Abstract
The proliferative life span of human cells is limited by telomere shortening, but the specific telomeres responsible for determining the onset of senescence have not been adequately determined. We here identify the shortest telomeres by the frequency of signal-free ends after in situ hybridization with telomeric probes and demonstrate that probes adjacent to the shortest ends colocalize with γH2AX-positive DNA damage foci in senescent cells. Normal BJ cells growth arrest at senescence before developing significant karyotypic abnormalities. We also identify all of the telomeres involved in end-associations in BJ fibroblasts whose cell-cycle arrest at the time of replicative senescence has been blocked and demonstrate that the 10% of the telomeres with the shortest ends are involved in >90% of all end-associations. The failure to find telomeric end-associations in near-senescent normal BJ metaphases, the presence of signal-free ends in 90% of near-senescent metaphases, and the colocalization of short telomeres with DNA damage foci in senescent interphase cells suggests that end-associations rather than damage signals from short telomeres per se may be the proximate cause of growth arrest. These results demonstrate that a specific group of chromosomes with the shortest telomeres rather than either all or only one or two sentinel telomeres is responsible for the induction of replicative senescence.
INTRODUCTION
Telomere shortening provides the counting mechanism for replicative senescence, the process by which normal diploid human cells limit the total number times they are able to divide (Hayflick and Moorhead, 1961; Harley et al., 1990; Wright and Shay, 2002). Replicative aging is thought to provide an initial barrier to the formation of cancer by preventing cells from having enough divisions to accumulate the many mutations needed to become malignant. Most cancer cells have escaped these limits by activating a mechanism to maintain telomere length, usually through the upregulation of telomerase (Kim et al., 1994; Shay and Bacchetti, 1997) or the activation of a recombination-based alternative lengthening of telomeres (ALT) pathway (Bryan et al., 1995; Dunham et al., 2000).
One of the major functions of telomeres is to hide the ends of the chromosomes from being recognized as double-strand breaks needing repair (de Lange, 2002), leading to the hypothesis that a DNA damage signal from too-short telomeres causes cellular senescence (Greider, 1990; Harley, 1991). This is consistent with earlier observations that viral oncoproteins that block p53 or antisense p53 extended the life span of human fibroblasts (Hara et al., 1991; Shay et al., 1991). The recent demonstrations that at senescence DNA sequences adjacent to telomeres coprecipitate with γH2AX antibodies (d'Adda di Fagagna et al., 2003) and that induced telomere dysfunction can cause telomere colocalization with DNA damage foci (Takai et al., 2003) reinforces this concept.
Using cell biological approaches, we have previously shown that the shortest telomeres determine the onset of replicative senescence in human cells (Ouellette et al., 2000; Steinert et al., 2000), an observation later confirmed with respect to end-fusions in murine cells (Hemann et al., 2001). It is not yet known if specific telomere(s) produce this result. Using quantitative in situ analysis of telomere lengths, no correlation between individual or even groups of short telomeres and replicative aging was reported (Martens et al., 2000). Indeed, several observations argue that it might be difficult to “program” one or two chromosomes to have the shortest ends and function as sentinel telomeres that regulate replicative aging. After replication, parental and daughter DNA strands have unequal lengths due a combination of the end-replication problem and end-processing events (Levy et al., 1992; Lingner et al., 1995). Depending on which strand is used as the template, daughter cells inherit telomeres of slightly different lengths, and this produces a distribution of sizes of several kb after 50 doublings, even for the progeny of a single telomere of defined length (Levy et al., 1992). This theoretical prediction has been confirmed by a PCR technique that measures the size of individual molecules of telomeric DNA at a specific allele of the Xp/Yp chromosome (Baird et al., 2003). This report showed that a several kilobase broad distribution of telomere lengths is present in multiple cell strains and is rapidly generated after subcloning. Thus even if two telomeres started out at the same length, their relative sizes could vary in individual cells after many cell divisions. An additional factor contributing to size variability is oxidative damage, which preferentially damages telomeres in a stochastic manner and can accelerate telomere shortening (von Zglinicki et al., 1995, 2002). The combination of the production of size heterogeneity during cell division and stochastic events due to oxidative damage might make it difficult to have a sentinel telomere regulating the onset of replicative aging unless it was dramatically shorter than the rest. Studies of the relative lengths of human telomeres have found a relatively continuous distribution of sizes without very short outliers (Martens et al., 2000; Graakjaer et al., 2003).
The rationale for the present study was to obtain direct evidence about the role of specific short telomeres in senescence. We identified the specific telomeres involved in chromosomal end-associations at senescence in order to determine whether or not a sentinel telomere, a subgroup of telomeres, or random telomeres were used to monitor telomere shortening. We show that there is not a sentinel telomere. The ten shortest telomeres accounted for >90% of the observed end-associations that occur at the time of M1, although in any given cell only a few telomeres are involved. We further show that telomere-adjacent probes from the shortest telomeres colocalize with DNA damage foci in near-senescent cells. As expected given the variability of telomere sizes between individuals, the 10 ends with the shortest telomeres identified in the BJ strain of fibroblasts used here only partially overlaps with reports of telomere lengths in fibroblasts from other donors. These observations are consistent with the concept that replicative aging functionally monitors groups of telomeres in order to accommodate the possible variability of germline transmission, heterogeneity among daughter cells, and stochastic events during their proliferative life span.
MATERIALS AND METHODS
Cell Culture
BJ foreskin fibroblasts were grown in a 4:1 mixture of DMEM and medium 199 containing 10% iron-supplemented calf serum (Hyclone, Logan, UT) and gentamicin (25 μg/ml; Sigma, St. Louis, MO) at 37°C in 5% CO2. Approximately 30 doublings before senescence, some cells were infected with a pLXSN retrovirus expressing HPV16 E6 and E7 proteins (Halbert et al., 1992) and selected on G418 (400 μg/ml) for 10–14 days.
Metaphase Spread Preparation and Cytogenetics Analysis
Cells were incubated with colcemid (Invitrogen, Carlsbad, CA) for 4 h, trypsinized, treated with hypotonic KCl buffer (0.075 M) for 30 min at 37°C, and washed several times with methanol-acetic acid (3:1) until a clean white cell pellet was obtained. Pellets were stored at -20°C until being dropped onto slides and GTG-banded using standard methods (Gustashaw, 1997). Metaphase images were analyzed using CytoVision software (Applied Imaging, San Jose, CA) at the cytogenetics laboratory, University of Texas Southwestern Medical Center.
