Abstract
Centromeric protein-E (CENP-E) is a kinesin-like motor protein required for chromosome congression at prometaphase. Functional perturbation of CENP-E by various methods results in a consistent phenotype, i.e., unaligned chromosomes during mitosis. One unresolved question from previous studies is whether cells complete mitosis or sustain mitotic arrest in the presence of unaligned chromosomes. Using RNA interference and video-microscopy, we analyzed the dynamic process of mitotic progression of HeLa(H2B)-GFP cells lacking CENP-E. Our results demonstrate that these cells initiated anaphase after a delayed mitotic progression due to the presence of unaligned chromosomes. In some dividing cells, unaligned chromosomes are present during anaphase, causing nondisjunction of some sister chromatids producing aneuploid daughter cells. Unlike in Xenopus extract, the loss of CENP-E in HeLa cells does not impair gross checkpoint activation because cells were arrested in mitosis in response to microtubule-interfering agents. However, the lack of CENP-E at kinetochores reduced the hyperphosphorylation of BubR1 checkpoint protein during mitosis, which may explain the loss of sensitivity of a cell to a few unaligned chromosomes in the absence of CENP-E. We also found that presynchronization with nocodazole sensitizes cells to the depletion of CENP-E, leading to more unaligned chromosomes, longer arrest, and cell death.
INTRODUCTION
The kinetochore is an essential component of the chromosome, required for correct transmission of cells' genetic materials during mitosis (Maney et al., 2000). It consists of a number of proteins that are assembled at the centromeric region of condensed mitotic chromosomes. One function of the kinetochore is to establish sister chromatid attachment to a bipolar spindle before chromosome segregation at anaphase. The spindle checkpoint machinery monitors the process at the kinetochores and relays a “wait-anaphase” signal to a cell when chromosomes are not properly attached or aligned (reviewed in Musacchio and Hardwick, 2002). Mutations causing defects in loss of checkpoint function can result in missegregation of chromosomes, leading to chromosomal instability and possibly cancer (Pihan and Doxsey, 1999).
The spindle checkpoint machinery consists of several proteins that are well conserved in various species. To date, two checkpoint proteins are known to directly mediate the waitanaphase signal, i.e., Mad2 and BubR1 (Yu, 2002; Zhou et al., 2002). These checkpoint proteins are recruited and activated at the kinetochores of unattached and/or unaligned chromosomes. These activated checkpoint components subsequently inhibit the anaphase-promoting complex/cyclosome (APC/C) and prevent the ubiquitination of substrates whose destruction is required for advance to anaphase (for review see Cleveland et al., 2003). It has been shown that a single unaligned chromosome is sufficient to inhibit anaphase onset, correlating with the presence of an activated checkpoint at the kinetochore (Rieder et al., 1994; Li and Nicklas, 1995).
CENP-E is a kinesin-like motor protein localized on the kinetochore. It has an apparent molecular mass of 312 kDa, with an ATP-dependent motor domain located at the N terminus (Yen et al., 1992). It is required for efficient capture and attachment of spindle microtubules by kinetochores, a necessary step in chromosome alignment during prometaphase (Lombillo et al., 1995; Yao et al., 1997; McEwen et al., 2001; Putkey et al., 2002). Functional disruption of CENP-E by various methods consistently resulted in the appearance of some unaligned chromosomes at metaphase (Schaar et al., 1997; Wood et al., 1997; Yao et al., 2000; Putkey et al., 2002). However, the effects on mitotic progression and cell fate have not been thoroughly investigated. Previous studies using either microinjection or the antisense approach showed that cells had prolonged mitotic arrest, and some reported that cells initiated apoptosis (Schaar et al., 1997; Yao et al., 2000). On the other hand, a study on mouse cells in the conditional knockout of CENP-E did not reveal significant long-term mitotic arrest in spite of the presence of unaligned chromosomes (Putkey et al., 2002). This apparent discrepancy has not been further addressed until the present study.
In Xenopus, CENP-E is essential for the localization and activation of the mitotic checkpoint proteins at the kinetochores. Depletion of CENP-E from Xenopus extracts resulted in loss of mitotic arrest in the presence of microtubule-depolymerizing agent (Abrieu et al., 2000). The lack of mitotic arrest in mouse knockout cells also supported that CENP-E may be involved in a checkpoint-sensing mechanism (Putkey et al., 2002). In contrast, activation and kinetochore localization of mitotic checkpoint proteins in cultured mammalian cells seem to be unaffected by CENP-E reduction (Yao et al., 2000; McEwen et al., 2001). However, the findings of CENP-E colocalization and its association with the checkpoint protein BubR1 at the kinetochores suggest that the spindle checkpoint activation may be sensed through CENP-E kinetochore activity (Chan et al., 1998; Yao et al., 2000).
The aim of the study is to investigate the fate of mitotic HeLa cells with reduced levels of CENP-E by using RNA interference and to further probe the possibility of CENP-E's involvement in the mitotic checkpoint mechanism.
MATERIALS AND METHODS
Small Interfering RNA (siRNA) Design
In this study, we designed three siRNA duplexes to knock down CENP-E: 1) 5′-AAGCAGAGAGAAGGGUGAACC, 2) 5′-AACACGGAUGCUGGUGACCUC, and 3) 5′-AACGAAGAGUUACUUGGUGCC. We also used the following published siRNA duplexes to knockdown CENP-E (Harborth et al., 2001), Lamin A/C (Elbashir et al., 2001) and Mad2 (Martin-Lluesma et al., 2002). All duplexes were chemically synthesized and purchased from Dharmacon (Lafayette, CO).
Cell Culture and Transfection
HeLa cells and HeLa(H2B)-GFP cells (Kanda et al., 1998) were cultured in Dulbecco's modified Eagle's media containing 10% fetal bovine serum. Transfections were carried out with siRNA duplexes and LipofectAMINE 2000 transfection reagent (Invitrogen, Carlsbad, CA) mixture. These components were mixed in Opti-MEM I per manufacturer's instructions. The final concentrations of siRNA and LipofectAMINE 2000 used in each transfection were 50 nM and 1.6 μg/ml, respectively. For mock transfections, cells were treated with LipofectAMINE 2000 alone.
Quantitative Polymerase Chain Reaction (QPCR) Analysis
Total RNA were isolated from adherent cells using the RNeasy mini kit (QIAGEN, Valencia, CA). First-strand cDNA was synthesized from 3 μg of total RNA by using random hexamers and SuperScript II (Invitrogen). All reverse transcription-polymerase chain reaction reactions were performed on an Applied Biosystems 7000 sequence detection system, by using 50 ng of cDNA per Sybr Green reaction (Applied Biosystems, Foster City, CA). The primers used for quantitating CENP-E mRNA are 5′-GTGGGACCAGTTCAGCCTGATA (forward) and 5′-CCAAGTGATTCTTCTCTGCTGTTC (reverse). The probes against Lamin were used to normalize CENP-E mRNA (forward, 5′-CTCGTCGTCCTCAACCACAGT; and reverse, 5′-TGCGTACGGCTCTCATCAACT).
