Abstract
Precursor-mRNA splicing removes the introns and ligates the exons to form a mature mRNA. This process is carried out in a spliceosomal complex containing >150 proteins and five small nuclear ribonucleoproteins. Splicing protein hSlu7 is required for correct selection of the 3′ splice site. Here, we identify by bioinformatics and mutational analyses three functional domains of the hSlu7 protein that have distinct roles in its subcellular localization: a nuclear localization signal, a zinc-knuckle motif, and a lysine-rich region. The zinc-knuckle motif is embedded within the nuclear localization signal in a unique functional structure that is not required for hSlu7's entrance into the nucleus but rather to maintain hSlu7 inside it, preventing its shuttle back to the cytoplasm via the chromosomal region maintenance 1 pathway. Thus, the zinc-knuckle motif of hSlu7 determines the cellular localization of the protein through a nucleocytoplasmic-sensitive shuttling balance. Altogether, this indicates that zinc-dependent nucleocytoplasmic shuttling might be the possible molecular basis by which hSlu7 protein levels are regulated within the nucleus.
INTRODUCTION
Precursor-mRNA splicing removes the introns and ligates the exons to form a mature mRNA. This process is carried out in a spliceosomal complex containing >150 proteins and five small nuclear ribonucleoproteins (snRNP) (Burge et al., 1999; Liu et al., 2002; Rappsilber et al., 2002; Jurica and Moore, 2003). The exon-intron borders, identified by the spliceosome with a remarkable fidelity, are defined by the 5′ and 3′ splice sites (ss) as well as the polypyrimidine tract and branch-site sequence (Brow, 2002). Combinatorial control of different nuclear protein concentrations varying among tissues (Hanamura et al., 1998) and cell types (Jensen et al., 2000) or during development (Mahe et al., 2000) was shown to determine exon choice (Wang and Manley, 1995; Smith and Valcarcel, 2000; Lallena et al., 2002), and these can be altered upon physiological stimuli (van der Houven van Oordt et al., 2000), environmental effects (Meshorer et al., 2002), or phosphorylation state (Xie et al., 2003). Aberrant regulation of splicing has been implicated in an increasing number of human diseases, including cancer (Hastings and Krainer, 2001; Modrek and Lee, 2002; Stoilov et al., 2002; Changela et al., 2003).
In a previous study, we showed the possible role of zinc, or a metalloprotein, in the second step of splicing in vitro (Shomron et al., 2002; Shomron and Ast, 2003). Zinc is an essential element for all organisms as a catalytic and/or structural cofactor for many zinc-dependent enzymes and proteins. Zinc homeostasis in higher animals and humans is a process that requires cells to move minute amounts of zinc ions into and out of cells by a series of transport proteins or transporters with the main purpose of obtaining zinc from the environment, protecting cells against zinc toxicity, and maintaining ample supplies of zinc for metabolic purposes (Harris, 2002). The hSlu7 protein is the only known secondstep splicing protein that possesses a potential zinc binding domain in the form of a zinc-knuckle motif, which led us to examine the role of zinc in hSlu7. At first, the yeast Slu7 protein (SLU; Synergistic Lethal with U5 snRNA) was isolated based on its interaction with U5 (Frank et al., 1992) and was shown to be required for the second catalytic step of splicing (Jones et al., 1995). Yeast Slu7 binds before the second catalytic step to U5 snRNP and to Prp18 in the spliceosome; Slu7's interaction can be carried out even without a 3′ ss and then it remains bound to the spliceosome after the second catalytic step (Frank and Guthrie, 1992; Frank et al., 1992; Jones et al., 1995; Umen and Guthrie, 1995; Jiang et al., 2000). The human homolog hSlu7 was isolated by Chua and Reed (1999a), and human nuclear extract immunodepleted of hSlu7 reduces the affinity of the 5′ exon to the spliceosome, leading to different selection of AGs at the 3′ ss. This suggested that hSlu7 is required for holding the 5′ exon in the spliceosome for the correct 3′ ss attack (Chua and Reed, 1999b).
