Abstract
The Aspergillus nidulans proteome possesses a single formin, SepA, which is required for actin ring formation at septation sites and also plays a role in polarized morphogenesis. Previous observations imply that complex regulatory mechanisms control the function of SepA and ensure its correct localization within hyphal tip cells. To characterize these mechanisms, we undertook a screen for mutations that enhance sepA defects. Of the mutants recovered, mesA1 causes the most dramatic defect in polarity establishment when SepA function is compromised. In a wild-type background, mesA1 mutants undergo aberrant hyphal morphogenesis, whereas septum formation remains unaffected. Molecular characterization revealed that MesA is a novel fungal protein that contains predicted transmembrane domains and localizes to hyphal tips. We show that MesA promotes the localized assembly of actin cables at polarization sites by facilitating the stable recruitment of SepA. We also provide evidence that MesA may regulate the formation or distribution of sterol-rich membrane domains. Our results suggest that these domains may be part of novel mechanism that directs SepA to hyphal tips.
INTRODUCTION
Fungal hyphae represent one of the most polarized cellular structures found in nature. Hyphae grow solely by apical extension, whereby cell surface expansion and cell wall deposition are confined to a discrete region at the hyphal tip (Gooday, 1971). This mode of growth is supported by extreme polarization of the cytoskeleton and endomembrane network, which permits the long-range transport of vesicles containing precursors required for apical extension to the tip region (Grove and Bracker, 1970; Bourett and Howard, 1991; Roberson and Vargas, 1994). Although hyphae are a defining feature of filamentous fungi, the molecular mechanisms underlying hyphal morphogenesis remain poorly understood. However, a general paradigm for these mechanisms has emerged from the detailed study of polarity establishment in yeast cells (Pruyne and Bretscher, 2000a, 2000b; Chang and Peter, 2003). This paradigm is based on the notion that positional cues bias intrinsic cellular asymmetries to generate an axis of polarity that is subsequently stabilized by localized activation of Rho-related GTPase signaling modules. Polarization of the cytoskeleton and other components of the morphogenetic machinery follow in response to the GTPase signals. Functional studies have revealed that elements of the fungal cytoskeleton and endomembrane network play conserved roles in the establishment and maintenance of hyphal polarity (reviewed by Wendland, 2001). For example, type I myosin appears to direct localized vesicle transport along microfilaments in the vicinity of the hyphal tip (McGoldrick et al., 1995), whereas the microtubule motors kinesin and dynein regulate long-range vesicle transport (Seiler et al., 1997; Inoue et al., 1998; Wu et al., 1998). In contrast, the role of Rho-related GTPases in polarized growth may not be strictly conserved between filamentous fungi and yeast (Boyce et al., 2001; Wendland and Philippsen, 2001). Moreover, several well-characterized positional cues from yeast, such as Bud8p, Bud9p, and Bud10p, appear to be absent from the fungal proteome (Harris and Momany, 2004). Accordingly, additional genetic approaches are needed to understand how positional information is generated and relayed to the morphogenetic machinery in filamentous fungi.
Ultrastructural analysis and live cell imaging have revealed the importance of the Spitzenkorper in hyphal morphogenesis. The Spitzenkorper is a dynamic body that consists of an aggregate of vesicles located immediately behind the hyphal tip (Lopez-Franco and Bracker, 1996). Mathematical models and experimental observations support the notion that the Spitzenkorper regulates hyphal extension by functioning as a vesicle distribution center (Bartnicki-Garcia et al., 1989). For example, changes in the direction of hyphal extension clearly correlate with alterations in the position of the Spitzenkorper (Riquelme et al., 1998). Functional microtubules appear to be required for both the formation and proper localization of the Spitzenkorper (Riquelme et al., 2000). In addition, vesicle transport to and from the Spitzenkorper is largely controlled by kinesins and dynein (Seiler et al., 1999; Riquelme et al., 2000).
The filamentous fungus Aspergillus nidulans produces dormant conidiospores that germinate in response to glucose signals (Osherov and May, 2000). During germination, spores undergo an initial period of isotropic swelling, which is then followed by the establishment of an axis of hyphal polarity and emergence of a germ tube (reviewed by d'Enfert, 1997). Once new hyphae grow to a sufficient extent, they are partitioned by the formation of septa near the spore/germ tube junction (reviewed by Harris, 2001). Additional polarity establishment events lead to the formation of lateral branches from primary hyphae, and also underlie the emergence of secondary and tertiary hyphae from the original spore (reviewed by Momany, 2002).
Formins are large proteins that function as molecular scaffolds to integrate cellular signals with localized reorganization of the actin cytoskeleton (reviewed by Evangelista et al., 2003). On interaction with activated Rho-related GTPases, formins are capable of directing the formation of actin cables (Evangelista et al., 2002; Sagot et al., 2002a). The FH1 and FH2 domains, which are characteristic features of most formins, are necessary and sufficient for this activity (Pruyne et al., 2002; Sagot et al., 2002b). In contrast, the amino-terminal GTPase-binding domain and the FH3 domain regulate formin activation and localization (Petersen et al., 1998; Ozaki-Kuroda et al., 2001). In yeast, the formin Bni1p is the central component of a large multiprotein complex known as the polarisome (Sheu et al., 1998), which mediates the formation of actin cables at presumptive bud sites in yeast. Other components of the polarisome, such as Bud6p and Spa2p, appear to facilitate proper Bni1p activation and localization, respectively (Sagot et al., 2002a).
The A. nidulans proteome contains a single formin, SepA, which is required for septum formation and also plays a role in the establishment and maintenance of hyphal polarity (Harris et al., 1997, 1999). SepA localizes to septation sites, where it forms a dynamic ring structure, and to hyphal tips, where it appears as a thin crescent subtended by a bright spot that likely corresponds to the Spitzenkorper (Sharpless et al., 2002). Because these structures can exist simultaneously in hyphal tip cells, their formation is presumably subject to precise temporal and spatial regulatory mechanisms. To gain insight into the nature of these mechanisms, we used a genetic screen to identify mutations that enhance sepA defects. Here, we present the functional characterization of a novel gene product, MesA, which was identified in this screen. We propose that MesA acts as a spatial determinant to stabilize axes of polarized hyphal extension in A. nidulans.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions
All A. nidulans strains used in this study are described in Table 1. Media used for the growth of A. nidulans include the following: CM (1% dextrose, 0.2% peptone, 0.1% yeast extract, 0.1% casamino acids, nitrate salts, vitamins, and trace elements; pH 6.5), MAG (2% dextrose, 2% malt extract, 0.2% peptone, trace elements, and vitamins), YGV (2% dextrose, 0.5% yeast extract, and vitamins), and MNV (1% dextrose, 5% nitrate salts, trace elements, and vitamins, pH 6.5). MNV-G and MNV-E were identical to MNV, except that glycerol (50 mM) and ethanol (1.2%) were used, respectively, in place of dextrose as the sole carbon source. Nitrate salts, trace elements, and vitamins were added as described in the Appendix to Kafer (1977). Arginine (1 mM), uridine, (5 mM), and uracil (10 mM) were added as needed. Media were solidified using 1.5% agar. Additional media supplements and protocols for mitotic mapping are provided in the Appendix to Kafer (1977).
Table 1.
Strain list
| Strain | Relevant genotype | Source |
|---|---|---|
| GR5 | pyrG89; pyroF4; wA3 | FGSCa |
| ASH162 | pyrG89; pabaA6 yA2 | Lab stock |
| ASH630 | sepA1 pyrG89; wA3 | Lab stock |
| ASH35 | sepA1; argB2; yA2 | Lab stock |
| AKS3 | alcA(p)::sepA::pyr-4 pyrG89; wA3; pyroA4 | This study |
| AKS70 | sepA::GFP::pyr-4 pyrG89 pabaA6 yA2 | Lab stock |
| ACP20 | mesA1 wA3; alcA(p)::sepA::pyr-4 pyrG89; pyroF4 | This study |
| ACP21 | mesA1 wA3; pyrG89 | This study |
| ACP65 | sepA1 yA2; mesA1 | This study |
| ACP79 | mesA::FLAG::pyr-4 mesA1; pyrG89 | This study |
| ACP86 | tpmA::GFP::pyr-4; pyrG89; mesA1 | This study |
| ACP94 | mesA1; sepA::GFP::pyr-4 pyrG89 | This study |
| ACP108 | mesA::FLAG::pyr-4 wA3; pyrG89 | This study |
| ACP115 | tpmA::GFP::pyr-4; pyrG89; wA3 | This study |
Fungal Genetic Stock Center, Department of Microbiology, University of Kansas Medical Center, Kansas City, KS 66160-7420
Identification and Genetic Analysis of mes Mutants
Strain AKS3 was obtained by transforming GR5 (Table 1) with pKES4, which was obtained by cloning a 916-base pair SpeI-XbaI fragment from sepA (encompassing 58-base pairs of upstream sequence plus the first 286 amino acids of SepA) into pAL3x (Waring et al., 1989). Homologous integration at the sepA locus resulted in a strain possessing a single functional copy of sepA driven by the alcohol-inducible alcA promoter. Accordingly, on inducing (MNV-E) or nonrepressing (MNV-G) media, growth and hyphal morphology of AKS3 were indistinguishable from wild type. In contrast, on repressing media (YGV or MAG), AKS3 colonies displayed restricted growth, and hyphae appeared similar in morphology to sepA4 mutants incubated at restrictive temperature (Harris et al., 1997). Furthermore, as in sepA4 mutants (Harris et al., 1997), the morphological defects were exacerbated by growth at 42°C, and AKS3 hyphae eventually septated under all conditions. Therefore, growth of AKS3 on repressing media appears to approximate the phenotypes caused by the hypomorphic sepA4 mutation (Sharpless and Harris, 2002).