Western Blotting
For CENP-E protein level analysis, cells were lysed with M-PER cell lysis buffer (Pierce Chemical, Rockford, IL) supplemented with protease inhibitors. The lysates were cleared by centrifugation at 14,000 rpm for 15 min at 4°C and quantitated by DC Protein Assay (Bio-Rad, Hercules, CA). The cell lysates were separated on 6% SDS-PAGE, and then transferred onto Immobilon polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membrane was probed with affinity-purified rabbit polyclonal antibody against the C-terminal peptide of human CENP-E, followed by anti-rabbit horseradish peroxidase-conjugated secondary antibody. After washing, the membrane was incubated in ECL Plus reagent before exposure to x-ray film. To quantitate the intensity of the bands, the membrane was scanned on Typhoon 9400 variable mode imager and analyzed on ImageQuant (Amersham Biosciences, Piscataway, NJ). For confirming the mitotic status, the extracts were separated on 4–20% gradient gel and transferred to Immobilon membrane as described above. The membrane was probed with rabbit polyclonal antibodies against phosphorylated histone H3 (Upstate Biotechnology, Lake Placid, NY).
Nocodazole or Taxol Treatment
The following experiments were carried out with cells treated with microtubule-interfering agents. For enrichment of mitotic population for Western blot analysis, HeLa(H2B)-GFP cells were transfected with CENP-E siRNA or mock as described above. Six to 10 h later, cells were treated with 50 ng/ml nocodazole for 18 h. Mitotic cells were collected by mechanical shake-off 24 h posttransfection. Subsequently, cell lysates were prepared for Western blot analysis. For presynchronization for time lapse analysis, HeLa(H2B)-GFP cells were treated with nocodazole at 100 ng/ml for 16–18 h. Mitotic cells collected by mechanical shake-off were seeded onto six-well culture dishes and allowed to attach and exit from mitosis. Three hours later, they were transfected with siRNA duplexes and incubated for 24 h. Subsequently, the culture medium was replaced and the cells were placed on a fluorescent microscope for time-lapse analysis. For mitotic checkpoint study, 24 h after transfection with CENP-E, Mad2, and Lamin siRNA duplexes, cells were treated with either nocodazole (100 ng/ml) or Taxol (50 nM) for 16 h. We also stained cells with an antibody against the mitotic marker MPM-2 for quantitation of mitotic index by flow cytometry. Cells were transfected with various siRNA duplexes for 24 h and treated with nocodazole or Taxol as described above before the cytometry analysis.
Immunoprecipitation
HeLa(H2B)-GFP cells were transfected with either control siRNA or CENP-E siRNA for 24 h and subsequently treated with 100 ng/ml nocodazole for 16 h. Mitotic cells were shaken off and lysed in radioimmunoprecipitation assay buffer supplemented with protease and phosphatase inhibitor mix. Cycling extracts were processed in the same way as mitotic extracts. The lysate were cleared and quantitated as described for CENP-E immunoblot. Cell lysates containing a total of 200 μg of proteins from each experimental condition were immunoprecipitated by incubation with 0.8 μg/ml polyclonal rabbit anti-human BubR1 and protein A-beads overnight. For alkaline phosphatase experiment, the beads were washed in phosphatase buffer and incubated with 20 U of alkaline phosphatase for 90 min at 37°C. The samples were separated on 4–20% SDS-PAGE, transferred to a nitrocellulose membrane (Pierce Chemical), and probed with mouse anti-human BubR1 antibody (BD Biosciences, San Jose, CA).
Time-Lapse Microscopy
HeLa(H2B)-GFP cells were transfected with either CENP-E or Lamin (control) siRNA duplexes. For cell fate analysis, transfected cells were placed on an Axiovert S-100 inverted microscope (Carl Zeiss, Thornwood, NY) 24 h after transfection with a 32× (0.4 numerical aperture [NA]) objective in an environmental chamber maintained at 37°C with 5% CO2. For each siRNA transfection, at least five positions were selected and imaged every 15 min. On average, 20–30 cells within a position were imaged to view the unaligned chromosomes during mitosis. The mitotic progression of each cell was visually examined using Axiovision version 3.1 software (Carl Zeiss). To examine unaligned chromosomes at anaphase onset, HeLa(H2B)-GFP cells were seeded at 55 × 103 cells in a chamber slide and transfected as described above. Twenty-four hours after transfection, the cells were placed on a DMIRE-2 confocal microscope (Leica Microsystems, Deerfield, IL) equipped with a 63× objective (0.7 NA) in an environmental chamber maintained at 37°C with 5% CO2. The images were captured with a charge-coupled device digital camera every 2–4 min and analyzed with Leica confocal software version 2. Selected images were processed with Adobe Photoshop version 6.0.
Immunofluorescence Microscopy
Immunofluorescence of CENP-E, BubR1, Mad2, and tubulin were carried out in HeLa cells as described by Liu et al., (2003). The following primary antibodies were used at the various dilutions indicated: mouse anti-CENP-E monoclonal antibody (1.8 μg/ml; a kind gift from Dr. T. Yen, Fox Chase Cancer Center. Philadelphia, PA), CREST-human anti-centromere autoantibody (1:1000; Cortex Biochem, San Leandro, CA), rabbit anti-BubR1 (1:250) and rabbit anti-Mad2 (1:250) (Covance Research Products, Berkeley, CA), and mouse anti-α-tubulin (1:1000) and γ-tubulin (1:500) (Sigma-Aldrich, St. Louis, MO). Secondary antibodies included Texas-Red–conjugated goat anti-rabbit antibodies, Alexa Fluor 488- and Alexa Fluor 594-conjugated goat anti-mouse, and Texas-Red–conjugated rabbit anti-human antibodies were used at 1:1000. DNA was stained with 4,6-diamidino-2-phenylindole contained in the mounting medium (Vector Laboratories, Burlingame, CA). Slides were examined with a Nikon Eclipse E800 fluorescence microscope with 100× objectives (1.4 NA).
Flow Cytometry Analysis
To examine the cell cycle profile, HeLa(H2B)-GFP cells were harvested and fixed in 70% ethanol overnight, washed once in phosphate-buffered saline, and incubated for at least 15 min in propidium iodide/RNase staining solution (BD Biosciences PharMingen, San Diego, CA) before analysis. For MPM-2 analysis, ethanol-fixed cells were washed in phosphate-buffered saline buffer containing 0.3 mg/ml saponin and incubated with a mouse anti-MPM-2 antibody (Upstate Biotechnology). The flow cytometry analysis was conducted on FACScalibur by using CellQuest software (BD Biosciences).