Nuclear localization signals (NLS) and nuclear export signals (NES) are the essential elements for a protein subjected to nucleocytoplasmic transportation. The typical NLS consensus sequence is either monopartite, consisting of a single, short stretch of basic amino acids, or bipartite, containing two separated clusters of basic residues divided by a spacer region of ∼10 amino acids (Boulikas, 1993). These sequences bind to specific receptors that allow selective passage though the nuclear pore complex (Fahrenkrog and Aebi, 2003).
Here, we identify by bioinformatics and mutational analyses three functional domains of the hSlu7 protein that have distinct roles in its subcellular localization: a nuclear localization signal, a zinc-knuckle motif, and a lysine-rich region. The zinc-knuckle motif is embedded within the NLS in a unique functional structure that is not required for hSlu7's entrance into the nucleus but rather to maintain hSlu7 inside it, preventing its shuttle back to the cytoplasm via the chromosomal region maintenance 1 (CRM1) pathway. Thus, the zinc-knuckle motif of hSlu7 determines the cellular localization of the protein through a nucleocytoplasmic sensitive shuttling balance. Altogether, this indicates that zinc-dependent nucleocytoplasmic shuttling might be the possible molecular basis by which hSlu7 protein levels are regulated within the nucleus.
MATERIALS AND METHODS
Plasmid Construction and Site-directed Mutagenesis
To generate fusion constructs with the green fluorescent protein (GFP) and glutathione S-transferase (GST) the hSlu7 entire coding region (provided by Robin Reed, Harvard Medical School, Boston, MA) was cloned into pEGFP-C1 (BD Biosciences Clontech, Palo Alto, CA) and pGEX-6P (Amersham Biosciences, Piscataway, NJ). Mutations were introduced using overlapping oligonucleotide primers containing the desired mutation as described in Lev-Maor et al. (2003). List of primers used is added as Supplementary Material.
Cell Line Maintenance, Transfection, and Treatments
HeLa and 293T cells were grown on coverslides in 24-well plates or 100-mm culture dish and maintained in DMEM at 37°C in a humidified atmosphere of 5% CO2. Cell media also were supplemented with 10% fetal calf serum, 0.29 mg/ml l-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 1 U/ml nystatin (Biological Industries, Beit HaEmek, Israel). Transfection was performed using FuGENE6 (Roche Diagnostics, Indianapolis, IN) as described by manufacturer's protocol. Cell lines were subjected to the following treatments. Leptomycin B (LMB; Sigma-Aldrich, St. Louis, MO) was added for 3 h at 2, 5, 15, and 25 ng/ml to inhibit nuclear export through the CRM1-mediated pathway. Divalent metals (ZnCl2, MnCl2, and CuCl2; Sigma-Aldrich) were added onto the medium of the growing cells at 50–200 μM concentrations for 3–18 h.
Recombinant Protein and Antibody Production
Recombinant GST-hSlu7 protein was produced as described in Shomron and Ast (2003). This protein was used for polyclonal antibody production in rabbits according to standard procedures.
Inductively Coupled Plasma-Atomic Emission Spectrometry (ICP-AES)
One hundred micrograms of purified recombinant wild-type hSlu7 and a zinc-knuckle mutant version of the same protein (CCHC>SSGS) were inserted into Spectra/Por molecular porous membrane tubing (Spectrum Medical Industries, Los Angeles, CA; molecular weight cut-off 12,000–14,000) and dialyzed at >3000 volumes against dialysis solution (50 mM Tris, pH 7.5, 50 mM NaCl in metal-free H2O) for 12 h at 4°C. Samples were then transferred to reading Buffer (5% HNO3 in metal-free H2O), boiled at 100°C for 1 h, and introduced into an ICP-AES (Spectro Modula E; Spectro, Kleve, Germany) for measurement of zinc atom levels.
Splicing Substrates, Reactions, and Spliceosome Complex Analysis
Standard splicing reactions were performed on adenovirus major late transcript as described in Shomron et al. (2002). Briefly, uniformly labeled RNA transcript was incubated in in vitro splicing conditions with the addition of recombinant hSlu7 at 200 M excess compared with the endogenous hSlu7. RNA was extracted and analyzed by 8% denaturing PAGE as described previously (Ast and Weiner, 1986). Spliceosome complexes were analyzed in 4% native gels as described previously (Konarska and Sharp, 1986) (with 0.5 mg/ml heparin in the loading buffer).
