δ atg21Δ (PSY286) cells expressing Atg5-GFP from the chromosomal locus and RFP-prApe1 from an integrating plasmid were grown to early mid-log phase in YPD before being viewed by fluorescence microscopy as described in the legend to Figure 6. DIC, differential interference contrast.
Correct localization and stability of Atg8-PE has been shown to depend on the presence of the Atg12-Atg5 conjugate (Suzuki et al., 2001). We therefore investigated the effect of removing Atg21 on the conjugation of Atg5. The majority of Atg5 is conjugated to Atg12 under all conditions in wild-type cells, and we were not able to detect any changes in the conjugation level in cells lacking Atg21 (Figure 9B). Atg5 also has been proposed to play a direct role in the formation of the autophagosome in mammalian cells, and Atg5-GFP has been shown to label the autophagic membranes before vesicle completion (Mizushima et al., 2001). In yeast cells, a fraction of Atg5-GFP has been shown to be present at the PAS (Suzuki et al., 2001). A significant amount of Atg5-GFP can be seen associated with the PAS marker RFP-prApe1 in cells lacking Atg8 (Figure 9C) (Kim et al., 2002). This punctate Atg5-GFP is probably associated with the sequestering membrane (Mizushima et al., 2001). However, in atg8Δ atg21Δ double mutant cells, the amount of Atg5-GFP associated with RFP-prApe1 was almost undetectable. A very faint Atg5-signal could sometimes still be detected in these cells, suggesting that a small fraction of Atg5 is still able to bind to the PAS.
DISCUSSION
ATG21 was originally described as a gene required for the maturation of prApe1 in rich media, but not for starvation-induced autophagy or for maturation of prApe1 during starvation (Barth et al., 2002). atg21Δ cells therefore seemed to be in the same category as several Cvt pathway mutants, including atg11, atg20, and atg24. However, unlike these mutants, atg21 cells are fully capable of degrading peroxisomes (Figure 1). Atg21 belongs to a family of proteins consisting of Atg18, Atg21, and Ygr223c. Atg18 is required for all types of autophagy, whereas a strong phenotype has not yet been found for Ygr223c. An intriguing feature of these proteins is that they all seem to be associated with the vacuole and perivacuolar structures (Figure 2). This could suggest that they all contain a common localization determinant binding to vacuolar-associated proteins or lipids. The overall homology between the three proteins is relatively low, with the exception of the N terminus and the central portion of the proteins, which therefore are candidates for harboring such a domain. Whereas we so far have not been able to detect protein interactions that could be responsible for localization of these proteins, our testing of the proteins in vitro suggests that they all bind preferentially to the phosphoinositide monophosphate PtdIns(3)P (Figure 4). The preferred binding of Atg21 to PtdIns(3)P was not altered by the presence of magnesium (our unpublished data), which is reported to increase the specificity of Atg18 for PtdIns(3,5)P2 (Dove et al., 2004). While this work was in revision, two reports were published indicating that the human homologue of Atg18 also binds to PtdIns(3)P (Jeffries et al., 2004) and that yeast Atg18 furthermore binds PtdIns(3,5)P2 (Dove et al., 2004). The latter work proposed that Atg18 is a PtdIns(3,5)P2 effector; however, PtdIns(3,5)P2 is not required for autophagy or the Cvt pathway (Dove et al., 2004). It is also clear that Atg21 and at least some Atg18 are still associated with the vacuole in cells lacking PtdIns(3,5)P2 altogether (Figure 3B). Further analysis is required to resolve the binding specificity as well as to determine the exact location of the binding domain. However, the localization of the proteins to the vacuole could suggest that in vivo, binding of the proteins to PtdIns(3)P could be important for the function of Atg18 and Atg21 in autophagy and the Cvt pathway. The PtdIns 3-kinase complex I, which consists of Vps34, Vps15, Vps30/Atg6, and Atg14, has been shown to be required for all types of autophagy (Kihara et al., 2001). Indeed, by using strains lacking either Vps15 or Vps34 or a temperature-sensitive allele of Vps34 (Figure 3A) (Stack et al., 1995), we were able to verify that PtdIns(3)P is required for the association of Atg18, Atg21, and Ygr223c with the vacuole.