Fluorescence In Situ Hybridization and Comparative Genomic Hybridization Analysis
One-day-old slides were rehydrated in phosphate-buffered saline (PBS, pH 7.5) for 15 min at room temperature, fixed in 4% formaldehyde in PBS for 2 min, and then washed in PBS three times for 5 min. Slides were treated with pepsin (1 mg/ml, pH 2.0) at 37°C for 10 min and washed twice for 2 min in PBS. The slides were again fixed in formaldehyde for 2 min, washed in PBS three times, and then dehydrated by 2-min serial incubations in 70, 90, and 100% ethanol and air-dried. The slides were denatured for 10 min at 78°C in a hybridization mixture (20 μl) containing 70% formamide, 15 ng 3′-Cy3-conjugated (CCCTAA)3 2′-deoxyoligonucleotide N3′-P5′ phosphoramidate telomeric probe, 0.25% (wt/vol) blocking reagent (Roche Molecular Biochemicals, Indianapolis, IN) and 5% MgCl2 in 10 mM Tris, pH 7.2, and then annealed for 2 h at room temperature. After two washes with 70% formamide, 0.1% bovine serum albumin, and 10 mM Tris, pH 7.2, and two washes with 0.15 M NaCl, 0.05% Tween-20, and 0.05 M Tris, the slides were dehydrated by 2-min serial incubations in ethanol and air-dried in the dark. The slides were then annealed without denaturation in the same hybridization solution as above but containing the complementary telomeric oligonucleotide (3′-Cy3-conjugated (TTAGGG)3 2′-deoxyoligonucleotide N3′-P5′ phosphoramidate telomeric probe, for 2 h at room temperature. The slides were washed again using the same wash steps as above, dehydrated by an ethanol series, and air-dried in the dark. Chromosomes were counterstained with Vectashield containing 4,6-diamidino-2-phenylindole-dihydrochloride (DAPI, 0.6 μg/ml final concentration, Vector Laboratories, Burlingame, CA) for chromosome identification.
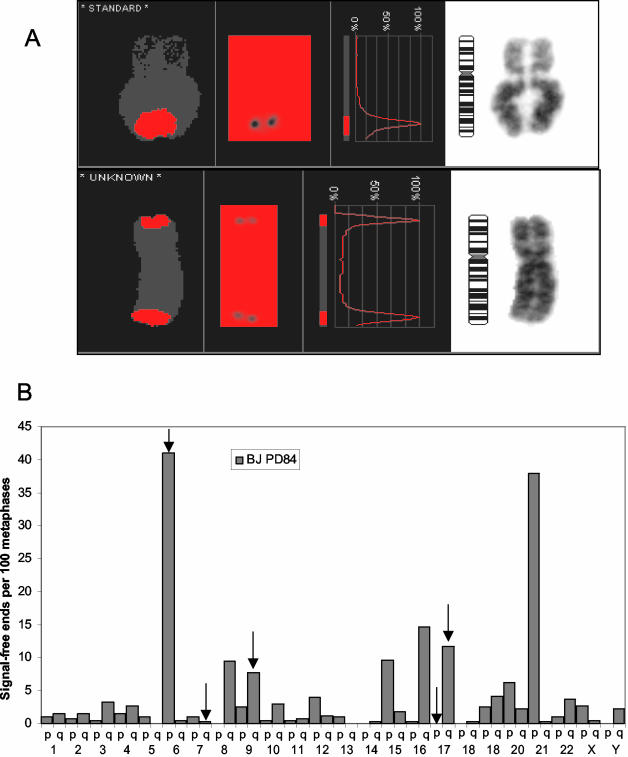
The slides were scanned and metaphase spreads were automatically found using the Msearch mode of the Metafer software system (MetaSystem Hard and Software, Altlussheim, Germany). The metaphase images were autocaptured as 90 stacks separated by 0.2 μm by a Zeiss Axioplan 2 microscope (63×, 1.4 NA; Plan-Apochromat oil immersion objective; Thornwood, NY) with Cy3 and DAPI single-pass filter sets using the AutoCapt mode of the Metafer software system. The inverted DAPI image of each metaphase spread was karyotyped using the ISIS digital image analysis system (MetaSystem) and the intensity of telomere signal was measured by modified CGH analysis software (MetaSystem). Telomere ends were interpreted as telomere signal-free ends when the red (telomere) to blue (DAPI) ratio was zero (Figure 1).
Figure 1.
Identification of the shortest telomeres. BJ metaphases at PD84 were serially hybridized to both C- and G-rich telomeric probes to maximize the detection of small telomeric regions. (A) Telomeres so short that they failed to give an observable signal (signal-free ends) were identified by CGH analysis. (B) The frequency of signal-free ends per 100 metaphases is shown for all telomeric ends from an analysis of 403 metaphases. BACs within 100 kb of the ends of the telomeres indicated by vertical arrows were used in the analysis shown in Figure 2.
For dual color fluorescence studies of telomeric variant repeats, 20 ng of the telomeric sequence variant probe (either Cy3-conjugated (TCAGGG)3 or (TGAGGG)3 2′-deoxyoligonucleotide N3′-P5′ phosphoramidate) was combine with 15 ng 3′-FITC-conjugated (TTAGGG)3 2′-deoxyoligonucleotide N3′-P5′ phosphoramidate probe and processed as above with a 10-min denaturation step and a 16-h 37°C hybridization. After washing and DAPI staining, metaphase spreads were digitally captured with precision Cy3/FITC/DAPI bandpass filter sets.
Immunofluorescence and Fluorescence In Situ Hybridization Analysis
Cells were grown on glass chamber slides for at least 96 h before immunostaining for γH2AX and 53BP1 proteins to minimize stress arising during trypsinization. Cells were briefly washed with PBS and fixed with freshly prepared 4% paraformaldehyde in PBS for 15 min at room temperature. After washing three times with PBS, cells were permeabilized for 15 min with PBS containing 0.2% Triton X-100, blocked for 1 h with 1.5% goat serum (Vector Laboratories) in PBS, and double-stained with 50 ng/μl mouse monoclonal anti-53BP1 (kindly provided by Dr. Chen, Mayo Clinic) and 10 ng/μl rabbit antiphospho-H2AX (Ser 139; Upstate Biotechnology, Lake Placid, NY) at 37°C for 1 h. Cells were subsequently washed three times for 5 min with PBS and incubated for 1 h with Alexa Fluor 568–conjugated anti-rabbit (1:200) and Alexa 488–conjugated anti-mouse antibodies (1:200; Jackson ImmunoResearch, West Grove, PA; Molecular Probes, Eugene, OR) at room temperature. After washing the cells three times for 5 min with PBS, the cells were dehydrated by an ethanol series (70, 90, and 100%) for 1 min each and counterstained with Vectashield containing DAPI for nuclear identification. The slides were scanned, and positive nuclei were automatically found and captured by a Zeiss Axioplan 2 microscope (63×, 1.4 NA; Plan-Apochromat oil immersion objective) with Texas Red, FITC, and DAPI single-pass filter sets using the AutoCapt mode of the Metafer software system. The images were saved as ninety z-stacks separated by 0.2 μm.
