Abstract
The adaptor protein Numb is necessary for the cell fate specification of progenitor cells in the Drosophila nervous system. Numb is evolutionarily conserved and previous studies have provided evidence for a similar functional role during mammalian development. The Numb protein has multiple protein-protein interaction regions including a phosphotyrosine binding (PTB) domain and a carboxy-terminal domain that contains conserved interaction motifs including an EH (Eps15 Homology) domain binding motif and α-adaptin binding site. In this study we identify the EHD/Rme-1/Pincher family of endocytic proteins as Numb interacting partners in mammals and Drosophila. The EHD/Rme-1 proteins function in recycling of plasma membrane receptors internalized by both clathrin-mediated endocytosis and a clathrin-independent pathway regulated by ADP ribosylation factor 6 (Arf6). Here we report that Numb colocalizes with endogenous EHD4/Pincher and Arf6 and that Arf6 mutants alter Numb subcellular localization. In addition, we present evidence that Numb has a novel function in endosomal recycling and intracellular trafficking of receptors.
INTRODUCTION
Signaling from cell surface receptors is regulated, in part, by controlling the level of receptor protein at the plasma membrane. The rapid internalization and incorporation of receptors into intracellular vesicles facilitates their removal from the plasma membrane and subsequent downregulation. Internalization occurs through either constitutive or receptor-activation–dependent pathways that can be clathrin-dependent or clathrin-independent. During clathrin-dependent internalization, receptor proteins at the cell surface interact with endocytic proteins that target them into clathrin-coated pits. Sorting to clathrin-coated pits is largely dependant on sequence motifs within the cytoplasmic domain of the receptor. The two major classes of sorting signals, tyrosine- and dileucine-based signals, are recognized by the heterotetrameric adaptor complex AP2, which recruits clathrin and nucleates the formation of the clathrin-coated vesicle (Bonifacino and Traub, 2003). In contrast, little is known about the sorting mechanisms involved in clathrin-independent internalization pathways. Nonclathrin endocytic pathways include phagocytosis, macropinocytosis, and caveolae-mediated uptake (Nichols and Lippincott-Schwartz, 2001). Receptors such as major histocompatibilty complex (MHC) I, integrins, and E-cadherin are internalized independently of clathrin but require the small GTPase ADP ribosylation factor 6 (Arf6; Donaldson, 2003). Both clathrin-coated vesicles and vesicles internalized by clathrin-independent pathways are trafficked to the tubulo-vesiclular early endosome (Bonifacino and Traub, 2003). Within this compartment several groups have noted that vesicles internalized by clathrin-independent pathways converge or fuse with clathrin-derived vesicle populations (Naslavsky et al., 2003; Sharma et al., 2002). From the early endosome, receptors are then either routed through the recycling endosome back to the cell surface or retained and sorted into the multivesicular body and ultimately degraded in the lysosome (Katzmann et al., 2002). Sorting decisions are directed by sequences intrinsic to the receptors themselves and by posttranslational modifications such as phosphorylation and ubiquitination that provide docking sites for endocytic proteins that subsequently coordinate receptor sorting and trafficking (Hicke, 1999; Katzmann et al., 2001).
The adaptor protein Numb interacts with proteins involved in protein ubiquitination (Juven-Gershon et al., 1998; Susini et al., 2001; Nie et al., 2002; McGill and McGlade, 2003) and receptor-mediated endocytosis (Santolini et al., 2000; Berdnik et al., 2002). Numb was originally identified in Drosophila (dNumb) as a mutation that caused the development of extra hair cells at the expense of sensory neurons during development (Uemura et al., 1989). The Numb protein functions as an intrinsic cell fate determinant that is asymmetrically localized in neuronal precursor cells where it influences cell fate by antagonizing signaling from the Notch receptor (Frise et al., 1996). Although the exact mechanism by which Numb mediates its neurogenic effect is unknown, evidence suggests that asymmetric cell divisions may involve partitioning of the endocytic machinery allowing differential control of cell surface receptor levels. Indeed, a report by Berdnik et al. (2002) demonstrated that the clathrin adaptor α-adaptin homologue in Drosophila is also asymmetrically localized and α-adaptin mutants show cell fate transformations similar to the Numb phenotype (Berdnik et al., 2002).
Studies suggest that mammalian Numb (mNumb) functions in a similar manner to its Drosophila counterpart. Transgenic expression of mNumb in Drosophila rescues the Numb null phenotype (Zhong et al., 1996), and its ectopic overexpression results in cell fate transformations identical to overexpression of dNumb (Verdi et al., 1996). Mammalian Numb is asymmetrically localized in dividing neuroepithelial cells in the developing vertebrate brain (Zhong et al., 1996; Wakamatsu et al., 1999) and retina (Cayouette et al., 2001), and homozygous deletion of Numb results in severe defects during neurogenesis, resulting in embryonic lethality around day 11.5 in utero (Zhong et al., 2000; Zilian et al., 2001; Petersen et al., 2002). Furthermore, recent work has demonstrated that Numb overexpression antagonizes Notch receptor signaling (Berezovska et al., 1999; French et al., 2002; McGill and McGlade, 2003).
Both mammalian and Drosophila Numb proteins possess an amino terminal phosphotyrosine binding, or PTB, domain followed by a carboxy-terminal region that contains conserved sequence motifs including aspartate-proline-phenylalanine (DPF) and asparagine-proline-phenylalanine (NPF) motifs. The DPF motif is a consensus binding site for the clathrin adaptor α-adaptin, whereas NPF motifs associate with proteins containing Eps15 Homology (EH) domains. Both motifs are conserved in all vertebrate Numb isoforms, in the related mammalian protein Numblike, and in the Drosophila homologue.
To identify Numb-binding proteins with conserved function, we conducted a parallel screen of embryonic fly and mammalian expression libraries to identify targets of the conserved sequence motifs in the Numb carboxy-terminal domain. Here we report the identification of an evolutionarily conserved interaction between Numb and the EHD/Rme-1/Pincher family of endocytic proteins and describe a novel role for Numb in endocytic recycling.
MATERIALS AND METHODS
Screening of Expression Libraries
Drosophila embryo (0–24 h) and mouse embryo (16-days) cDNA expression libraries (Novagen, Madison, WI) were plated and protein expression was induced according to the manufacturer's instructions. The nitrocellulose filters with immobilized proteins were washed several times in TBST (20 mM Tris-base, pH 7.5, 150 mM NaCl, 0.05% [vol/vol] Tween-20 and blocked overnight in blocking buffer (5% BSA [wt/vol], 1 mM DTT in TBST). The filters were then incubated overnight with blocking buffer containing biotinylated peptides (25 pmol/ml; Drosophila Numb peptide: ARHSTNPFISP and mammalian Numb peptide: NPSPTNPFSSDAQKAFEIEL) that were preconjugated to streptavidin-alkaline phosphatase (Bio-Rad, Richmond, CA). The filters were washed five times with TBST and detection was performed with a colorimetric reaction with NBT/BCIP (Roche Molecular Biochemicals, Indianapolis, IN). Positive plaques were isolated, and two rounds of plaque purification were performed. The pExLox plasmids were isolated according to the manufacturer's directions and sequenced.
Antibodies and Constructs
Anti-mNumbA was generated in rabbits with a synthetic peptide (TTHPQSPSLAKQQTFPQYE) corresponding to amino acids 538–557 as previously described (Cat. no. 07147; Upstate Biotechnology, Lake Placid, NY; Dho et al., 1999). Mouse anti-Prospero was a generous gift from Gabrielle Boulianne. Rabbit anti-dEHD was raised against a GST fusion protein of the EH-domain of dEHD (amino acids 422–533). Rabbit anti-dNumb was raised against a peptide corresponding to the carboxy terminus of dNumb (sequence ISPPKAPAQSFQVQL). Other antibodies used in this study are commercially available and include the following: anti-Flag M2 (Sigma-Aldrich, St. Louis, MO); anti-HA (Covance, Madison, WI); anti-Arf6 (Santa Cruz Biotechnology, Santa Cruz, CA); and anti-interleukin (IL) 2α (Santa Cruz). Full-length expression constructs of EHD4, wild-type Numb, and Numb mutants were generated by PCR and subcloned into pFlag (Sigma-Aldrich) or pcDNA3.1–2HA in frame with the amino-terminal epitope tag. Similarly, the GST fusions were generated using PCR and cloned into pGex-4T3 (Clontech, Palo Alto, CA) with an amino terminal GST sequence. All constructs were sequenced in both directions to ensure sequences were correct (ACTG Corporation, Toronto, Ontario, Canada).