AKS3 conidia were mutagenized with 4-nitroquinoline-1-oxide as described previously (Harris et al., 1994) and then plated on MNV-G supplemented with 0.01% Triton X-100 to restrict colony growth. After 3 days' growth at 28°C, surviving colonies were replica plated onto MAG plates and incubated at 42°C for 36–48 h. Colonies unable to grow on MAG were subsequently picked and retested for growth on MNV-G at 42°C (to eliminate general temperature sensitive mutants) and MAG at 28°C (to eliminate mutants with general defects in glucose metabolism). Finally, candidates incubated on MAG plates at 42°C were screened for obvious defects in hyphal morphology that were at least partially rescued by growth on MNV-G at 42°C or MAG at 28°C. Standard approaches were used for the genetic analyses of all mutants (Harris et al., 1994).
Cloning and Characterization of the mesA Gene
Strain ACP21 was transformed with the pRG3-AMA1 plasmid-based genomic library (Osherov and May, 2000; kindly provided by G. May). Transformations were performed using a standard protocol with the following modifications. First, protoplasts were generated from germinating conidia using a lytic mix containing 6 mg/ml Driselase (Sigma, St. Louis, MO), 6 mg/ml lysing enzymes (Sigma), and 14 mg/ml β-d-glucanase (Interspex, Foster City, CA). Second, protoplasts were recovered by centrifugation after they were layered on 20 ml of 1 M sucrose. Third, protoplasts were left on ice overnight at 4°C before transformation. Fourth, transformed protoplasts were plated in 8 ml of molten YGV agar supplemented with 1 M sucrose and left at room temperature overnight. Finally, 25 ml of molten YGV agar was poured onto each plate the following morning, and the plates were transferred to 42°C (with appropriate controls left at 28°C).
pCP1 is the original pRG3-AMA1–based complementing plasmid recovered from mesA1 transformants. To localize complementing activity, pCP1 was digested with KpnI or PstI and religated to produce pCP2 and pCP3, respectively. To construct pCP6 and pCP7, a 3-kb EcoRI fragment derived from pCP1 was subcloned into pBS-KS- (Stratagene, La Jolla, CA) or pRG3 (Waring et al., 1989), respectively. cDNA fragments were obtained by RT-PCR using the RETROscript RT-PCR kit (Ambion, Austin, TX). DNA sequencing was performed with custom oligonucleotide primers using the Molecular Core Facility at the University of Connecticut Health Center. The DNA sequence of mesA is deposited in GenBank (accession no. AAP31022.1).
Restriction fragment–based cotransformation experiments using pRG3-AMA1 were used to localize the mesA1 mutation to a 1.1-kb KpnI fragment derived from the 5′ end of the predicted coding region. Using genomic DNA from ACP21 as a template, three independent PCR products derived from this region were obtained and, along with wild-type control samples, sequenced using the Genomics Core Facility of the University of Nebraska Center for Biotechnology.
Standard approaches were used for the preparation of genomic DNA from A. nidulans (Oakley and Osmani, 1993). Total RNA was prepared from lyophilized mycelia using Trizol reagent (Invitrogen, Carlsbad, CA). Oligonucleotides were obtained from IDT, Inc. (Coralville, IA; sequences will be provided upon request).
Bioinformatic Analysis of MesA
MacVector software (Oxford Molecular Sciences, Inc., Hunt Valley, MD) was used for processing and analysis of DNA sequences. Potential transmembrane regions were identified using TM-PRED. Fungal homologues were identified by performing BLAST searches in the following public databases: http://www.broad.mit.edu/annotation/fungi/fgi/, http://www.tigr.org/tdb/fungal/, http://www-sequence.stanford.edu/group/candida/, http://genome.jgi-psf.org/whiterot/, and http://cogeme.ex.ac.uk/.
Construction of mesA::FLAG and tpmA::GFP
The mesA::FLAG vector, pCP30, was constructed in two steps. First, a 2.53-kb fragment, encompassing the entire mesA coding region plus 54 base pairs of upstream sequence, was amplified from pCP1 using the PCR and cloned into pCR2.1-TOPO (Invitrogen) to create pCP28. Second, pCP28 was digested with EcoRI and BamHI to release mesA::FLAG, which was then cloned into pRG3 to generate pCP30. Strain ACP108 was obtained by transforming GR5 with pCP30. Southern analysis was used to identify transformants in which mesA::FLAG had undergone homologous integration at the mesA locus, such that its expression is controlled by endogenous promoter sequences. ACP108 possesses a single integrated copy of mesA::FLAG at the mesA locus, plus three additional ectopic copies. However, when compared with a wild-type control, or to a transformant containing no additional ectopic copies of mesA::FLAG, the kinetics of spore polarization, septation, and general growth are identical. The ability of pCP30 to complement mesA1 growth defects in strain ACP79 confirmed that the mesA::FLAG construct is functional. Expression of mesA::FLAG was verified using standard protocols for protein extraction, SDS-PAGE, Western blotting, and chemiluminescent detection. MesA-FLAG was detected using the anti-FLAG M2 mAb (Sigma) at 1 μg/ml. The secondary antibody was peroxidase-conjugated goat anti-mouse (Sigma) diluted to 1:10,000.
To facilitate the construction of protein fusions to the N-terminus of GFP, the GFP coding region was amplified from pMCB32 (Fernandez-Abalos et al., 1998) and cloned into pCR2.1-TOPO to create pCP15. Digestion of pCP15 with BamHI and SphI released a 750-base pair fragment that was cloned into pRG3 to generate pCP19.
The tpmA::GFP fusion was constructed in two steps. First, a 1-kb fragment, encompassing the entire coding region of tpmA, was amplified from genomic DNA and cloned into pCR2.1-TOPO to create pCP29. Second, pCP29 was digested with EcoRI and BamHI to release tpmA, which was cloned into pCP19 to generate pCP32. Strains ACP86 and ACP115 were obtained by transforming ACP21 and GR5, respectively, with pCP32. Because of the lack of a viable tpmA mutant, it was not possible to determine if TpmA-GFP is fully functional. However, when the spore polarization kinetics of ACP86 and ACP115 were compared with their respective wild-type and mesA1 parents, no dramatic differences were observed. For example, after 10 h in YGV media, 99.5% of ACP115 (tpmA::GFP) spores had produced a detectable germ tube, compared with 96.5% of its wild-type parent strain (n = 200). Similarly, 70% of ACP86 (mesA1 tpmA::GFP) spores had polarized, compared with 86% of its mesA1 parent. Because TpmA-GFP does not modify mesA1 morphogenetic defects, we conclude that it is likely to be functional. In addition, we verified that TpmA-GFP detects actin cables by showing that exposure of growing ACP115 hyphae to cytochalasin A (CA; 2 μg/ml; Sigma) for 10 min abolished all detectable signal (Supplemental Figure 1).
All amplification steps were done using Expand High Fidelity Taq polymerase (Roche, Indianapolis, IN) and PCRx enhancer buffers (Invitrogen).
Microscopy
Coverslips with adherent cells were processed for microscopy and stained with Hoechst 33258 (Somerville, NJ) and Calcofluor (Molecular Probes, Eugene, OR) as described previously (Harris et al., 1994). To examine patterns of chitin deposition, strains were grown at 42°C in YGVUU for 6–12 h and then stained for 5 min with wheat germ agglutinin (Sigma) that had been diluted (1/1000) in prewarmed media. To visualize proteins fused to GFP, hyphae were grown on coverslips in YGV, briefly rinsed in sterile water, and mounted. The effect of sphingolipid depletion on SepA-GFP, TpmA-GFP, and MesA-FLAG localization was assessed by shifting hyphae grown in YGV for 10 h at 28°C into YGV + 80 μg/ml myriocin (Sigma). This compound inhibits serine palmitoyltransferase, the first committed step in sphingolipid biosynthesis, and blocks spore polarization without adversely affecting growth or nuclear division (Cheng et al., 2001). Samples were examined for up to 120 min after the shift.
To examine sterol distribution, strains were grown at 28°C or 42°C in YGVUU for 12 h and then stained for no more than 5 min with filipin (Sigma) at a concentration of 25 μg/ml in prewarmed growth media. They were then washed in growth media and mounted. Lower concentrations of filipin (i.e., 12.5 μg/ml) produced an identical, but much fainter, staining pattern (Supplemental Figure 2). Recent observations using yeast question the reliability of filipin as a marker for sterol distribution in living cells (Valdez-Taubas and Pelham, 2003). Accordingly, we also examined the pattern of filipin staining in formaldehyde-fixed hyphae (Supplemental Figure 2). Although background staining was much higher, a prominent, but somewhat smaller, patch was still observed at hyphal tips.
To determine how disruption of the cytoskeleton affects the localization of MesA-FLAG, ACP108 hyphae (plus appropriate wild-type controls) were grown at 28°C in YGV for 16 h and then treated with CA (Sigma; 2 μg/ml) or benomyl (Sigma; 5 μg/ml). Hyphae treated with CA were monitored at 10-min intervals for 30 min, whereas benomyl-treated hyphae were examined at 2 h after treatment.
