Abstract
Background:
A cluster of genes are involved in the pathogenesis and adhesion of Candida albicans to mucosa and epithelial cells in the vagina, the important of which is agglutinin-like sequence (ALS) genes. As well as vaginitis is a significant health problem among women, the antifungal resistance of Candida species is continually increasing. This cross-sectional study investigates the expression of ALS1 and ALS3 genes and biofilm formation in C. albicans isolate isolated from vaginitis.
Materials and Methods:
Fifty-three recognized isolates of C. albicans were collected from women with recurrent vulvovaginal candidiasis in Iran, cultured on sabouraud dextrose agar, and then examined for gene expression. Total messenger RNA (mRNA) extracted from C. albicans isolates and complementary DNA synthesized using reverse transcriptase enzyme. Reverse transcription-polymerase chain reaction (RT-PCR) using specific primer was used to evaluate the expression of ALS1 and ALS3 through housekeeping (ACT1) genes. 3-(4,5-dimethyl-2-thiazyl)-2,5-diphenyl-2H-tetrazolium bromide assay was performed to assess adherence capacity and biofilm formation in the isolated.
Results:
Forty isolates (75.8%) expressed ALS1 and 41 isolates (77.7%) expressed ALS3 gene. Moreover, 39 isolates (74%) were positive for both ALS1 and ALS3 mRNA by the RT-PCR. Adherence capability in isolates with ALS1 or ALS3 genes expression was greater than the control group (with any gene expression), besides, it was significantly for the most in the isolates that expressed both ALS1 and ALS3 genes simultaneously.
Conclusion:
The results attained indicated that there is an association between the expression of ALS1 and ALS3 genes and fluconazole resistance in C. albicans. A considerable percent of the isolates expressing the ALS1 and ALS3 genes may have contributed to their adherence to vagina and biofilm formation.
Keywords: Agglutinin-like sequence proteins, biofilm formation, Candida albicans, vulvovaginal candidiasis
INTRODUCTION
Candida albicans is an opportunistic fungal pathogen which causes a broad spectrum of diseases such as oral, vagino-mucosal, and systemic infections.[1]
Vaginal candidiasis occurs in three-fourth of women during their life and C. albicans is the etiological agent in over 80% of the cases.[2]
C. albicans has a large number of virulence factors causing disease, including phenotypic switching, filamentation, adherence and secreted hydrolyses. Some of these pathogeneses are associated with gene families, particularly the agglutinin-like sequence (ALS) (6), secreted aspartyl proteinase, and lipase families.[3,4,5,6,7]
Among these, the ALS gene family that encodes cell wall glycoprotein is related to adherence to host surfaces. ALS genes are a family of adhesions recognized to play a role in adherence and early biofilm formation. Since biofilm formation contributes to drug resistance, ALS genes appear to be responsible for fluconazole resistance.[8,9,10,11] Each ALS gene has a similar three-domain structure, including a 5’ domain of 1299–1308 bp that is 55–90% identical across the family; a central domain of variable numbers of tandemly are repeated copies of a 108-bp motif; and a 3′ domain that is relatively variable in length and sequence across the family.[12] Although they distribute a similar three-domain structure, sequence differences between the ALS proteins can be so large that the proteins may have different functions.[13] Since adherent isolate of Candida is more pathogenic, there is a theory that ALS1 and ALS3p may be responsible for the pathogenesis of C. albicans.[14] The expression of each ALS1 and ALS3 gene can be detected by reverse transcription-polymerase chain reaction (RT-PCR) and confirmed by using this method for the analysis of ALS gene expression in the C. albicans genome that is correlated with its virulence. An RT-PCR assay investigates a specific gene expression in a variety of clinical specimens and in different host and model conditions.[15,16] The objective of our research was to determine the expression of ALS1 and ALS3 genes, its correlation with fluconazole resistance and finally biofilm formation in C. albicans, isolated from women with symptoms of recurrent vaginitis.