Cell Culture and Transfections
CHO cells were grown in Ham's minimal media (Wisent, St. Bruno, Quebec) supplemented with 10% fetal bovine serum and transfected with LipofectAMINE 2000 reagent (Invitrogen, Carlsbad, CA) in OptiMEM (Invitrogen) according to the manufacturer's instructions. Cells were cultured at 37°C for 24–48 h before lysis. HeLa cells were grown in DMEM (Wisent) supplemented with 10% fetal bovine serum. For siRNA experiments, a 21-nucleotide oligomer (Dharmacon, Boulder, CO) was designed to a region homologous to Numb (nucleotides 39–57). As a control, the Scramble II duplex from Dharmacon (D-001205–20) was used.
In Vitro Binding Experiments
Recombinant GST-fusion proteins were expressed and purified as previously described (Pelicci et al., 1992). Purified proteins bound to glutathione-Sepharose 4B beads (Amersham Pharmacia Biotechnol, Piscataway, NJ) were suspended in an equal volume of Nonidet-P40 (NP40) lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 100 μM ZnCl2, 1% [vol/vol] NP40, 10% [vol/vol] glycerol) containing Complete protease inhibitors (Roche Molecular Biochemicals) and 1 mM DTT. Each of the purified GST-fusion proteins were quantitated by SDS-PAGE and Coomassie staining and compared with BSA standards. Lysates of transfected HEK293T cells were prepared in NP40 lysis buffer and incubated with ∼1 μg of GST-fusion protein coupled to glutathione-Sepharose 4B beads for 2 h at 4°C. The beads were washed five times with cold NP40 lysis buffer and suspended in SDS-Laemmli sample buffer. The proteins were resolved on a polyacrylamide gel and transferred to Immobilon-P (Millipore, Bedford, MA) membrane.
Immunoprecipitations and Western Blotting
Cell lysates were prepared from transiently transfected cultured HEK293T cells by scraping 90% confluent 10-cm dishes into 1 ml lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1% [vol/vol] Triton X-100, 10% [vol/vol] glycerol, and protease inhibitors; Roche Molecular Biochemicals) and centrifuged at 14,000 rpm to pellet the insoluble material. The protein concentrations of cell lysates were determined using the MicroBCA kit (Pierce, Rockford, IL) according to the manufacturer's directions. One milligram of total cell lysate was incubated with 1 μg antibody and 50 μl of 20% (wt/vol) protein G-Sepharose bead slurry. The immune complexes were washed five times in NP40 lysis buffer (described above) and eluted in boiling SDS-Laemmli sample buffer. Proteins were separated by SDS-PAGE, transferred onto PVDF membrane (Immobilon-P, Millipore, Billerica, MA), and immunoblotted with primary antibodies overnight at 4°C. Bound antibodies were visualized using HRP-conjugated protein A (Bio-Rad) or goat anti-mouse in conjunction with the ECL system (Amersham Pharmacia Biotechnol).
Immunohistochemistry
For Drosophila antibody staining, mixed-staged embryos were fixed in 5% formaldehyde in PEMS buffer (1 M PIPES, pH 7.2, 2 mM MgSO4, 50 mM EGTA), stained following standard procedures, and mounted in antifade (70% glycerol + 2.5% DABCO [Sigma-Aldrich] in PBS). The following primary antibodies were used: rabbit anti-dNumbC, 1:200; rabbit anti-dEHD, 1:400; and mouse anti-Prospero, 1:40. Cy3- and Alexa fluor 488–labeled secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at 1:400.
For mammalian immunostaining experiments, cells were cultured on glass coverslips. The cells were fixed in 4% paraformaldehyde (freshly prepared in PBS containing 30 mM sucrose) for 30 min at room temperature, washed with 10 mM glycine in PBS, and then permeabilized with either 0.05% saponin or 0.1% Triton X-100 for 5 min. After several short washes, the cells were blocked with either 5% BSA or 5% normal goat serum in PBS and then incubated for 30 min at 37°C with primary antibodies diluted in the blocking medium as follows: rabbit anti-NumbA 1:200, mouse anti-Arf6 1:100 (Santa Cruz) and rabbit anti-EHD4 1:100. Cy3-labeled donkey anti-mouse IgG (1:500; Jackson ImmunoResearch Laboratories) or Alexa fluor 488 goat anti-rabbit IgG (1:500; Molecular Probes, Eugene, OR) were used to indirectly label the primary antibodies. The cells were washed with PBS and mounted in DAKO mounting medium.
A modified protocol was used to costain cells with anti-NumbA and anti-EHD4, which are both rabbit polyclonal antibodies. After blocking with 5% BSA, cells were incubated with anti-NumbA, washed, and then incubated with an excess of monovalent goat anti-rabbit IgG Fab fragment (1:100; Jackson ImmunoResearch Laboratories) to block all anti-rabbit binding sites with goat IgG. The first primary antibody was then visualized with Alexa fluor 488 donkey anti-goat (1:500; Molecular Probes). The second primary antibody, anti-EHD4, was then added, followed by incubation with Cy3-labeled anti-rabbit IgG. To ensure that there was no binding of the second antibody to unoccupied sites on the first primary antibody, control slides were prepared identically except that no second primary antibody was added. Cells were examined and images taken with a scanning laser confocal microscope (Zeiss, Thornwood, NY) and processed in Adobe Photoshop or Illustrator (San Jose, CA).
Surface Biotinylation and Trafficking Assays
Surface biotinylation was adapted to measure the recycling of the IL-2α receptor (Tac) in CHO cells as described previously (Le et al., 2002). Briefly, cells that had been cotransfected with 1 μg Tac and double-stranded Numb specific or scrambled RNA or GFP and GFP-T27N were cooled on ice for 40 min. Cells were washed with PBS and biotinylated at 4°C with EZ-Link Sulfo-NHS-SS-Biotin (Pierce) in biotinylation buffer (154 mM NaCl, 10 mM HEPES, 3 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 10 mM glucose, pH 7.6) for 45 min. Cells were washed with Ham's F12 media containing 100 mM glycine for 5 min to quench free biotin and then washed several times more with PBS. At this point one sample was removed and lysed in lysis buffer in order to measure total surface biotinylation. In addition, one sample was reserved to determine the effectiveness of MESNA stripping of biotinyl groups from the surface proteins with two 20-min incubations at 4°C in stripping buffer (50 mM 2-mercaptoethanesulfonic acid). The remaining samples were then incubated at 18°C in Ham's F12 media containing 10% FBS for 60 min to allow the cells to accumulate internalized biotinylated receptors and then MESNA-stripped as described above to remove any remaining biotinyl groups remaining at the surface. After PBS washes, the cells were then rapidly warmed to 37°C in Ham's-F12 media containing 10% FBS for 0, 3, 5, or 10 min to permit recycling to the surface and then stopped by chilling on ice in PBS maintained at 4°C. The samples were then stripped of all surface biotinylated proteins in MESNA stripping buffer. The samples were then lysed in RIPA buffer, and protein concentrations were determined. Between 600 and 750 μg of protein lysates were then mixed overnight at 4°C with streptavidin sepharose beads (Pierce) to collect biotinylated proteins sequestered inside the cell and therefore protected from MESNA treatment. The proteins were then collected by centrifugation and washed four times in NP40 buffer, and bed volume removed with a 30-gauge needle before resuspending in SDS-Laemmli sample buffer and analyzing by SDS-PAGE and immunoblotting with anti-Tac (Santa Cruz).