Indirect immunofluorescence was performed as described previously (Harris et al., 1999). Because of the apparent accumulation of cell wall polymers, we were unable to establish conditions for cell wall removal that permitted optimal antibody accessibility in mesA1 mutants grown at 42°C. Accordingly, immunofluorescence experiments utilized cultures grown at 28°C, a temperature at which the mesA1 mutant still displays obvious polarity defects. The anti–α-tubulin mAb DM1A (Sigma) was used at a 1/200 dilution to detect microtubules. The anti-actin mAb C4 (ICN Biomedicals, Inc, Costa Mesa, CA) was used at a 1/500 dilution to detect actin. The anti-FLAG mAb M2 (Sigma) was used at 10 μg/ml to detect MesA-FLAG. A 1/200 dilution of FITC-conjugated rabbit anti-mouse IgG (Sigma) was used as the secondary antibody.
Slides were viewed using the 60× or 100× objectives (PlanApo) of an Olympus BX51 fluorescent microscope. Images were captured with a Photometrics CoolSnap HQ CCD camera (Roper Scientific, Tucson, AZ), and processed using IPLab software (Scanalytics, Inc., Billerica, MA). Images were converted from 16–8 bits and saved as TIF files. Additional processing was performed using Adobe PhotoShop 6.0 (San Jose, CA). Live cell and confocal images were obtained using the 60× water immersion lens (PlanApo) of an Olympus BX50 microscope equipped with a Laser Sharp MRC-1024 confocal scanning system (v3.2; Bio-Rad, Richmond, CA). Images were captured by direct acquisition with Z step ranging from 0.5 to 2 μm. They were subsequently processed using ImageJ (National Institutes of Health, Bethesda, MD; maximum pixel intensity), Imaris (Bitplane Inc., St. Paul, MN; for display of 3D images in Figures 5 and 6; maximum pixel intensity for projection of Z series), and Adobe PhotoShop 6.0.
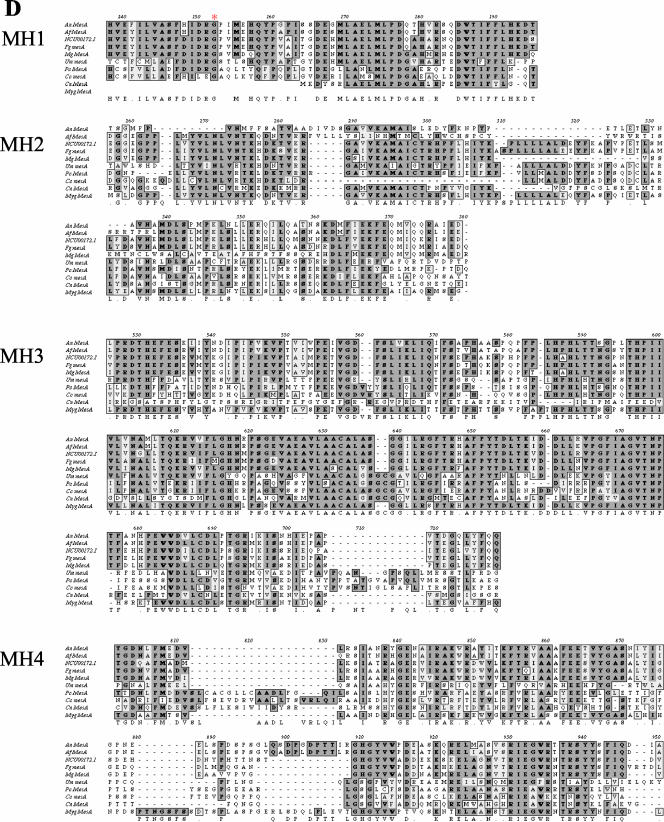
Figure 5.
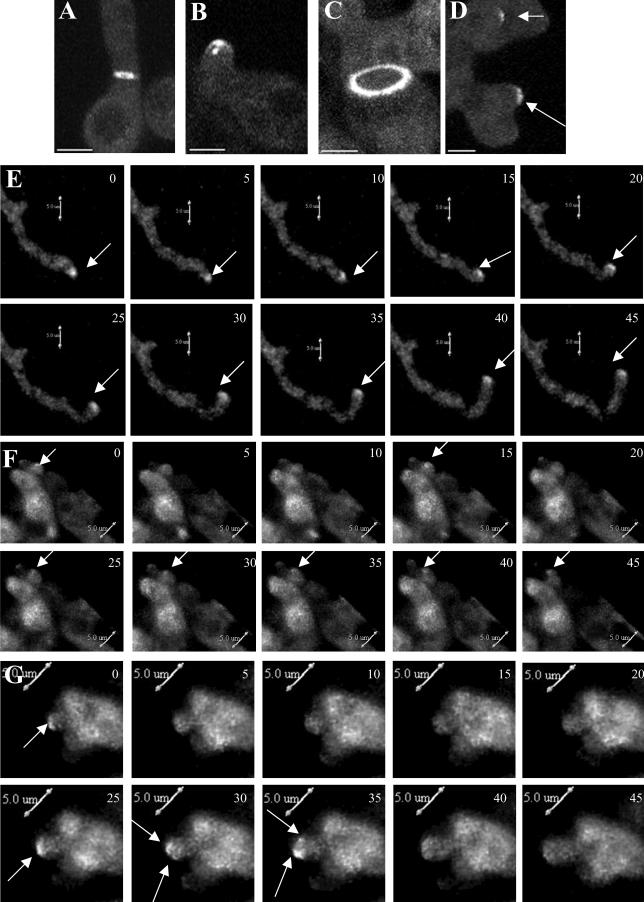
The mesA1 mutation affects the stabilization of SepA-GFP at hyphal tips. Wild-type (AKS70; A, B, and E) and mesA1 mutant (ACP94; C, D, F, and G) hyphae were grown in YGV at 28°C for 12 h, and live cell images were taken at 5-min intervals. Images were captured by confocal microscopy with a Z stop of 2 μm, 60× immersion lens (Kalmon 2) and processed with Imaris (maximum pixel intensity). (A) SepA-GFP ring at a septation site in a wild-type hypha. (B) SepA-GFP crescent with subtending spot at the tip of a wild-type hypha. (C) SepA-GFP ring at a septation site in a mesA1 hypha. (D) SepA-GFP patches (arrows) at the tips of a mesA1 hypha. (E) Persistent localization of SepA-GFP (arrows) at the extending tip of a wild-type hypha. Numbers in the top right corner indicate time in minutes. (F) Sporadic localization of SepA-GFP (arrows) at the tip of a mesA1 hypha. A weak patch is evident at time zero that disappears, reappears at 15 min, disappears again, and then reappears and persists from 25 through 45 min. (G) Splitting of a SepA-GFP patch (arrows) at the tip of a mesA1 hypha. The patch, which disappears between 5 and 20 min, reappears at 25 min before splitting into two discrete patches at 30 min. Bars, 5 μm.
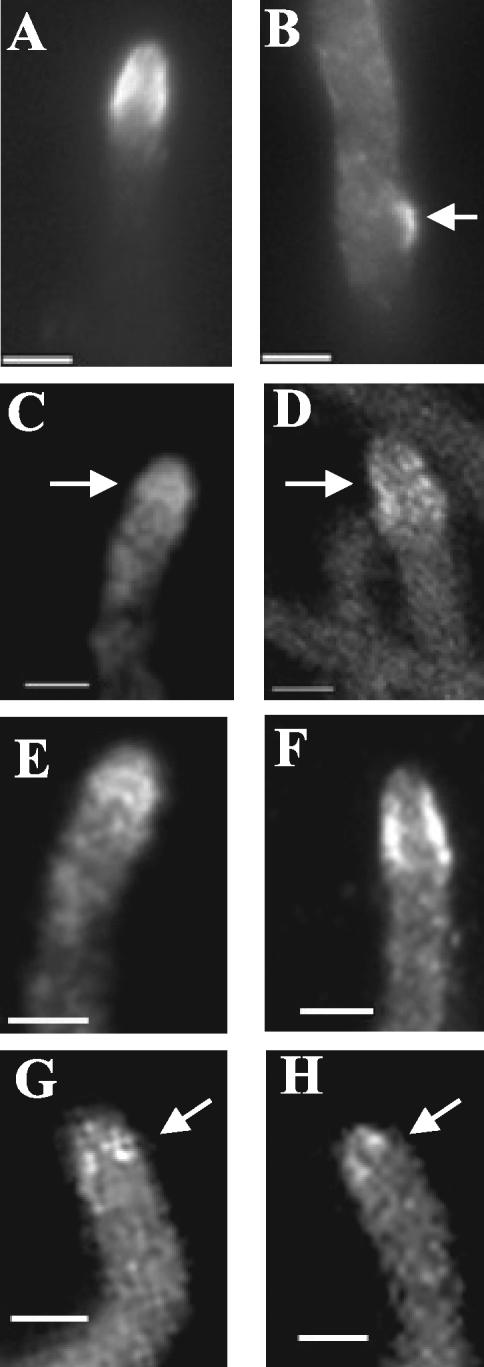
Figure 6.
