Abstract
Tetrahymena telomeres usually consist of ∼250 base pairs of T2G4 repeats, but they can grow to reach a new length set point of up to 900 base pairs when kept in log culture at 30°C. We have examined the growth profile of individual macronuclear telomeres and have found that the rate and extent of telomere growth are affected by the subtelomeric region. When the sequence of the rDNA subtelomeric region was altered, we observed a decrease in telomere growth regardless of whether the GC content was increased or decreased. In both cases, the ordered structure of the subtelomeric chromatin was disrupted, but the effect on the telomeric complex was relatively minor. Examination of the telomeres from non-rDNA chromosomes showed that each telomere exhibited a unique and characteristic growth profile. The subtelomeric regions from individual chromosome ends did not share common sequence elements, and they each had a different chromatin structure. Thus, telomere growth is likely to be regulated by the organization of the subtelomeric chromatin rather than by a specific DNA element. Our findings suggest that at each telomere the telomeric complex and subtelomeric chromatin cooperate to form a unique higher order chromatin structure that controls telomere length.
INTRODUCTION
Telomeres from most organisms exhibit a characteristic mean length that results from a balance between addition of telomeric DNA by telomerase or recombination and loss of DNA due to incomplete replication or nuclease activity (Greider, 1996; McEachern et al., 2000). If this balance is perturbed, telomeres grow or shrink until a new length set point is reached. Factors that control the balance, and hence regulate telomere length, include telomere and telomerase components, replication and repair proteins, and environmental conditions. In Saccharomyces cerevisiae, more than a dozen different proteins have been shown to affect telomere length (Bourns et al., 1998; Blackburn, 2001), whereas rapid proliferation and increased culture temperature induce telomere growth in Tetrahymena, trypanosomes, and Candida albicans (Bernards et al., 1983; Larson et al., 1987; McEachern and Hicks, 1993).
In organisms such as Tetrahymena and S. cerevisiae that have relatively short telomeres (250–350 nt), the entire telomeric tract can be packaged into a nonnucleosomal complex (Blackburn and Chiou, 1981; Wright et al., 1992; Cohen and Blackburn, 1998), but in organisms with longer telomeres, the telomeric DNA is bound by a combination of nucleosomes and specialized telomere proteins (Tommerup et al., 1994). Both short and long telomeres seem to be subject to a second level of packaging that involves folding or looping of the telomeric tract to form a more compact higher order structure (Grunstein, 1997; Griffith et al., 1999). In S. cerevisiae, this folding is mediated by protein–protein interactions with the telomere protein Rap1 interacting with SIR proteins bound to nucleosomes along the subtelomeric region (Grunstein, 1997). In vertebrate and plant cells, the folding seems to be mediated by nucleic acid interactions. The single-strand overhang that is present on the telomeric G-strand invades and pairs with an internal portion of the telomeric tract to form a large loop on the end of the chromosome (Griffith et al., 1999; Cesare et al., 2003).
Both the composition of the telomeric complex and telomerase access to the DNA terminus have been shown to change during the cell cycle (Diede and Gottschling, 1999; Marcand et al., 2000; Taggart et al., 2002; Smith et al., 2003). These findings suggest that telomeres cycle between a closed state where the telomeric DNA is heavily protected and an open, less compact state (Blackburn, 2001). Switching between states is thought to reflect opening and closing of the higher order folded structure. Conditions that promote telomere growth are thought to increase access to telomerase by increasing the time that the telomere remains in the open state and/or decreasing the overall compaction of the terminal DNA–protein complex.
Although higher order packaging of the telomere can involve interactions with the subtelomeric region, for most organisms the extent to which the subtelomeric DNA or chromatin contributes to telomere length regulation is unclear. Most information is available for S. cerevisiae where telomere length has been shown to depend on the subtelomeric sequence. In certain genetic backgrounds, chromosomes that lack Y' elements are regulated differently from those that have both X and Y' elements (Craven and Petes, 1999). Alteration of the subtelomeric chromatin also can affect length regulation as removal of Sir3 and Sir4 correlates with a decrease in telomere length (Palladino et al., 1993). We now add to these studies by showing that, in Tetrahymena, the subtelomeric region directly affects telomere growth and final telomere length.
Tetrahymena thermophila is useful for studying many aspects of telomere biology because its unusual genomic organization results in an abundance of telomeres and telomerase. Like other ciliates, it has a transcriptionally active macronucleus and a germline, transcriptionally quiescent micronucleus (Jahn and Klobutcher, 2002). The macronucleus is formed from a copy of the micronucleus during sexual reproduction through a process that involves subdivision of the five micronuclear chromosomes into ∼200 smaller molecules (Coyne et al., 1996; Yao et al., 2002). Most of these molecules are 200–400 kb and are present at a copy number of ∼45; however, the ribosomal DNA exists as a 21-kb minichromosome that is amplified to a copy number of ∼10,000. Each of the ∼20,000 macronuclear chromosomes have telomeres composed of 250- to 300-base pair T2G4 repeats. Most of the telomeres exist as a nonnucleosomal DNA–protein complex that extends <50 base pairs beyond the end of the telomeric tract, but ∼10% have a smaller telomeric complex, so part of the telomeric tract is packaged into nucleosomes (Blackburn and Chiou, 1981; Cohen and Blackburn, 1998). The telomeric tract can be induced to grow to ∼900 base pairs if the cells are maintained in continuous log culture at 30°C (Larson et al., 1987; Ahmed et al., 1998). The underlying cause of this change in length set point is unknown, but the telomeres shrink to their original length when the culture is returned to room temperature. The current study examines the rate and extent of telomere growth at both rDNA and non-rDNA telomeres. We show that the growth profile is heavily influenced by the adjacent subtelomeric region, indicating that this nontelomeric segment of the chromosome cooperates with the telomeric complex to regulate telomerase access to the DNA terminus.
MATERIALS AND METHODS
Tetrahymena Growth and Transformation
Tetrahymena thermophila cell lines CU428, CU427, B2086, and D1 were grown in PPYS medium (2% protease peptone, 0.2% yeast extract, 0.003% sequestrin) at room temperature or 30°C. Strain D1 was derived from a cross between CU427 and CU428. To promote telomere growth, cultures were maintained in logarithmic growth (maximum cell density of 2.5 × 105 cells/ml) at 30°C by diluting daily by using prewarmed 1–2% PPYS medium. The cultures of D1 CU428 and B2086 used to compare telomere lengths were not grown in parallel. Clones containing the 11- and 14-kb nonpalindromic rDNA molecules were generated by exconjugant tranfection by using the rDNA vectors D-500 and D-500A that are defective in palindrome formation (Yao et al., 1990; Yasuda and Yao, 1991). Cell lines CU428 and B2086 or CU427 and B2086 were mated and transformed by particle bombardment 10 h later (Cassidy-Hanley et al., 1997). Transformants with 11- or 14-kb rDNA molecules were obtained by selection in 2 mg/ml paramomycin. Clones that lacked the native rDNA were identified by Southern hybridization after growth for several days in the absence of drug.