MATERIALS AND METHODS
Isolates
In this cross-sectional study, there were used 53 isolates of C. albicans obtained from women with recurrent vulvovaginal candidiasis that referred to clinical centers, Tehran, Iran. The patients were enrolled in terms of clinical symptoms of vaginitis and resistance to fluconazole therapy during the treatment process. Out of 53 patients, 57.69% would consume several antibiotics, 28.84% took an oral contraceptive, and 15.38% were diabetic. The isolates were confirmed by phenotypic and genotypic assays such as CHROM agar Candida and PCR-restriction fragment length polymorphism technique. Fluconazole susceptibility test had already been determined in our previous study.[17]
Reverse transcriptase-polymerase chain reaction analysis of ALS1 and ALS3 genes
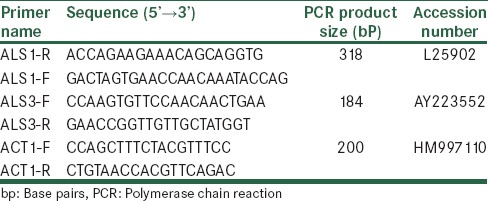
The confirmed C. albicans isolates are used to analyze the expression of ALS1 and ALS3 genes with minor modification. Expression of ALS1 and ALS3 genes was analyzed in planktonic cells of Candida isolates. For RNA extraction, a 24-h colony on sabouraud dextrose agar (Merck) was transferred to a 1.5 ml eppendorf tube and then 200 μl of lysis buffer containing (200 mM Tris-Hcl, pH 7.5, 10 mM EDTA, 0.5 M NaCl), 200 μl of phenol-chloroform-isoamillalchol (25:24:1) and glass beads, were added, and the tube was strongly vibrate for 60 min. After centrifuging for 3 min at 5000 rpm, the supernatant was removed to a new tube and 300 μl of chloroform was added. After centrifuging, the aqueous phase was moved to a clean tube and then 1 volume of cold isopropanol were added and was reserved at −20°C for 15 min. After that, the sample was washed by 70% ethanol, 30 μl distilled water was added, and the sample was kept at −20°C.[18] The RNA samples were treated with 1U of DNaseI (Fermentas) per 10 µl of RNA at 37°C for 1 h, and then its quality checks by electrophoresis on agarose gel (Merck). The first strand complementary DNA (cDNA) was synthesized using 2-step RT-PCR kit (Vivantis, Malaysia) according to the manufacturer's instruction and directly used in RT-PCR assay. Briefly, 2 μl 10X Viva Buffer A, 18.5 μl distilled water, 1–2 μl cDNA, 0.35 μl forward primer and 0.35 μl reverse primer for each gene. In continue, the microtube put into thermocycling device: Initial denaturation step at 94°C for 5 min followed by 35 cycles consisting of denaturation (94°C for 1 min), annealing (58°C for 1 min), and extension (72°C for 1 min) were followed by a final extension step at 72°C for 3 min. Finally, 25 µl of PCR product should be run out on a 1% agarose gel. ACT1 was used ACT1 (actin) gene was used as a control housekeeping gene. Primers using in this study are shown in Table 1.
Table 1.
ALS1, ALS3, and ACT1 specific primers for reverse transcription-polymerase chain reaction assay
Biofilm formation assay
Candida biofilm formation of isolates was performed on a 96-well plate in four groups including the isolates with ALS1 expression (Group 1), ALS3 expression (Group 2), ALS1 and ALS3 expression (Group 3) and those with any expression (Group 4). Eighty microliters of C. albicans cell suspension from each group was inoculated onto each well. The yeast cells were allowed to adhere to the bottom of the plate at 37°C for 90 min. Afterward, the unattached cells were gently washed with phosphate-buffered saline (PBS) and then added 4 ml of yeast nitrogen base (YNB) medium with 50 mM glucose. The plates were incubated at 37°C for 48 h, and the biofilm was made by Candida. Subsequently, the YNB medium was detached and washed with 4 ml of PBS.
3-(4,5-dimethyl-2-thiazyl)-2,5-diphenyl-2H-tetrazolium bromide assay
For this, 100 μl 3-(4,5-dimethyl-2-thiazyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) (5 mg/ml in PBS, Sigma) MTT was added to cells at 37°C for 3 h dimethyl sulfoxide (sigma) was added and the plates incubated for 15 min to stop the reaction and to dissolve the insoluble purple formazan. Optical absorbance was measured at the wavelength of 520 nm using an ELISA reader (Memmert, Germany).[19] The results were evaluated with regard to the percentage of viable cells in comparison to the negative control (isolates without ALS gene expression). All tests were performed in three independent experiments.