Actin cables are absent from the tips of mesA1 hyphae. Wild-type (ACP115; A and C–F) and mesA1 mutant (ACP86; B and G–J) hyphae were grown in YGV at 28°C for 12 h. Images were captured by confocal microscopy with a Z stop of 2 μm, 60× immersion lens (Kalmon 2) and processed with Imaris (maximum pixel intensity). (A) In wildtype hyphae, TpmA-GFP localizes to tips and to rings at septation sites. A tip-associated actin cable is shown in the inset. (B) In mesA1 mutants, TpmA-GFP localizes to rings at septation sites, but no longer appears at tips. (C–F) Higher resolution images showing TpmA-GFP localization at wild-type hyphal tips. Prominent (D, red arrow) and faint (C, white arrow) actin cables are indicated. (G–J) Higher-resolution images showing of TpmA-GFP localization at mesA1 hyphal tips. At best, faint patches are observed (G, white arrow). Bars, 3 μm.
Characterization of Yeast MesA Homologues
Haploid and diploid Saccharomyces cerevisiae Δyor129c strains were obtained from Research Genetics (Huntsville, AL). Schmoo formation, cell fusion, and budding pattern were scored using previously described protocols (Gammie et al., 1998; Ni and Snyder, 2001).
The following S. pombe strains were used in this study; TE365 (h+, ura4-D18), TE366 (h-, ura4-D18), TE236 (h-, leu1-322, ura4-D18), and TE237 (h+, leu1-322, ura4-D18). Standard protocols were used for the growth, transformation and analysis of mating in fission yeast (available from the following Web site: http://www.hsph.harvard.edu/wolflab/Protocols/Protocols.html). The SPBC776.06c coding region was precisely deleted using a previously described PCR-mediated approach (Bahler et al., 1998). Transformants possessing the correct gene replacement were identified by PCR using SPBC776.06c and URA4 gene-specific oligonucleotide primers. Cell lengths were obtained from cultures grown to logarithmic phase and stained with both Hoechst and Calcofluor. Measurements of cell length and width (n = 100) were obtained using IPLab software and were tabulated using Microsoft Excel.
RESULTS
Screen for Enhancers of sepA Defects
Our previous observations suggest that complex regulatory mechanisms control the function and localalization of sepA in hyphal tip cells (Sharpless and Harris, 2002). We reasoned that it would be possible to further understand the nature of these mechanisms by using an enhancer screen similar to that described by Efimov and Morris (1998). This screen requires a starting strain possessing a target gene (i.e., sepA) whose expression is regulated by the glucose-repressible alcA promoter. Accordingly, we constructed strain AKS3 (see MATERIALS AND METHODS) and showed that it displays wild-type growth on nonrepressing media (i.e., MNV-G). However, on repressing media (i.e., MAG), the strain exhibits morphological phenotypes in common with the hypomorphic sepA4 mutation (Harris et al., 1997). Nonetheless, AKS3 still forms small colonies on MAG plates. Taking advantage of this phenotype, we conducted a genetic screen for Ts mutations that abolish AKS3 colony formation on MAG and display associated morphological defects.
From a total of 48,000 mutagenized conidia, 97 candidate mutants were selected for subsequent genetic analysis. Of this group, 78 mutants failed to produce viable cleistothecia when backcrossed to strain ASH162. Of the remaining 19 mutants, ≥20 Pyr+ random ascospore progeny (i.e., containing the pyr-4 marked alcA::sepA construct) from the ASH162 backcross were analyzed to determine if failure to grow on MAG plates at 42°C cosegregated with the aberrant morphology and was likely caused by mutation of a single gene. Only four mutants, each of which defined a single linkage group (mesA–mesD), were retained for further characterization. Preliminary observations suggested that the mesA1 and mesD1 mutations enhance sepA defects, whereas mesB1 and mesC1 are Ts mutations that are partially suppressed by the modest increase in sepA expression conferred by growth on MNV-G. Because the mesA1 mutation appears to display the strongest interaction with sepA, we chose to characterize it in further detail.
Phenotypic Characterization of mesA1 Mutants
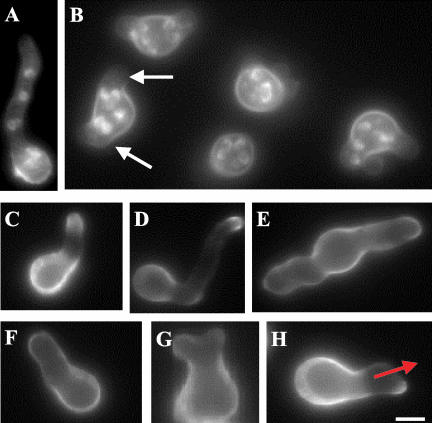
When incubated on repressing MAG media at 42°C, mesA1 alcA(p)::sepA::pyr-4 conidia displayed dramatic morphological defects indicative of problems in establishing hyphal polarity. Many spores (i.e., >80%) formed large, swollen, multinucleate (i.e., typically ≥32 nuclei) spheres that ultimately lysed (Figure 1). The remainder formed short, wide, apolar hyphae that also lysed. Even at 28°C, mesA1 alcA(p)::sepA::pyr-4 conidia produced abnormally wide hyphae that appeared to have no defined axis of extension. To verify that these synthetic phenotypes were not due to a unique interaction between mesA1 and the alcA(p)::sepA::pyr-4 allele, we constructed mesA1 sepA1 double mutants. As expected, because the sepA1 allele causes defects that are more severe than the “alcoholic” allele, the double mutant exhibited a stronger growth defects at 42°C and formed swollen, multinucleate spores that ultimately lysed. These observations suggest that mesA is required for polarity establishment when SepA function is compromised.
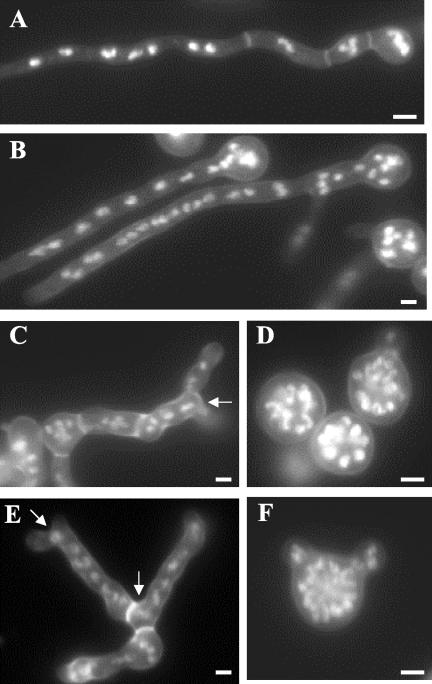
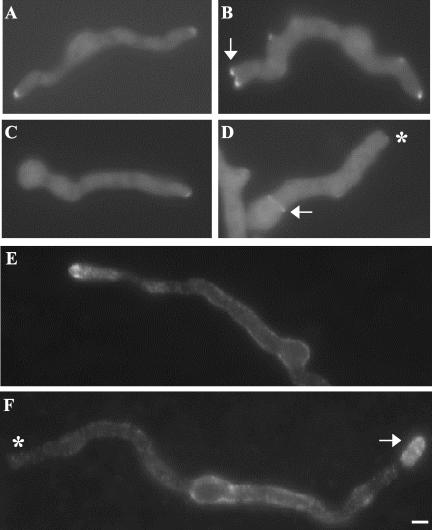
Figure 1.
Hyphal morphology of mesA1 sepA double mutants. Hyphae were grown for 12 h on repressing media (YGV) at 42°C. They were subsequently fixed and stained with Hoechst 33258 and Calcoflour to detect nuclei and cell wall material, respectively. (A) Wildtype (GR5) hyphae. (B) alcA::sepA (AKS3) hyphae. (C and E) mesA1 (ACP21) mutants. A characteristic feature of mesA1 mutants is apical branching leading to the formation of “split tips” (arrows). (D and F) alcA::sepA mesA1 (ACP20) double mutants. Note that most (i.e., 80%) sepA mesA1 double mutants fail to establish polarity at 42°C (D), whereas the remainder form abnormally wide germ tubes (F). Bar, 3 μm.
In an otherwise wild-type background, the mesA1 mutant produced a Ts colonial growth phenotype. The switch from isotropic expansion to apical growth was delayed in mutant conidia. Whereas wild-type conidiospores have uniformly produced germ tubes by the time they possess four nuclei (i.e., 97.0 ± 1.0%; n = 200), only 45.0 ± 1.0% of mesA1 spores possessing four nuclei had reached a similar stage despite having swollen to the same extent. In addition, when they did polarize, mesA1 conidiospores often produced multiple germ tubes (Figure 2, A and B), a condition that is rarely observed in wild-type spores. Once established, the axis of hyphal polarity was frequently altered in mesA1 mutants (Figure 2H), resulting in the formation of wide hyphae with an aberrant pattern of tip branching (Figure 1, C and E). Although the growth phenotype was more severe on plates at 42°C, the polarity defect was largely temperature independent. The effects of the mesA1 mutation appear to be restricted to polarity establishment and maintenance, because the timing and position of septum formation was indistinguishable from wild type. These observations demonstrate that MesA function is required for normal hyphal morphogenesis.