Telomere Length Analysis and Generation of Subtelomeric Probes
DNA was isolated as described previously (Brehm and Cech, 1983). Telomere length was analyzed by Southern hybridization of restriction-digested genomic DNA by using subtelomeric or telomeric probes. The median telomere length was determined by PhosphorImager analysis of each lane to find the position with maximum signal. SD was calculated from three different growth experiments for rDNA telomeres and from two experiments for non-rDNA telomeres. Length heterogeneity was estimated by eye using the PhorphorImager trace to identify the longest and shortest telomeres. To generate probes to 11- and 14-kb rDNAs, the minichromosomes were cloned by ligation of adaptors to the telomeric G-strand overhang and the subtelomeric regions were sequenced. Probes to individual non-rDNA chromosomes were generated by polymerase chain reaction by using the following primer sets. The numbers that precede the sequence identify the contigs in the Tetrahymena database that encode each chromosome end.
Tel-1; 1172290f1-TGTAGAAATGAAGATAGTTAGTATT, 1172290r1-ATGTTATTATATTTTAATTTTAGTTG
1172290f2-GGCAGTTAATATTTGATAATTCGTA, 1172290r2-TCCTGCTTGCCTTTAAAAGCTCC
Tel-2; 1173116f1-GTAATAACCATTAATAAGTGCTTAAA, 1173116r1-TTTAAAAGCTATCAATAAATGATGC
Tel-3; 1173090f1-ATTTGGTTGGTCAATCTATGTAAG, 1173090r1-GTAATTAATTATTTTATTCCTCACC
Tel-4; 1173014f1-GTTAATTCTAAAAACTTTAACTTAAC, 1173014r1-TTGTGGCCTATAAAGTTAATTCC
1173014f2- TGCAGTAAATGTAATAAATAGTTAG, 1173014r2-CCATTACTTCT C CTTCTATAAC
Tel-5; 1173021f1-GATGCATAAATGAGACACGCTGGTA, 1173021r1-CAACTAATTTATAACCCCAATTCTC
Tel-7; 1173338f1-CCATTAAGATTCCATTGCTGATAAA, 1173338r1-CTTCTATGAATTAACTTTGAGAGG
Tel-8; 1173200f1-TGGCTAATGGATAGAGTAGATAAAG, 1173200r1-AAGCTTCTACTCCCGCAACTAAGT
Tel-9; 1173226f1-TTGATTTTATAACAATTAATTAGATG, 1173226r1-GCTTTACGAATTTATATTTTGGCTA
Tel-10; 1172432f1-GCAAGTAAATCTATGAATAATATGAG, 1172432r1-CAGATATTAACCCCTATGTTTATTG
Tel-11; 1173097f1-ATAGGACTGAATATATCCTCACTTG, 1173097r1-CAAGCTTTTAAGATTTGATAAAGAAG
Tel-12; 1171891f1-AAATATGAAGGAAATTAGGACAAGAG, 1171891r1-GAAGCTTTATTATGTCTATCTGGAA
Tel-13; 1173043f1-GCTAATAAAATTAAAAACAACTAGG, 1173043r1-AATAAATTGAAAGTGTTTCTTTAAG
1173043f3-TAAAATTAAAAACAACTAGGGACC, 1173043r3-CTTTAAGTATTTTTACTAATAGATC
Isolation and Nuclease Digestion of Macronuclei
Macronuclei were isolated essentially as described by Karrer and VanNuland (1999). Briefly, 400 ml of cells grown to a density of 3 × 105 cells/ml were pelleted and lysed in 60 ml of TMS (10 mM Tris, pH 7.5, 10 mM MgCl2, 3 mM CaCl2, 250 mM sucrose, 1 mM dithiothreitol) and 0.16% Igepal CA-630 (NP-40) at 4°C. Sucrose was added to a concentration of 0.816 g/ml and stirred until completely dissolved. The lysate was centrifuged at 9000 × g for 30 min at 4°C, and the pelleted nuclei were washed with TMS before micrococcal nuclease (MNase) digestion.
For MNase digestion, nuclei were resuspended in buffer A (15 mM Tris 7.5, 60 mM KCl, 15 mM NaCl, 2 mM CaCl2, 0.05% spermidine phosphate, 1 mM dithiothreitol) at a concentration of 200 μg/ml and MNase (Worthington Biochemicals, Freehold, NJ) was added at a concentration of 30 U/mg nuclei for 2–60 min at 30°C. The reaction was stopped by adding EGTA to 14 mM, and the DNA was purified. Purified macronuclear DNA was digested with 8 U of MNase/mg DNA for 1, 2, and 4 min at 30°C. The digestion products were purified and further digested with HindIII or EcoRI. Subtelomeric restriction fragments were visualized by indirect end-labeling by using oligonucleotides (for the rDNAs) or polymerase chain reaction products (for the non-rDNAs) that hybridized adjacent to the HindIII or EcoRI site.
RESULTS
Effect of Subtelomeric Sequence on Telomere Growth
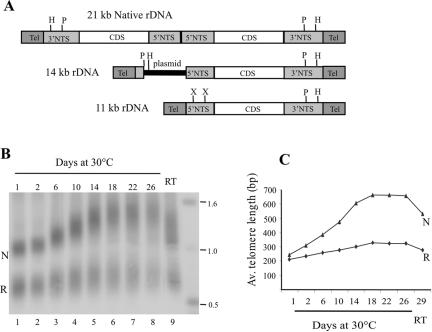
Although the Tetrahymena rDNA is normally a 21-kb palindrome that has two transcription units and two identical telomeres (Figure 1A), it is possible to generate cells that contain recombinant, nonpalindromic rDNAs (Yao et al., 1990; Yasuda and Yao, 1991). These molecules have only one transcription unit and two different telomeric restriction fragments. We were using 11- and 14-kb nonpalindromic rDNAs to study telomeric DNA structure when we observed an unexpected difference in the growth rate of telomeres lying adjacent to natural and recombinant subtelomeric regions of the chromosome. This observation suggested that the subtelomeric DNA might be influencing telomere length regulation.
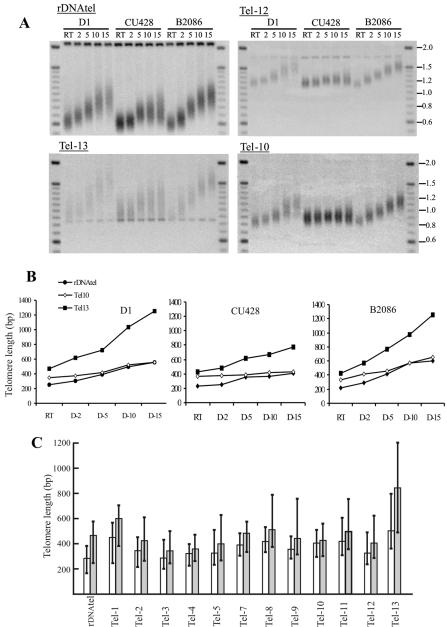
Figure 1.