The slides were then processed for fluorescence in situ hybridization (FISH) analysis using the BAC probes RP3-416J7 (6pter), RP11-1197K16 (17qter), RP4-764O12 (7qter), RP11-1260E13 (17pter), RP11-974F22 (9qter), and RP11-81L3 (9q21, 62 Mb from 9qter; Children's Hospital Oakland Research Institute). All of the BACs except 9q21 were within 100 kb of the telomere. BAC DNAs were extracted using a large-construct kit (Qiagen, Chatsworth, CA), and labeled with either Spectrum-orange or Spectrum-green conjugated dUTP after a nick translation kit protocol (Vysis, Downers Grove, IL). After washing three times for 5 min with PBS, the cells were dehydrated through an ethanol series as above and air-dried. DNA was denatured for 10 min at 70°C in hybridization buffer containing 70% formamide (Sigma) and 2× SSC solutions (pH 7.0). After denaturation, the cells were dehydrated by a cold ethanol series and air-dried. The cells were put through another two denaturation and dehydration steps above to fully remove γ-H2AX and 53BP1 fluorescent antibodies. A mix of 100 ng of one Spectrum-orange– and one Spectrum-green–conjugated BAC probes in a hybridization buffer containing 50% formamide, 10% dextran sulfate, and 1× SSC was denatured at 78°C for 5 min and then hybridized to the cells for 16 h at 37°C in a humidified chamber. The slides were washed for 2 min with 0.4× SSC/0.3% NP-40 at 70°C, and 1 min in 2× SSC/0.1% NP-40 at room temperature. After being air-dried, the cells were counterstained with DAPI. The same nuclei identified for γH2AX/53BP staining above were automatically found and captured with Spectrum-orange, Spectrum-green, and DAPI single-pass filter sets using the AutoCapt mode of the Metafer software system. The images were saved as 90 z-stacks separated by 0.2 μm. The nuclei were analyzed using the ISIS digital image analysis system (MetaSystem). The same slide was then reprobed with a different set of BAC probes after again removing the previous signals with the denaturation steps described above. The physical mapping sites of these BACs was confirmed using regular FISH analysis of at least 20 metaphase spreads of normal fibroblast cells per BAC, and no polymorphism was observed.
RESULTS
The Shortest Telomeres Colocalize with γH2AX/53BP1 Foci
We used quantitative in situ hybridization to identify the shortest telomeres in near-senescent BJ foreskin fibroblasts. Several studies (Londono-Vallejo et al., 2001; Baird et al., 2003; der-Sarkissian et al., 2004) have demonstrated that the telomere length on homologous chromosomes can differ. Rather than measuring the strength of the telomere signal, which would provide the average of both homologues, we identified the shortest telomeres as those lacking a detectable signal. Only telomeres lacking a signal on both chromatids when serially hybridized with both (TTAGGG)4 and (CCCTAA)4 probes were scored as signal-free ends. The chromosomes were then scanned with a Comparative Genomics Hybridization program (Figure 1A) in order to identify ends lacking a detectable signal. No signal-free ends were found at any telomere in young BJ fibroblasts (PD 15, 59 metaphases, unpublished data). However, in near senescent BJ cells (PD 84), only 10% of the metaphases had no signal-free ends, whereas 6% of the metaphases had one, 75% had 2–3 and 9% had 4–5 signal-free ends. Figure 1B shows the distribution of very short telomeres (% of metaphases exhibiting a signal free end at each chromosome end) for 403 metaphases at PD84, approximately three doublings before senescence. Chromosome ends 6p and 21p were the shortest, and 80% of all of the signal-free ends could be accounted for by the 10 shortest telomeres.
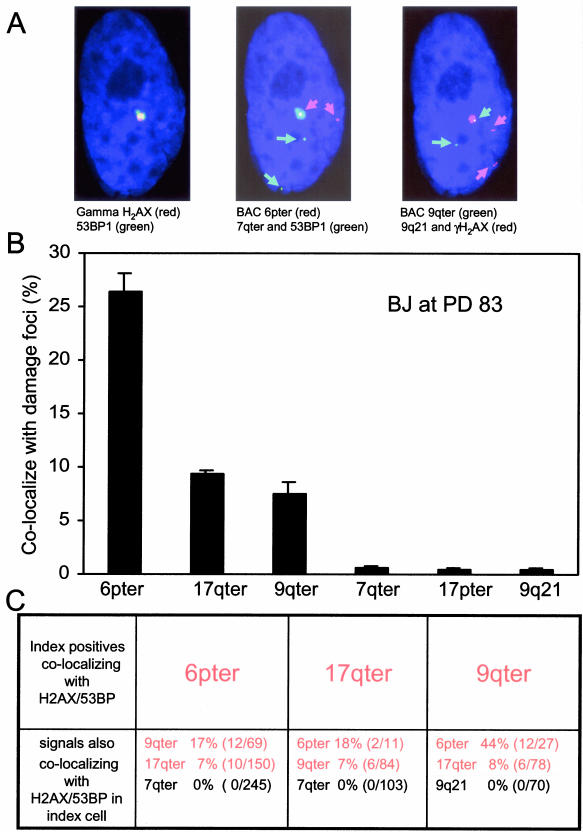
Although telomere dysfunction through removal of a telomeric protein can induce DNA damage foci in mammalian cells (Takai et al., 2003), and γH2AX has been found near telomeres in senescent cells by chromatin immunoprecipitation analysis (d'Adda di Fagagna et al., 2003), the specific association of the shortest telomeres in human senescent cells with these foci has not been demonstrated. In our near senescent BJ, 79% of 2000 interphase cells analyzed contained γH2AX/53BP1 DNA damage foci, and the vast majority of these (80%) had only one or two foci. We obtained BACs within 100 kb of three of the short telomere ends (6p [41% signal-free ends], 17q [11.7% signal-free ends] and 9q [7.7% signal-free ends]), two from ends having much longer telomeres (7q [0.2% signal-free ends] and 17p [0% signal-free ends]), and one that was 62 Mb from the telomere (9q21).