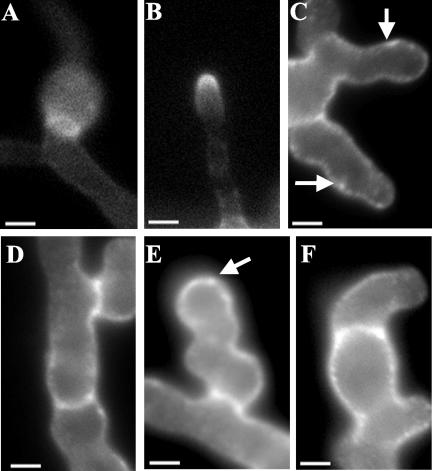
Figure 2.
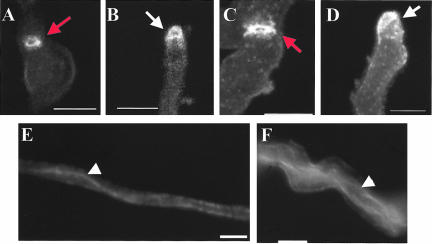
Morphological defects caused by the mesA1 mutation. Conidiospores were allowed to germinate in YGV for 7 h. They were subsequently fixed and stained with Hoechst 33258 and Calcofluor (A and B), or stained with FITC-conjugated WGA and fixed (C–H). (A) Wild-type (GR5) hyphae. (B) mesA1 (ACP21) mutants. Note that mesA1 mutants, despite possessing an equivalent number of nuclei, have only formed short germ tubes. Also, they frequently produce multiple germ tubes (white arrows). (C and D) Wild-type (GR5) hyphae, in which chitin deposition occurs at a discrete patch at the hyphal tip. (E–H) mesA1 (ACP21) mutants. Note that chitin appears to be deposited in a more random manner in mesA1 mutants. In H, although chitin deposition is properly localized to the hyphal tip, the polarity axis (red arrow) appears to be unstable and has been redirected. Bar, 4 μm.
To further characterize the morphogenetic defects caused by the mesA1 mutation, the pattern of cell wall deposition was examined in hyphae incubated at 42°C. In wild-type A. nidulans hyphae, FITC-conjugated wheat germ agglutinin stains a discrete patch at the hyphal tip that corresponds to active sites of chitin deposition (Figure 2, C and D). Although a similar patch was observed in mesA1 mutants, it was more diffuse and was accompanied by an increase in background staining (Figure 2, E–G). This observation suggests that cell wall deposition may be mislocalized in mesA1 mutants.
The polarity phenotypes caused by the mesA1 mutation could reflect an underlying defect in cytoskeletal organization. To address this possibility, the integrity of both microtubules and the actin cytoskeleton in hyphae grown at 28°C was examined by indirect immunofluorescence using the appropriate monoclonal antibodies. Despite their aberrant morphology, mesA1 mutants displayed a normal pattern of actin localization (Figure 3, A–D), including prominent staining at hyphal tips and rings at septation sites. Furthermore, cytoplasmic microtubules were intact and generally organized in longitudinal arrays (Figure 3, E and F). Therefore, the abnormal morphology of mesA1 hyphae is not caused by gross cytoskeletal defects.
Figure 3.
Actin and microtubule localization. Wild-type (GR5; A, B, and E) and mesA1 mutant (ACP21; C, D, and F) hyphae were grown for 12 h. They were subsequently fixed and processed for immunofluorescence microscopy to detect actin (A–D) or microtubules (E and F). Red arrows, actin rings at septation sites. White arrows, actin patch at hyphal tips; white arrowheads, cytoplasmic microtubules. Bars, 6 μm.
Molecular Characterization of MesA
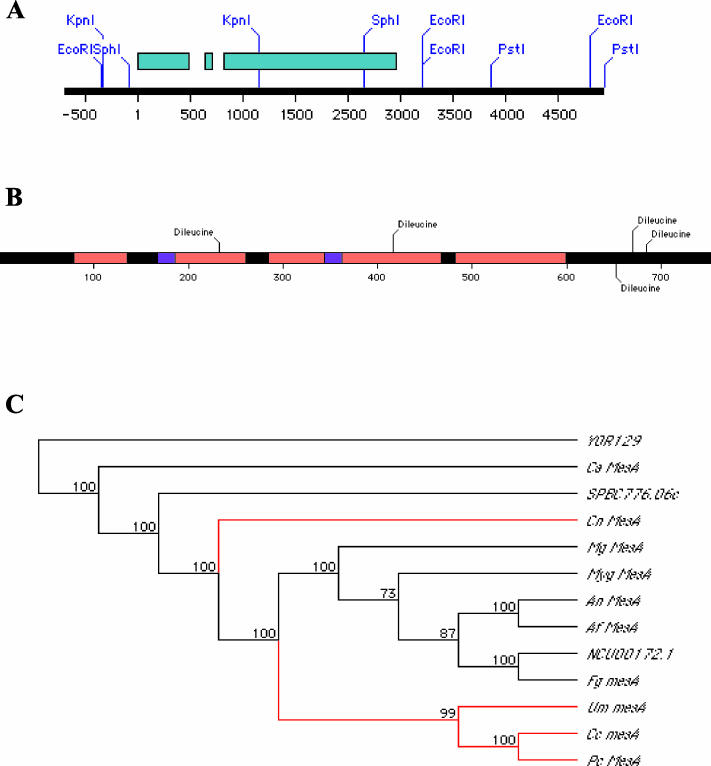
The mesA gene was cloned from the pRG3-AMA1 plasmid library by complementation of the Ts colonial growth phenotype. Analysis of subclones derived from the original insert localized complementing activity to the ∼4.0-kb PstI/SphI fragment cloned in pCP3 (Figure 4A). Two observations suggest that this fragment encompasses the bona fide mesA gene. First, it was able to complement mesA defects when present in low copy. Second, a smaller 1.1-kb KpnI/KpnI subclone (pCP21; Figure 4A) consistently yielded a low number of transformants, consistent with the notion that it was repairing the mesA1 mutation.
Figure 4.
Molecular characterization of MesA. (A) Schematic diagram of the insert from plasmid pCP1. Exons are depicted in blue. Coordinate number 1 refers to the A in the start codon. (B) Schematic depiction of MesA, showing conserved regions (red), potential transmembrane domains (purple), and dileucine motifs (black). (C) A cladogram depicting the evolutionary relationship between fungal MesA homologues. Ascomycete and basidiomycete lineages are shown in black and red, respectively. (D) Alignments of conserved domains (MH1–4) from MesA family members. The mesA1 mutation is located in the MH1 domain and is indicated by a red asterisk.
The DNA sequence of the 4.0-kb PstI/SphI fragment revealed the presence of a single open reading frame capable of encoding an 83-kDa protein with as estimated pI of 6.5. RT-PCR was used to confirm the location of two short introns at nucleotide positions 637–696 and 756–803 relative to the predicted start site (Figure 4A). A variety of bioinformatic tools were used to screen the predicted MesA sequence for potentially informative features. Multiple programs strongly suggested the presence of two potential transmembrane domains (Figure 4B). MesA appears to be oriented such that it possesses an extracellular loop, which is rich in serine and threonine residues, flanked by cytoplasmic N- and C-termini. The C-terminus includes a cluster of dileucine motifs that are typically involved in the endocytic retrieval of membrane proteins (Figure 4B).
BLAST searches using the predicted MesA sequence as the query failed to detect homology to any known animal or plant sequence. However, predicted homologues of MesA were found in the filamentous fungus Neurospora crassa (NCU00172.1; 53% identity, E = 0.0) and the fission yeast Schizosaccharomyces pombe (SPBC776.06c; 27% identity, E = 5e - 61). In addition, through the use of multiple iterative PSI-BLAST searches, weak homology to a predicted S. cerevisiae protein (YOR129c; 21% identity, E = 2e - 07) was observed. The function of the putative N. crassa and S. pombe homologues has not been characterized. In S. cerevisiae, YOR129c is not required for viability (Winzeler et al., 1999), but the observation that its expression is induced by mating pheromone in a Ste12p-dependent manner suggests that it may play a role in mating (Ren et al., 2000).
Systematic searches of several public databases (see MATERIALS AND METHODS) revealed that MesA is likely to be conserved throughout much of the fungal kingdom (Figure 4C). Common features of MesA homologues include the presence of at least two predicted transmembrane domains as well as four blocks of homology that we refer to as MH1–MH4 (Figure 4D). Strikingly, phylogenetic analyses demonstrated that homologues from filamentous ascomycetes and basidiomycetes cluster together, whereas homologues from ascomycete yeasts are more divergent (Figure 4C). This observation suggests that MesA function may be particularly important for the filamentous mode of fungal growth.
The mesA1 mutation maps to the first exon and consists of a G:C to C:G transversion that results in the substitution Gly93→Arg (Figure 4D). Notably, Gly93 is a strongly conserved residue that lies within the MH1 homology block. However, mesA1 is most likely a hypomorphic allele, because a null mutation constructed by gene replacement (deleting the entire mesA coding region plus 56-base pairs of upstream sequence) could not be recovered in haploid segregants derived from heterokaryotic transformants (C. Pearson and S. Harris, unpublished data).