Growth of rDNA telomeres. (A) Organization of native and recombinant rDNA molecules. Coding sequences (CDS) are represented as open boxes, 5′ and 3′ NTS are light-shaded boxes, and telomeres are dark boxes. HindIII (H), PstI (P), and XbaI (X) sites are marked. (B and C) Differential growth of 14-kb rDNA telomeres with natural and recombinant subtelomeric regions. (B) Native (N) and recombinant (R) telomeric restriction fragments from the 14-kb rDNA were detected by Southern hybridization with probe for telomeric DNA. Cells were grown at 30°C for 1–26 d then at room temperature (RT) for 3 d. (C) Average length of the telomeric tract from the native and recombinant telomere is plotted against time in culture.
To examine the growth of telomeres with natural and recombinant subtelomeric sequences more closely, we isolated DNA from cells maintained in continuous log culture at 30°C for up to 26 d and determined the length of the telomeric restriction fragments. The 11- and 14-kb molecules used in this study both have the natural subtelomeric sequence at one end (the native telomere, Figure 1A) and recombinant sequence at the other end (the recombinant telomere). In the 14-kb molecule, the 3 kb of recombinant sequence is derived from the cloning vector used to generate the nonpalindromic molecule. This plasmid DNA is removed during formation of the 11-kb rDNA, so the telomere lies directly adjacent to the 5′ nontranscribed spacer (NTS) of the single remaining transcription unit.
In initial experiments, we examined telomeric restriction fragments from cells carrying the 14-kb rDNA (Figure 1, B and C). Hybridization of a telomeric DNA probe to PstI-digested rDNA revealed that the length of the telomeric tract from the recombinant telomere was on average ∼50 base pairs shorter than that of the native telomere when cells were maintained under conditions that did not cause telomere growth (e.g., room temperature or 1 d at 30°C). The fragment from the native telomere was 1.1–1.2 kb and contained ∼230- to 380-base pair T2G4 repeat and 835-base pair subtelomeric DNA, whereas the fragment from the recombinant telomere was 700–800 base pairs and contained 200- to 340-base pair T2G4 repeat and 472 base pairs subtelomeric sequence (Figure 1B, lane 1).
On continuous passage at 30°C, the length of the native telomere became more heterogeneous and increased by up to six base pairs (1 telomeric repeat) per population doubling (Figure 1B, lanes 2–5) as had been observed previously for wild-type cells (Larson et al., 1987). By 10–11 d of growth, the overall length had increased by 250–400 base pairs to give a total telomere length of 500–800 base pairs. The length of the recombinant telomere also became more heterogeneous; however, the average growth rate was considerably slower with an average increase of only approximately one base pair per population doubling. Thus, after 10–11 d growth they were only 250–450 base pairs in length. Both telomeres gradually returned to their original size when the cultures were transferred back to room temperature or kept in nonlogarithmic growth (Figure 1B, lane 9).
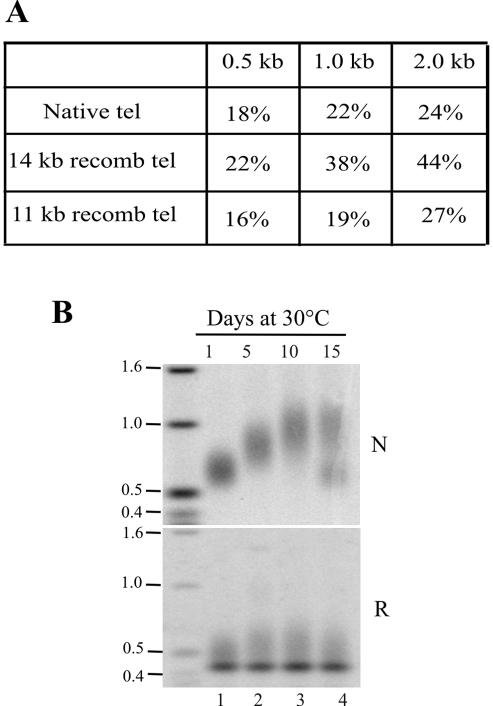
One difference between the native and the 14-kb rDNA is that the vector sequence incorporated into the subtelomeric region of the 14-kb molecule has an ∼44% GC base content compared with 24% GC in the native subtelomeric region and ∼30% GC in the total Tetrahymena genome (Figure 2A). To determine whether this change in base composition contributed to the altered telomere growth, we examined the growth profile of telomeres from the 11-kb rDNA molecule because it has a comparable GC base composition in both subtelomeric regions. DNA isolated from clones carrying the 11-kb rDNA was digested with XbaI or HindIII to release the telomeric restriction fragments and hybridized with subtelomeric probes. XbaI cuts the subtelomeric DNA of the recombinant telomere 269 base pairs internal to the telomeric tract, whereas HindIII cuts the native telomere 360 nt from the telomeric tract. As observed for the 14-kb rDNA, the recombinant telomere from the 11-kb rDNA initially had a slightly shorter telomeric tract than the native telomere, and upon continuous culture, the two telomeres again showed a difference in growth rate (Figure 2B). As before, the native telomere grew by as much as six base pairs per population doubling, whereas the recombinant telomere grew at a rate of ∼1.4 base pairs per doubling. We therefore conclude that alteration of the subtelomeric sequence can lead to changes in telomere growth; however, these changes do not correlate with the subtelomeric base composition in any simple way.
Figure 2.
Effect of subtelomeric base composition on telomere growth. (A) Table showing the percentage of G + C within the first 0.5, 1, or 2 kb of the natural and recombinant subtelomeric DNA. (B) Southern blot showing native and recombinant telomeric restriction fragments from the 11-kb rDNA after 1–15 d growth at 30°C. Hybridization was with subtelomeric probes. The sharp bands in the lower panel reflect cross-hybridization to an internal restriction fragment. The bimodal native telomere signal in lane 4 reflects outgrowth of a short telomere mutant (Ahmed et al., 1998).