RESULTS
Identification of EHD/Rme-1 Proteins That Interact with Numb Carboxy-terminus
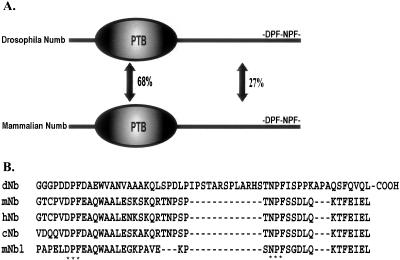
At the primary amino acid level, Drosophila and mammalian Numb proteins share weak sequence identity outside the PTB domain (27%; Figure 1A). However, alignment of the carboxy-terminal sequences of Numb from Drosophila, mouse, human, chicken, and the related mammalian protein Numblike revealed conserved DPF and NPF motifs (Figure 1B). In Drosophila, deletion of these motifs compromised Numb function in vivo (Frise et al., 1996). The presence of conserved endocytic protein-binding motifs within this functionally important region of dNumb prompted us to search for proteins that associate with the NPF-based motifs. Peptides encompassing the NPF motif of both mouse and Drosophila Numb (Figure 2A) were used to screen mouse and Drosophila cDNA expression libraries respectively. Multiple cDNAs encoding the EH-domain–containing protein, EHD1 (Mintz et al., 1999), also named Rme-1 (Lin et al., 2001), and a related gene, EHD4/Pincher (Pohl et al., 2000; Shao et al., 2002), were identified from the mammalian screen (BIND ID's 133328, 133329). Both are members of the EHD family of proteins and contain an amino-terminal phosphate-binding loop (or p-loop) and a central coiled-coil domain, followed by a carboxy-terminal EH-domain (Figure 2B). To date, the EHD protein family has been described in both Caenorhabditis elegans and mammalian systems, where they appear to function as components of the endocytic recycling machinery.
Figure 1.
Structure of Numb adaptor protein. (A) Schematic representation of Drosophila and mammalian Numb proteins. PTB-phosphotyrosine binding domain. (B) Multiple sequence alignment of Numb carboxy-terminal sequences in Drosophila (dNb), mouse (mNb), human (hNb), chicken (cNb), and mouse Numblike (mNbl). Asterisks mark the conserved DPF and NPF endocytic motifs.
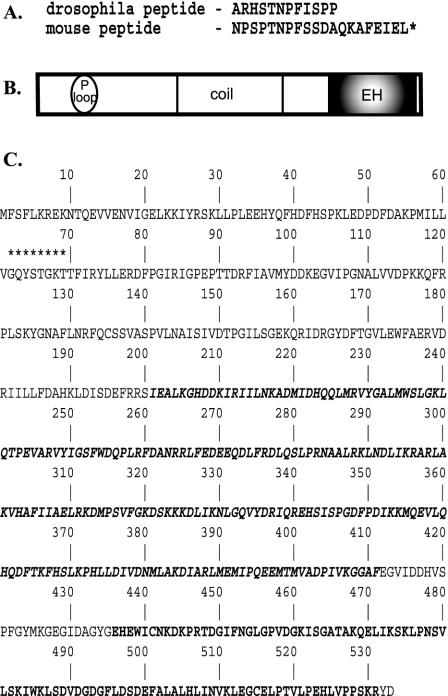
Figure 2.
Structure of the EHD/Rme-1 family of EH-domain–containing proteins. (A) Amino acid sequence of peptides used to screen expression libraries. Peptides were coupled to biotin at the amino terminus. (B) Schematic representation of domain organization of EHD protein family. P-loop, phosphate-binding motif; coil, coiled-coil domain; EH, Eps15-Homology domain. (C) Primary amino acid sequence of Drosophila EHD orthologue (accession no. AF473822). Asterisks indicate the p-loop consensus sequence. The coiled-coil domain is denoted by bold italic type, and the EH-domain is indicated by boldface type.
Our expression screen of the Drosophila library identified the mEHD1/mRme-1 orthologue, which we have named dEHD (accession no. AF473822; BIND ID 133327; Figure 2C). Drosophila EHD shares 69% identity with the mammalian EHD proteins. The full-length dEHD sequence was nearly identical to a GenBank entry for PAST-1 or Putative Achaete Scute Target, with the exception that the GenBank entry contains an error between nucleotides 728-1078, where the sequence is inverted. The isolation of the Drosophila and mouse EHD/Rme-1 homologues in two separate screens with two distinct peptides is a strong indication that EHD proteins are conserved components of a Numb protein complex.
Characterization of Drosophila EHD
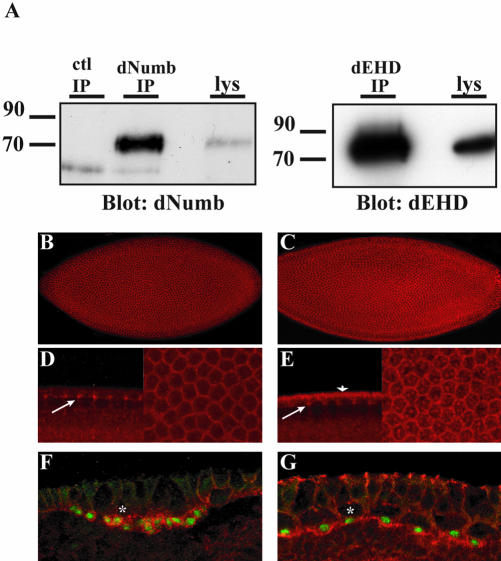
Our screen revealed a novel interaction between dNumb and Drosophila EHD (dEHD). To date, no characterization of the Drosophila orthologue of the EHD family of proteins has been reported. To analyze the expression patterns of dEHD and dNumb in Drosophila, polyclonal antibodies were generated. The antibodies generated recognized a single band corresponding to the expected molecular weights of dNumb and dEHD proteins in immunoprecipitation and Western analysis of whole fly lysates (Figure 3A). Whole-mount embryo preparations immunostained with dNumb and dEHD revealed that both dEHD and dNumb are expressed ubiquitously during early embryogenesis (unpublished data). We observed an enrichment of both proteins within the syncytial blastoderm and during cellularization (Figure 3, B and C). Both dNumb and dEHD were enriched at the apico-lateral cell borders, giving rise to the characteristic honey-comb pattern in en face views of the embryo. dNumb is predominantly membrane-associated (Figure 3D, right panel), whereas dEHD exhibits both membrane-associated and punctate cytosolic staining (Figure 3E, right panel). We also observed significant coexpression of dNumb and dEHD within structures identified as the progressing furrow canal, a region of active membrane expansion (Figure 3, D and E, arrows, left panels). In addition, we observed an apical accumulation of dEHD (Figure 3E, arrowhead), distinct from dNumb, which may be associated with apical membrane (egg membrane) or apical cytoplasm, and is consistent with the early membrane insertion site identified by Lecuit and Wieschaus (2000).
Figure 3.
Expression pattern of dNumb and dEHD during embryogenesis. (A) Whole flies were homogenized in lysis buffer, and immunoprecipitations with either affinity-purified anti-dNumb (left panel) or anti-dEHD (right panel) were performed. Western blotting revealed a single band in both immunoprecipitation and lysate (lys) lanes corresponding to the expected molecular weight of the dNumb or dEHD protein. (B--E) Whole-mount immunostaining of stage 4–5 Drosophila embryos during cellularization. dNumb expression (B) is found ubiquitously in all cells. dEHD expression (C) is similarly enriched at cell borders but is also found within large vesicular like structures. Lateral and face views of B and C at higher magnification demonstrating accumulation of dNumb (D) and dEHD (E) at the progressing furrow channel (arrows, left panel) and enrichment at apico-lateral cell borders (right panels). Apical staining of dEHD (arrowhead, E) corresponds to apical membrane or apical cytoplasm. (F and G) Lateral view of whole-mount immunostaining of stage 8–9 embryos double-stained with α-prospero (F and G, green) and α-dNumb (F, red), or α-dEHD (G, red). Asterisks demark apical neuroblast. Both dNumb and dEHD are coexpressed in Prospero-positive ganglion mother cells (GMC).