RESULTS
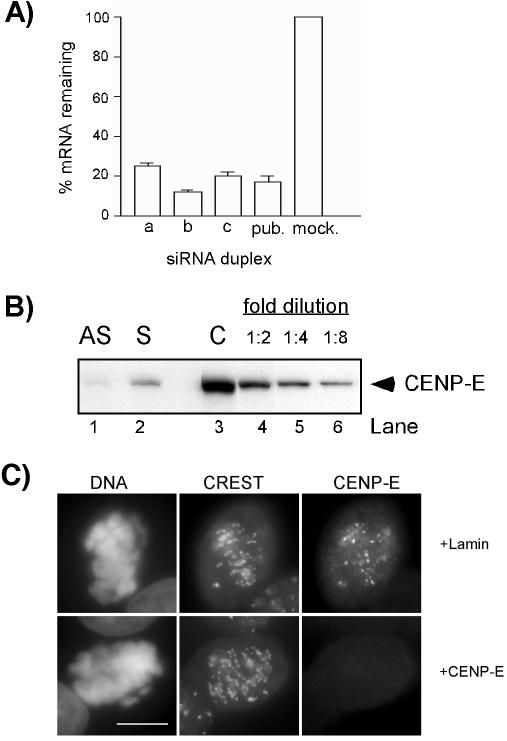
Transfection of HeLa(H2B)-GFP Cells with CENP-E siRNA Resulted in the Loss of CENP-E from Kinetochores We synthesized siRNA duplexes (a, b, and c) and the published duplex to CENP-E mRNA (Harborth et al., 2001), and compared their knockdown efficiencies in HeLa(H2B)-GFP cells. QPCR analysis consistently showed an ∼85–90% reduction of CENP-E mRNA 24 h after transfection of cells with duplex b, which was used for the remaining of this study (Figure 1A, b). Next, we examined the knockdown of CENP-E at the protein level by Western blotting. Because CENP-E accumulates only during mitosis (Brown et al., 1994), we compared the level of CENP-E protein in mitotic extracts of cells 24 h after transfection with CENP-E siRNA with those untransfected cells. Only a trace amount of CENP-E was detected in 75 μg of lysates of CENP-E siRNA-transfected cells. CENP-E protein levels, as measured by quantitative immunoblotting, were diminished by at least eightfold compared with the untransfected cells.
Figure 1.
siRNA-induced reduction of CENP-E mRNA and protein levels. (A) Reduction of CENP-E mRNA. HeLa(H2B)-GFP cells were transfected with various CENP-E siRNA duplexes as indicated, and the mRNA levels were measured 24 h posttransfection by QPCR. pub., published siRNA duplex; mock, LipofectAMINE 2000 alone. (B) Analysis of CENP-E protein levels by Western blot. Seventy-five micrograms of mitotic extracts derived from asynchronous cells (lane 1, AS) or presynchronized cells (lane 2, S) 24 h after transfection with CENP-E siRNA was compared with twofold serial dilutions of control mitotic extracts starting at 75 μg (lanes 3–5). (C) Immunofluorescence of CENP-E of HeLa cells 24 h posttransfection with Lamin (control) or CENP-E siRNA. Cells were double stained with CREST and CENP-E antibodies. Identical exposure times were used for imaging both control and CENP-E siRNA-transfected cells. DNA was visualized with 4,6-diamidino-2-phenylindole staining. Bar, 10 μm.
We also examined the presence of CENP-E at the single-cell level by indirect immunofluorescence assay of the kinetochores with CREST antiserum as the centromere marker. In Lamin siRNA-transfected cells (control), the signals corresponding to CENP-E were readily detected at the kinetochores of mitotic cells (Figure 1C, top). However, in CENP-E siRNA-transfected cells, no signal was detectable at the kinetochores in 85% of mitotic cells (Figure 1C, bottom).
Reduction of CENP-E Levels in HeLa(H2B)-GFP Cells Resulted in Delayed Mitotic Progression
Previously, it was demonstrated that the loss of CENP-E in mammalian cells reduced the efficiency of microtubule capture by kinetochores, causing unaligned chromosomes (Schaar et al., 1997; Yao et al., 2000; McEwen et al., 2001; Putkey et al., 2002). However, the outcome of this has not been fully investigated. In some studies, long-term mitotic arrest was observed; in others, apoptosis followed mitotic arrest (Schaar et al., 1997; Yao et al., 2000). In contrast, liver cells in CENP-E knockout mice showed no sign of mitotic arrest or cell death despite the presence of unaligned chromosomes (Putkey et al., 2002).
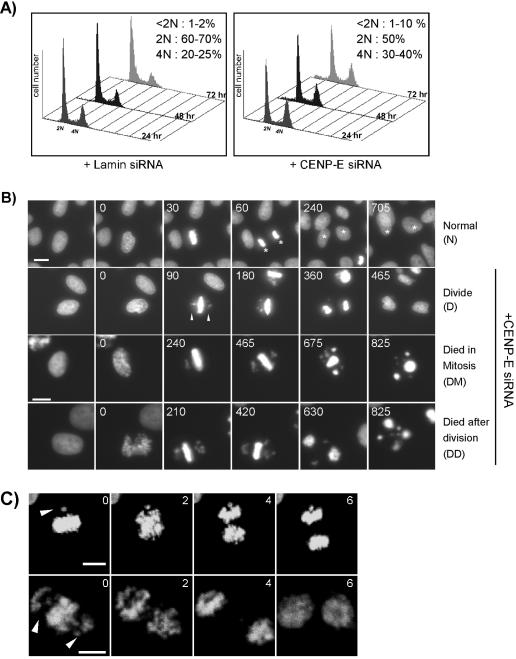
We wished to confirm which of the above-mentioned terminal phenotypes is associated with loss of CENP-E in HeLa(H2B)-GFP cells. By flow cytometry analysis, we expected that any effect of mitotic arrest would cause a change in the cell cycle profile. Compared with control siRNA-transfected cells, the cells transfected with CENP-E siRNA revealed a slight increase of 4N population (10–15% above control cells) (Figure 2A). Furthermore, the loss of CENP-E did not cause significant cell death because there was only a slight difference in percentage of dead/dying cells (i.e., <2N DNA content) between the control and experimental samples (Figure 2A).
Figure 2.