MesA Is Required for the Formation of Actin Cables at Hyphal Tips
The recovery of the mesA1 mutation in a screen for enhancers of sepA defects suggested that MesA might regulate the activity and/or localization of SepA. To test this notion, we first examined SepA localization in mesA1 mutants. We previously reported that a functional SepA-GFP fusion protein localizes to hyphal tips, where it forms a crescent subtended by a bright spot that presumably corresponds to the Spitzenkorper (Sharpless and Harris, 2002). We generated mesA1 sepA::gfp::pyr-4 transformants and showed that, regardless of copy number, their growth and morphology was indistinguishable from mesA1 mutants. Three-dimensional confocal imaging (using Z-stacks) of an integrant possessing four copies of SepA-GFP revealed that it could form a broad crescent at hyphal tips, but the associated bright spot was never observed (Figure 5, A–D). Furthermore, when live cells were similarly imaged, SepA-GFP localization at the tips of mesA1 hyphae was unstable. In wild type, although the crescent and spot display rapid movement, they persist at the hyphal tip throughout an entire 45-min sequence of 4D confocal images (Figure 5E). In contrast, in mesA1 mutants, SepA-GFP frequently disappears within 5–10 min. In some cases, SepA-GFP reappeared at the same hyphal tip (59.7%; 43 of 72 tips analyzed; Figure 5F), whereas other SepA-GFP crescents appeared to split into two patches (16.7%; 12 of 72 tips analyzed; Figure 5G). These observations suggest that MesA may play a role in stabilizing SepA localization at hyphal tips.
By analogy to other formins (Evangelista et al., 2003), SepA is presumably required for the formation of actin cables. Therefore, the failure to stably recruit SepA to hyphal tips suggests that mesA1 mutants may not possess tip-associated actin cables. Because the antibody that was originally used to localize actin in wild-type and mesA1 hyphae does not discriminate between actin patches and cables, we constructed a GFP-labeled probe that would allow us to specifically detect cables (note that exhaustive attempts to stain A. nidulans actin structures with phalloidin have repeatedly failed; our unpublished results). Fusions of tropomyosin, an actin filament-stabilizing protein, to GFP have been effectively used for this purpose in yeast (Evangelista et al., 2002), although tropomyosin also associates with patches in S. pombe (Arai et al., 1998). Accordingly, we fused the A. nidulans tropomyosin gene, (AN5686.1, renamed as tpmA), to the N-terminus of GFP, such that its expression was under the control of endogenous promoter sequences. When transformed into wild-type or mesA1 strains, multiple copies of TpmA-GFP (i.e., 1–5 copies) did not alter the timing or pattern of morphogenetic events such as spore polarization or septation (see MATERIALS AND METHODS). As expected, TpmA-GFP localized to the tips of wild-type hyphae, where longer cables were occasionally observed and, unlike antiactin antibodies (Harris et al., 1994), actin patches were not detected (compare Figure 6, A and C–F, with Figure 3). In addition, it also formed a circumferential ring at septation sites (Figure 6A). Notably, TpmA-GFP localization was completely abolished by treatment with CA (Supplemental Figure 1), thereby suggesting that it is an effective probe for the detection of actin cables. By contrast, we were surprised to detect faint TpmA-GFP localization at the tips of sepA1 hyphae shifted to restrictive temperature (Supplemental Figure 1; see DISCUSSION). Although TpmA-GFP was found at septation sites in mesA1 mutants (Figure 6B), it was generally absent from hyphal tips, where, at best, punctate spots that may correspond to actin patches were infrequently seen (Figure 6, B and G–J). These observations suggest that the morphological defects caused by the mesA1 mutation may reflect the inability to assemble actin cables at incipient hyphal tips.
MesA Localizes to Hyphal Tips
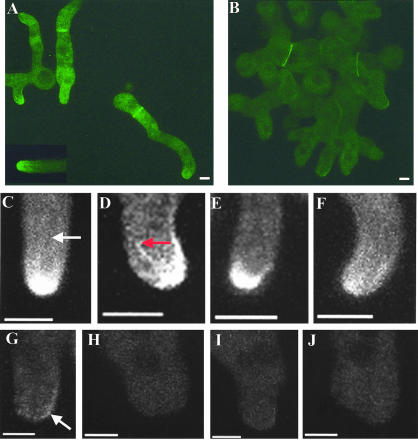
Because MesA appears to stabilize SepA localization at sites of polarized growth, we presumed that, like SepA, it would localize to hyphal tips. Because initial attempts to localize a functional MesA-GFP fusion protein were unsuccessful, we resorted to the use of a functional MesA-FLAG fusion that was expressed under the control of endogenous promoter sequences. As expected, MesA-FLAG localized to hyphal tips and sites of branch emergence (Figure 7, A and B). When examined by confocal microscopy (Figure 7, C–F), MesA-FLAG appeared to form a circumferential band that localized to the cell surface. Strikingly, the band is found in the subapical zone just behind the hyphal tip, and MesA seems to be largely excluded from the tip itself (Figure 7). By contrast, at an incipient polarization site (i.e., the branch site shown in Figure 7B), MesA appears to form a cell surface patch that will ultimately become a new tip. Consistent with the observation that the mesA1 mutation has no effect on septum formation, MesA-FLAG did not localize to septation sites.
Figure 7.
MesA localizes to hyphal tips in an actin-dependent manner. Strain ACP108 was grown in YGV at 28°C for 12 h. (A and B) Hyphae were fixed and processed for immunofluorescence microscopy using anti-FLAG antibodies. MesA-FLAG localization to a hyphal tip (A) and a branch site (B) are shown. (C–H) Images were captured by confocal microscopy, 100× objective, Z step of 0.5 μm, and processed with ImageJ (maximum pixel intensity). (C and D) Single sections showing MesA-FLAG localization at hyphal tips. (E and F) Stacked Z series of the sections shown in C and D, respectively. MesA-FLAG appears to form a circumferential band just behind the apex of the hyphal tip. (G and H) Hyphae were exposed to cytochalasin A (2 μg/ml) for 20 min before fixation. MesA-FLAG collapses into a series of punctate spots in the absence of actin filaments. Arrows, MesA-FLAG. Bars, 3 μm.
We previously reported that the maintenance of SepA-GFP localization at hyphal tips depended on the integrity of the actin cytoskeleton (Sharpless and Harris, 2002). To determine if filamentous actin has a similar role in MesA-FLAG localization, hyphae were examined after treatment with CA. A dose of 2 μg/ml CA, which was sufficient to eliminate all tip-associated TpmA-GFP signal (Supplemental Figure 1), disrupted organization of the subapical band of MesA-FLAG such that only a series of punctate spots remained (Figure 7, G and H). These spots bear some resemblance to the patch observed at an incipient polarization site (Figure 7B), which may reflect the unstable nature of these MesA structures. By contrast, as observed for SepA-GFP (Sharpless and Harris, 2002), microtubule depolymerization had no obvious affect on the localization of MesA-FLAG at hyphal tips. These observations suggest that actin cables may stabilize the localization of both SepA and MesA at hyphal tips.
Relationship between Sterol-rich Membrane Domains and SepA Localization
In yeast and animal cells, lipid microdomains play a key role in polarity establishment by allowing signaling and morphogenetic proteins to cluster within specific membrane regions (Simons and Toomre, 2000; Bagnat and Simons, 2002). Recently, sterol-rich membrane domains were implicated in the establishment and maintenance of hyphal polarity in A. nidulans (Cheng et al., 2001). This result could conceivably reflect a role for these domains in the localized recruitment of SepA. To test this notion, localization of SepA-GFP and TpmA-GFP was examined in hyphae treated with the chemical inhibitor myriocin, which blocks the first step in sphingolipid biosynthesis. Within 2 h of treatment, myriocin triggered the formation of multiple SepA-GFP patches and the apparent loss of polarity, as shown by the appearance of “split tips” (Figure 8, A and B). During this time, actin cables presumably disappeared, as TpmA-GFP localization to hyphal tips was largely abolished (Figure 8, C and D). Notably, localization of MesA-FLAG at hyphal tips was either lost or became diffuse (Figure 8, E and F). Inhibition of sphingolipid biosynthesis is thought to disrupt the formation of sterol-rich membrane domains (Bagnat et al., 2000). Accordingly, these observations suggest that sterolrich membrane domains may promote stable recruitment of SepA to hyphal tips and the subsequent formation of actin cables, by regulating the localization of MesA.
Figure 8.
Inhibition of sphingolipid biosynthesis alters the localization patterns of SepA-GFP, TpmA-GFP, and MesA-FLAG. sepA::gfp (AKS70) (A and B), tpmA::gfp (ACP115) (C and D), and mesA::FLAG (ACP108) (E and F) spores were germinated in YGV at 28°C for 10 h. One sample was left untreated (A, C, and E), whereas the other was shifted into YGV + 80 μg/ml myriocin (B, D, and F). Samples were incubated an additional 120 min and viewed by epifluorescence microscopy either directly (A–D) or after fixation and exposure to anti-FLAG antibodies (E and F). (B) Multiple SepA-GFP patches are present at the splitting hyphal tip (arrow). (D) TpmA-GFP appears to be absent from the hyphal tip (asterisk), although it can still form rings at septation sites (arrow). (F) MesA-FLAG is either absent (asterisk) or displays a diffuse localization pattern (arrow) at hyphal tips. Bar, 4 μm.