The proteins so far characterized as having a role in the transport of prApe1 to the vacuole have been shown to have a diverse localization pattern, but most of them show some punctate localization that corresponds to the PAS (Suzuki etal., 2001; Kim et al., 2002; Shintani et al., 2002). It therefore seems likely that even though most of Atg21 does not seem to localize to the PAS, a fraction of the protein may reside at this site where the protein exerts its function. The Atg20 and Atg24 proteins bind to PtdIns(3)P through a PX domain (Nice et al., 2002; Yu and Lemmon, 2001), and binding of the two proteins to the PAS requires Atg14 (Nice et al., 2002). This, together with the observation that a fraction of Atg14 is found at the PAS, suggests that PtdIns(3)P is present at this structure. Atg21 could therefore be attracted to the PAS due to its affinity to PtdIns(3)P.
Although we could not detect any interaction of Atg21 with other Atg proteins, and none of the atg mutants investigated showed adverse localization of Atg21, removal of Atg21 had a strong effect on two Atg proteins, namely, Atg5 and Atg8. Atg5 is transiently located at the PAS; the localization can be enhanced in mutants defective in the progression of the Cvt/Atg pathways. We found, however, that the Atg5 localization to the PAS seen in an atg8Δ mutant strain was disrupted by the loss of Atg21 (Figure 9C). Atg8 is normally represented strongly at the PAS and has even been regarded as the best marker protein for this compartment (Suzuki et al., 2001). In the atg21Δ mutant, however, Atg8 seems to be largely absent from the PAS. Along these lines, in the atg21Δ mutant some Atg8 is going to the vacuole, but at a much lower rate (Figure 6C). Our data furthermore show that Atg21 is required for efficient lipidation of Atg8 (Figure 7). A lack of Atg8 at the PAS also has been described for cells lacking Atg14 (Suzuki et al., 2001), suggesting a possible functional link between Atg21 and the PtdIns 3-kinase complex I. However, the reduced lipidation seen with the loss of Atg21 suggests that Atg21 may function at a different step than the PtdIns 3-kinase complex I. The activity of this complex, but not Atg21, is required for the localization of Atg2 to the PAS (Shintani et al., 2001). Together, these data suggest that the PtdIns 3-kinase activity of Vps34 is required at two independent steps of the Cvt pathway. The first of these steps involves the generation of PtdIns(3)P at the PAS and is required for the recruitment of Atg2 as well as Atg20-Atg24 as part of the vesicle-forming machinery. The second step requiring Vps34 activity involves Atg21 and may be independent of the first step.
Although localization of Atg5 and Atg8 at the PAS was disrupted in the atg21Δ mutant, formation of the Atg12-Atg5 conjugate seemed to be normal and lipidation of Atg8 still occurred at a low level. Similarly, analysis of atg8Δ atg21Δ double mutants suggested that Atg21 exerted some function independent of its effect on Atg8 (Figure 9). These results, combined with the unique location of Atg21, lead us to propose that this protein may be involved in membrane recruitment to, or generation at, the site of sequestering vesicle formation. A defect in membrane recruitment would result in a defect in Cvt vesicle formation and also could cause the observed defects in Atg5 and Atg8 localization. A recent study suggests that Atg18 and the homologous proteins contain β-propeller folds, and it also was suggested that Atg18 is required for recycling of membrane proteins from the vacuole to the late endosome (Dove et al., 2004). Additional studies will be needed to determine whether Atg18 and Atg21 similarly could be involved in transport from the vacuole or late endosomes to the PAS by a mechanism involving PtdIns(3)P but not PtdIns(3,5)P2.
Acknowledgments
We thank Dr. Scott D. Emr and members of the Klionsky laboratory for helpful discussions. This work was supported by Public Health Service grant GM-53396 from the National Institutes of Health (to D.J.K.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-02-0147. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-02-0147.