Effect of CENP-E depletion on mitotic progression. (A) Flow cytometry analysis of asynchronous HeLa(H2B)-GFP cells transfected with Lamin A/C (control) or CENP-E siRNA at 24, 48, and 72 h posttransfection. Cellular DNA content was determined by staining with propidium iodide. The percentage ranges of DNA contents at the three different time points are indicated on the upper right corner of each plot. (B) Live recordings of cells progressing through mitosis. HeLa(H2B)-GFP cells were transfected with Lamin (control) or CENP-E siRNA duplex, 24 h posttransfection. The morphological consequences were assessed by green fluorescent-chromosome/nucleus by using fluorescent time-lapse microscopy. Four different series of mitotic progression (i.e., normal [N], divide [D], died in mitosis [DM], and died after division [DD]) were observed and six images are shown for each series. Numbers in each panel denote the time (minutes) after the nuclear envelope breakdown (0 min). Asterisks denote the same daughter cells observed at different time points. Bar, 10 μm. (C) Anaphase occurs in the presence of unaligned chromosomes. HeLa(H2B)-GFP cells were transfected with CENP-E siRNA for 24 h and examined under a confocal fluorescent time-lapse microscope. The images were taken at 2-min intervals as indicated and show a cell with a pair of (top) or many unaligned chromosomes (bottom). Arrowheads denote unaligned chromosomes. Bar, 10 μm.
Two potential outcomes could explain the slight increase of 4N population observed. 1) Cells continued to divide after a short mitotic delay. 2) Only a small percentage of cells showed the mitotic defect. Using fluorescent time-lapse microscopy, we examined the mitotic progression in live HeLa(H2B)-GFP cells (Kanda et al., 1998). We defined the length of mitotic progression as the moment of nuclear envelope breakdown to chromosome segregation at anaphase. It typically took 45–60 min for control cells to go through this process (Figure 2B, N). In contrast, cells transfected with CENP-E siRNA took longer to reach anaphase (varying from 2.5 to 8 h) (Figure 2B, D). Our live cell recordings consistently revealed up to 80% of cells showing a few unaligned chromosomes close to spindle poles at metaphase, whereas the majority of chromosomes in these cells aligned at the metaphase plate. The majority of the cells initiated anaphase to produce viable daughter cells (Figure 5A, +CENP-E siRNA, D). These results indicate that the slight increase in 4N population observed in flow cytometry analysis is due to a delayed mitotic progression.
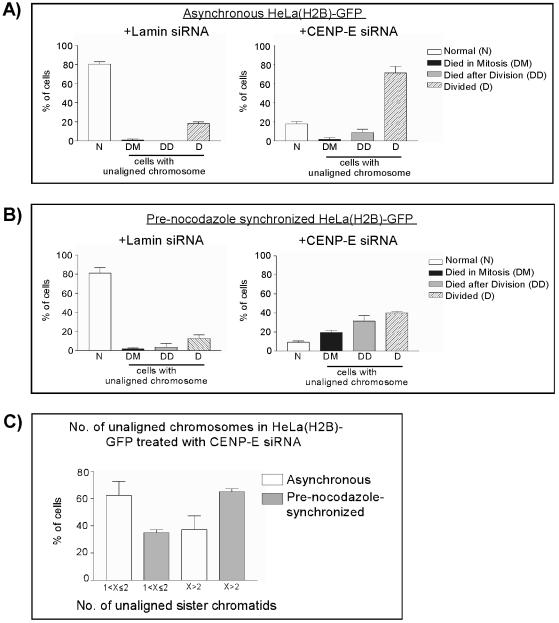
Figure 5.
Summary of the fate of mitotic cells with reduced levels of CENP-E. (A) Cell fate analysis of asynchronous HeLa(H2B)-GFP cells transfected with Lamin (control) or CENP-E siRNA duplex by using fluorescent videomicroscopy as described in Figure 2B. The number of cells without (N) or with unaligned chromosomes during mitosis were counted, and their fates were followed. (B) Same analysis as in A except that these cells were presynchronized with nocodazole before transfection. (C) Number of unaligned chromosomes in asynchronous and prenocodazole synchronized HeLa(H2B)-GFP cells 24 h after transfection with CENP-E siRNA duplex. 1 <X ≤2 denotes one to two pairs of unaligned sister chromatids. X>2 denotes more than two pairs unaligned sister chromatids. The data presented in A and B are the average of three independent experiments in each protocol. The data presented in C are the sum of all six experiments.
Cells with Reduced Levels of CENP-E Had a Grossly Intact Mitotic Checkpoint but Did Not Prevent Anaphase Onset in the Presence of a Few Unaligned Chromosomes
The lack of prolonged mitotic arrest in cells with reduced CENP-E levels is consistent with the result from a study of the conditional knockout mouse (Putkey et al., 2002). In that study, liver cells initiated anaphase in the presence of unaligned chromosomes. To examine whether anaphase occurred in the presence of unaligned chromosomes in HeLa(H2B)-GFP cells with reduced levels of CENP-E, we applied shorter intervals for live cell recording to examine carefully some of the cells progressing through anaphase. We observed that among the cells that initiated anaphase, ∼40% had at least one unaligned chromosome, resulting in uneven chromosome segregation (Figure 2C). A similar aneuploidy frequency was observed in primary mouse fibroblasts with nonfunctional copies of CENP-E (Weaver et al., 2003).
The initiation of anaphase in the presence of unaligned chromosomes in cells with reduced levels of CENP-E has raised the question whether the loss of CENP-E impairs checkpoint activation in mammalian cells. The work from Xenopus extract supports this possibility (Abrieu et al., 2000). To investigate this possibility in mammalian cells, we examined whether HeLa(H2B)-GFP cells transfected with CENP-E siRNA would escape mitotic arrest in the presence of microtubule-interfering agents. HeLa(H2B)-GFP cells transfected with CENP-E siRNA were incubated with the microtubule-depolymerizing agent nocodazole to inhibit spindle assembly. The mitotic index was scored by direct visualization of live cells with green-fluorescent chromosomes. Our Western blotting (Figure 1B) and immunofluorescence assay (Figure 1C) showed that CENP-E protein was significantly reduced.
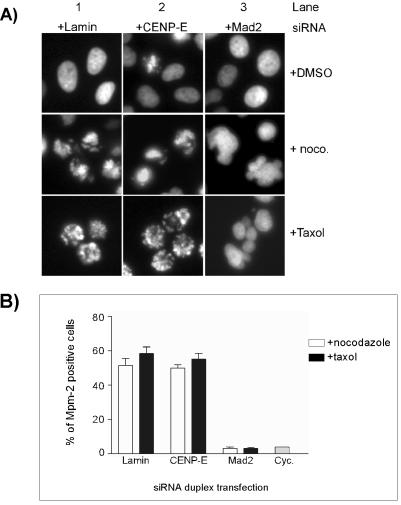
Similar to control siRNA-transfected cells, treatment of CENP-E siRNA-transfected HeLa(H2B)-GFP cells with nocodazole for 16 h resulted in accumulation of cells with condensed DNA, indicative of mitotic arrest (Figure 3A). We used Mad2 siRNA transfection as a positive control. As expected, Mad2-reduced cells exited mitosis when challenged with nocodazole, as was shown by the lack of condensed nuclei and the formation of micronucleated cells (Figure 3A). A similar mitotic arrest in both control siRNA and CENP-E siRNA-transfected cells also was observed after Taxol-induced suppression of microtubule dynamics (Figure 3A).