One of the hallmark expression patterns of dNumb occurs within neuroblasts, the progenitor cells of the Drosophila CNS. The neuroectoderm of stage 8–9 embryos contains neuroblasts that divide into a large apical daughter cell that retains neuroblast characteristics and a smaller basal ganglion mother cell (GMC). During neuroblast cell division, dNumb is asymmetrically localized to the basal side of the neuroblast and the dNumb protein is preferentially delivered to the GMC (Knoblich et al., 1995). Another cell fate determinant, the transcription factor Prospero is also asymmetrically segregated with dNumb into the GMC and then translocated to the nucleus and serves as a marker for the GMCs. We examined the expression of dNumb and dEHD in the embryonic neuroectoderm. As previously reported, dNumb expression was observed in Prospero-positive GMC's (Figure 3F; Knoblich et al., 1995). In addition, Prospero positive GMC's were found to be enriched for dEHD (Figure 3G) and interestingly, dEHD accumulated basally in these cells.
Numb and EHD Proteins Associate In Vitro through an EH-domain–mediated Interaction
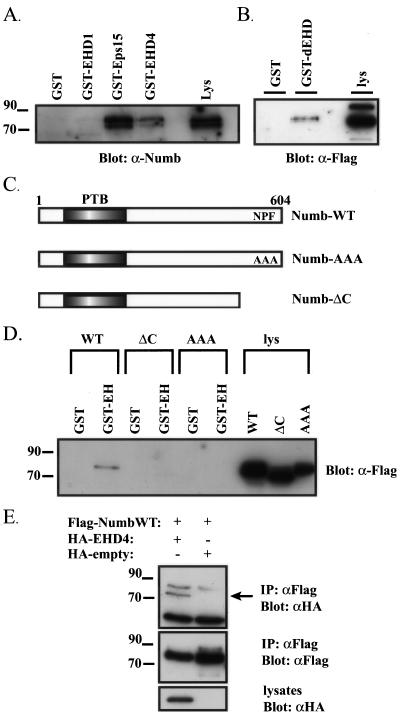
To further investigate the interaction between mammalian Numb and EHD family proteins, GST fusion proteins of the EH-domain of EHD1 and EHD4 were produced, and Numb binding was examined. In addition, a GST fusion of the second EH-domain of Eps15 was used as a positive control. Eps15 was previously shown to associate with Numb in vitro and in vivo (Salcini et al., 1997). GST-EH or GST proteins were immobilized on glutathione agarose beads and incubated with HeLa cell lysates. Endogenous Numb precipitated with the EH-domains of EHD4 and Eps15 but not with GST alone (Figure 4A). The association of the EHD1 EH-domain with endogenous Numb was barely detectable, and given that Numb binding to EHD4 was considerably stronger, subsequent experiments focused on the Numb-EHD4 interaction.
Figure 4.
Numb and EHD proteins interact in vitro and in vivo. (A) HEK293T whole cell lysates were mixed with GST, or GST fusion with the EH-domains of EHD1 (GST-EHD1), EHD4 (GST-EHD4), or the second EH-domain of Eps15 (GST-Eps15). The complexes were washed, analyzed by SDS-PAGE, and immunoblotted with anti-NumbA (lys, whole cell lysate). (B) Cell lysates from HEK293T cells transfected with Flag-epitope–tagged Drosophila Numb were mixed with immobilized GST or dEHD EH-domain fusion proteins (GST-dEHD). The complexes were washed, analyzed by SDS-PAGE, and immunoblotted with anti-Flag. (lys, whole cell lysate). (C) Schematic representation of Numb and Numb mutants used to map the region of Numb responsible for interaction with EHD4. Numb-AAA contains a triple point mutation of amino acids 590–592, where the NPF (Asn-Pro-Phe) motif is mutated to AAA (Ala-Ala-Ala). Numb-ΔC is a truncation mutant that removes amino acids 567–604. (D) Immobilized GST or EHD4 EH-domain fusion proteins (GST-EH) were mixed with lysates prepared from HEK293T cells expressing Flag epitope–tagged wild-type Numb p66 (WT), carboxy-terminal deletion mutant (ΔC), or triple point mutant (AAA). Protein complexes were washed and analyzed by SDS-PAGE and immunoblotted for Flag epitope. (E) HEK293T cells were transiently cotransfected with HA-EHD4 and Flag NumbWT. Empty vector plasmids were used in all combinations and served as internal controls to ensure binding was specific. Flag immunoprecipitations were performed and immunoblotted with α-HA (top panel). α-Flag immunoprecipitates is nonspecificand cross reacts with α-HA antibodies. The membranes were stripped and reblotted with α-Flag to confirm the presence of Numb in the immunoprecipitations (middle panel). Similarly, whole cell lysates were immunoblotted with α-HA to confirm the expression of HA-EHD4 (bottom panel).
The interaction of Drosophila Numb and the EH-domain of dEHD was also confirmed using in vitro binding assays. Full-length wild-type dNumb was transiently expressed in HEK293T cells, and its association with GST or the GST-dEHD was examined. Wild-type dNumb bound specifically to GST-dEHD and not control GST alone (Figure 4B), suggesting that the interaction of EHD proteins with Numb is evolutionarily conserved.
To localize the region of Numb important for the interaction with the EH-domain of EHD4, we generated a truncation mutant of Numb in which a region of the carboxy-terminal domain (amino acids 567–604) was deleted (Numb-ΔC), and a triple point mutant that converted the NPF motif (amino acids 590–592) to alanine residues (Numb-AAA; Figure 4C). GST-EH or GST proteins were immobilized on glutathione agarose beads and incubated with cell lysates containing FLAG epitope–tagged wild-type Numb or Numb mutants. Wild-type Numb bound to GST-EH; however, neither of the carboxy-terminal Numb mutants interacted with GST-EH (Figure 4D). This confirms that the formation of a Numb-EHD4 complex is mediated by an EH-domain interaction with the NPF motif in Numb.
To determine if Numb and full-length EHD4 form a complex in vivo, HEK293T cells were cotransfected with Flag-tagged wild-type Numb (FLAG-NumbWT) and either HA-tagged full-length EHD4 (HA-EHD4) or vector only (HA-empty). HA-EHD4 coimmunoprecipitated with FLAG-NumbWT (Figure 4E, top panel). Expression of FLAG-NumbWT was confirmed by stripping the membrane and immunoblotting with anti-FLAG antibodies (Figure 4E, middle panel). Similarly, whole cell lysates were immunoblotted with HA antisera to confirm the expression of HA-EHD4 in transfected lysates (Figure 4E, bottom panel).