Statistics analysis
Data analysis was performed by t-test method with SPSS Statistics version 17, chicago. The level of statistical significance was set at P < 0.05.
RESULTS
In this study, the expressions of ALS1 and ALS3 genes were uniformly assessed on 3 µg of total RNA extract of isolates. RT-PCR showed the expression of ALS1 in 75.3% and of ALS3 in 77.2% of the isolates. Moreover, 73.5% of the isolates expressed both ALS1 and ALS3 genes (14.9% of the isolates had no expression for any ALS1 and ALS3 genes). Figure 1 illustrates the result of RT-PCR analysis. Of 53 isolates, 50 isolates were resistant to fluconazole (inhibition zone <12 mm), whereas 3 isolates were sensitive (inhibition zone >19 mm). The relation between fluconazole resistance with ALS1 and ALS3 gene expression is shown in Table 2. 79% of isolates that expressed ALS1/ALS3 or both genes were resistant to fluconazole.
Figure 1.
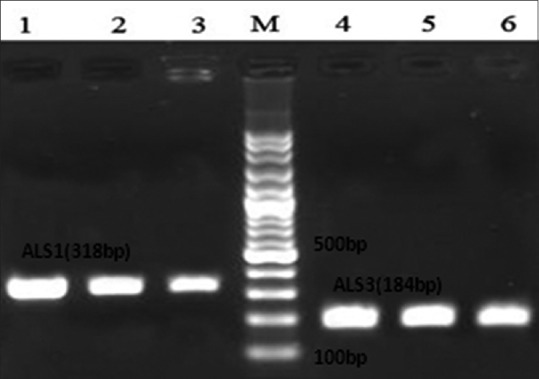
Messenger RNA of Candida albicans isolates extracted, and first strand complementary DNA was synthesized. Reverse transcriptase-polymerase chain reaction with ALS1 and ALS3 specific primers was done. Line 1−3, gene expression of ALS1 in isolates (318 bp); line 4−6, gene expression of ALS3 in isolates (184 bp). M: Marker molecular weight (100 bp)
Table 2.
Frequency of expression of ALS1 and ALS3 genes of Candida albicans isolates

Data from MTT assay showed the isolates with an expression of ALS1 (Group 1) or ALS3 genes (Group 2) had more adherence capacity than those lacking any gene expression (Group 4) and this difference was significant (P < 0.05). However, the isolates which had both ALS1 and ALS3 expressions had the highest adherence capability. Data were means ± standard deviations of three independent experiments (P < 0.05).
The result is shown in Figure 2.
Figure 2.
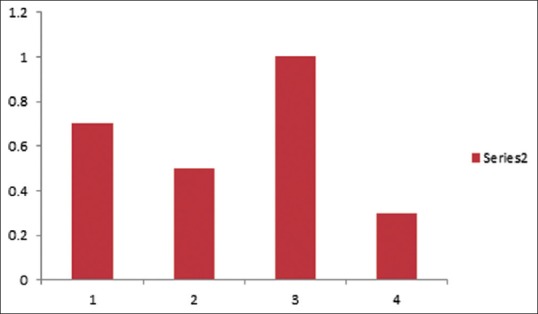
3-(4,5-dimethyl-2-thiazyl)-2,5-diphenyl-2H-tetrazolium bromide assay: Biofilm formations in four groups of Candida albicans isolates in yeast nitrogen base medium containing 50 mM glucose. The method used for biofilm quantification was 3-(4,5-dimethyl-2-thiazyl)-2,5-diphenyl-2H-tetrazolium bromide reduction. Data were means ± standard deviations of three independent experiments (P < 0.05). Group 1 (isolates with ALS1 expression), Group 2 (isolates with ALS3 expression), Group 3 (isolates with ALS1 and ALS3 expression), and Group 4 (control group without gene expression)
DISCUSSION
In this study, the expressions of ALS1 and ALS3 genes among the C. albicans isolates were analyzed using RT-PCR method. The presence of the genes has been reported in our previous study.[17] The simultaneous existence of both ALS1 and ALS3 genes was detected in 83% of the isolates, among which 74% expressed the two genes, summarized in Table 2. Our results are consistent with what reported by Cheng et al.[20] that evaluated the expression of ALS genes in C. albicans isolates using RT-PCR and showed a high frequency of ALS1, ALS2, ALS3, and ALS9 genes expression.