Figure 3.
CENP-E knockdown cells sustain the mitotic checkpoint in the presence of microtubule-interfering agents. (A) Images of green fluorescent-nuclei/chromosomes were taken after HeLa(H2B)-GFP cells were transfected with CENP-E, Lamin (negative control), and Mad2 siRNA (positive control) followed by either nocodazole or Taxol treatment for 16 h. Bar, 10 μm. (B) Flow cytometry profiles of HeLa cells with MPM-2 staining. HeLa cells were transfected with various siRNA duplexes and treated with nocodazole/Taxol as described in A. Subsequently, the cells were fixed, permeabilized, and stained with an antibody against the MPM-2 epitope, and the percentage of positive cells against fluorescent intensity was plotted. Cyc., untreated cycling cells.
To quantify the mitotic index, we carried out flow cytometry analysis on cells stained with MPM-2, a mitotic-specific proteins antibody (Yaffe et al., 1997). In normal cycling cells, <5% of cells are MPM-2 positive, reflecting the normal mitotic index. However, treatment of control siRNA-transfected cells with either nocodazole or Taxol increased the number of MPM-2–positive cells (55–60%) (Figure 3B). A similar percentage was observed in CENP-E siRNA-transfected cells when they are exposed to these microtubule-interfering agents (Figure 3B). On the contrary, cells transfected with Mad2 siRNA showed very few MPM-2–positive cells when treated with either nocodazole or Taxol (Figure 3B). These results are consistent with the chromosome morphology observed above with HeLa(H2B)-GFP cells. Together, our data demonstrated that the loss of CENP-E at the kinetochore did not impair the gross activation of the spindle checkpoint as cells were arrested in response to microtubule-interfering agents.
Both Mad2 and BubR1 Were Recruited to the Kinetochores of Unaligned Chromosomes, but Hyperphosphorylation of BubR1 Was Reduced in CENP-E Knockdown Cells
All the results presented so far have raised a paradox. On the one hand, cells with reduced levels of CENP-E initiated anaphase in the presence of unaligned chromosomes, suggesting that CENP-E is involved in the checkpoint-sensing mechanism. On the other hand, perturbation of spindle function with microtubule-interfering agents resulted in mitotic arrest in the absence of CENP-E, suggesting that it is not involved in spindle checkpoint activation. We questioned whether the spindle checkpoint components are recruited to the kinetochores on these unaligned chromosomes, another indicator for mitotic checkpoint activation. To address this, we carried out double immunofluorescences of BubR1 or Mad2 with CENP-E in HeLa cells transfected with CENP-E siRNA.
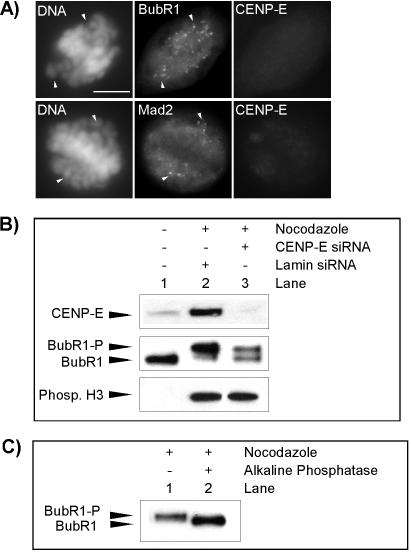
In control siRNA-transfected cells, both BubR1 and Mad2 antibodies stained unaligned chromosomes in prometaphase cells. Similarly, in CENP-E siRNA-transfected cells, both BubR1 and Mad2 also were detected on unaligned chromosomes in cells with no detectable CENP-E at the kinetochores (Figure 4A). In both control and CENP-E siRNA-transfected cells, Mad2 was not detected, and only low levels of BubR1 were observed on fully aligned chromosomes, indicating that the majority of BubR1 and Mad2 left the kinetochores when the checkpoint was silenced (Figure 4A). This indicates that these checkpoint proteins are still recruited to the kinetochores in cells with reduced levels of CENP-E, consistent with previous studies using either an antisense oligo approach or antibody injection (Yao et al., 2000; McEwen et al., 2001).
Figure 4.
Effect of CENP-E reduction on checkpoint activation. (A) Immunofluorescence analysis for BubR1 (top) or Mad2 (bottom) and CENP-E in HeLa cells 24 h after transfection with CENP-E siRNA. Arrowheads denote unaligned chromosomes and fluorescent signal of BubR1 or Mad2 on these locations. Bar, 10 μm. (B) Phosphorylation of BubR1 is reduced in mitotic extracts of CENP-E siRNA-transfected cells. Middle, comparison of hyperphosphorylation status of hBubR1 in cycling extracts (lane 1), mitotic extracts of Lamin siRNA-transfected cells (lane 2), and mitotic extracts of CENP-E siRNA-transfected cells (lane 3). The samples also were examined for CENP-E (top) and phosphorylated histone H3 (bottom). (C) hBubR1 immunoprecipitated from mitotically blocked cell lysates was incubated without (lane 1) or with alkaline phosphatase (lane 2) (see MATERIALS AND METHODS). BubR1-P, hyperphosphorylated form; BubR1, unphosphorylated form.
Using purified components and Xenopus egg extracts, Mao et al., (2003) showed that CENP-E is involved in the activation of the essential checkpoint kinase BubR1. We wondered whether reduction of CENP-E in HeLa cells impaired the ability of BubR1 to activate the mitotic checkpoint, which might explain why cells were insensitive to a few unaligned chromosomes in CENP-E knockdown cells. It was previously shown that hyperphosphorylation of BubR1 was related to the mitotic checkpoint (Chan et al., 1999). We decided to probe whether CENP-E knockdown by siRNA would lower the degree of BubR1 phosphorylation of the mitotically blocked cells. As shown in Figure 4B, BubR1 mobility was reduced in mitotically blocked cells relative to the cycling cells (compare lanes 1 and 2). This upshift was due to phosphorylation of BubR1 as the electrophoretic mobility was no longer retarded after phosphatase treatment (Figure 4C, compare lanes 1 and 2). We found that BubR1 was partially phosphorylated in mitotically blocked cells with reduced levels of CENP-E (Figure 4B, lane 3). Western blotting by using antibodies against phosphorylated histone H3 confirmed that cells collected by mitotic shake-off were indeed arrested in mitosis (Figure 4B, bottom). These results suggest that the ability of BubR1 to activate mitotic checkpoint seemed to be partially impaired in CENP-E knockdown cells.