These probes were serially hybridized to cells after first identifying DNA damage foci by γH2AX/53BP1 staining. Figure 2A shows an example of colocalization of DNA damage foci with the shortest but not longer telomere ends or internal signals. Figure 2B shows that the frequency of colocalization approximates the frequency of signal-free ends shown in Figure 1B, with only background levels for the internal and longer-telomere probes. Many cells exhibited more than one short telomere colocalizing with foci, often with the same γH2AX/53BP1 focus (Figure 2, A and C). For example, about one-sixth of the cells showing colocalization of 6ptel with foci also showed colocalization of 9qtel (Figure 2C).
Figure 2.
Colocalization of short telomeres with γH2AX/53BP1 foci at senescence. (A) BJ cells at PD 83 were first stained with γH2AX/53BP1 and then serially hybridized in pairs to four different BAC probes, The probes to 6pter (41% signal-free ends), 9qter (7.7% signal-free ends), and 7qter (0.2% signal-free ends) were within 100 kb of the telomere, whereas that to 9q21 was 62 Mb from the telomere. Colored arrows highlight the position of the small but intense BAC signals. The same cell appears in all three panels and shows colocalization of both 6pter (middle) and 9qter (right) to the same γH2AX/53BP1 focus (left, digitally overlaid onto the other two images). (B) The frequency of colocalization with markers of DNA damage foci. The ends having shorter telomeres (6pter, 17qter, and 9qter) but not the ends having longer telomeres (7qter and 17pter) or the internal locus (9q21) showed frequent colocalization with markers of DNA damage foci. Data are for at least 900 DNA damage foci per BAC on three slides, ±SD between slides. The % colocalization was calculated as the percent of all γH2AX/53BP1 foci that colocalized with the specific BAC probe. The γH2AX/53BP1 positive foci in near-senescent BJ cells were distributed as follows: 43% of the cells had a single focus, 35% had two, 8% had three, 6% had four, and 8% had five or more foci. Correction for the number of colocalized foci per cell would result in a doubling of the % colocalization if expressed per cell instead of the data per focus as shown. (C) Presence of multiple short ends colocalizing with damage foci in single cells. The data in B have been reorganized to show the frequency with which a cell showing colocalization of one particular short telomere (e.g., 6pter) with a DNA damage focus also showed colocalization of other probes to DNA damage foci within the same cell. The short telomeres are shown in red.
End-associations Involving Short Telomeres
A high frequency of polyploidy and multiple chromosomal abnormalities have been reported as cells approach senescence (Saksela and Moorhead, 1963; Thompson and Holliday, 1975; Benn, 1976; Sherwood et al., 1988). However, previous reports were generally performed in embryonic lung fibroblasts. These cells undergo an oxidative crisis when grown in room oxygen (Atamna et al., 2000; Forsyth et al., 2003), and the inability of telomerase to immortalize these cells under ambient oxygen conditions in spite of elongating telomeres (Forsyth et al., 2003) strongly implicates global rather than just telomere-specific effects of oxidative damage. BJ foreskin fibroblasts have been shown to have much greater oxygen protective mechanisms than embryonic lung fibroblasts (Lorenz et al., 2001), and they immortalize directly after ectopic expression of telomerase (Bodnar et al., 1998). They thus provided an opportunity to examine the consequences of telomere shortening with fewer complications from other clastogenic processes. We confirmed that WI-38 embryonic lung fibroblasts exhibited many chromosomal abnormalities as they approached senescence (unpublished data). In stark contrast, in spite of the abundance of signal-free ends in metaphases from PD 84 BJ cells, the frequency of chromosomal abnormalities and end-associations in metaphase spreads from nearly senescent cells was too low to be easily quantitated. Tetraploid metaphases were also very rare (<1%). These cells were only three population doublings before senescence, and their growth rate had decreased from three doublings per week to one doubling per several weeks, so they clearly were in the period of culture where many cells were already senescent and only the longest-lived subpopulations were continuing to divide. This implies that most of the previously reported chromosomal abnormalities might reflect the combined results of oxidative crisis/cultured stress and telomere shortening rather than just the consequences of short telomeres. It also implies that in BJ fibroblasts, the presence of telomeres sufficiently short to appear in γH2AX foci is not sufficient to cause the appearance of gross chromosomal abnormalities.
The absence of abnormal metaphases suggested that either the short telomeres were inducing growth arrest before their formation or that the very first abnormality prevented the cells from dividing again so they dropped out of the mitotic population. We thus introduced factors to block cell-cycle checkpoint functions (human papilloma virus 16 proteins E6 and E7 (Scheffner et al., 1990; Werness et al., 1990; Barbosa et al., 1991) in order to obtain metaphases exhibiting the abnormalities due to short telomeres. HPV 16 E6+E7 does not activate telomerase in BJ cells but permits the bypass of senescence and confers an extended life span to the population.
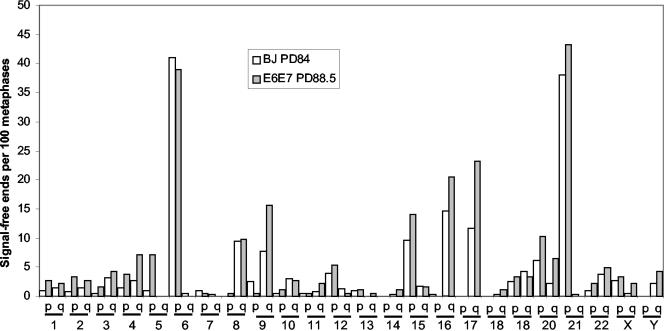
Figure 3 compares the frequency of signal-free ends in 485 metaphases from E6+E7-expressing BJ fibroblasts at PD 88 with those observed in PD 84 parental BJ cells. There is a very close correspondence to the frequency of signal-free telomeres in the uninfected cells, indicating that ∼30 doublings in the presence of E6+E7 has not significantly altered the telomere-shortening rate or affected the specific chromosomes having the shortest telomeres. The few minor alterations in frequency that are present may reflect that at PD 88 the E6+E7 cells are still dividing vigorously and continue to represent the clonal diversity in the original population, whereas at PD 84 most clonal lineages have become senescent and only a few of the longest-lived clones are continuing to divide.