Nas et al.,[21] evaluated the expression of ALS1 gene in C. albicans isolated from vaginal candidiasis and showed its expression in 69% of total isolates.
In this study, 42 (79.5%) of isolates with an expression of at least one ALS gene were resistant to fluconazole and 3 (5.5%) of them without ALS gene expression were absolutely sensitive to fluconazole. A significant relationship was found between the ALS1 and ALS3 genes and biofilm formation in C. albicans isolates in MTT assay. Our finding hypothesized that there is a positive correlation between the expressions of ALS1, ALS3 genes, and fluconazole resistance. Since all of C. albicans isolates expressing the ALS1 or ALS3 were resistant to fluconazole, and their adherence was stronger than the control group (without any expression) and it seems reasonable to conclude that biofilm contributes to fluconazole resistance in isolates. This finding is in agreement with the results of other studies according to which the ALS1 was introduced as the major over-expressed gene in biofilm formation and adherence which in turn play an important role in drug inhibition to gain access to the fungi microcolonies.[22,23,24] Similar findings by other investigators which show pathogenic Candida species established well-developed biofilm and are resistant to drug therapy and act as a source of reinfections. More studies are required to realize the exact mechanisms involved.[25,26] According to our result, 9.4% isolates of C. albicans did not contain ALS1 and ALS3 genes, though they were resistant to fluconazole. This may occur due to other mechanisms involved in fluconazole resistance which affect phenotype. Moreover, the role of Med31, Med20, and Srb9/Med13 transcriptional regulator genes in the expression of the relevant virulence genes including ALS gene family and biofilm formation should not be ignored.[27]
It has been shown that C. albicans Tor1 gene plays a crucial role in the regulation of expression of several virulence-associated genes such as ALS1 and ALS3 and other adhesions. In addition, as a regulatory complex, Tec1, Bcr1, Efg1, Nrg1, and Tup1 affect the expression of several adhesions and ALS genes family.[28] Many other studies show the significance of regulatory genes in the expression of ALS gene family that explain the nonexpressed ALS1 and/or ALS3 genes in the isolates.[29,30]
CONCLUSION
The findings of this study demonstrate that the ALS genes expression contributes to adherence and fluconazole resistance in clinical isolates of Candida to the vagina. Regulatory network genes play a significant role in whether ALS genes are actively expressed or kept silence. The expression of ALS genes on biofilm cells is suggested in future studies.
Financial support and sponsorship
We would like to thank Tarbiat Modares University for financial support.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Naglik JR, Fostira F, Ruprai J, Staab JF, Challacombe SJ, Sundstrom P. Candida albicans HWP1 gene expression and host antibody responses in colonization and disease. J Med Microbiol. 2006;55:1323–7. doi: 10.1099/jmm.0.46737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves CT, Wei XQ, Silva S, Azeredo J, Henriques M, Williams DW. Candida albicans promotes invasion and colonisation of Candida glabrata in a reconstituted human vaginal epithelium. J Infect. 2014;69:396–407. doi: 10.1016/j.jinf.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Brockert PJ, Lachke SA, Srikantha T, Pujol C, Galask R, Soll DR. Phenotypic switching and mating type switching of Candida glabrata at sites of colonization. Infect Immun. 2003;71:7109–18. doi: 10.1128/IAI.71.12.7109-7118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun JN, Solis NV, Phan QT, Bajwa JS, Kashleva H, Thompson A, et al. Host cell invasion and virulence mediated by Candida albicans. PLoS Pathog. 2010;6:1–14. doi: 10.1371/journal.ppat.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gropp K, Schild L, Schindler S, Hube B, Zipfel PF, Skerka C. The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol Immunol. 2009;47:465–75. doi: 10.1016/j.molimm.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, Oh SH, Hoyer LL. Unequal contribution of ALS9 alleles to adhesion between Candida albicans and human vascular endothelial cells. Microbiology. 2007;153(Pt 7):2342–50. doi: 10.1099/mic.0.2006/005017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hube B, Stehr F, Bossenz M, Mazur A, Kretschmar M, Schäfer W. Secreted lipases of Candida albicans: Cloning, characterisation and expression analysis of a new gene family with at least ten members. Arch Microbiol. 2000;174:362–74. doi: 10.1007/s002030000218. [DOI] [PubMed] [Google Scholar]