Western Blotting
Total protein harvesting was performed by incubating the cells in lysis buffer (50 mM Tris 7.5, 1% NP-40, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholic acid, protease inhibitor cocktail and phosphatase inhibitor cocktail I and II; Sigma-Aldrich) for 15 min on ice. Supernatant containing proteins was collected after centrifugation for 30 min at 14,000 rpm. Total protein concentrations were measured using Bio-Rad protein assay (Bio-Rad, Hercules, CA). Proteins were separated in 12% SDS-PAGE and then electroblotted onto a Protran membrane (Schleicher & Schuell, Keene, NH). The membranes were probed with anti-hSlu7, anti-GST, or anti-GAPDH followed by the appropriate secondary antibody. Immunoblots were visualized by enhanced chemiluminescence (Lumi-Light Western blotting substrate; Roche Diagnostics) and autoradiography.
Immunofluorescence
To detect subcellular localization of native and GFP fusion proteins cells cultured on coverslips were transfected with plasmids as described above. After 48 h, cells were washed with phosphate-buffered saline (PBS) and fixed for 10 min in 4% paraformaldehyde. Permeabilization was performed with 0.5% Triton X-100 in PBS for 10 min, and then cells were incubated for 5 min with 0.1 mg/ml 4,6-diamidino-2-phenylindole (DAPI) to distinguish nuclear and cytoplasmic localization. To observe hSlu7 and Sm proteins, the cells were incubated first with anti-hSlu7 or anti-Sm antibodies for 1 h and then reacted with secondary antibodies either rhodamine-conjugated anti-IgG or fluorescein isothiocyanate-conjugated anti-IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) in blocking solution (5% fetal calf serum, 5% bovine serum albumin; PBS) for 0.5 h. Fluorescent images were observed under a 410 confocal laser-scanning microscope (CLSM410; Carl Zeiss, Thornwood, NY) at 365-, 488-, and 568-nm laser lines. Images of the same antibody staining were obtained using the same parameters (e.g., brightness, contrast, pinhall).
Image Processing
For determination of hSlu7 localization multiple fields were examined to count at least 100 positive cells. Images were acquired under identical conditions, ensuring that the maximal signal was not saturated. On some cells, random test lines also were drawn over the cell nucleus, and the absolute intensity value of the test points was measured. Acquisition of images and measurement of speckles intensity was performed using Adobe Photoshop and TINA software. All results represent values obtained from at least two separate experiments, and the results are average values.
Structural Modeling
The structural model of the hSlu7 zinc-knuckle domain was built and optimized with Swiss-PDB viewer and RasMol by using the coordinates of viral Gag polyprotein (PDB ID code 2znf) as the template because it produced the most significant score in the Prosite search. Based on this structure the same software was used to build the mutant forms of hSlu7.
RNA Isolation and Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Amplification
Cells were grown in 100-mm culture dish to 50% confluence, and transfection was performed using 4 μg of the indicated plasmid DNA as described above. After 48 h total cytoplasmic RNA was extracted using TRI Reagent (Sigma-Aldrich) followed by treatment with 2 U of DNase (RNase-Free; Ambion, Austin, TX). First-strand oligo(dT)-primed cDNA synthesized with reverse transcriptase of avian myelobstosis virus (Roche Diagnostics) from 2 μg of total RNA was amplified with High Fidelity Taq polymerase (Roche) and hSlu7, ADAR2 and E1A primers for 30 cycles, consisting of 94°C for 30 s, 55–65°C for 45 s and 72°C for 1 min. The products were separated in 1% agarose gel and confirmed by sequencing.
Web-based Software
The URLs for Web-based software are as follows: MotifScan, motif scan in a protein sequence, http://hits.isb-sib.ch/cgi-bin/PFSCAN Prosite; database of protein families and domains, http://www.expasy.org/prosite/Pfam; protein families database, http://www.sanger.ac.uk/Software/Pfam/PDB; a browser for structure/function, http://pdb.tau.ac.il/Swiss-PDB viewer; and molecular visualization, http://www.usm.maine.edu/spdbv/RasMol and http://www.umass.edu/microbio/rasmol/.