Figure 3.
The same telomeres are short in normal BJ and BJ cells expressing E6+E7. BJ cells infected with a retrovirus expressing human papilloma virus 16 proteins E6+E7 at about PD 65 were analyzed for the presence of telomeres with signal-free ends at PD 88.5 using the same techniques as for BJ cells in Figure 1. The results of both analyses demonstrate that E6+E7 has neither affected the telomere shortening rate nor the telomeres showing signal-free ends in cells with abrogated cell-cycle checkpoints, because both cell types show the same frequencies and distribution.
The frequency of signal-free ends at the Xp/Yp telomere was 3% for both normal BJ at PD 84 and BJ with E6+E7 at PD 88. To determine the approximate sensitivity of detecting signal-free ends, we used STELA (Baird et al., 2003) to amplify individual Xp/Yp telomeres. One hundred seventy-seven individual bands (90 from BJ and 87 from BJ E6+ E7) were analyzed. After subtracting the 400 base pairs of subtelomeric sequences amplified with each product (Baird et al., 2003), the shortest five bands (3% of 177) contained <100 base pairs of C-strand telomeric sequences. Telomeres identified as signal-free thus had very few remaining telomeric repeats.
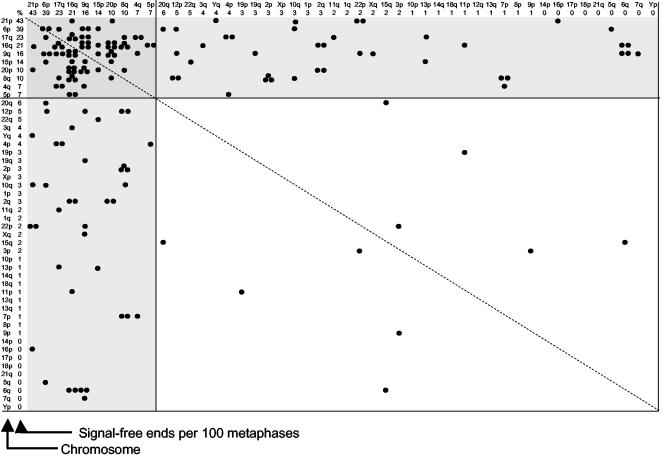
Telomere associations appeared in the presence of E6+E7 at approximately the same population doubling level as when senescence occurred in control cultures (Table 1). The frequency of end-associations at PD 84–91 is plotted in Figure 4 as a function of the percentage of signal-free ends for each telomere from Figure 3. Each end-association is plotted twice, i.e., an association between 20p and 21p is considered equivalent to one between 21p and 20p. The vast majority (94%) of the end-associations occurring at the time of M1 involved the 10 shortest telomeres. Forty-four percent of the associations occurred within members of this group, whereas most of the remainder (49%) occurred between,one of these 10 and other chromosomes. There is a wide distribution of end-associations, indicating that the events are polyclonal and do not represent the overgrowth of a cell with one particularly short telomere. This distribution demonstrates that the end-associations are not due to the “programming” of one or two “sentinel” telomeres that monitor divisions, but rather the cell is using the shortening of roughly 10% of its telomeres as a measure of replicative history.
Table 1.
Frequency of end-associations involving the 10 shortest telomeres
| Telomeric end-associations
|
|||||
|---|---|---|---|---|---|
| PDL | One short | Both short | Total with short | Total | % involving short |
| 73 | 0 | 0 | 0 | 0 | — |
| 81 | 0 | 2 | 2 | 2 | 100 |
| 84 | 32 | 20 | 52 | 56 | 93 |
| 87 | 6 | 9 | 15 | 16 | 94 |
| 91 | 2 | 6 | 8 | 8 | 100 |
| 96 | 7 | 10 | 17 | 19 | 89 |
| 101 | 10 | 6 | 16 | 19 | 84 |
| 103 | 19 | 6 | 25 | 39 | 64 |
M1 (senescence) occurred at approximately PD 85 in control cultures without E6+E7 and M2 (crisis) occurred at PD 103–105. At least 20 metaphases were scored at each population doubling level (PDL).
Figure 4.
Chromosomal end-associations in BJ E6+E7 cells. The frequency of end-associations for specific telomeres is shown for BJ foreskin fibroblasts expressing E6+E7 at PD 84–91, the approximate time when M1 would have occurred in parallel cultures without E6+E7. The axes are arranged according to the frequency of signal-free ends per 100 metaphases shown in Figure 3. Each event is plotted twice, because an association between telomeres A and B is equivalent to an association between B and A. The dotted diagonal line reflects this line of symmetry. Almost all of the associations either involve short-short associations (darker gray box at the top left) or associations between the 10 shortest and other chromosomes (lighter gray boxes). There is no significant bias of one or a few specific ends. The distribution as plotted does not distinguish between homologues of the 22 autosomal chromosomes, so there are only 48 ends along each axis. If separated into homologues, the 10 shortest would represent 10 of 92 ends (see Figure 5).
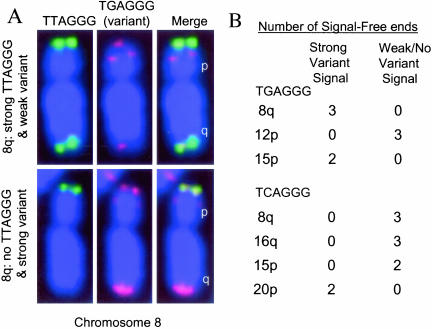
Consistent with previous observations of the size heterogeneity of homologous chromosomes (Londono-Vallejo et al., 2001; Baird et al., 2003; der-Sarkissian et al., 2004), we found little correlation between the telomere lengths of the two parental chromosomes (e.g., metaphase spreads in which there was no telomeric FISH signal on one chromosome 8q exhibited good doublet signals on the other chromosome 8q). In some cases, the parental chromosomes could be distinguished by the presence of very different signals when hybridized to telomeric sequence variants (such as (TGAGGG)3 or (TCAGGG)3), which can be present in a polymorphic pattern at the base of the telomeres (Allshire et al., 1989; Baird et al., 2000). This confirmed that the signal-free ends were either on the maternal or paternal chromosome but not both (Figure 5, p = 0.0005). Consequently, the 10 shortest telomeres represent the 10 shortest among 92 rather than among 46 telomeres. The expected fraction of short-short associations by random chance is 5% (assuming 10/46 ends) or 1% (assuming 10/92 ends), very different from the 44% observed (p < 0.0001 by the one sample proportions test). The results in Figure 4 are thus highly significant.