CENP-E Knockdown after Synchronization with Nocodazole Induced More Unaligned Chromosomes and Cell Death
We were curious about why, in our hands, the HeLa(H2B)-GFP cells transfected with CENP-E siRNA did not display long-term mitotic arrest, which had been previously observed in HeLa cells transfected with CENP-E antisense oligonucleotides (Yao et al., 2000). However, in that study, cells were presynchronized with nocodazole before transfection. We reasoned that presynchronization may sensitize cells to the loss of CENP-E, resulting in the observed longterm mitotic arrest. To investigate this possibility, we presynchronized HeLa(H2B)-GFP cells with nocodazole before siRNA transfection and analyzed their fate by fluorescent time-lapse microscopy. By Western blotting, the level of CENP-E was reduced slightly more than eightfold compared with untransfected cells. However, the knockdown level was less than in asynchronous cells transfected with CENP-E siRNA.
As expected, the majority of presynchronized cells transfected with control siRNA showed profiles similar to asynchronous cells transfected with control siRNA (Figure 5, A and B, +Lamin siRNA). Most of these cells did not show unaligned chromosomes and divided normally to produce viable daughter cells. In contrast, transfection of the cells with CENP-E siRNA by using a presynchronization protocol resulted in the majority having unaligned chromosomes during mitosis (Figure 5B, +CENP-E siRNA). Of those with unaligned chromosomes, ∼20% of the cells never reached anaphase and died during division, as indicated by a collapsed metaphase plate and further condensation of chromosomes (Figure 2B, DM, T = 675, 825; Figure 5B, +CENP-E siRNA, DM). Another 30% of cells died after they completed mitosis (Figure 2B, DD, T = 825; Figure 5B, +CENP-E siRNA, DD), whereas the remaining 40% managed to complete cell division and produced viable daughter cells during 4 h of observation after the division (Figure 5B, +CENP-E siRNA, D). To rule out the possibility of cell death caused by phototoxicity, within each time-lapse analysis we imaged the control siRNA-transfected cells for the same duration of time as CENP-E siRNA-transfected cells. The results indicate that <5% of cells transfected with control siRNA died either during mitosis or after mitosis (Figure 2B, normal; Figure 5B, +Lamin siRNA, DM, DD). On the other hand, the fate of asynchronous cells transfected with CENP-E siRNA was significantly different from presynchronized cells. The majority of these cells with unaligned chromosomes initiated anaphase and produced viable daughter cells (Figure 5A, +CENP-E siRNA, D) and no significant increase of cell death during mitosis compared with control cells (Figure 5A, compare +Lamin siRNA and +CENP-E siRNA, DM). There was a slight increase in the percentage of cells that died after mitosis (9% above control cells) (Figure 5A, +CENP-E siRNA, DD).
Next, we questioned whether the number of unaligned chromosomes contributed to the different fates observed between the asynchronous and presynchronization methods. By the resolution of our time-lapse microscope, we are able to differentiate between one to two pairs of unaligned sister chromatids and more than two pairs of unaligned sister chromatids. It seemed that presynchronization with nocodazole increased the number of unaligned chromosomes in CENP-E siRNA-transfected cells. Compared with approximately 40% of asynchronous cells with CENP-E knockdown showing more than two unaligned sister chromatids, 65% of cells presynchronized with nocodazole displayed more than two unaligned chromosomes (Figure 5C). We also found that cells with more than two pairs of unaligned sister chromatids tended to spend more time in mitosis (Figure 2B, DM and DD, and Table 1). Generally, cells with fewer than two pairs of unaligned chromosomes spent 2.5–3 h, on average, between nuclear envelope breakdown and anaphase onset, whereas cells with more than two pairs of unaligned chromosomes spent between 6 and 8 h in this process. We interpret that the variations in time spent by the groups of cells with more than two pairs of unaligned chromosomes are due to the mixed population of cells with various numbers of unaligned chromosomes.
Table 1.
Relationship between the number of unaligned sister chromatids and the length of mitotic progression (minutes)
| No. of unaligned sister chromatids | 1 <X ≤2 | X>2 |
|---|---|---|
| Asynchronous | 155 ± 75 | 495 ± 288 |
| Prenocodazole synchronized | 185 ± 75 | 375 ± 168 |
We also directly followed the fate of cells with more unaligned chromosomes and found that most of these cells died either during mitosis or soon after cell division (our unpublished data). Hence, our data indicate that presynchronization with nocodazole increases the number of unaligned chromosomes in cells with reduced levels of CENP-E, subsequently leading to prolonged mitotic arrest and cell death in some cells.
Depletion of CENP-E Caused Defective Chromosome Congression but Had No Effect on Mitotic Spindle Assembly
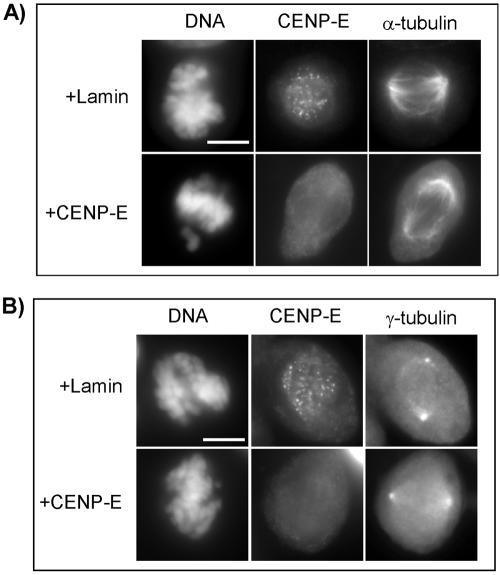
Our results presented above have confirmed that CENP-E is involved in chromosome congression and that the depletion of CENP-E resulted in a few unaligned chromosomes at metaphase. However, there has been a controversy with regard to spindle morphology when the function of CENP-E is perturbed. Most studies have shown normal or close to normal spindles in the cells with CENP-E perturbation (Brown et al., 1996; Schaar et al., 1997; Yao et al., 2000). In contrast, a report has shown that adenocarcinoma cells microinjected with anti-CENP-E antibody developed multiple spindle poles during mitosis (McEwen et al., 2001). To determine whether the spindle abnormalities occur in HeLa cells transfected with CENP-E siRNA, we carried out immunofluorescence of α- and γ-tubulin to examine the spindle morphology.
As expected, cells transfected with CENP-E siRNA have unaligned chromosomes that are monopolar and located close to the spindle poles (Figure 6, A and B, bottom), consistent with a previous report (Yao et al., 2000). However, immunofluorescence of α-tubulin and γ-tubulin of all cells from three independent experiments (a total of 50 cells examined for each antibody staining) showed that these cells have bipolar spindles. The morphology of the spindles is indistinguishable from that of control cells (Figure 6, A and B. top vs. bottom).
Figure 6.
CENP-E knockdown does not affect spindle morphology. Images show immunofluorescence assay for CENP-E and α-tubulin (A) and CENP-E and γ-tubulin (B) in HeLa cells 24 h after transfection with Lamin or CENP-E siRNA. Bars, 10 μm.