Submitted Sequences
The submitted sequence to National Center for Biotechnology Information/European Molecular Biology Laboratory/DNA Database of Japan sequence databases was as follows: AY207315: Rattus norvegicus second-step splicing protein SLU7 (Slu7) mRNA, complete cds gi 29126644 gb AY207315.2 [22885214].
RESULTS
hSlu7 Possesses Evolutionarily Conserved and Semiconserved Motifs
With the aid of Web-based software (Prosite, MotifScan, and Pfam), we identified three putative motifs in the hSlu7 protein sequence: 1) a zinc-knuckle motif (only this motif was previously described in hSlu7; Chua and Reed, 1999a); 2) a bipartite NLS; and 3) a lysine-rich domain. Interestingly, the zinc-knuckle is located within the NLS domain, which is a unique structural organization (Figure 1A; see below).
Figure 1.
Protein motifs and phylogenetic conservation of hSlu7. (A) Schematic representation of the hSlu7 protein motifs. The respective location of each motif is indicated: NLS (diagonal bared box), zinc-knuckle (zinc knk; black box), four leucine repeats (Leu 1–4; gray shaded boxes), and lysine-rich region (Lys rich; doted box). The protein image is not drawn exactly to scale but is drawn proportionally. (B) Slu7 from 10 different organisms [human (AAD13774); rat (R. norvegicus; sequenced in our laboratory and deposited in GenBank/EMBL/DDBJ; NP_776208); mouse (Mus musculus; XP_126206); Drosophila (Drosophila melanogaster; NP_651659); Anopheles (Anopheles gambiae; AAAB01008968); nematode (Caenorhabditis elegans; NP_505661); Fugu (annotated from www.fugubase.org), Arabidopsis (Arabidopsis thaliana CAB82819); Yeast S.P. (Schizosaccharomyces pombe; NP_596034); and Yeast S.C. (Saccharomyces cerevisiae; A46229)] were aligned along their sequence. Phylogenetic conservation is high within the putative zinc-knuckle amino acids (marked in bold on the first row) but less in the potential NLS regions (single and double underlined on the first row). Semi-conservation is seen in the four leucine-rich regions (bold and underlined “L” in the two center rows). Weak conservation is observed in the lysine-rich region (underlined in the bottom row; the lysine amino acids–K–also are shown in bold). The most conserved amino acids are marked with an asterisk below the alignments. Two dots or one indicate decreasing conservation, respectively. The yeast S. cerevisiae (Yeast S.C.) sequence is written below the main alignment because it possesses the least conserved Slu7 out of the group. Two alignment results are shown; with and without the Yeast S.C. Slu7 sequence. Numbers on the right indicate amino acid location.
The zinc-knuckle domain is characterized by a zinc atominterconnecting cysteine (C) and histidine (H) residues forming a finger-like structure with a consensus sequence of CX2CX4HX4C (where X denotes any amino acid). This motif was reported in other proteins to bind single-stranded nucleic acids (Lopato et al., 1999) or to determine nuclear localization (mutations at this motif caused cytoplasmic localization; Dye and Patton, 2001; Braem et al., 2002; Pandya and Townes, 2002). Based on software analysis, we first identified an NLS of the bipartite type that harbors the zinc-knuckle motif of hSlu7 (Figure 1B, single underlined on the top row). We searched databases for Slu7 sequences from other species, by using whole and subregions of the protein. Alignment of hSlu7 derived from 10 different organisms (one sequenced by us; see MATERIALS AND METHODS) shows high conservation of the zinc-knuckle domain, partial conservation of the bipartite NLS, and additional regions containing highly conserved basic amino acids (R/K) that might serve as an NLS as well (Figure 1B, double underlined on the top row). Four putative NES signals, characterized by a stretch of several leucine amino acids (Henderson and Eleftheriou, 2000), but not necessarily (Brown et al., 2003), also were identified (Figure 1B, bold and underlined “L” in the center rows). A lysine-rich domain that can be associated with several cellular functions, such as binding to nucleic acids (Weeks and Crothers, 1992; Frugier et al., 2000; Stros, 2001), facilitating interactions between two proteins (Chakraburtty, 1999; Das and Maitra, 2001), or nuclear translocation (Kitagawa et al., 1996), was identified in the C terminal of the protein (Figure 1B, bold and underlined “K” on the bottom row).