Telomere Growth and Subtelomeric Chromatin Structure
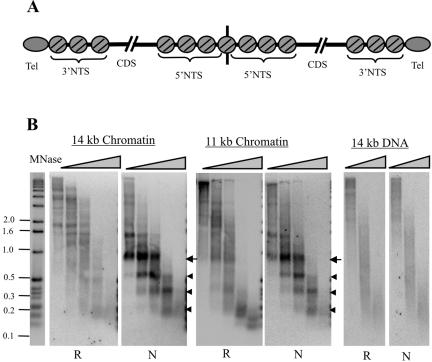
The native Tetrahymena rDNA has a very characteristic chromatin structure that consists of three positioned nucleosomes along each 3′ NTS and seven positioned nucleosomes along the 5′ NTS at the center of the palindrome (Figure 3A) (Blackburn and Chiou, 1981; Palen and Cech, 1984; Budarf and Blackburn, 1986; Cohen and Blackburn, 1998). Although the cause of the nucleosome positioning in the 3′ NTS is unknown, it is likely to be determined either by the underlying DNA sequence or by the telomeric complex serving as a boundary element. If positioning depends on the DNA sequence, we might expect it to be disrupted along the recombinant subtelomeric region of the 11- and 14-kb rDNAs. Such an alteration in chromatin structure might, in turn, cause the lower growth rate of the recombinant telomere. Given this possibility, we decided to use micrococcal nuclease (MNase) digestion to compare the chromatin structure within the recombinant and natural subtelomeric regions.
Figure 3.
Subtelomeric chromatin structure. (A) Chromatin structure of the native 21-kb rDNA. Positioned nucleosomes are shown as circles, and the telomeric complexes are ovals. CDS, coding sequence. (B) Southern blots showing MNase sensitivity of the 11- and 14-kb rDNA subtelomeric chromatin. Macronuclei were digested with 30 U of MNase/mg chromatin for 2, 4, 6, 16, or 60 min. For the DNA control, deproteinized DNA from the 14-kb rDNA clone was digested with 8 U of MNase/mg DNA for 1, 2, or 4 min. Each blot was probed first with an oligonucleotide that hybridized 400 base pairs from the telomeric tract within the recombinant subtelomeric region (R) and then with an oligonucleotide that hybridized at the same position within the native subtelomeric region (N). Arrowheads mark bands corresponding to positioned nucleosomes, and arrow marks band corresponding to positioned nucleosome + telomeric complex.
The 11- and 14-kb rDNAs have different restriction sites at the two telomeres, so we first examined the MNase digestion pattern by using probes that hybridized a set distance (40 or 400 nt) in from the junction between the telomeric tract and the subtelomeric DNA. Macronuclei were isolated from cultures carrying 11- or 14-kb rDNA molecules, digested with MNase, and the subtelomeric digestion products were identified by Southern blotting. As expected, a very distinct pattern of products was observed with probes that hybridized to the native subtelomeric region (Figure 3B; our unpublished data). This pattern was identical for the 11- and 14-kb molecules and corresponded to the three positioned nucleosomes in the 3′ NTS. However, the pattern obtained with probes to the recombinant subtelomeric region was very different. For the 14-kb rDNA, the banding pattern close to the telomeric tract was indistinct, but the chromatin was more resistant to digestion than the deproteinized DNA controls. This indicates that nucleosomes were probably still present along the subtelomeric region, but they were no longer at the same position on each molecule. For the 11-kb rDNA, a faint banding pattern could be observed; however, it was different from that observed at native telomeres and probably corresponded to a partial positioning of the nucleosomes normally present along the 5′ NTS.
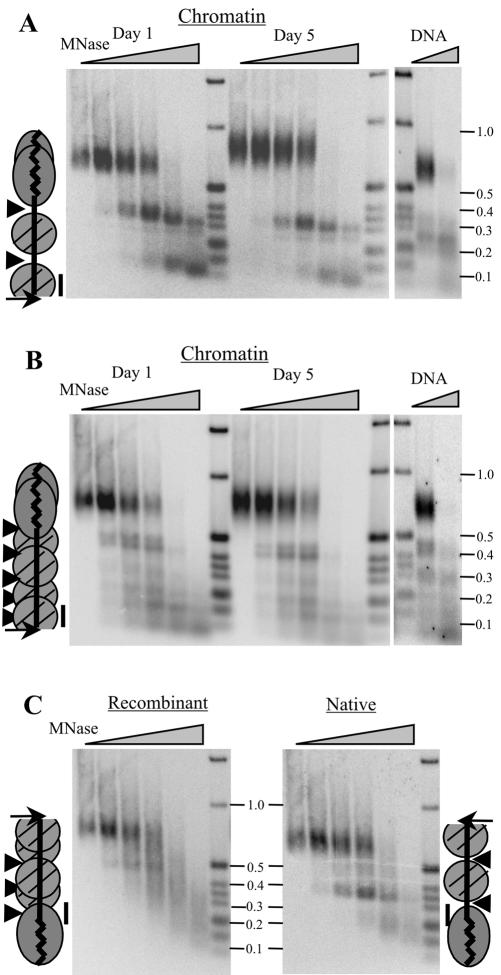
To look more closely at nucleosome positioning on the 14-kb rDNA, we used indirect end-labeling to map the MNase digestion sites. Macronuclei were isolated from cells after 1 or 5 d growth at 30°C, treated with MNase, and the purified DNA was digested with HindIII. Fragments extending toward the telomere were then identified with probes that hybridized adjacent to the cut site (Figure 4). The native and recombinant telomeres have HindIII sites 360 base pairs or 420 base pairs internal to the telomeric tract. The probe for the native subtelomeric region identified bands of ∼100 and 300 nt, which corresponded to the two most terminal nucleosomes in the 3′ NTS (Budarf and Blackburn, 1986). The banding pattern was unchanged by the continuous culture, confirming a previous observation that the rDNA subtelomeric chromatin structure is unaffected by conditions that promote telomere growth (Cohen and Blackburn, 1998). As in the previous experiment, the probe for the recombinant subtelomeric region revealed a less distinct digestion pattern that was composed of multiple bands. The pattern was unchanged during telomere growth and many of the same bands were seen in the naked DNA control, indicating that cleavage occurred preferentially at certain sequences within the subtelomeric DNA. Thus, the digestion pattern gave no indication of nucleosome positioning. Nonetheless, the sensitivity to digestion was considerably lower in the chromatin samples, indicating that although the subtelomeric region lacks positioned nucleosomes, it is unlikely to be nucleosome free.
Figure 4.
Nucleosome positioning on the 14-kb rDNA. MNase digestion products from macronuclei or deproteinized DNA were digested with HindIII, and the subtelomeric restriction fragments were identified by Southern hybridization by using a probe that hybridized adjacent to the restriction site (A and B), fragments protected by the telomeric complex were identified using a probe that hybridized 40 nt internal to the telomeric tract (C). The cartoons show the relative positions of the HindIII site (arrow), MNase hypersensitivity (arrowheads), hybridization probes (bar), telomeric complex on days 1 and 5 (overlapping ovals), and nucleosomes (circles). (A) Digestion products from the native rDNA telomere after 1 or 5 d culture at 30°C. Left, macronuclei were digested with 30 U of MNase/mg chromatin for 0, 2, 4, 6, 16, or 60 min. Right, deproteinized DNA controls; digested with 8 U of MNase/mg DNA for 1 or 2 min. (B) Same blot hybridized with the probe to the recombinant subtelomeric sequence. (C) Day 1 blot from A hybridized with probes 40 base pairs internal to the telomeric complex from the native telomere (right) or the recombinant telomere (left).