If sterol-rich membrane domains mediate the localization of SepA and MesA to hyphal tips, then these domains themselves should localize to polarization sites. To investigate this possibility, living hyphae were briefly stained with filipin, a fluorescent polyene antibiotic that forms complexes with free sterols. In wild type, filipin stained a prominent patch at hyphal tips and a ring at septation sites that appears to constrict (Figure 9, A and B). Staining of hyphal tips was also observed in sepA1 mutants incubated at restrictive temperature (Supplemental Figure 2), but was dramatically altered in mesA1 mutants. Although septal rings were still apparent, staining was generally delocalized at varying intensities over the entire hyphal surface (Figure 9, C–F). In addition, randomly distributed bright spots were occasionally observed in some hyphae (Figure 9, C and E). Similarly, localized filipin staining of hyphal tips was not observed in mesA1 sepA1 double mutants incubated at restrictive temperature (Supplemental Figure 2). These observations suggest that sterol-rich membrane domains localize to the hyphal tip, which places them in the appropriate location to mediate the localization of MesA and SepA. Moreover, they also support the possibility that MesA itself may contribute to the organization of these domains.
Figure 9.
The mesA1 mutation alters the distribution of sterol-rich membrane domains. Wild-type (GR5) and mesA1 (ACP21) hyphae were grown in YGV at 28°C for 14 h. They were subsequently stained with filipin (25 μg/ml) for 5 min. (A and B) Sterol-rich membrane domains localize to a ring structure at septation sites (A) and to a patch at the tips (B) of wild-type hyphae. (C–F) Sterol-rich membrane domains fail to form a discrete patch at the tips of mesA1 hyphae. Instead, they appear randomly distributed around the surface of hyphal tips cells, with occasional bright spots (arrows) observed at both apical and subapical sites. Localization to septation sites is not affected. Bars, 5 μm.
Characterization of Yeast MesA Homologues
To obtain further insight into the general function of MesA family members, we examined the phenotype of yeast cells in which the mesA homologue had been deleted. In S. cerevisiae, characterization of YOR129c expression suggested that it might play a role in mating (Ren et al., 2000). However, in both unilateral and bilateral mutant crosses, yor129c mutants displayed no obvious defects in the formation of mating projections, in cell fusion, or in mating efficiency (our unpublished results). In addition, neither haploid nor homozygous diploid mutants exhibited an altered budding pattern. These observations suggest that Yor129c either has a subtle role in cellular morphogenesis or performs a function that is shared with another protein.
We also examined the phenotype of S. pombe mutants in which the SPCB0776.06c gene had been deleted using a PCR-based approach. Although they retained their normal rod-shaped morphology, spcb0776.06c mutants were slightly shorter than wild-type cells (Δspcb0776.06c, 11.7 ± 0.8 μm, n = 40; wild type, 13.3 ± 0.8 μm, n = 34). In addition, they displayed a modest defect in mating efficiency (wild type h+× wild-type h-, 21.0% mated; Δspcb0776.06c h+× Δspcb0776.06c h-; 3.7% mated; n = 400). However, the mutants still formed colonies of normal size and did not show obvious sensitivity to agents that induce cell wall stress. Moreover, expression of the SPCB0776.06c gene under alcA(p) control did not complement the growth defects of mesA1 mutants (our unpublished results). These results are consistent with the notion that the pivotal function of MesA in hyphal morphogenesis is not conserved in yeast cells.
DISCUSSION
Our screen for mutations that enhance sepA defects resulted in the identification of MesA, a novel fungal protein required for hyphal morphogenesis. Functional characterization of MesA shows that it is required for the stabilization of polarity axes. Our observations suggest that MesA accomplishes this task by promoting the stable recruitment of SepA to hyphal tips.
MesA Is a Novel Protein with a Unique Function in Filamentous Fungi
Analysis of the predicted MesA sequence revealed a lack of obvious homology to any known animal or plant gene product. In contrast, homologues were identified in all sequenced fungal genomes. Notably, the MesA homologues of filamentous ascomycetes are more similar to those identified in filamentous basidiomycetes than they are to the homologues from ascomycete yeasts. Because the ascomycetes and basidiomycetes are thought to have diverged ∼500 million years ago (Berbee and Taylor, 2001), this observation suggests that MesA function has been more strongly conserved in filamentous fungi than it has in yeasts. This notion is supported by the phenotypic effects of mutations in the respective homologues; in particular, the morphogenetic defects caused by the mesA1 mutation are far more severe than those caused by deletion of either yeast homologue.
Several results obtained from our characterization of MesA suggest that it functions as a cell surface protein to direct the axis of hyphal extension in A. nidulans. First, MesA is predicted to possess at least one transmembrane domain. Second, MesA appears to localize to the cell surface at sites of polarized morphogenesis (i.e., hyphal tips, branch sites). Finally, mesA1 mutants display dramatic defects in the maintenance of hyphal polarity, including an aberrant pattern of cell wall deposition and delocalization of sterol-rich membrane domains. The localization pattern of MesA is particularly intriguing. Its appearance as a circumferential band just behind the apex of the hyphal tip indicates that it might function as a cortical marker that specifies the boundaries of the apical growth zone. In S. cerevisiae, the septins perform a similar function marking cortical domains (Barral et al., 2000). Because A. nidulans possesses a large family of septins (Momany et al., 2001), it is tempting to speculate that one or more of these may interact with MesA to control the establishment of a specialized cortical domain that defines the hyphal tip and facilitates the generation of positional information.
The Role of Membrane Lipds in Hyphal Morphogenesis
Recent results implicate lipid microdomains in the regulation of polarized morphogenesis in filamentous fungi. In A. nidulans, the characterization of sphingolipid biosynthetic mutants suggested that sterol-rich membrane domains are required for polarity establishment (Cheng et al., 2001). Similarly, in N. crassa, mutants defective in synthesis of ergosterol or specific phosphoinositides display dramatic polarity defects (Seiler and Plamann, 2003). Our observations suggest that sterol-rich membrane domains localize to the hyphal tip, where they may promote polarity maintenance by ultimately regulating the localization of SepA. A key intermediate in this process appears to be the interaction between these domains and MesA. Although additional studies are needed to clarify the nature of this interaction, their mutual dependency for proper localization at the hyphal tip is consistent with the notion that MesA and sterol-rich membrane domains function in a cooperative manner to stabilize polarity axes.
MesA Is Required for Actin Cable Formation at Hyphal Tips
Like wild type, SepA-GFP localized to crescents at the tips of mesA1 hyphae. However, the subtending spot, which likely corresponds to the Spitzenkorper, was absent. Furthermore, while SepA-GFP was relatively stable at the tips of wild-type hyphae, it generally failed to persist for longer than 15 min at mesA1 hyphal tips. These observations suggest that MesA is not involved in the initial recruitment of SepA to polarization sites, but is required for its subsequent retention. Because formins nucleate the formation of actin cables (Evangelista et al., 2003), the failure to retain SepA would ultimately preclude the assembly of these structures, thereby preventing the transport of factors needed to stabilize the nascent polarity-axis. Presumably, this would cause the axis to collapse or split, as was observed in the timelapse images of mesA1 hyphae.
How are mesA1 mutants able to establish hyphal polarity in the apparent absence of actin cables? One possible explanation is that polarization may become reliant on Arp2/3-dependent branched actin filaments (reviewed by Pollard and Borisy, 2003). This possibility is consistent with the observation that actin structures could still be detected at mesA1 hyphal tips using anti-actin antibodies. Alternatively, because cytoplasmic microtubules remained intact in mesA1 mutants, they may become important for polarization in the absence of actin cables. In accord with this notion, microtubules are required for polarity establishment in sepA mutants (our unpublished results).
Surprisingly, faint TpmA-GFP structures could still be seen in sepA1 hyphae shifted to restrictive temperature. Although the nature of these structures remains unknown, their presence provides a potential explanation for the ability of the mesA1 mutation to enhance sepA1 defects. For example, assembly of the faint structures may be controlled by other scaffolding proteins that function in parallel with SepA to direct organization of the actin cytoskeleton (i.e., WASP; annotated as AN8715.2 in the A. nidulans genome). By affecting the localized recruitment of these proteins in addition to SepA, the mesA1 mutation would presumably eliminate all detectable actin cables from hyphal tips.
Stabilization of Polarity Axes in Filamentous Fungi
The role of feedback loops in the amplification of polarization signals and subsequent stabilization of polarity axes has recently been described in S. cerevisiae (Nern and Arkowitz, 2000; Irazoqui et al., 2003; Wedlich-Soldner et al., 2003). Our observations suggest that MesA stabilizes polarity axes in A. nidulans by promoting the localized assembly of actin cables. Moreover, we found that MesA localization to hyphal tips is dependent on the presence of actin cables. On the basis of these results, we propose that MesA is a key element of a feedback loop that stabilizes polarity axes in filamentous fungi. In this model, MesA triggers the localized assembly of actin cables, which in turn reinforce the polarization signal by consolidating MesA localization at hyphal tips. The presence of such a feedback loop may provide filamentous fungi with a dynamic mechanism for adjusting the direction of hyphal extension in response to local signals (Riquelme et al., 1998).
Supplementary Material
Acknowledgments
We thank Christian Elowsky and Joe Zhao (Microscopy Core, University of Nebraska Center for Biotechnology) as well as Ann Cowan (Center for Biomedical Imaging Technology, University of Connecticut Health Center) for invaluable assistance with confocal microscopy and image processing. We also thank Tom Wolkow (University of Colorado-Colorado Springs) for the gift of S. pombe strains and Greg May (The University of Texas M.D. Anderson Cancer Center) for providing the pRG3-AMA1 genomic library. This project was supported by National Science Foundation and funds provided by the University of Nebraska Research Foundation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-11-0803. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-11-0803.