We conclude that hSlu7 possesses four potential motifs that may impact its cellular localization: an overlapping zinc-knuckle and NLS region, four stretches of leucine repeats, and a lysine-rich domain. Given that hSlu7 is an integral component of the spliceosome, and the evolutionary conservation of these regions (Figure 1B) can only suggest functional importance, an experimental validation was needed to confirm their purpose. Thus, we set to determine the effect of each on the intracellular localization of hSlu7.
The Cellular Localization of hSlu7
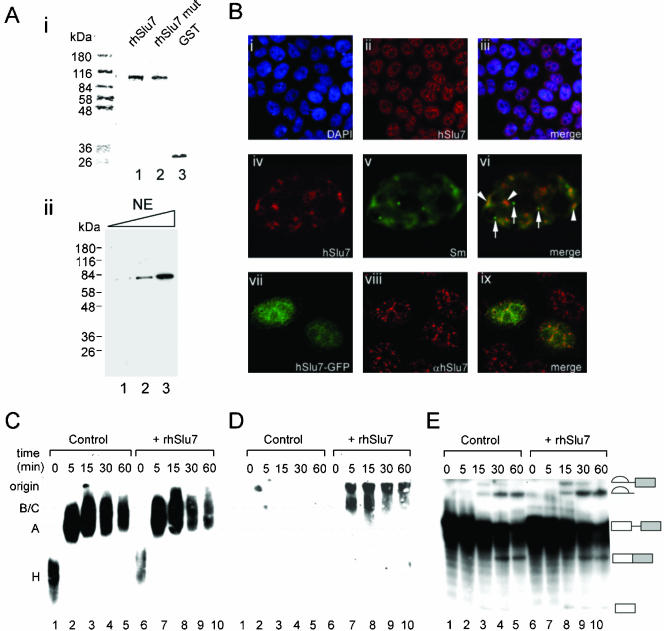
To study the cellular localization of hSlu7, we expressed and purified a recombinant hSlu7 protein fused to a GST tag (rhSlu7; Figure 2Ai, lane 1). This protein was used for antibody production in rabbits. Mutant rhSlu7 (in which the CCHC amino acids of the zinc-knuckle motif were mutated to SSGS) and GST-only proteins also were produced (Figure 2Ai, lanes 2 and 3, respectively) and will be discussed below (see Figure 7). Western blot analysis, by using the anti-hSlu7 polyclonal antibody serum, recognized one polypeptide in HeLa nuclear extract at ∼70 kDa, which was confirmed by mass spectroscopy to be hSlu7 (Figure 2Aii, lanes 1–3; our unpublished data). The hSlu7 polyclonal antibody serum, hereafter termed anti-hSlu7, was first used for immunofluorescence studies to locate hSlu7's staining pattern within the cells. HeLa cells were grown on cover slides for 48 h, fixed, and then stained for their nuclear DNA (using DAPI) and stained with anti-hSlu7 as well. Cellular localization of endogenous hSlu7 exhibits a distinct nuclear staining pattern known as speckles, which is common to other splicing factors, including snRNPs (such as in SF2/ASF; (Caceres et al., 1997; Mintz and Spector, 2000; Lamond and Spector, 2003), in addition to a diffusible nucleoplasmic signal and a lack of cytoplasmic and nucleolar staining (Figure 2Bi–iii; DAPI nuclear staining, anti-hSlu7, and a merged image, respectively). We therefore wanted to observe the localization of hSlu7 in comparison with snRNPs. HeLa cells were fixed and then stained with anti-hSlu7 and anti-Sm (Sm proteins are bound to the snRNPs). Superimposed images of the endogenous hSlu7 (Figure 2Biv, marked red) with snRNP proteins (Figure 2Bv, anti-Sm, marked green) result in yellow color at colocalization sites seen in 84% of the nuclear stained regions (marked by arrowheads in Figure 2Bvi), but not in the Cajal bodies (marked by arrows on the same image). Cajal bodies are nuclear organelles that contain factors required for splicing, ribosome biogenesis, and transcription (for review, see Lamond and Spector, 2003) and also are enriched in common box C/D small nucleolar ribonucleoprotein proteins (Leary et al., 2003). Similar results were obtained using different cell lines (our unpublished data). Thus, we conclude that hSlu7 is a distinct nuclear protein that localizes to the nucleoplasm in a speckles pattern and colocalizes with snRNPs.