- 8.Hoyer LL, Green CB, Oh SH, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family – A sticky pursuit. Med Mycol. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, et al. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150:2415–28. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- 10.Du H, Guan G, Xie J, Sun Y, Tong Y, Zhang L, et al. Roles of Candida albicans Gat2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS One. 2012;7:e29707. doi: 10.1371/journal.pone.0029707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F, Svarovsky MJ, Karlsson AJ, Wagner JP, Marchillo K, Oshel P, et al. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot Cell. 2007;6:931–9. doi: 10.1128/EC.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murciano C, Moyes DL, Runglall M, Tobouti P, Islam A, Hoyer LL, et al. Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PLoS One. 2012;7:e33362. doi: 10.1371/journal.pone.0033362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoyer LL, Hecht JE. The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast. 2001;18:49–60. doi: 10.1002/1097-0061(200101)18:1<49::AID-YEA646>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell. 2011;10:168–73. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaller M, Bein M, Korting HC, Baur S, Hamm G, Monod M, et al. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect Immun. 2003;71:3227–34. doi: 10.1128/IAI.71.6.3227-3234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stehr F, Felk A, Gácser A, Kretschmar M, Mähnss B, Neuber K, et al. Expression analysis of the Candida albicans lipase gene family during experimental infections and in patient samples. FEMS Yeast Res. 2004;4:401–8. doi: 10.1016/S1567-1356(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 17.Roudbary M, Roudbarmohammadi SH, Bakhshi B, Farhadi Z. Relation of ALS 1 and ALS3 genes and fluconazole resistance in Candida albicans isolated from vaginal candidiasis. Inter J Mol Clin Microbiol. 2012;2:170–4. [Google Scholar]
- 18.Ausubel FM, Brent R, Kingston RE, More D. Current Protocols in Molecular Biology. John wiley & sons Inc; ringbou edition. 2003 [Google Scholar]
- 19.Kavanagh K. New Insight in Medical Mycology. Netherland: Springer; 2007. [Google Scholar]
- 20.Cheng G, Wozniak K, Wallig MA, Fidel PL, Jr, Trupin SR, Hoyer LL. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect Immun. 2005;73:1656–63. doi: 10.1128/IAI.73.3.1656-1663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nas T, Kalkanci A, Fidan I, Hizel K, Bolat S, Yolbakan S, et al. Expression of ALS1, HWP1 and SAP4 genes in Candida albicans strains isolated from women with vaginitis. Folia Microbiol (Praha) 2008;53:179–83. [PubMed] [Google Scholar]
- 22.Dhamgaye S, Bernard M, Lelandais G, Sismeiro O, Lemoine S, Coppée JY, et al. RNA sequencing revealed novel actors of the acquisition of drug resistance in Candida albicans. BMC Genomics. 2012;13:396. doi: 10.1186/1471-2164-13-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Sánchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d’Enfert C. Candida albicans biofilms: A developmental state associated with specific and stable gene expression patterns. Eukaryot Cell. 2004;3:536–45. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.İnci M, Atalya MA, Ozer B, Evirgen O, Duran N, Nedretkoc A, et al. Investigations of ALS1 and HWP1 genes in clinical isolates of Candida albicans. Turk J Med Sci. 2013;43:125–30. [Google Scholar]
- 25.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–94. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumamoto CA. Candida biofilms: An update. Eukaryot Cells. 2005;4:633–638. doi: 10.1128/EC.4.4.633-638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uwamahoro N, Qu Y, Jelicic B, Lo TL, Beaurepaire C, Bantun F, et al. The functions of Mediator in Candida albicans support a role in shaping species-specific gene expression. PLoS Genet. 2012;8:e1002613. doi: 10.1371/journal.pgen.1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modrzewska B, Kurnatowski P. Adherence of Candida sp. to host tissues and cells as one of its pathogenicity features. Ann Parasitol. 2015;61:3–9. [PubMed] [Google Scholar]
- 29.Argimón S, Wishart JA, Leng R, Macaskill S, Mavor A, Alexandris T, et al. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot Cell. 2007;6:682–92. doi: 10.1128/EC.00340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastidas RJ, Heitman J, Cardenas ME. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 2009;5:e1000294. doi: 10.1371/journal.ppat.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]