Because both digestion experiments revealed that the positioned nucleosomes in the 3′ NTS were lost when the DNA sequence was altered, our results indicate that the chromatin structure within the natural subtelomeric region of the rDNA must be determined by the underlying DNA sequence. Our experiments also demonstrated that the reduced growth of the recombinant rDNA telomeres correlates directly with an alteration in the subtelomeric chromatin structure. Thus, our findings support a role for the subtelomeric chromatin in regulating the rate and extent of telomere growth in Tetrahymena.
Size and Organization of the Telomeric Complex
One way that altered subtelomeric chromatin might affect telomere growth is by causing a change in the size or structure of the terminal DNA–protein complex. For example, loss of positioned nucleosomes might allow the terminal complex to extend beyond the end of the telomeric tract to make a larger more stable complex that reduces telomerase access to the DNA terminus. To determine whether this had occurred, we calculated the size of the telomeric complex at native and recombinant telomeres by subtracting the size of the largest MNase digestion product (second band down in Figure 4, A and B) from the average size of the HindIII restriction fragment. The resulting value provides an estimate for the maximum size of the complex because it includes any MNase-sensitive linker DNA that lies between the telomeric complex and the first nucleosomes on the subtelomeric sequence.
Measurements from several experiments indicated that before telomere growth, the complex at the recombinant telomere protected, on average, 210–250 base pairs DNA, whereas the complex at the native telomere protected 300–330 base pairs. Because the recombinant telomere had ∼50 base pairs less telomeric DNA than the native telomere, this indicated that the altered chromatin structure did not cause the telomeric complex to expand into the recombinant subtelomeric region. To the contrary, it seemed to cover less of the junction between the telomeric tract and the subtelomeric DNA. This lack of expansion upon disruption of the phased nucleosomes in the 3′ NTS indicates that the size of the complex is likely to be dictated by the length of the telomeric tract rather than by boundary nucleosomes limiting its expansion. Interestingly, the decrease in size of the complex at the recombinant telomere did not lead to a change in MNase sensitivity because the size of the largest band remained the same with increased digestion. Thus, the complexes at the native and recombinant telomeres both prevent cleavage within the telomeric tract, suggesting that their gross structure is not significantly different.
We attempted to examine the size of each telomeric complex more precisely using probes that hybridized to the subtelomeric DNA either 10 or 40 nucleotides internal to the telomeric tract (Figure 4C; our unpublished data). Both probes identified a series of products that corresponded to telomeric DNA that had been protected by the telomeric complex; however, as previously reported, no limit digestion product was observed (Blackburn and Chiou, 1981; Cohen and Blackburn, 1998), so it was not possible to determine the minimum size of the complex at either telomere. Each probe also identified one discrete digestion product (340 nt at the native telomere and 450 nt at the recombinant telomere) that probably came from rDNA molecules where the telomeric complex did not extend into the beginning of the subtelomeric region. These molecules may correspond to the population that has part of the telomeric tract packaged by nucleosomes (Cohen and Blackburn, 1998).
Tetrahymena telomeres have a very well defined terminal DNA structure (Jacob et al., 2001) where the G-strands are extended to form 3′ overhangs that are mostly 14–15 or 20–21 nt in length and end with the sequence 5′-G4T. Because telomerase adds new telomeric repeats directly to these overhangs, any change in their structure could make them a better or worse telomerase substrate and hence alter the ability of a telomere to grow. Although it seemed unlikely that the recombinant and native telomeres would have a different overhang structure, we tested this possibility by using an oligonucleotide ligation and primer extension procedure developed previously in the laboratory (Jacob et al., 2001). As expected, the length and sequence of the overhangs was identical at the native and recombinant telomeres (our unpublished data).
Together, our findings indicate that altering the subtelomeric chromatin has a relatively minor effect on either the size or the gross structure of the telomeric complex. Thus, the reduced growth of the recombinant telomere cannot be explained by the presence of a larger more stable telomeric complex. The decrease in size of the complex might instead have been expected to render the recombinant telomere more accessible to telomerase, and hence more prone to growth. Because this does not occur, our results suggest that the telomeric complex and subtelomeric chromatin may cooperate to regulate access to the DNA terminus
Growth of non-rDNA Telomeres
If growth of rDNA telomeres is affected by the subtelomeric chromatin, we would expect this to also be true for telomeres on other macronuclear chromosomes because rDNA and non-rDNA telomeres are structurally very similar (Jacob et al., 2001). Although previous studies of telomere growth demonstrated that growth during continuous culture is not limited to rDNA telomeres (Larson et al., 1987), preliminary analysis of the subtelomeric DNA from non-rDNA molecules indicated that each chromosome end has a different DNA sequence (Orias, personal communication). Thus, it seemed likely that each chromosome might have its own characteristic subtelomeric chromatin structure and hence growth profile. To determine whether this is the case, we set out to examine both the growth profile and subtelomeric chromatin structure from a selection of macronuclear chromosomes.
The ends of non-rDNA chromosomes were identified by searching the Tetrahymena macronuclear DNA database for contigs that ended with telomeric sequence T2G4. Further analysis revealed a subset of 12 chromosome ends that gave restriction fragments of a suitable size for telomere length analysis (≥1.5 kb). All the subtelomeric sequences were AT rich (19–26% GC within the first 2 kb), and they each had a different sequence. They did not contain copies of native or variant telomeric repeats or the type VI or V repeats that are found in the rDNA subtelomeric region. They also seemed to lack conserved complex sequence elements analogous to the X or Y' elements found at S. cerevisiae telomeres or the complex repeats found at human telomeres (Mefford and Trask, 2002)
To examine the growth profile of each chromosome end, we made probes that hybridized to the sequence adjacent to the restriction site on each end. We then used these probes to detect the telomeric restriction fragments from cells grown for 1–15 d at 30°C. As shown in Figure 5, each telomere exhibited a unique but characteristic growth pattern in terms of both rate and extent of growth, and generation of length heterogeneity. Some showed a fairly large increase in size (e.g., rDNAtel and Tel-13), whereas others showed a more modest increase. In all cases, the growth rate was fastest initially but then slowed as the telomeres approached a new length set point. When we determined the size of the telomeric restriction fragments before telomere growth, we observed that the initial length of the telomeric tract varied considerably between chromosomes ends (Figure 5). However, the rate and extent of growth did not depend on the initial length of the telomeric tract because chromosome ends with the same initial length showed very different growth patterns (compare rDNAtel and Tel-3 or Tel-12).
Figure 5.