DISCUSSION
Presynchronization with Nocodazole Sensitizes Cells to CENP-E Gene Silencing by siRNA
We have shown that reduction of CENP-E expression by siRNA transfection in HeLa(H2B)-GFP cells delays mitotic progression. The majority of cells exhibited a few unaligned chromosomes at metaphase, consistent with what has been reported previously in both cellular knockdown and mouse knockout studies (Schaar et al., 1997; Yao et al., 2000; Putkey et al., 2002). The fact that only a few unaligned chromosomes present in CENP-E knockdown/knockout cells supports previous hypothesis that the controlling mechanism for chromosome alignment is functionally redundant (McEwen et al., 2001).
However, the lack of prolonged mitotic arrest in HeLa(H2B)-GFP cells transfected with CENP-E siRNA is contradictory to previous observations of cellular studies (Schaar et al., 1997; Yao et al., 2000). Our quantitative immunoblotting indicates that there is more than an eightfold reduction of CENP-E protein in siRNA-transfected cells. Moreover, there seems to be longer mitotic arrest in presynchronized cells, even though the knockdown is less than in asynchronous cells, which strongly argues that mitotic exit is not due to incomplete protein reduction. We attempted to quantify the immunofluorescence intensity of CENP-E at the single-cell level but encountered technical difficulties (such as saturation of antibody binding at the kinetochores, resulting in an extremely narrow dynamic range and high background staining causing poor signal-to-noise ratio). Therefore, we were not able to take accurate measurements.
We have found that presynchronization of cells with nocodazole sensitizes cells to the reduction of CENP-E; cells displayed an overall increase of unaligned chromosomes and a prolonged delay. This may explain the discrepancy between our results and previous observations. It is plausible that the residual amount of nocodazole is still present in the cells, although we do not have absolute proof. Low concentrations of nocodazole has been shown to reduce microtubule dynamics (Vasquez et al., 1997), which may synergize with loss of CENP-E tethering activity, thereby enhancing the failure of chromosome congression. Alternatively, we speculate that the presynchronization procedure per se introduces additional stress to sensitize cells to the CENP-E siRNA knockdown. Further characterizations are needed to understand the nature of this potential synergy. Interestingly, the study of CENP-E by the antibody microinjection also was carried out with synchronized cells (Schaar et al., 1997), further demonstrating that additional stress sensitizes cells to depletion of CENP-E. We believe that the effect is specific to CENP-E gene silencing because the control siRNA-transfected cells after presynchronization do not reveal abnormal effects.
CENP-E and Mitotic Spindle Checkpoint
It has been demonstrated that a single unattached kinetochore is sufficient to prevent progression to anaphase due to checkpoint activation (Rieder et al., 1994; Li and Nicklas, 1995). Our videorecordings of mitotic progression have revealed that cells with reduced CENP-E do not prevent progression to anaphase in the presence of unattached kinetochores. This has raised a question of whether CENP-E is involved in the mitotic checkpoint-sensing mechanism. It has been shown that Xenopus extracts in which all kinetochores are unattached no longer recruit Mad1 and Mad2 to their kinetochores and fail to arrest after immunodepletion of CENP-E (Abrieu et al., 2000).
First, we investigated whether HeLa cells transfected with CENP-E siRNA would exit mitosis when they are challenged with microtubule-interfering agents. Our results showed that CENP-E knockdown cells sustained the mitotic checkpoint after incubation with either nocodazole or Taxol both by morphological observation of condensed chromosomes and quantitative measurement of the mitotic marker MPM-2.
Second, we examined whether CENP-E knockdown would affect recruitment of essential checkpoint components Mad2 and BubR1 to unattached kinetochores. Both Mad2 and BubR1 are recruited and activated at the kinetochores of unattached and/or unaligned chromosomes (Musacchio and Hardwick, 2002). Our results showed that both Mad2 and BubR1 were recruited to unaligned chromosomes in CENP-E siRNA-transfected cells, consistent with previous observations of both cellular and mouse knockout studies (Yao et al., 2000; McEwen et al., 2001; Putkey et al., 2002). While we were revising our manuscript, Weaver et al., (2003) reported that BubR1 and Mad2 recruitment to unaligned chromosomes were reduced in MEF cells isolated from CENP-E knockout mouse. Our initial quantitative immunofluorescence of BubR1 in CENP-E siRNA transfected HeLa cells also supports their finding (our unpublished data).
BubR1 kinase activity is essential for the checkpoint activation in mammalian cells (Chan et al., 1999). Using Xenopus extract, Mao et al., (2003) showed that CENP-E is required for BubR1 activation. Previous work on comparison of hyperphosphorylation status of BubR1 in normal mitotic HeLa cells, nocodazole-blocked mitotic cells, or cells exiting mitosis showed that the hyperphosphorylation of BubR1 is related to an activated checkpoint (Chan et al., 1999). Therefore, we analyzed whether hyperphosphorylation status of BubR1 was compromised in CENP-E knockdown cells. We used an antibody that recognizes both hyperphosphorylated and unphosphorylated forms of BubR1 to probe whether the phosphorylation of BubR1 is reduced in nocodazole-treated cells. Indeed, BubR1 gets partially phosphorylated in CENP-E siRNA-transfected cells, suggesting that its activity may be impaired. In the study of Weaver et al., (2003), they showed that not only CENP-E bound and directly stimulated BubR1 kinase activity in vitro but also that the BubR1 kinase activity was compromised in MEF cells depleted of CENP-E.
Thus, the paradox of CENP-E's involvement in mitotic checkpoint may be explained by the following hypothesis, as proposed previously by the Salmon and the Yen laboratories (Shannon et al., 2002; Liu et al., 2003). Our studies as well as those by Weaver et al., (2003) support the hypothesis. A certain threshold level of the wait-anaphase signal may be needed for a cell to induce mitotic arrest. When cells with reduced levels of CENP-E (our study) or CENP-E knockout cells (Weaver et al., 2003) are challenged with microtubule-interfering agents, the sum of the signals generated at each kinetochore are still sufficient to reach the threshold required for mitotic arrest, despite having reduced BubR1 activity. However, in normal mitotic progression of cells with knockdown/knockout of CENP-E, the recruitment and activation of BubR1 at the few unaligned chromosomes are not enough to generate a sufficiently robust checkpoint signal to prevent anaphase onset and the consequent development of aneuploidy. The result from cells presynchronized with nocodazole further strengthens the hypothesis, because we have discovered that more unaligned chromosomes are needed to induce a longer arrest.
Effects of CENP-E Reduction on the Fate of Cells
We have observed that cells with gene silencing of CENP-E by using RNAi undergo three major fates: die in mitosis, exit mitosis and then die, or exit mitosis and become viable with partially missegregated chromosomes. We also have discovered that presynchronization of cells with nocodazole before CENP-E siRNA transfection causes more cell death, either at mitosis or after cell division. Because loss of CENP-E reduces the fidelity of chromosome segregation, the loss of critical genes may impair proper cellular functions in the next cell cycle, thereby triggering apoptosis. This process may occur randomly in asynchronous cells with knockdown of CENP-E, because most daughter cells are viable. Furthermore, by using a presynchronization protocol, the increased number of missegregated chromosomes is expected to increase the loss of essential genes and nonviable progeny. As expected, a significant percentage of cell death is observed in that population.