Figure 5.
Telomeric sequence variants distinguish parental chromosomes. (A) Telomeres were hybridized with probes for the telomeric repeat [(TTAGGG)3-FITC] or a Cy3-labeled sequence variant (TGAGGG)3 often found at the very base of the telomere. The top chromosome 8 shows a strong telomeric signal on the q arm but a weak variant repeat signal, whereas the lower chromosome 8q from the same cell exhibits no TTAGGG fluorescence (thus a signal-free end) but a strong sequence variant signal, thus allowing the parental homologues to be determined. (B) In cases where the parental homologues could be determined by polymorphisms in the amount of sequence variants, the signal-free ends were always present on one but not both parental telomeres. The probability of finding this pattern in 3 metaphases is 0.25, in 2 metaphases is 0.5, and for all 18 metaphases on 7 ends shown is 0.0005.
Telomere Associations during the Extended Lifespan
The specific chromosomes involved in end-associations were monitored as cells progressed beyond M1 until they reached crisis (Table 1). The predominance of end-association events involving the 10 shortest chromosomes continued for an additional 10–15 doublings, consistently representing ∼90% of all telomere associations. As the cells approached M2 (PD103–105) their relative abundance diminished slightly as other telomeric ends become sufficiently short to initiate additional events.
DISCUSSION
The process by which cells regulate the onset of cellular senescence represents an important question for understanding replicative aging and the mechanisms by which tumor cells escape these limits and become immortal. We have previously demonstrated that the timing of senescence is dependent on the shortest telomeres (Ouellette et al., 2000; Steinert et al., 2000), but these studies did not distinguish between one or a few short sentinel telomeres or a more general monitoring of short telomeres and did not identify the specific telomeres involved. The present results show that the shortest telomeres are found in γH2AX/53BP1 foci at senescence and that in a cell type with strong oxidative protection mechanisms the frequency of chromosomal abnormalities at senescence are very low. If checkpoint arrest is blocked by the papilloma virus proteins E6+E7, then ∼10% of the telomeres are found to be responsible for >90% of the telomeric end-association events at the population-doubling level when senescence would have occurred. Thus, a substantial subset rather than one or two telomeres is being used to time the onset of replicative arrest in the population of cells. These results also indicate that although blocking p53/pRB checkpoints with E6+E7 bypasses senescence, it does not prevent end-associations involving the shortest telomeres. der-Sarkissian et al. (2004) have shown that the shortest telomeres drive karyotypic changes in transformed human embryonic kidney cells. Our results are consistent with their observations.
Approximately 85% of metaphase spreads from near-senescent cultures contained more than one signal-free end. This establishes that normal cells are still able to divide in the presence of telomeres sufficiently short to fail to give a hybridization signal (which may or may not be short enough to initiate a DNA damage response). Furthermore, many interphase nuclei from near-senescent cultures had γH2AX/53BP foci colocalizing with more than one of the shortest telomeres (Figure 2). It is as yet unknown whether the initial failure to growth arrest is due to a graded response from individual telomeres (the strength of the damage signal is initially low when the telomere is sufficiently short to initiate any signal and increases as that telomere gets progressively shorter) or if more than one short telomere is required to initiate growth arrest (for example, a short telomere has to find a partner and form an end-association before growth arrest occurs). Telomeric end-associations were not seen in metaphases from near-senescent BJ cultures. The fact that end-associations first appear in E6+E7 expressing BJ cells at the same population doubling level when senescence occurred in control cultures suggests that end-associations were correlated with growth arrest in the normal cells (and thus failure to form metaphases). This supports the hypothesis that cells can divide in the presence of signal-free telomeres until an end-association occurs.
Although there is a general correlation between chromosome size and telomere length (Suda et al., 2002), the fact that there is variation between maternal and paternal telomere length on any given chromosome (Londono-Vallejo et al., 2001) suggests that the control of telomere length in the germline is imprecise. A 3-kb variation in size between a 15-kb maternal and an 18-kb paternal telomere would make little difference in the germline, but would make an enormous difference in which specific telomere became limiting when shortening reduced the respective lengths to 0.5 and 3.5 kb. Furthermore, if telomere shortening/processing of C-strands is much more extensive than G-strands, a several kilobase distribution of sizes develops over time even for a single telomere of defined length (Levy et al., 1992). Given the importance of counting cell divisions in order for replicative aging to function as a brake against the progression of cancer, it thus makes biological sense for cells to monitor a group of short telomeres to control the timing of replicative aging as found here, rather than attempting to have one or two specified telomeres function as “sentinels” that count divisions and determine when senescence should occur.
Although there is a statistically significant difference in the length of telomeres at particular chromosome ends when multiple individuals are analyzed (Martens et al., 2000; Graakjaer et al., 2003), there is considerable variability between individuals. This is consistent with the variability in telomere sizes between maternal and paternal chromosomes (Figure 5; Londono-Vallejo et al., 2001). We would thus predict that the 10 shortest telomeres that regulate replicative aging in BJ foreskin fibroblasts should be enriched for ends that have been found to be short in other studies, but that the overlap should not be complete and a different subset of ends would produce senescence in other cell strains. This analysis is complicated by the fact that the published studies on chromosome-specific telomere length have averaged maternal and paternal chromosomes, whereas our results and those of others (Figure 5; Londono-Vallejo et al., 2001; der-Sarkissian et al., 2004) suggest that only one of each pair is among the 10 shortest. Nonetheless, 8 of our 10 shortest telomeres were found in the shortest 50% of chromosome ends from an analysis of the average of 20 older individuals (Graakjaer et al., 2003). This study also showed that although there is an overall pattern of telomere length on different chromosome ends, there is an individual pattern superimposed on the average pattern such that the particular telomeres that are the shortest will vary between individuals.