Growth profile and average length of individual macronuclear telomeres. (A) Southern blots showing telomeric restriction fragments from different chromosome ends after growth at room temperature (RT) or continuous culture at 30°C for 2–15 d. DNA was isolated from three Tetrahymena strains D1, CU428, and B2086. (B) Median length of the telomeric tract from three different chromosome ends (rDNAtel, Tel-10, and Tel-13; A) is plotted against time in culture. (C) Histogram showing the median telomere length at different chromosome ends in strain CU428 after growth at room temperature (open bars) or 30°C for 15 d (shaded bars). The lines that extend above and below each bar indicate the length heterogeneity by showing the maximum and minimum length of the telomeric restriction fragments. Data are from one representative experiment.
To determine whether individual telomere growth profiles always remained the same, we recultured Tetrahymena strain CU428 and grew an additional two strains (D1 and B2086; Figure 5, A and B) for 15 d at 30°C. As observed previously for rDNA telomeres (Cohen and Blackburn, 1998) (N.K.J. and C.P., unpublished observations), reanalysis of CU428 showed that the extent of telomere growth varied slightly when the same strain was analyzed on different occasions. However, the relative tendency of each telomere to grow remained the same, i.e., telomere 13 and the rDNA telomere always grew the most and telomere 10 and 4 grew the least (our unpublished data). The median telomere length for rDNAtel in strain CU428 was 261 base pairs ± 44 base pairs at room temperature and 358 base pairs ± 36 after 10 d of growth. The median telomere length Tel-10 was 411 base pairs ± 14 base pairs at room temperature and 420 base pairs ± 5 base pairs after 10 d of growth.
Comparison of the three different strains revealed that there were some strain specific differences in the extent of telomere growth and generation of length heterogeneity (Figure 5A). However, the relative growth potential of the individual telomeres and the initial length of their telomeric tract was strikingly similar (Figure 5B). Because the three strains have similar subtelomeric sequences (N.K.J. and C.P., unpublished observations), this finding further supports the importance of the subtelomeric region in determining both the length set point and growth potential for an individual telomere.
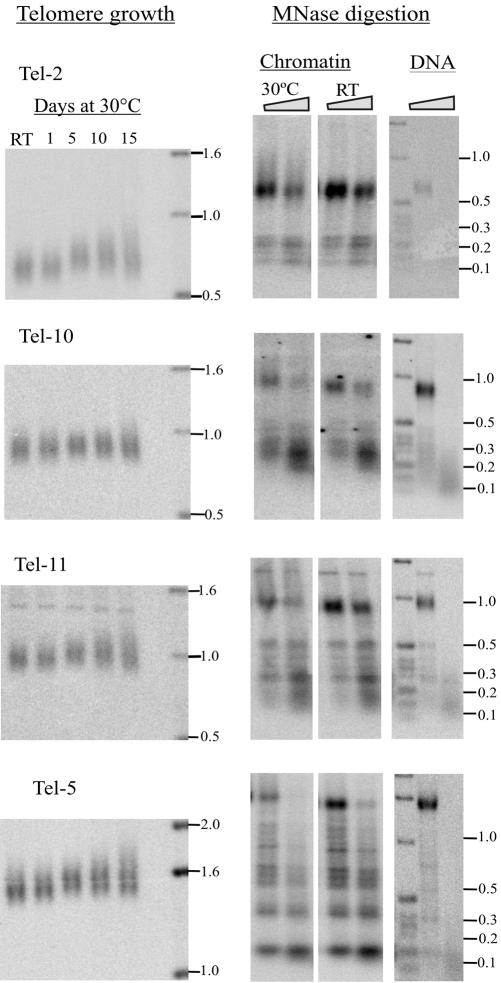
We next examined the chromatin structure within the subtelomeric region of each chromosome end. Freshly isolated macronuclei were digested with MNase as described above, and then the chromosome-specific probes were used to indirectly end-label the subtelomeric digestion products. Strikingly, each chromosome end gave a different MNase digestion pattern (Figure 6, right; our unpublished data). Some displayed a banding pattern that was similar to that observed for the naked DNA control, suggesting a lack of nucleosome positioning. Others had discrete bands that were absent in the DNA control, indicating a well defined chromatin structure and the possibility of positioned nucleosomes. In all cases, the banding pattern and hence the nucleosome placement was very different from that observed for the rDNA 3′ NTS. The unique organization of the chromatin within each subtelomeric region indicates that subtelomeric chromatin structure is likely to be the underlying cause of the characteristic growth profile exhibited by individual telomeres. Interestingly, there was no obvious correlation between any one type of MNase digestion pattern (e.g., discrete vs. indistinct) and the extent of telomere growth. Thus, telomere growth does not seem to be determined by the exact positioning of nucleosomes within the subtelomeric region, but rather by a different level of subtelomeric chromatin structure. Because the telomeric complex that packages the telomeric tract also contributes to telomere length regulation, one possibility is that the subtelomeric chromatin from each telomere interacts with the telomeric complex to form a unique higher order telomeric chromatin structure that determines the growth potential and telomere length set point for that telomere
Figure 6.
Telomere growth and chromatin structure at non-rDNA telomeres. Left, telomeric restriction fragments from different chromosome ends after growing strain CU428 at room temperature (RT) or in continuous culture at 30°C. Middle, corresponding MNase digests of subtelomeric chromatin. Macronuclei were isolated from cells grown at RT or for 30°C for 15 d. Digestion was with 30 U of MNase/mg chromatin for 4 and 6 min. Right, deproteinized DNA controls digested with 8 U of MNase/mg DNA for 1 and 2 min. Purified MNase digestion products were restriction digested and analyzed by Southern hybridization with a probe that hybridized adjacent to the restriction site.
DISCUSSION
Although various studies have shown that proper telomere function depends on the telomeric tract being packaged into a higher order chromatin complex (Grunstein, 1997; Lowell and Pillus, 1998; Smogorzewska et al., 2000; Chan and Blackburn, 2002), the contribution of the subtelomeric region to the organization of this complex, and hence to telomere length regulation, remains largely unexplored. In this study, we show that the subtelomeric region has a direct impact on both the rate and extent of telomere growth in Tetrahymena cells. We have been able to alter the final length set point and amount of length heterogeneity at a telomere by simply changing the subtelomeric sequence. This finding indicates that the subtelomeric region somehow controls accessibility of the DNA terminus to the enzymes (telomerase and nucleases) that alter the length of the telomeric tract. We also have shown that individual telomeres from different macronuclear chromosomes each have a unique basal telomere length and growth profile. This basal length and growth tendency is highly reproducible and is conserved in different Tetrahymena strains. Because every chromosome end has a different subtelomeric sequence, the unique behavior of individual telomeres confirms the importance of the subtelomeric region in telomere length regulation.
Given the known contribution of the telomeric complex to telomere length regulation (Lowell and Pillus, 1998; Smogorzewska et al., 2000), our results suggest that the capacity of a telomere to grow may be determined by a higher order chromatin structure that is composed of both the subtelomeric chromatin and the telomeric complex. Because the subtelomeric chromatin at each telomere is different, the precise organization of this higher order structure, and hence telomere growth potential, would be telomere specific. Alteration of the subtelomeric chromatin at a particular telomere would change this structure, and hence either increase or decrease telomerase or nuclease access to the DNA terminus.