Alternatively, these presynchronized cells with reduced CENP-E exit mitosis with an activated spindle checkpoint in the subsequent G1 phase of the cell cycle, which can initiate apoptosis, as suggested previously (Taylor and McKeon, 1997). Chemotherapeutics such as Taxol and the vinca alkaloids perturb kinetochore–microtubule attachment, which consequently activates a checkpoint pathway and induces mitotic arrest. Prolonged mitotic arrest is thought to induce cell death through proapoptotic pathways (Sorger et al., 1997). Apoptosis is most likely a safeguard mechanism used by a cell to prevent improper mitotic exit, leading to daughter cells with abnormal DNA content.
Our results indicate that chromosome missegregation upon CENP-E knockdown seems to be universal between CENP-E gene deletion in mouse and gene silencing by RNA interference in a human tumor cell line. Our results indicate that disruption of CENP-E function is undesirable because this causes chromosome instability; potentially, it may promote tumorigenesis in the long run. On the other hand, studying CENP-E-depleted cells in vivo may serve as a model to understand chromosome instability and tumor development.
Acknowledgments
We thank Drs. Emma Lees, David Parry, Ronald Herbst, Wolfgang Seghezzi, Scott Frank, and Dovie Wylie for critically reading the manuscript. We also thank Dr. Yongke Zhang for polyclonal antibodies against CENP-E, and we are grateful for the gift of monoclonal CENP-E antibody from Dr. Tim Yen. DNAX Research Institute is supported by Schering-Plough Corporation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0482. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0482.
References
- Abrieu, A., Kahana, J.A., Wood, K.W., and Cleveland, D.W. (2000). CENP-E as an essential component of the mitotic checkpoint in vitro. Cell 102, 817-826. [DOI] [PubMed] [Google Scholar]
- Brown, K.D., Coulson, R.M., Yen, T.J., and Cleveland, D.W. (1994). Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J. Cell Biol. 125, 1303-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K.D., Wood, K.W., and Cleveland, D.W. (1996). The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J. Cell Sci. 109, 961-969. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., Jablonski, S.A., Sudakin, V., Hittle, J.C., and Yen, T.J. (1999). Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146, 941-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G.K., Schaar, B.T., and Yen, T.J. (1998). Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143, 49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y., and Sullivan, K.F. (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407-421. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Bechert, K., Tuschl, T., and Weber, K. (2001). Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557-4565. [DOI] [PubMed] [Google Scholar]
- Kanda, T., Sullivan, K.F., and Wahl, G.M. (1998). Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377-385. [DOI] [PubMed] [Google Scholar]
- Li, X., and Nicklas, R.B. (1995). Mitotic forces control a cell-cycle checkpoint. Nature 373, 630-632. [DOI] [PubMed] [Google Scholar]
- Liu, S.T., Hittle, J.C., Jablonski, S.A., Campbell, M.S., Yoda, K., and Yen, T.J. (2003). Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 5, 341-345. [DOI] [PubMed] [Google Scholar]
- Lombillo, V.A., Nislow, C., Yen, T.J., Gelfand, V.I., and McIntosh, J.R. (1995). Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J. Cell Biol. 128, 107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney, T., Ginkel, L.M., Hunter, A.W., and Wordeman, L. (2000). The kinetochore of higher eucaryotes: a molecular view. Int. Rev. Cytol. 194, 67-131. [DOI] [PubMed] [Google Scholar]
- Mao, Y., Abrieu, A., and Cleveland, D.W. (2003). Activating and silencing the mitotic checkpoint through CENP-E dependent activation/inactivation of BubR1. Cell 114, 87-98. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma, S., Stucke, V.M., and Nigg, E.A. (2002). Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297, 2267-2270. [DOI] [PubMed] [Google Scholar]
- McEwen, B.F., Chan, G.K., Zubrowski, B., Savoian, M.S., Sauer, M.T., and Yen, T.J. (2001). CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell 12, 2776-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio, A., and Hardwick, K.G. (2002). The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell. Biol. 3, 731-741. [DOI] [PubMed] [Google Scholar]
- Pihan, G.A., and Doxsey, S.J. (1999). The mitotic machinery as a source of genetic instability in cancer. Seminars in Cancer Biology 9, 289-302. [DOI] [PubMed] [Google Scholar]
- Putkey, F.R., Cramer, T., Morphew, M.K., Silk, A.D., Johnson, R.S., McIntosh, J.R., and Cleveland, D.W. (2002). Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell 3, 351-365. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., Schultz, A., Cole, R., and Sluder, G. (1994). Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 127, 1301-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar, B.T., Chan, G.K., Maddox, P., Salmon, E.D., and Yen, T.J. (1997). CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139, 1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, K.B., Canman, J.C., and Salmon, E.D. (2002). Mad2 and BubR1 function in a single checkpoint pathway that responds to a loss of tension. Mol. Biol. Cell 13, 3706-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger, P.K., Dobles, M., Tournebize, R., and Hyman, A.A. (1997). Coupling cell division and cell death to microtubule dynamics. Curr. Opin. Cell Biol. 9, 807-814. [DOI] [PubMed] [Google Scholar]
- Taylor, S.S., and McKeon, F. (1997). Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89, 727-735. [DOI] [PubMed] [Google Scholar]
- Vasquez, R.J., Howell, B., Yvon, A.M., Wadsworth, P., and Cassimeris, L. (1997). Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell 8, 973-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, B.A., Bonday, Z.Q., Putkey, F.R., Kops, G.J., Silk, A.D., and Cleveland, D.W. (2003). Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162, 551-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, K.W., Sakowicz, R., Goldstein, L.S., and Cleveland, D.W. (1997). CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell 91, 357-366. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.B., et al. (1997). Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278, 1957-1960. [DOI] [PubMed] [Google Scholar]
- Yao, X., Abrieu, A., Zheng, Y., Sullivan, K.F., and Cleveland, D.W. (2000). CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484-491. [DOI] [PubMed] [Google Scholar]
- Yao, X., Anderson, K.L., and Cleveland, D.W. (1997). The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, T.J., Li, G., Schaar, B.T., Szilak, I., and Cleveland, D.W. (1992). CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 359, 536-539. [DOI] [PubMed] [Google Scholar]
- Yu, H. (2002). Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14, 706-714. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Yao, J., and Joshi, H.C. (2002). Attachment and tension in the spindle assembly checkpoint. J. Cell Sci. 115, 3547-3555. [DOI] [PubMed] [Google Scholar]