mNumb and mEHD Proteins Colocalize In Vivo
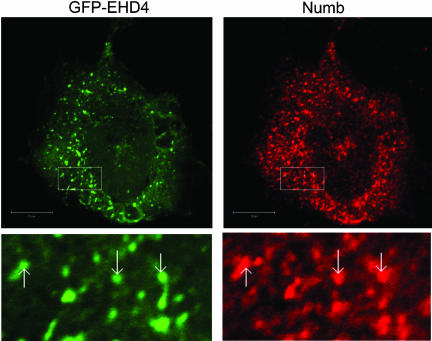
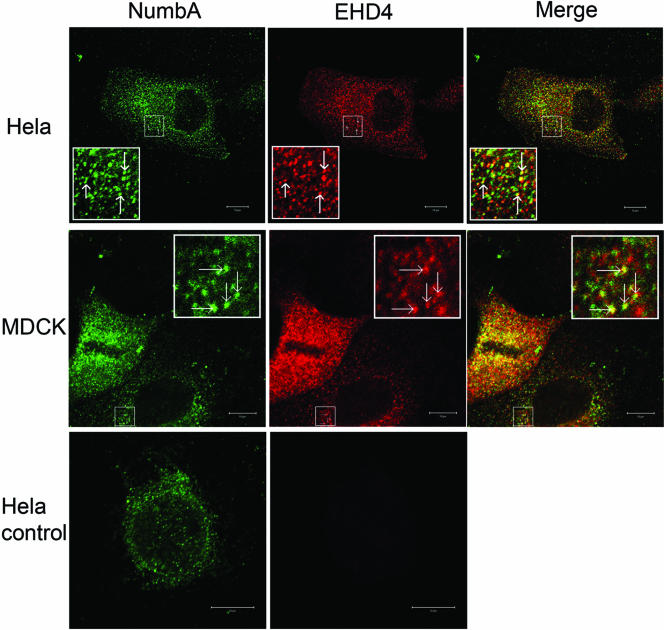
To directly assess if Numb and EHD4 are located in the same intracellular compartments, HeLa cells were cotransfected with GFP-EHD4 and Numb and examined by immunocytochemistry. Overexpressed Numb and EHD4 were observed in both overlapping and distinct intracellular compartments. However, the degree of overlap varied across the population of transfected cells. Some cells showed very little colocalization, whereas others demonstrated marked colocalization as shown in Figure 5. Examination of optical sections near the plasma membrane did not reveal Numb and EHD4 colocalization (unpublished data). However, when midcell optical sections were analyzed, colocalization of Numb and EHD4 was observed on intracellular vesicles and tubular structures (Figure 5). In this representative field we determined that ∼51% of Numb-positive vesicles contained EHD4. To examine the distribution of endogenous Numb and EHD4 proteins, MDCK and HeLa cells were costained using a modified immunohistochemical protocol. Briefly, after incubation with rabbit α-NumbA, cells were incubated with excess goat α-rabbit monovalent Fab fragments, followed by Alexa 488 donkey α-goat secondary antibody. The second primary antibody, rabbit α-EHD4, was then applied and used in conjunction with a Cy3-labeled donkey α-rabbit secondary antibody. Efficient blockage of the first primary was confirmed in control preparations treated identically except for the addition of the second primary antibody. Colocalization of the endogenous proteins was observed on intracellular vesicles in both HeLa (Figure 6, top) and MDCK cells (Figure 6, middle). We noted that many of these vesicles were distinct and did not show overlapping staining; however, a proportion of vesicles were labeled by both α-NumbA and α-EHD4 antibodies (Figure 6). Control immunostaining experiments performed in parallel confirmed that the observed colocalization was not due to the use of two rabbit polyclonal antibodies (Figure 6, bottom).
Figure 5.
Overexpressed Numb and EHD4 colocalize in HeLa cells. HeLa cells were transfected with pEF-Numb p66 and GFP-EHD4 and fixed 24 h after transfection, and processed for immunofluorescent localization of Numb (red) and GFP-EHD4 (green). Boxed regions have been digitally enlarged and represented as inserts in order to show more clearly examples of colabeling (arrows). In this representative section, Zeiss colocalization software compatible with Zeiss Axiovert 100 with LSM510 estimates there is ∼51% colocalization of Numb and EHD4. Scale bar, 10 μm.
Figure 6.
Endogenous Numb and EHD4 colocalize in intracellular vesicles. HeLa (top and bottom panels) and MDCK (middle panels) cells were costained with rabbit anti-NbA plus goat anti-rabbit Fab (green), followed by rabbit anti-EHD4 (red) as described in MATERIALS AND METHODS. The bottom HeLa cell image is a representative cell taken from a control coverslip that was treated identically to the HeLa and MDCK cells shown above it except that no anti-EHD4 was added. The parameters used for image acquisition were the same. Confocal sections taken through the middle region of the cell are presented. The red and green images were merged to show regions of colocalization of mNumb and mEHD4 (yellow). Scale bar, 10 μm.
Endogenous Numb and EHD4 Proteins Colocalize with Arf6
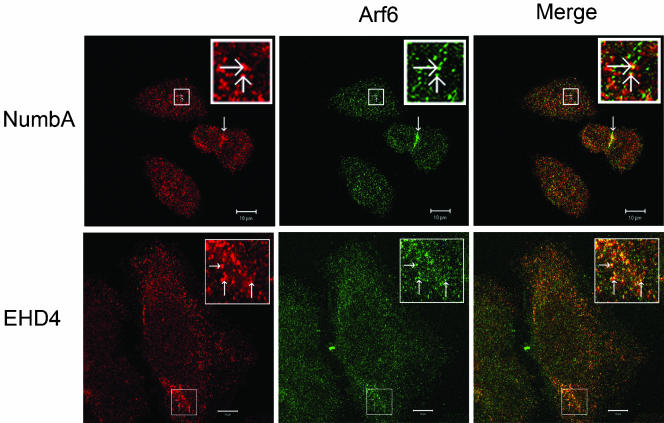
Our immunostaining experiments showed that Numb localizes to multiple endocytic structures at the plasma membrane and intracellular vesicles. To characterize the identity of Numb positive vesicles, cells were costained with markers for known subcellular compartments including α-EEA1 (early endosomes), α-KDEL (endoplasmic reticulum), α-GM130 (trans-Golgi network [TGN]), and α-Lamp1 (lysosomes). No significant colocalization of these markers with either Numb nor EHD4 was observed (unpublished data). Recently, EHD proteins have been implicated in the ADP ribosylation factor 6 (Arf6) regulated membrane recycling pathway (Caplan et al., 2002). Arf6 belongs to a subfamily of the ras-related class of small GTPases, and cycles between the active GTP bound and the inactive GDP-bound form. This dynamic cycle is thought to mediate the regulation of a membrane recycling pathway. Previous immunocytochemistry studies demonstrated that Arf6 localized at the plasma membrane and to a distinct intracellular vesicle population which, similar to Numb, did not colocalize with known endocytic markers (Radhakrishna and Donaldson, 1997; D'Souza-Schorey et al., 1998). We therefore examined whether Numb localized within the Arf6 recycling pathway, by using HeLa cells in which the Arf6 subcellular localization has been extensively characterized (Radhakrishna and Donaldson, 1997). HeLa cells were costained with anti-Numb and anti-Arf6 antisera. Endogenous Numb and Arf6 colocalized on a subset of intracellular vesicles (∼14–20%), suggesting Numb is distributed within the Arf6 recycling pathway (Figure 7, top panels). In addition, Numb and Arf6 colocalized to what appears to be the cleavage furrow (Figure 7, top panels, thickened arrow) where Arf6 was shown by Schweitzer and D'Souza-Schorey (2002) to concentrate during telophase.
Figure 7.
Endogenous Numb and EHD4 colocalize with Arf6. HeLa cells were fixed and processed for immunofluorescence localization of endogenous Numb (top panel, red) or EHD4 (bottom panel, red), and Arf6 (green). The green and red images were merged to demonstrate colocalization of Arf6 and Numb or EHD4 (yellow). Numb and Arf6 were enriched at what appears to be the cleavage furrow (thickened arrow; top panel). Boxed regions have been digitally enlarged and represented as inserts in order to show more clearly examples of colabeling (arrows). Scale bar, 10 μm.
Previously, Caplan et al. (2002) showed that Arf6 and transfected EHD1 proteins colocalize within the tubulo-vesiclular recycling compartment. We therefore examined whether EHD4 shared a similar expression pattern. Similar to our observations with Numb and Arf6, EHD4 and Arf6 colocalized to intracellular vesicles in HeLa cells (Figure 7, bottom panels). Taken together these results suggest that both Numb and EHD4 are resident proteins of the Arf6-regulated membrane recycling pathway.