The chromatin structure of an individual telomere could dictate its growth rate in a number of ways. First, it might determine how much DNA telomerase or nucleases add or remove during any one cell cycle. Second, it might determine the frequency with which telomerase gains access to the DNA terminus. Both the extent and frequency of elongation also could cause the increase in length heterogeneity that occurs at many telomeres during continuous culture. In yeast, telomerase does not extend each telomere at every cell cycle, and it is the frequency of telomerase extension, rather than the amount of DNA added, that promotes growth of very short telomeres (Teixeira et al., 2004). Thus, it seems likely that growth of Tetrahymena telomeres and the generation of length heterogeneity also may be dictated by the frequency of telomerase extension.
Early studies of chromosome healing suggested that subtelomeric sequences were not important for telomere function because when a broken chromosome is healed by the addition of telomeric repeats, the new telomere is maintained normally despite the lack of natural subtelomeric sequence (Gottschling et al., 1990; Barnett et al., 1993). However, more recent work has shown that if normal telomere structure is perturbed, the subtelomeric region can provide a backup structure that allows certain aspects of telomere function to continue (Chan and Blackburn, 2002). This is apparent in Drosophila and budding yeast where the subtelomeric region can be used to form a telomeric cap when the natural telomeric DNA is absent. S. cerevisiae that lack telomerase can amplify the Y' subtelomeric repeats to form large arrays that provide sufficient capping function to allow continued growth (Lundblad and Blackburn, 1993). The capping function is thought to result from formation of a nucleosome based heterochromatin structure along the amplified Y' elements (Chan and Blackburn, 2002). Likewise, Drosophila strains that have lost the natural telomeric HeT-A and TART elements form a functional cap that contains telomere-associated sequences and the heterochromatin proteins HP1 and HOAP (Fanti et al., 1998; Cenci et al., 2003). Subtelomeric chromatin also can promote chromosome segregation. In fission yeast, accurate meiotic chromosome segregation only occurs if the chromosomes cluster and associate with the spindle pole body. This process requires Taz1, a telomere protein that normally binds to telomeric DNA. However, in cells that lack a telomeric tract, Taz1 can bind to a subtelomeric element, thus allowing an epigenetically regulated association with the spindle pole body (Sadaie et al., 2003).
In addition to providing backup functions at dysfunctional telomeres, subtelomeric chromatin also seems to contribute to telomere length regulation at functionally normal telomeres. Because cells can maintain telomeres at an acceptable length in the absence of natural subtelomeric sequences (Gottschling et al., 1990; Barnett et al., 1993), the native subtelomeric chromatin seems to serve to fine-tune the length regulation process. Mice that lack histone methytransferases and hence have no H3 lysine 9 methylation or heterochromatin protein HP1 in the telomeric region undergo large increases in telomere length on a subset of their chromosomes (Garcia-Cao et al., 2004). These findings suggest that the subtelomeric chromatin is important for preventing abnormal telomere lengthening, perhaps through the ALT pathway. Subtelomeric chromatin also helps determine the length of individual telomeres in S. cerevisiae, Plasmodium falciparum, and probably also Drosophila melanogaster (Craven and Petes, 1999; Figueiredo et al., 2002; Mason et al., 2003), whereas in Tetrahymena it seems to dictate the new length set point when telomeres are induced to grow. Because the effect of subtelomeric chromatin on telomere length is usually not apparent until the subtelomeric region is somehow altered, it is likely this component of the telomere length regulation machinery is a normal feature of telomere biology that has not yet been characterized in many organisms. Indeed, recent studies indicate that subtelomeric regions may be responsible for the conserved telomere length profile that exists on human chromosomes. Each arm of a human chromosome has a characteristic relative telomere length that is maintained both throughout the life of an individual and between individuals (Martens et al., 1998; Graakjaer et al., 2003). Interestingly, the characteristic length of a specific telomere is dependent on a factor that is located in the distal region of the same chromosome arm (Graakjaer et al., 2003). Because human subtelomeric sequences can encompass hundreds of kilobases (Wilkie et al., 1991), these factors may well correspond to the subtelomeric DNA or chromatin.
Acknowledgments
We thank Meng-Chao Yao for providing the constructs containing the nonpalindromic rDNAs, Ed Orias for providing information about subtelomeric sequences on non-rDNA chromosome, and The Ohio State University Center for Plant Biotechnology for use of the particle gun. We are grateful to Marty Gorovsky, Jeff Kapler, Iain Cartwright, and members of the Price laboratory for helpful advice and discussions. This work was supported by grant GM-41803 from the National Institutes of Health to C.M.P.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0237. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0237.
References
- Ahmed, S., Sheng, H., Niu, L., and Henderson, E. (1998). Tetrahymena mutants with short telomeres. Genetics 150, 643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett, M.A., Buckle, V.J., Evans, E.P., Porter, A.C., Rout, D., Smith, A.G., and Brown, W.R. (1993). Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res. 21, 27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards, A., Michels, P.A.M., Lincke, C.R., and Borst, P. (1983). Growth of chromosome ends in multiplying trypanosomes. Nature 303, 592-597. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2001). Switching and signaling at the telomere. Cell 106, 661-673. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H., and Chiou, S.S. (1981). Non-nucleosomal packaging of a tandemly repeated DNA sequence at termini of extrachromosomal DNA coding for rRNA in Tetrahymena. Proc. Natl. Acad. Sci. USA 78, 2263-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourns, B.D., Alexander, M.K., Smith, A.M., and Zakian, V.A. (1998). Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol. 18, 5600-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm, S.L., and Cech, T.R. (1983). Fate of an intervening sequence ribonucleic acid: excision and cyclization of the Tetrahymena ribosomal ribonucleic acid intervening sequence in vivo. Biochemistry 22, 2390-2397. [DOI] [PubMed] [Google Scholar]
- Budarf, M.L., and Blackburn, E.H. (1986). Chromatin structure of the telomeric region and 3′-nontranscribed spacer of Tetrahymena ribosomal RNA genes. J. Biol. Chem. 261, 363-369. [PubMed] [Google Scholar]