Abbreviations used: GFP, green fluorescent protein; Ts, temperature sensitive.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Arai, R., Nakano, K., and Mabuchi, I. (1998). Subcellular localization and possible function of actin, tropomyosin and actin-related protein 3 (Arp3) in the fission yeast Schizosaccharomyces pombe. Eur. J. Cell Biol. 76, 288-295. [DOI] [PubMed] [Google Scholar]
- Bagnat, M., Keranen, S., Shevchenko, A., Shevchenko, A., and Simons, K. (2000). Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97, 3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat, M., and Simons, K. (2002). Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA 99, 14183-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., III, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Barral, Y., Mermall, V., Mooseker, M.S., and Snyder, M. (2000). Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5, 841-851. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia, S., Hergert, F., and Gierz, G. (1989). Computer simulation of fungal morphogenesis and the mathematical basis for (hyphal tip) growth. Protoplasma 153, 46-57. [Google Scholar]
- Berbee, M.L., and Taylor, J.W. (2001). Fungal molecular evolution: gene trees and geologic time. In: The Mycota VII Part B: Systematics and Evolution, ed. D.J. McLaughlin, E.J. McLaughlin, and P. Lemke, Heidelberg: Springer-Verlag, 229-243.
- Bourett, T.M., and Howard, R.J. (1991). Ultrastructural immunolocalization of actin in a fungus. Protoplasma 163, 199-202. [Google Scholar]
- Boyce, K., Hynes, M., and Andrianopoulos, A. (2001). The CDC42 homolog of the dimorphic fungus Penicillium marneffei is required for correct cell polarization during growth but not development. J. Bacteriol. 183, 3447-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F., and Peter, M. (2003). Yeasts make their mark. Nat. Cell Biol. 5, 294-299. [DOI] [PubMed] [Google Scholar]
- Cheng, J., Park, T.-S., Fischl, A.S., and Ye, X.S. (2001). Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol. Cell. Biol. 21, 6198-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert, C. (1997). Fungal spore germination: insights from the molecular genetics of Aspergillus nidulans and Neurospora crassa. Fungal Genet. Biol. 21, 163-172. [Google Scholar]
- Efimov, V., and Morris, N.R. (1998). A screen for dynein synthetic lethals in Aspergillus nidulans identifies spindle assembly checkpoint genes and other genes involved in mitosis. Genetics 149, 101-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D.C., Boone, C., and Bretscher, A. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 32-41. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Zigmond, S., and Boone, C. (2003). Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116, 2603-2611. [DOI] [PubMed] [Google Scholar]
- Fernandez-Abalos, J.M., Fox, H., Pitt, C., Wells, B., and Doonan, J.H. (1998). Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 27, 121-130. [DOI] [PubMed] [Google Scholar]
- Gammie, A., Brizzio, V., and Rose, M.D. (1998). Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9, 1395-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooday, G.W. (1971). An autoradiographic study of hyphal growth of some fungi. J. Gen. Microbiol. 132, 125-133. [Google Scholar]
- Grove, S.N., and Bracker, C.E. (1970). Protoplasmic organization of hyphal tips among fungi: vesicles and Spitzenkorper. J. Bacteriol. 104, 989-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.D. (2001). Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4, 736-739. [DOI] [PubMed] [Google Scholar]
- Harris, S.D., Morrell, J.L., and Hamer, J.E. (1994). Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136, 517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.D., Hamer, L., Sharpless, K.E., and Hamer, J.E. (1997). The Aspergillus nidulans sepA gene encodes an FH1/2 protein required for cytokinesis and the maintenance of cellular polarity. EMBO J. 16, 3474-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.D., Hofmann, A.F., Tedford, H.W., and Lee, M.P. (1999). Identification and characterization of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics 151, 1015-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.D., and Momany, M. (2004). Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41, 391-400. [DOI] [PubMed] [Google Scholar]
- Inoue, S., Turgeon, B.G., Yoder, O., and Aist, J.R. (1998). Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J. Cell Sci. 111, 1555-1566. [DOI] [PubMed] [Google Scholar]
- Irazoqui, J.E., Gladfelter, A.S., and Lew, D.J. (2003). Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5, 1062-1070. [DOI] [PubMed] [Google Scholar]
- Kafer, E. (1977). Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19, 33-131. [DOI] [PubMed] [Google Scholar]
- Lopez-Franco, R., and Bracker, C.E. (1996). Diversity and dynamics of the Spitzenkorper in growing hyphal tips of higher fungi. Protoplasma 195, 90-111. [Google Scholar]
- McGoldrick, C.A., Gruver, C., and May, G.S. (1995). myoA of Aspergillus nidulans encodes an essential myosin required for secretion and polarized growth. J. Cell Biol. 128, 577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany, M. (2002). Polarity in filamentous fungi: establishment, maintenance, and new axes. Curr. Opin. Microbial. 5, 580-585. [DOI] [PubMed] [Google Scholar]
- Momany, M., Zhao, J., Lindsey, R., and Westfall, P.J. (2001). Characterization of the Aspergillus nidulans septin (asp) gene family. Genetics 157, 969-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern, A., and Arkowitz, R.A. (2000). G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell 5, 853-864. [DOI] [PubMed] [Google Scholar]
- Ni, L., and Snyder, M. (2001). A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 2147-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B.R., and Osmani, S.A. (1993). Cell cycle analysis using the filamentous fungus Aspergillus nidulans. In: The Cell Cycle: A Practical Approach, ed. P. Fantes and R. Brooks, New York, NY: IRL Press, 127-142.
- Osherov, N., and May, G.S. (2000). Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155, 647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki-Kuroda, K., Yamamoto, Y., Nohara, H., Kinoshita, M., Fujiwara, T., Irie, K., and Takai, Y. (2001). Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 827-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, J., Nielsen, O., Egel, R., and Hagan, I.M. (1998). FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141, 1217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., and Borisy, G.G. (2003). Cellular motility driven by the assembly and disassembly of actin filaments. Cell 112, 453-465. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000a). Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113, 365-375. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000b). Polarization of cell growth in yeast. J. Cell Sci. 113, 571-585. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A., and Boone, C. (2002). Role of formins in actin assembly: nucleation and barbedend association. Science 297, 612-615. [DOI] [PubMed] [Google Scholar]
- Ren, B. et al. (2000). Genome-wide location and function of DNA binding proteins. Science 290, 2306-2309. [DOI] [PubMed] [Google Scholar]
- Riquelme, M., Reynaga-Pena, C.G., Gierz, G., and Bartnicki-Garcia, S. (1998). What determines growth direction in fungal hyphae? Fungal Genet. Biol. 24, 101-109. [DOI] [PubMed] [Google Scholar]
- Riquelme, M., Gierz, G., and Bartnicki-Garcia, S. (2000). Dynein and dynactin deficiencies affect the formation and function of the Spitzenkorper and distort hyphal morphogenesis of Neurospora crassa. Microbiol. 146, 1743-1752. [DOI] [PubMed] [Google Scholar]
- Roberson, R.W., and Vargas, M.M. (1994). The tubulin cytoskeleton and its sites of nucleation in hyphal tips of Allomyces macrogynus. Protoplasma 182, 19-31. [Google Scholar]
- Sagot, I., Klee, S.K., and Pellman, D. (2002a). Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4, 42-50. [DOI] [PubMed] [Google Scholar]
- Sagot, I., Rodal, A.A., Moseley, J., Goode, B.L., and Pellman, D. (2002b). An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4, 626-631. [DOI] [PubMed] [Google Scholar]
- Seiler, S., Nargang, F.E., Steinberg, G., and Schliwa, M. (1997). Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16, 3025-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., Plamann, M., and Schliwa, M. (1999). Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 9, 779-785. [DOI] [PubMed] [Google Scholar]
- Seiler, S., and Plamann, M. (2003). The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14, 4352-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless, K.E., and Harris, S.D. (2002). Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13, 469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y.J., Santos, B., Fortin, N., Costigan, C., and Snyder, M. (1998). Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18, 4053-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K., and Toomre, D. (2000). Lipid rafts and signal transduction. Nature Rev. Mol. Cell. Biol. 1, 31-39. [DOI] [PubMed] [Google Scholar]
- Valdez-Taubas, J., and Pelham, H.R.B. (2003). Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 13, 1636-1640. [DOI] [PubMed] [Google Scholar]
- Waring, R.B., May, G.S., and Morris, N.R. (1989). Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-encoding genes. Gene 79, 119-130. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner, R., Altschuler, S., Wu, L., and Li, R. (2003). Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299, 1231-1235. [DOI] [PubMed] [Google Scholar]
- Wendland, J. (2001). Comparison of morphogenetic networks of filamentous fungi and yeast. Fungal Genet. Biol. 34, 63-82. [DOI] [PubMed] [Google Scholar]
- Wendland, J., and Philippsen, P. (2001). Cell polarity and hyphal morphogenesis are controlled by multiple Rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics 157, 601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E.A. et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- Wu, Q., Sandrock, T.M., Turgeon, B.G., Yoder, O., Wirsel, S.G., and Aist, J.R. (1998). A fungal kinesin required for organelle motility, hyphal growth, and morphogenesis. Mol. Biol. Cell 9, 89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.