Numb Localization Is Altered by an Arf6 GDP/GTP Cycling Mutant
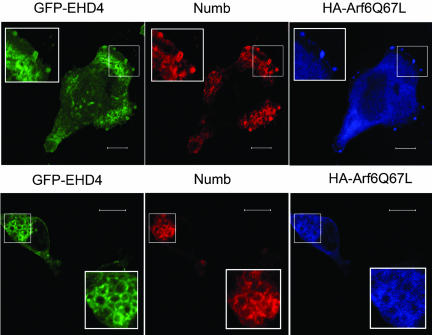
The use of dominant-acting Arf6 mutants that mimic either activated GTP-bound Arf6 (Arf6-Q67L) or inactive GDP-bound Arf6 (Arf6-T27N) have been instrumental in the study of Arf6-regulated membrane trafficking events. Arf6-Q67L localizes almost exclusively to the plasma membrane and its expression results in the formation of extensive plasma membrane protrusions (D'Souza-Schorey et al., 1995; Radhakrishna and Donaldson, 1997). We examined the effects of Arf6-Q67L on the intracellular localization of both Numb and EHD4 coexpressed within the same cell. GFP-EHD4, Arf6-Q67L-HA, and Numb were cotransfected into HeLa cells, and their expression pattern was examined by immunocytochemistry. In cells expressing Arf6-Q67L, EHD4 colocalized with Numb and Arf6-Q67L at plasma membrane protrusions (Figure 8, top panels). In addition, Numb, EHD4, and Arf6-Q67L accumulated within macropinosomes and intracellular vacuolar clusters (Figure 8, bottom panels). In contrast, expression of the Arf6 GTP-binding defective mutant, Arf6-T27N, did not alter Numb localization (unpublished data). These observations suggest that at sites of Arf6 activation, colocalization of Numb and EHD4 is induced.
Figure 8.
Activation of Arf6 alters Numb subcellular localization. HeLa cells cotransfected with pEF-Numb p66, GFP-EHD4, and Arf6-Q67L-HA were fixed, permeabilized, and processed for immunofluorescence. Expression of Arf6-Q67L-HA (blue) induces localization of Numb (red) and EHD4 (green) to Arf6-positive membrane protrusions (top panels) and macropinosomes/vacuolar clusters (bottom panels). Boxed regions have been digitally enlarged and represented as inserts in order to show more clearly examples of colabeling. Scale bar, 10 μm.
Numb Functions in the Arf6-regulated Recycling Pathway
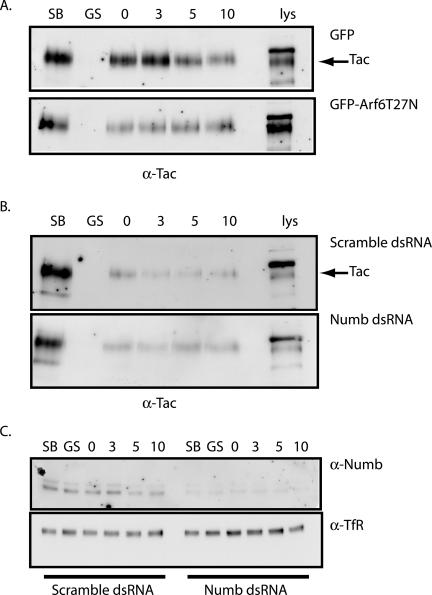
Given the intracellular distribution of Numb and its interaction with proteins of the Arf6 recycling pathway, we tested whether Numb plays a functional role in endocytic recycling. A number of cell surface receptors are constitutively internalized and recycled via the Arf6 membrane recycling pathway including cadherins, MHC class I, and IL-2α receptor (Tac). The kinetics of Tac receptor recycling has been previously characterized and was therefore chosen as a model receptor to investigate the influence of Numb on recycling (Radhakrishna and Donaldson, 1997). The Arf6-T27N mutant has been previously shown by immunocytochemistry to inhibit recycling of receptors back to the cell surface, and we assayed the effect of this mutant to validate our biochemical assay of Tac recycling in the Arf6-dependent pathway. CHO cells were cotransfected with pTac and either GFP-Arf6-T27N or GFP empty vector and recycling of the Tac receptor assessed by surface biotinylation (Le et al., 2002). Cells were biotinylated at 0°C and then incubated at 18°C to accumulate internalized, biotin-labeled Tac receptor. The cells were MESNA stripped to remove any biotin label remaining at the cell surface and returned to 37°C to resume trafficking. At the indicated time points, cells were MESNA stripped again to remove any biotin label from endocytosed proteins that had returned to the cell surface. The cells were lysed and the unrecycled biotinylated Tac receptor was analyzed by immunoblot. After 10 min, a loss of biotinylated Tac receptor from the intracellular pool was observed in GFP-expressing cells, indicating that the Tac receptor was recycled (Figure 9A, top). As predicted, expression of the Arf6-T27N mutant severely inhibited recycling, as shown by retention of intracellular Tac receptor over time (Figure 9A, bottom).
Figure 9.
Numb functions in regulation of Tac recycling. (A) CHO cells cotransfected with pTac and either GFP-Arf6-T27N or pEGFP control vector were surface-biotinylated on ice for 40 min. One plate was removed to depict total surface biotinylation (lane SB), and another was stripped with glutathione to ensure efficient removal of biotin label (lane GS). The cells were then allowed to internalize for 60 min at 18°C and returned to ice to strip any label remaining at the surface. After glutathione stripping, cells were returned to 37°C for chase periods 0, 3, 5, and 10 min. At each chase time point, cells were glutathione-stripped, and remaining biotinylated proteins were recovered from equal amounts of protein lysates using streptavidin beads and were analyzed by SDS-PAGE. Tac receptor was detected by immunoblotting. Arrow indicates band corresponding to the Tac receptor. (B) Similar to experiment above, CHO cells were cotransfected with pTac and either double-stranded RNA duplex specific for Numb (bottom panel) or with a scrambled RNA duplex (top panel), and intracellular Tac receptor levels were monitored by surface biotinylation. Arrow indicates band corresponding to the Tac receptor. (C) Equal amounts of protein lysates were analyzed by SDS-PAGE to confirm depletion of endogenous Numb levels by immunoblotting for Numb (top panel). The blots were then stripped and reprobed for endogenous transferrin receptor to confirm equal loading in of each condition (α-TfR, bottom panel).
We next assessed whether reduction of endogenous Numb levels by RNA-mediated interference (RNAi) had an effect on Tac recycling. CHO cells were transfected with pTac and either control-scrambled or Numb-specific double-stranded RNA duplexes (Dharmacon), and the recycling of the Tac receptor was examined. In control-scrambled transfected cells the amount of internalized Tac receptor decreased when chased at 37°C, with maximum recycling between 3 and 10 min (Figure 9B, top). In contrast, in cells with at least 50% reduced Numb protein levels (Figure 9C, top), the amounts of labeled Tac within the protected intracellular pool remained unchanged even after 10 min, suggesting minimal recycling (Figure 9B, bottom). Taken together these results are consistent with a role for Numb in the regulation of Tac receptor trafficking in the Arf6-dependent membrane recycling pathway.
DISCUSSION
Our study utilized an expression screen to identify a novel interaction between Numb and EHD family of endocytic proteins in both Drosophila and mammals. The four members of the mammalian EHD family are highly conserved (69–72% identity) and have differential tissue expression patterns, which may define the intracellular binding partners of the individual EHD proteins (Mintz et al., 1999; Pohl et al., 2000). Despite extensive functional characterization of the EHD proteins, in vivo targets of the EH-domain have, until now, remained elusive. We have confirmed the interaction in vitro and in vivo and mapped the interaction site to the NPF motif of Numb. NPF-EH-domain interactions occur with relatively low affinity (Santolini et al., 1999), and the in vivo interactions mediated by the EH-domain of EHD proteins are likely transient and highly regulated. Caplan et al. (2002) demonstrated that EHD proteins are continuously cycling on and off membranes, which is indicative of a dynamic regulatory process (Caplan et al., 2002). This may in part explain why we were able to observe a direct association in vitro, but were unable to isolate an endogenous Numb-EHD4 complex by coimmunoprecipitation. Furthermore, colocalization of endogenous Numb and EHD4 was variable across a given cell population, suggesting that the level of Arf6 activation within the cell induces the functional association of Numb and EHD4. In support of this, we observed that expression of the constitutively active Arf6 mutant induced a dramatic increase in Numb, EHD4 and Arf6 colocalization.