- Cassidy-Hanley, D., Bowen, J., Lee, J.H., Cole, E., VerPlank, L.A., Gaertig, J., Gorovsky, M.A., and Bruns, P.J. (1997). Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146, 135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci, G., Siriaco, G., Raffa, G.D., Kellum, R., and Gatti, M. (2003). The Drosophila HOAP protein is required for telomere capping. Nat. Cell Biol. 5, 82-84. [DOI] [PubMed] [Google Scholar]
- Cesare, A.J., Quinney, N., Willcox, S., Subramanian, D., and Griffith, J.D. (2003). Telomere looping in P. sativum (common garden pea). Plant J. 36, 271-279. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., and Blackburn, E.H. (2002). New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21, 553-563. [DOI] [PubMed] [Google Scholar]
- Cohen, P., and Blackburn, E.H. (1998). Two types of telomeric chromatin in Tetrahymena thermophila. J. Mol. Biol. 280, 327-344. [DOI] [PubMed] [Google Scholar]
- Coyne, R.S., Chalker, D.L., and Yao, M.C. (1996). Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu. Rev. Genet. 30, 557-578. [DOI] [PubMed] [Google Scholar]
- Craven, R.J., and Petes, T.D. (1999). Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics 152, 1531-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede, S.J., and Gottschling, D.E. (1999). Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99, 723-733. [DOI] [PubMed] [Google Scholar]
- Fanti, L., Giovinazzo, G., Berloco, M., and Pimpinelli, S. (1998). The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2, 527-538. [DOI] [PubMed] [Google Scholar]
- Figueiredo, L.M., Freitas-Junior, L.H., Bottius, E., Olivo-Marin, J.C., and Scherf, A. (2002). A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 21, 815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao, M., O'Sullivan, R., Peters, A.H., Jenuwein, T., and Blasco, M.A. (2004). Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36, 94-99. [DOI] [PubMed] [Google Scholar]
- Gottschling, D.E., Aparicio, O.M., Billington, B.L., and Zakian, V.A. (1990). Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63, 751-762. [DOI] [PubMed] [Google Scholar]
- Graakjaer, J., Bischoff, C., Korsholm, L., Holstebroe, S., Vach, W., Bohr, V.A., Christensen, K., and Kolvraa, S. (2003). The pattern of chromosome-specific variations in telomere length in humans is determined by inherited, telomerenear factors and is maintained throughout life. Mech. Ageing Dev. 124, 629-640. [DOI] [PubMed] [Google Scholar]
- Greider, C.W. (1996). Telomere length regulation. Annu. Rev. Biochem. 65, 337-365. [DOI] [PubMed] [Google Scholar]
- Griffith, J.D., Comeau, L., Rosenfield, S., Stansel, R.M., Bianchi, A., Moss, H., and de Lange, T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503-514. [DOI] [PubMed] [Google Scholar]
- Grunstein, M. (1997). Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9, 383-387. [DOI] [PubMed] [Google Scholar]
- Jacob, N.K., Skopp, R., and Price, C.M. (2001). G-overhang dynamics at Tetrahymena telomeres. EMBO J. 20, 4299-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, C.L., and Klobutcher, L.A. (2002). Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56, 489-520. [DOI] [PubMed] [Google Scholar]
- Karrer, K.M., and VanNuland, T.A. (1999). Nucleosome positioning is independent of histone H1 in vivo. J. Biol. Chem. 274, 33020-33024. [DOI] [PubMed] [Google Scholar]
- Larson, D.D., Spangler, E.A., and Blackburn, E.H. (1987). Dynamics of telomere length variation in Tetrahymena thermophila. Cell 50, 477-483. [DOI] [PubMed] [Google Scholar]
- Lowell, J.E., and Pillus, L. (1998). Telomere tales: chromatin, telomerase and telomere function in Saccharomyces cerevisiae. Cell. Mol. Life Sci. 54, 32-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad, V., and Blackburn, E.H. (1993). An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73, 347-360. [DOI] [PubMed] [Google Scholar]
- Marcand, S., Brevet, V., Mann, C., and Gilson, E. (2000). Cell cycle restriction of telomere elongation. Curr. Biol. 10, 487-490. [DOI] [PubMed] [Google Scholar]
- Martens, U.M., Zijlmans, J.M., Poon, S.S., Dragowska, W., Yui, J., Chavez, E.A., Ward, R.K., and Lansdorp, P.M. (1998). Short telomeres on human chromosome 17p. Nat. Genet. 18, 76-80. [DOI] [PubMed] [Google Scholar]
- Mason, J.M., Konev, A.Y., and Biessmann, H. (2003). Telomeric position effect in Drosophila melanogaster reflects a telomere length control mechanism. Genetica 117, 319-325. [DOI] [PubMed] [Google Scholar]
- McEachern, M.J., and Hicks, J.B. (1993). Unusually large telomeric repeats in the yeast Candida albicans. Mol. Cell. Biol. 13, 551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern, M.J., Krauskopf, A., and Blackburn, E.H. (2000). Telomeres and their control. Annu. Rev. Genet. 34, 331-358. [DOI] [PubMed] [Google Scholar]
- Mefford, H.C., and Trask, B.J. (2002). The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 3, 91-102. [DOI] [PubMed] [Google Scholar]
- Palen, T.E., and Cech, T.R. (1984). Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell 36, 933-942. [DOI] [PubMed] [Google Scholar]
- Palladino, F., Laroche, T., Gilson, E., Axelrod, A., Pillus, L., and Gasser, S.M. (1993). SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75, 543-555. [DOI] [PubMed] [Google Scholar]
- Sadaie, M., Naito, T., and Ishikawa, F. (2003). Stable inheritance of telomere chromatin structure and function in the absence of telomeric repeats. Genes Dev. 17, 2271-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.D., Smith, D.L., DeRisi, J.L., and Blackburn, E.H. (2003). Telomeric protein distributions and remodeling through the cell cycle in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 556-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M.R., Schnapp, G., and de Lange, T. (2000). Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20, 1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart, A.K., Teng, S.C., and Zakian, V.A. (2002). Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297, 1023-1026. [DOI] [PubMed] [Google Scholar]
- Teixeira, M.T., Arneric, M., Sperisen, P., and Lingner, J. (2004). Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117, 323-335. [DOI] [PubMed] [Google Scholar]
- Tommerup, H., Dousmanis, A., and de Lange, T. (1994). Unusual chromatin in human telomeres. Mol. Cell. Biol. 14, 5777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie, A.O., Higgs, D.R., Rack, K.A., Buckle, V.J., Spurr, N.K., Fischel-Ghodsian, N., Ceccherini, I., Brown, W.R., and Harris, P.C. (1991). Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell 64, 595-606. [DOI] [PubMed] [Google Scholar]
- Wright, J.H., Gottschling, D.E., and Zakian, V.A. (1992). Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 6, 197-210. [DOI] [PubMed] [Google Scholar]
- Yao, M.C., Duhacourt, S., and Chalker, D.L. (2002). Genome-wide rearrangements of DNA in ciliates. In: Mobile DNA II, ed. R. Craig, M. Craigie, M. Gellert, and A. Lambowitz, New York: Academic Press, 730-758.
- Yao, M.C., Yao, C.H., and Monks, B. (1990). The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell 63, 763-772. [DOI] [PubMed] [Google Scholar]
- Yasuda, L.F., and Yao, M.C. (1991). Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell 67, 505-516. [DOI] [PubMed] [Google Scholar]