References
- Abeliovich, H., Dunn, W.A., Jr., Kim, J., and Klionsky, D.J. (2000). Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151, 1025-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, H., Meiling-Wesse, K., Epple, U.D., and Thumm, M. (2001). Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 508, 23-28. [DOI] [PubMed] [Google Scholar]
- Barth, H., Meiling-Wesse, K., Epple, U.D., and Thumm, M. (2002). Mai1p is essential for maturation of proaminopeptidase I but not for autophagy. FEBS Lett. 512, 173-179. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove, S.K., et al. (2004). Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. (2004). EMBO J. 23, 1922-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary, J.D., Wurmser, A.E., Bonangelino, C.J., Weisman, L.S., and Emr, S.D. (1998). Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143, 65-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, J., Stromhaug, P.E., George, M.D., Habibzadegah-Tari, P., Bevan, A., Dunn, Jr., W.A., and Klionsky, D.J. (2001). Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell 12, 3821-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, P.K., and Emr, S.D. (1990). Characterization of Vps34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 6742-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins, M.U., Veenhuis, M., and Klionsky, D.J. (1999). Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112, 4079-4087. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries, T.R., Dove, S.K., Michell, R.H., and Parker, P.J. (2004). PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol. Biol. Cell 15, 2652-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Huang, W.-P., and Klionsky, D.J. (2001a). Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152, 51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Huang, W.-P., Stromhaug, P.E., and Klionsky, D.J. (2002). Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277, 763-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kamada, Y., Stromhaug, P.E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S.V., Ohsumi, Y., Dunn, Jr., W.A., and Klionsky, D.J. (2001b). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., Ohsumi, M., Takao, T., Noda, T., and Ohsumi, Y. (2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J., et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539-545. [DOI] [PubMed] [Google Scholar]
- Klionsky, D.J., Cueva, R., and Yaver, D.S. (1992). Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119, 287-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., III, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Yamamoto, A., Hatano, M., Kobayashi, Y., Kabeya, Y., Suzuki, K., Tokuhisa, T., Ohsumi, Y., and Yoshimori, T. (2001). Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice, D.C., Sato, T.K., Strømhaug, P.E., Emr, S.D., and Klionsky, D.J. (2002). Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277, 30198-30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Kim, J., Huang, W.-P., Baba, M., Tokunaga, C., Ohsumi, Y., and Klionsky, D.J. (2000). Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Matsuura, A., Wada, Y., and Ohsumi, Y. (1995). Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126-132. [DOI] [PubMed] [Google Scholar]
- Noda, T., and Ohsumi, Y. (2004). Macroautophagy in yeast. In: Autophagy, ed. D.J. Klionsky, Georgetown, TX: Landes Bioscience, 70-83.
- Ohsumi, Y. (2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell. Biol. 2, 211-216. [DOI] [PubMed] [Google Scholar]
- Paz, Y., Elazar, Z., and Fass, D. (1999). Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 275, 25445-25450. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., Tucker, K.A., Stromhaug, P.E., and Klionsky, D.J. (2004). The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79-90. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., Wang, C.-W., Stromhaug, P.E., Shintani, T., and Klionsky, D.J. (2003). Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 278, 5009-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.S., Klionsky, D.J., Banta, L.M., and Emr, S.D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., Huang, W.-P., Stromhaug, P.E., and Klionsky, D.J. (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., Suzuki, K., Kamada, Y., Noda, T., and Ohsumi, Y. (2001). Apg2p functions in autophagosome formation on the perivacuolar structure. J. Biol. Chem. 276, 30452-30460. [DOI] [PubMed] [Google Scholar]
- Stack, J.H., DeWald, D.B., Takegawa, K., and Emr, S.D. (1995). Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129, 321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug, P.E. and Klionsky, D.J. (2004). Cytoplasm to vacuole targeting. In: Autophagy, ed. D.J. Klionsky, Georgetown, TX: Landes Bioscience, 84-106.
- Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., DeWald, D.B., Boronenkov, I.V., Anderson, R.A., Emr, S.D., and Koshland, D. (1995). Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell 6, 525-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.W., and Lemmon, M.A. (2001). All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276, 44179-44184. [DOI] [PubMed] [Google Scholar]