Figure 2.
Cellular distribution of hSlu7. (A) i, recombinant hSlu7 protein (rhSlu7), mutant recombinant hSlu7 protein (rhSlu7 mut), and GST-only protein (GST) were expressed and purified in bacteria (see MATERIALS AND METHODS). The purified proteins were run in SDS-PAGE followed by staining of total proteins by using Coomassie. Protein size marker is indicated on the left. ii, polyclonal antibodies raised against hSlu7 detect a distinct polypeptide of ∼70 kDa on a Western blot of increasing concentrations of HeLa cells nuclear extract (NE; 1, 5, and 10 μg of total proteins). Mass spectroscopy confirmed that this protein is hSlu7 (our unpublished data). Protein size marker is indicated on the left. (B) HeLa cells were fixed, and the cellular localization of hSlu7 was determined by indirect immunofluorescence with anti-hSlu7. The same slides were also stained with DAPI for nuclear DNA and anti-Sm. The hSlu7 protein localizes in a speckled nuclear pattern and is also diffusely distributed throughout the nucleoplasm (i–iii). Colocalization of hSlu7 (iv; red) and Sm (v; green) can be seen at several nuclear locations (vi; yellow, indicated by arrowheads) but not in others (vi; such as in the Cajal bodies, indicated by an arrow). Cells also were transfected with hSlu7-GFP plasmids (vii). Colocalization with endogenous hSlu7 is seen in several of the nuclear regions (viii–ix; see text). (C) HeLa nuclear extract was incubated with a radiolabeled Adeno pre-mRNA transcript for the indicated time under splicing conditions (lanes 1–10). To some reactions recombinant hSlu7 was added (rhSlu7; at 200 mol excess than the wild-type hSlu7; lanes 6–10). The splicing reactions were separated in a 4% native polyacrylamide gel and proteins/RNA were transferred to nylon membrane, and then visualized by autoradiography. (D) After radioactive decay, the same membrane described in C was probed with anti-GST. (E) Splicing products, extracted from the same experiment, were separated on a denaturing gel. RNA intermediates, and products are schematically represented on the right. Exons are drawn as boxes and introns as lines.
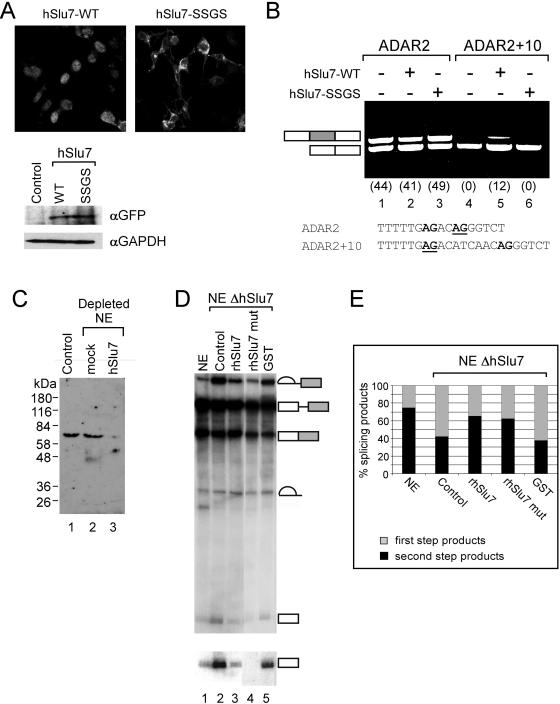
Figure 7.