Recently several groups have established that EHD/Rme1 proteins function within a tubulo-vesicular endocytic recycling compartment (Caplan et al., 2002; Galperin et al., 2002). Both the C. elegans and mammalian orthologues have been characterized and were found to promote recycling of plasma membrane receptors such as the transferrin receptor in CHO cells (Lin et al., 2001) and the yolk receptors in C. elegans (Grant et al., 2001). In addition to a role in recycling back to the cell surface, EHD/Rme-1 proteins also regulate traffic from the recycling compartment to the TGN (Caplan et al., 2002). This suggests that the EHD proteins may be involved in the formation of transport vesicles within the recycling compartment. EHD/Rme-1 was also found to promote recycling of receptors internalized by the clathrin-independent Arf6 endocytic pathway (Caplan et al., 2002).
Previously, mammalian Numb was shown to partially colocalize with AP2 and Eps15 in clathrin-coated pits and early endosomes (Santolini et al., 2000). In addition, mNumb has been shown to interact with components of the endocytic internalization machinery, AP2 and Eps15 and fragments of Numb inhibited internalization of the EGF and transferrin receptors, implicating Numb in clathrin-dependent endocytosis (Salcini et al., 1997; Santolini et al., 2000). We have also observed a subset of Numb colocalized at the plasma membrane with AP2 (unpublished data). However, we have found that a significant proportion of Numb is localized to intracellular vesicles that do not contain AP2 and that a subset of this vesicle population contains Arf6 and EHD4. Furthermore, we have shown that knock down of Numb by RNAi inhibits recycling of Tac; a receptor trafficked via a clathrin-independent pathway regulated by Arf6. This indicates that Numb functions in multiple steps of the endocytic pathway and reveals a role for Numb in trafficking events subsequent to internalization.
Arf6 has been predominantly associated with clathrin-independent trafficking events; however, it has also been shown to affect clathrin-dependent internalization of the transferrin receptor, in a cell type–dependent manner (D'Souza-Schorey et al., 1995). Furthermore, proteins internalized by clathrin-dependent endocytosis and the Arf6 pathway have been shown to converge within the early sorting endosome (Naslavsky et al., 2003). The EHD family of proteins, like Arf6 and Numb, are also found to be involved in both clathrin-dependent and -independent processes. For example, expression of EHD1 mutants disrupt transferrin receptor recycling (Lin et al., 2001), but not its internalization in TRVb1 cells, whereas EHD4/Pincher was previously shown to promote clathrin-independent trafficking of the TrkA receptor in PC12 cells (Shao et al., 2002). Therefore Numb, like Arf6 and EHD proteins, likely functions in multiple locations along the endocytic pathway and may represent a common component of both the clathrin-dependent and -independent vesicle-routing pathways that coordinates the trafficking of cargo between these two compartments.
Although we have shown that perturbation of Numb protein levels can affect recycling or trafficking of receptors within the Arf6-regulated membrane recycling pathway, it is not yet clear if Numb has a general or cargo-specific trafficking function. Members of the Disabled family of adaptor proteins, Dab1 and Dab2, share structural features with Numb proteins, including an amino-terminal PTB domain and a carboxy-terminal domain containing DPF motifs that interact with AP2 (Morris and Cooper, 2001). Disabled proteins have been described as cargo-specific adaptors that endocytose members of the LDL receptor family (Gotthardt et al., 2000; Mishra et al., 2002). There is also evidence that Dab proteins may modulate the trafficking of other transmembrane receptors. For example, Dab2 was shown to link the transforming growth factor β (TGF-β) receptor to the downstream Smad pathway (Hocevar et al., 2001). Interestingly Smad2/3 activation and signaling occurs only following TGF-β receptor internalization into an endocytic compartment (Penheiter et al., 2002). In a similar manner, Numb proteins could exert its effects on Notch signaling by specifically modifying the interaction of Notch with the endocytic machinery. Therefore, Numb and Disabled proteins may be members of a distinct family of endocytic adaptors with specialized functions in the regulation of signaling pathways.
In this report, we identify the Drosophila orthologue of the EHD/Rme1 family of endocytic proteins as the first described target of the NPF motif in dNumb. Although further genetic analysis will be required to establish the function of Drosophila EHD, our results strongly suggest that Numb-mediated regulation of Notch signaling may involve an Arf6-regulated trafficking pathway. A Drosophila orthologue of Arf6 has recently been identified (Chen et al., 2003); however, its function with respect to endocytosis and vesicle trafficking has not been characterized. Given that the functional relationship between EHD and Arf6 proteins was recently confirmed in C. elegans (Dang et al., 2004), the identification of the Drosophila EHD protein will facilitate further studies of the Arf6-dependent membrane trafficking pathway.
Acknowledgments
We thank Shun Cheng Li for the Drosophila Numb peptide, Gabrielle Boulianne and Chris Doe for the anti-Prospero antibody, J. Bonifacino for Tac construct, and Melanie McGill for comments on the manuscript. C.A.S. is a recipient of a Terry Fox Studentship from the National Cancer Institute of Canada (NCIC). C.J.M. is a research scientist of the NCIC. This work was supported by a grant from the NCIC (C.J.M.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-01-0026. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-01-0026.
References
- Berdnik, D., Torok, T., Gonzalez-Gaitan, M., and Knoblich, J.A. (2002). The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3, 221-231. [DOI] [PubMed] [Google Scholar]
- Berezovska, O., McLean, P., Knowles, R., Frosh, M., Lu, F.M., Lux, S.E., and Hyman, B.T. (1999). Notch1 inhibits neurite outgrowth in postmitotic primary neurons. Neuroscience 93, 433-439. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J.S., and Traub, L.M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395-447. [DOI] [PubMed] [Google Scholar]
- Caplan, S., Naslavsky, N., Hartnell, L.M., Lodge, R., Polishchuk, R.S., Donaldson, J.G., and Bonifacino, J.S. (2002). A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 21, 2557-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette, M., Whitmore, A.V., Jeffery, G., and Raff, M. (2001). Asymmetric segregation of Numb in retinal development and the influence of the pigmented epithelium. J. Neurosci. 21, 5643-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E.H., Pryce, B.A., Tzeng, J.A., Gonzalez, G.A., and Olson, E.N. (2003). Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell 114, 751-762. [DOI] [PubMed] [Google Scholar]
- Dang, H., Li, Z., Skolnik, E.Y., and Fares, H. (2004). Disease-related myotubularins function in endocytic traffic in Caenorhabditis elegans. Mol. Biol. Cell 15, 189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dho, S.E., French, M.B., Woods, S.A., and McGlade, C.J. (1999). Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 274, 33097-33104. [DOI] [PubMed] [Google Scholar]
- Donaldson, J. (2003). Multiple roles for Arf 6, sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278, 41573-41576. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., Li, G., Colombo, M.I., and Stahl, P.D. (1995). A regulatory role for ARF6 in receptor-mediated endocytosis. Science 267, 1175-1178. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., van Donselaar, E., Hsu, V.W., Yang, C., Stahl, P.D., and Peters, P.J. (1998). ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J. Cell Biol. 140, 603-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, M.B., Koch, U., Shaye, R.E., McGill, M.A., Dho, S.E., Guidos, C.J., and McGlade, C.J. (2002). Transgenic expression of Numb inhibits Notch signaling in immature thymocytes but does not alter T cell fate specification. J. Immunol. 168, 3173-3180. [DOI] [PubMed] [Google Scholar]