Our results differ from those reported by Martens et al. (2000), which failed to find a correlation between the telomere length determined by Q-FISH and replicative aging. There are a variety of possible explanations. Their study averaged the telomere length between homologous chromosomes, thus the influence of particular parental telomeres would have been diminished. Furthermore, they used a strain of fetal rather than neonatal skin fibroblasts. We have found considerable differences in the culture requirements of fetal vs. adult lung fibroblasts. Fetal lung fibroblasts exhibited stasis (growth arrest due to inadequate culture conditions; Ramirez et al., 2001; Drayton and Peters, 2002; Wright and Shay, 2002) rather than pure telomere-based replicative senescence using standard tissue culture media in room oxygen (Forsyth et al., 2003). If fetal skin fibroblasts exhibit similar differences, the short telomeres in the above study may have failed to predict the onset of growth arrest because the cells never reached telomere-based replicative senescence.
The shortest telomeres preferentially formed end-associations with other short telomeres rather than random chromosome ends (Figure 4). Short telomeres might be metastable, spending progressively larger fractions of time in a partially unprotected configuration. The presence of one unprotected telomere could then trap a second metastable telomere in an end-association before it could be repackaged into a protected conformation, explaining the preponderance of short-short rather than short-random associations.
One report has suggested that M1 is caused by the disappearance of the G-rich 3′ telomeric overhang and that it is the global loss of this overhang on all telomeres with progressive cell divisions rather than telomere length that causes replicative aging (Stewart et al., 2003). Furthermore, this and a subsequent report (Masutomi et al., 2003) suggested that telomerase immortalizes cells by restoring the overhang rather than by elongating telomeres. Our results are largely incompatible with these reports because we establish that specific telomeres with a high fraction of signal-free ends become localized to γH2AX/53BP1 foci at the time of senescence, whereas telomeres with a very low fraction of signal-free ends do not (Figure 2). Furthermore, if cell cycle arrest is blocked, only a small subset of telomeres accounts for almost all of the end-association events that occur (Figure 4). Not only are these specifically the shortest telomeres, in any given cell it is only one or two of the shortest telomeres that are producing end-associations. One should thus not see an 85% reduction in overhangs in BJ cells (Stewart et al., 2003) using an assay that measures all 92 ends at once if only one or two telomeres per cell are limiting for growth. These authors also demonstrated that the induction of DNA damage in young fibroblasts could induce a shortening of the overhangs (Stewart et al., 2003). Given the known recruitment to telomeres of a large number of DNA repair factors, it would not be surprising if the activation of a DNA damage response influenced telomere end-processing. A reasonable hypothesis is that 1 of the 10 shortest telomeres induces a DNA damage signal, and any subsequent global shortening of overhangs then occurs as a simple consequence of signal-induced altered processing. Short overhangs would thus represent an epiphenomenon, a secondary consequence of short telomeres, rather than a proximate cause of senescence. We cannot exclude the possibility that the consequences of short overhangs contribute to a greater deprotection of the shortest telomeres or that there might be cis-acting effects so that overhang loss is greatest on the shortest telomeres.
Studies of telomere lengths on specific human chromosomes have shown a progression of sizes rather than one or a few individual telomeres that are dramatically shorter than the rest. Our increasing knowledge of stochastic processes that influence telomere lengths suggest that it would be difficult to have a few sentinel telomeres that could time replicative aging. The demonstration that relatively large groups of short telomeres are functionally monitored to determine the timing of cellular senescence now provides a sound biological rationale for understanding this process.
Acknowledgments
We thank Linda Hynan for help with the statistical analysis. This work was supported by National Institutes of Health Grant AG07992 (W.E.W. and J.W.S.). W.E.W. is an Ellison Foundation Senior Scholar.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0207. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0207.
References
- Allshire, R.C., Dempster, M., and Hastie, N.D. (1989). Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 17, 4611-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamna, H., Paler-Martinez, A., and Ames, B.N. (2000). N-t-butyl hydroxylamine, a hydrolysis product of alpha-phenyl-N-t-butyl nitrone, is more potent in delaying senescence in human lung fibroblasts. J. Biol. Chem. 275, 6741-6748. [DOI] [PubMed] [Google Scholar]
- Baird, D.M., Coleman, J., Rosser, Z.H., and Royle, N.J. (2000). High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution. Am. J. Hum. Genet. 66, 235-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, D.M., Rowson, J., Wynford-Thomas, D., and Kipling, D. (2003). Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 33, 203-207. [DOI] [PubMed] [Google Scholar]
- Barbosa, M.S., Vass, W.C., Lowy, D.R., and Schiller, J.T. (1991). In vitro biological activities of the E6 and E7 genes vary among human papillomaviruses of different oncogenic potential. J. Virol. 65, 292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn, P.A. (1976). Specific chromosome aberrations in senescent fibroblast cell lines derived from human embryos. Am. J. Hum. Genet. 28, 465-473. [PMC free article] [PubMed] [Google Scholar]
- Bodnar, A.G. et al. (1998). Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349-352. [DOI] [PubMed] [Google Scholar]
- Bryan, T.M., Englezou, A., Gupta, J., Bacchetti, S., and Reddel, R.R. (1995). Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14, 4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna, F., Reaper, P.M., Clay-Farrace, L., Fiegler, H., Carr, P., von Zglinicki, T., Saretzk, G., Carter, N.P., and Jackson, S.P. (2003). A DNA damage checkpoint-mediated response in telomere-initiated senescence. Nature 426, 194-198. [DOI] [PubMed] [Google Scholar]
- de Lange, T. (2002). Protection of mammalian telomeres. Oncogene 21, 532-540. [DOI] [PubMed] [Google Scholar]
- der-Sarkissian, H., Bacchetti, S., Cazes, L., and Londono-Vallejo, J.A. (2004). The shortest telomeres drive karyotype evolution in transformed cells. Oncogene 23, 1221-1228. [DOI] [PubMed] [Google Scholar]
- Drayton, S., and Peters, G. (2002). Immortalisation and transformation revisited. Curr. Opin. Genet. Dev. 12, 98-104. [DOI] [PubMed] [Google Scholar]
- Dunham, M.A., Neumann, A.A., Fasching, C.L., and Reddel, R.R. (2000). Telomere maintenance by recombination in human cells. Nat. Genet. 26, 447-450. [DOI] [PubMed] [Google Scholar]
- Forsyth, N.R., Evans, A.P., Shay, J.W., and Wright, W.E. (2003). Developmental differences in the immortalization of lung fibroblasts by telomerase. Aging Cell 2, 235-243. [DOI] [PubMed] [Google Scholar]
- Graakjaer, J., Bischoff, C., Korsholm, L., Holstebroe, S., Vach, W., Bohr, V.A., Christensen, K., and Kolvraa, S. (2003). The pattern of chromosome-specific variations in telomere length in humans is determined by inherited, telomere-near factors and is maintained throughout life. Mech. Ageing Dev. 124, 629-640. [DOI] [PubMed] [Google Scholar]
- Greider, C.W. (1990). Telomeres, telomerase and senescence. Bioessays 12, 363-369. [DOI] [PubMed] [Google Scholar]
- Gustashaw, K.M. (1997). Chromsome Stains. Philadelphia: Lippincott-Raven.
- Halbert, C.L., Demers, G.W., and Galloway, D.A. (1992). The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 66, 2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, E., Tsurui, H., Shinozaki, A., Nakada, S., and Oda, K. (1991). Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem. Biophys. Res. Commun. 179, 528-534. [DOI] [PubMed] [Google Scholar]
- Harley, C.B. (1991). Telomere loss: mitotic clock or genetic time bomb? Mutat. Res. 256, 271-282. [DOI] [PubMed] [Google Scholar]
- Harley, C.B., Futcher, A.B., and Greider, C.W. (1990). Telomeres shorten during ageing of human fibroblasts. Nature 345, 458-460. [DOI] [PubMed] [Google Scholar]
- Hayflick, L., and Moorhead, P.S. (1961). The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585-621. [DOI] [PubMed] [Google Scholar]
- Hemann, M.T., Strong, M.A., Hao, L.Y., and Greider, C.W. (2001). The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107, 67-77. [DOI] [PubMed] [Google Scholar]
- Kim, N.W. et al. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011-2015. [DOI] [PubMed] [Google Scholar]
- Levy, M.Z., Allsopp, R.C., Futcher, A.B., Greider, C.W., and Harley, C.B. (1992). Telomere end-replication problem and cell aging. J. Mol. Biol. 225, 951-960. [DOI] [PubMed] [Google Scholar]
- Lingner, J., Cooper, J.P., and Cech, T.R. (1995). Telomerase and DNA end replication: no longer a lagging strand problem? Science 269, 1533-1534. [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo, J.A., DerSarkissian, H., Cazes, L., and Thomas, G. (2001). Differences in telomere length between homologous chromosomes in humans. Nucleic Acids Res. 29, 3164-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M., Saretzki, G., Sitte, N., Metzkow, S., and von Zglinicki, T. (2001). BJ fibroblasts display high antioxidant capacity and slow telomere shortening independent of hTERT transfection. Free Radic. Biol. Med. 31, 824-831. [DOI] [PubMed] [Google Scholar]
- Martens, U.M., Chavez, E.A., Poon, S.S., Schmoor, C., and Lansdorp, P.M. (2000). Accumulation of short telomeres in human fibroblasts prior to replicative senescence. Exp. Cell Res. 256, 291-299. [DOI] [PubMed] [Google Scholar]
- Masutomi, K. et al. (2003). Telomerase maintains telomere structure in normal human cells. Cell 114, 241-253. [DOI] [PubMed] [Google Scholar]
- Ouellette, M.M., Liao, M., Herbert, B.S., Johnson, M., Holt, S.E., Liss, H.S., Shay, J.W., and Wright, W.E. (2000). Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem. 275, 10072-10076. [DOI] [PubMed] [Google Scholar]
- Ramirez, R.D., Morales, C.P., Herbert, B.S., Rohde, J.M., Passons, C., Shay, J.W., and Wright, W.E. (2001). Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 15, 398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksela, E., and Moorhead, P.S. (1963). Aneuploidy in the degenerative phase of serial cultivation of human cell strains. Proc. Natl. Acad. Sci. USA 50, 390-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner, M., Werness, B.A., Huibregtse, J.M., Levine, A.J., and Howley, P.M. (1990). The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63, 1129-1136. [DOI] [PubMed] [Google Scholar]
- Shay, J.W., and Bacchetti, S. (1997). A survey of telomerase activity in human cancer. Eur. J. Cancer 5, 787-791. [DOI] [PubMed] [Google Scholar]
- Shay, J.W., Pereira-Smith, O.M., and Wright, W.E. (1991). A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 196, 33-39. [DOI] [PubMed] [Google Scholar]
- Sherwood, S.W., Rush, D., Ellsworth, J.L., and Schimke, R.T. (1988). Defining cellular senescence in IMR-90 cells: a flow cytometric analysis. Proc. Natl. Acad. Sci. USA 85, 9086-9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert, S., Shay, J.W., and Wright, W.E. (2000). Transient expression of human telomerase extends the life span of normal human fibroblasts. Biochem. Biophys. Res. Commun. 273, 1095-1098. [DOI] [PubMed] [Google Scholar]
- Stewart, S.A., Ben-Porath, I., Carey, V.J., O'Connor, B.F., Hahn, W.C., and Weinberg, R.A. (2003). Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 33, 492-496. [DOI] [PubMed] [Google Scholar]
- Suda, T., Fujiyama, A., Takimoto, M., Igarashi, M., Kuroiwa, T., Waguri, N., Kawai, H., Mita, Y., and Aoyagi, Y. (2002). Interchromosomal telomere length variation. Biochem. Biophys. Res. Commun. 291, 210-214. [DOI] [PubMed] [Google Scholar]
- Takai, H., Smogorzewska, A., and de Lange, T. (2003). DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549-1556. [DOI] [PubMed] [Google Scholar]
- Thompson, K.V., and Holliday, R. (1975). Chromosome changes during the in vitro ageing of MRC-5 human fibroblasts. Exp. Cell Res. 96, 1-6. [DOI] [PubMed] [Google Scholar]
- von Zglinicki, T. (2002). Oxidative stress shortens telomeres. Trends Biochem. Sci. 27, 339-344. [DOI] [PubMed] [Google Scholar]
- von Zglinicki, T., Saretzki, G., Docke, W., and Lotze, C. (1995). Mild hyperoxia shortens telomeres and inhibits proliferation of fibroblasts: a model for senescence? Exp. Cell Res. 220, 186-193. [DOI] [PubMed] [Google Scholar]
- Werness, B.A., Levine, A.J., and Howley, P.M. (1990). Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248, 76-79. [DOI] [PubMed] [Google Scholar]
- Wright, W.E., and Shay, J.W. (2002). Historical claims and current interpretations of replicative aging. Nat. Biotechnol. 20, 682-688. [DOI] [PubMed] [Google Scholar]