- Frise, E., Knoblich, J.A., Younger-Shepherd, S., Jan, L.Y., and Jan, Y.N. (1996). The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl. Acad. Sci. USA 93, 11925-11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin, E., Benjamin, S., Rapaport, D., Rotem-Yehudar, R., Tolchinsky, S., and Horowitz, M. (2002). EHD 3, a protein that resides in recycling tubular and vesicular membrane structures and interacts with EHD1. Traffic 3, 575-589. [DOI] [PubMed] [Google Scholar]
- Gotthardt, M., Trommsdorff, M., Nevitt, M.F., Shelton, J., Richardson, J.A., Stockinger, W., Nimpf, J., and Herz, J. (2000). Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J. Biol. Chem. 275, 25616-25624. [DOI] [PubMed] [Google Scholar]
- Grant, B., Zhang, Y., Paupard, M.C., Lin, S.X., Hall, D.H., and Hirsh, D. (2001). Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 3, 573-579. [DOI] [PubMed] [Google Scholar]
- Hicke, L. (1999). Gettin'down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 9, 107-112. [DOI] [PubMed] [Google Scholar]
- Hocevar, B.A., Smine, A., Xu, X.X., and Howe, P.H. (2001). The adaptor molecule Disabled-2 links the transforming growth factor beta receptors to the Smad pathway. EMBO J. 20, 2789-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon, T., Shifman, O., Unger, T., Elkeles, A., Haupt, Y., and Oren, M. (1998). The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol. Cell. Biol. 18, 3974-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., Babst, M., and Emr, S.D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145-155. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Odorizzi, G., and Emr, S.D. (2002). Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3, 893-905. [DOI] [PubMed] [Google Scholar]
- Knoblich, J.A., Jan, L.Y., and Jan, Y.N. (1995). Asymmetric segregation of Numb and Prospero during cell division. Nature 377, 624-627. [DOI] [PubMed] [Google Scholar]
- Le, T.L., Joseph, S.R., Yap, A.S., and Stow, J.L. (2002). Protein kinase C regulates endocytosis and recycling of E-cadherin. Am. J. Physiol. Cell Physiol. 283, C489-C499. [DOI] [PubMed] [Google Scholar]
- Lecuit, T., and Wieschaus, E. (2000). Polarized insertion of new membrane from a cytoplasmic resevoir during cleavage of the Drosophila embryo. J Cell Biol. 150, 849-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.X., Grant, B., Hirsh, D., and Maxfield, F.R. (2001). Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat. Cell Biol. 3, 567-572. [DOI] [PubMed] [Google Scholar]
- McGill, M.A., and McGlade, C.J. (2003). Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278, 23196-23203. [DOI] [PubMed] [Google Scholar]
- Mintz, L., Galperin, E., Pasmanik-Chor, M., Tulzinsky, S., Bromberg, Y., Kozak, C.A., Joyner, A., Fein, A., and Horowitz, M. (1999). EHD1—an EH-domain-containing protein with a specific expression pattern. Genomics 59, 66-76. [DOI] [PubMed] [Google Scholar]
- Mishra, S.K., Keyel, P.A., Hawryluk, M.J., Agostinelli, N.R., Watkins, S.C., and Traub, L.M. (2002). Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor. EMBO J. 21, 4915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, S.M., and Cooper, J.A. (2001). Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2. Traffic 2, 111-123. [DOI] [PubMed] [Google Scholar]
- Naslavsky, N., Weigert, R., and Donaldson, J.G. (2003). Convergence of Nonclathrin- and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Mol. Biol. Cell 14, 417-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, B.J., and Lippincott-Schwartz, J. (2001). Endocytosis without clathrin coats. Trends Cell Biol. 11, 406-412. [DOI] [PubMed] [Google Scholar]
- Nie, J., McGill, M.A., Dermer, M., Dho, S.E., Wolting, C.D., and McGlade, C.J. (2002). LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 21, 93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci, G., Lanfrancone, L., Grignani, F., McGlade, J., Cavallo, F., Forni, G., Nicoletti, I., Pawson, T., and Pelicci, P.G. (1992). A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell 70, 93-104. [DOI] [PubMed] [Google Scholar]
- Penheiter, S.G., Mitchell, H., Garamszegi, N., Edens, M., Dore, J.J., Jr., and Leof, E.B. (2002). Internalization-dependent and -independent requirements for transforming growth factor beta receptor signaling via the Smad pathway. Mol. Cell. Biol. 22, 4750-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, P.H., Zou, K., Hwang, J.K., Jan, Y.N., and Zhong, W. (2002). Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929-934. [DOI] [PubMed] [Google Scholar]
- Pohl, U. et al. (2000). EHD2, EHD3, and EHD4 encode novel members of a highly conserved family of EH domain-containing proteins [In Process Citation]. Genomics 63, 255-262. [DOI] [PubMed] [Google Scholar]
- Radhakrishna, H., and Donaldson, J.G. (1997). ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 139, 49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcini, A.E., Confalonieri, S., Doria, M., Santolini, E., Tassi, E., Minenkova, O., Cesareni, G., Pelicci, P.G., and Di Fiore, P.P. (1997). Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 11, 2239-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini, E., Puri, C., Salcini, A.E., Gagliani, M.C., Pelicci, P.G., Tacchetti, C., and Di Fiore, P.P. (2000). Numb is an endocytic protein [In Process Citation]. J. Cell Biol. 151, 1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini, E., Salcini, A.E., Kay, B.K., Yamabhai, M., and Di Fiore, P.P. (1999). The EH network. Exp. Cell Res. 253, 186-209. [DOI] [PubMed] [Google Scholar]
- Schweitzer, J.K., D'Souza-Schorey, C. (2002). Localization and activation of the ARF6 GTPase during cleavage furrow ingression and cytokinesis. J. Biol. Chem. 277, 27210-27216. [DOI] [PubMed] [Google Scholar]
- Shao, Y., Akmentin, W., Toledo-Aral, J.J., Rosenbaum, J., Valdez, G., Cabot, J.B., Hilbush, B.S., and Halegoua, S. (2002). Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. J. Cell Biol. 157, 679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, D.K., Choudhury, A., Singh, R.D., Wheatley, C.L., Marks, D.L., and Pagano, R.E. (2002). Glycosphingolipids internalize via caveolar-related endocytosis rapidly merge with the clathrin pathway in early endosomes and form microdomains for recycling. J. Biol. Chem. 278, 7564-7572. [DOI] [PubMed] [Google Scholar]
- Susini, L. et al. (2001). Siah-1 binds and regulates the function of Numb. Proc. Natl. Acad. Sci. USA 98, 15067-15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T., Shepherd, S., Ackerman, L., Jan, L.Y., and Jan, Y.N. (1989). numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349-360. [DOI] [PubMed] [Google Scholar]
- Verdi, J.M., Schmandt, R., Bashirullah, A., Jacob, S., Salvino, R., Craig, C.G., Program, A.E., Lipshitz, H.D., and McGlade, C.J. (1996). Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 6, 1134-1145. [DOI] [PubMed] [Google Scholar]
- Wakamatsu, Y., Maynard, T.M., Jones, S.U., and Weston, J.A. (1999). NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron 23, 71-81. [DOI] [PubMed] [Google Scholar]
- Zhong, W., Feder, J.N., Jiang, M.M., Jan, L.Y., and Jan, Y.N. (1996). Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 17, 43-53. [DOI] [PubMed] [Google Scholar]
- Zhong, W., Jiang, M.M., Schonemann, M.D., Meneses, J.J., Pedersen, R.A., Jan, L.Y., and Jan, Y.N. (2000). Mouse numb is an essential gene involved in cortical neurogenesis [In Process Citation]. Proc. Natl. Acad. Sci. USA 97, 6844-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilian, O., Saner, C., Hagedorn, L., Lee, H.Y., Sauberli, E., Suter, U., Sommer, L., and Aguet, M. (2001). Multiple roles of mouse Numb in tuning developmental cell fates. Curr. Biol. 11, 494-501. [DOI] [PubMed] [Google Scholar]