The zinc-knuckle of hSlu7 is not essential for mRNA splicing. (A) Equal amounts of wild-type and mutant hSlu7 are expressed after transfection. Wild-type and zinc-knuckle mutant plasmids were transfected into 293T cells. After 48 h, GFP fluorescence was observed and total proteins were extracted, separated in SDS-PAGE, transferred to membrane, and probed with anti-GFP and GAPDH. (B) 293T cells were cotransfected with plasmids containing either wild-type hSlu7 (hSlu7-WT) or zinc-knuckle SSGS mutant (hSlu7-SSGS) and the ADAR2 mini-genes (ADAR2 and ADAR2 + 10; Lev-Maor et al., 2003). Sequence of the 3′ ss of the Alu exon is shown in the lower part of the panel, and the selected 3′ ss is underlined (proximal and distal 3′ ss are from left to right, respectively). Forty-eight hours posttransfection, cytoplasmic RNA was collected, and the splicing pattern was examined by RT-PCR by sing specific primers, after separation in agarose gel. A schematic representation of PCR products is indicated to the left of the gel. Alu exon is shaded gray. Numbers in brackets indicate percent of Alu exon inclusion. (C) Western blot analysis of total HeLa nuclear extract (lane 1), mock (lane 2), and hSlu7 depleted (lane 3) with anti-hSlu7. Protein size marker is indicated on the left. (D) In vitro splicing reaction of Adeno pre-mRNA by using mock depleted nuclear extract (NE; lane 1) and HeLa nuclear extract depleted of hSlu7 (presented in C; lanes 2–5). To the depleted extracts rhSlu7 (lane 3; also see Figure 2Ai), mutant rhSlu7 (SSGS mutant; lane 4; also see Figure 2Ai), and GST (lane 5, also see Figure 2Ai) were added as well. Total RNA was extracted and separated in a 10% denaturing gel. RNA intermediates and products are schematically represented on the right. The 5′ exon also is shown in a longer exposure. (E) Quantification of the splicing reaction presented in D. Total amount of splicing products were divided according to first (light shaded gray) or second step (black) splicing products.
The hSlu7 cDNA also was fused downstream to GFP to study the localization of mutant hSlu7. Wild-type hSlu7 and hSlu7-GFP colocalize to the same nuclear region (Figure 2Bvii–ix); however, speckles are not visible in the GFP-tagged protein, and it also is more diffused within the nucleus. To test whether this tag affects the function of the protein and thus impair our experimental results, we added the recombinant hSlu7 (rhSlu7) in excess amounts (200 mol excess) to an in vitro splicing assay. The GST and GFP tags are similar in size (∼27 kDa); therefore, it is possible that their hindrance to the protein, if present, might be similar. However, the large amount of competing rhSlu7 did not have any effect on spliceosomal complexes formation and splicing activity (Figure 2, C and E, respectively). Recombinant hSlu7 is within the spliceosomal complexes B/C as shown by Western blotting with anti-GST on the same membrane shown in Figure 2C (Figure 2D, lanes 7–10; as also shown by Chua and Reed, 1999a). This indicates that rhSlu7, similarly to endogenous hSlu7, joins the splicing reaction during complex B/C assembly (Chua and Reed, 1999a). We conclude that rhSlu7 participates in the splicing reaction.
The Zinc-Knuckle Motif of hSlu7 Binds Zinc
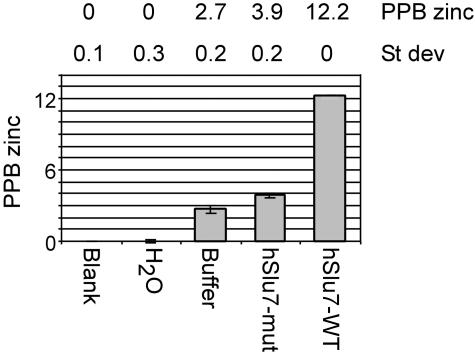
We wanted to determine whether the zinc-knuckle of hSlu7 binds zinc and whether mutations in this motif abrogate zinc binding. For this purpose, we introduced the purified recombinant wild-type hSlu7 (rhSlu7; Figure 2A) and a zinc-knuckle mutant version of the same protein (in which the CCHC amino acids of the zinc-knuckle motif were mutated to SSGS; rhSlu7 mut; Figure 2A) into an ICP-AES and measured the amount of zinc present in each sample (see MATERIALS AND METHODS). The blank and the water samples gave a zero reading. The wild-type recombinant hSlu7 protein occupied 12.2 ppb zinc atoms, whereas the zinc-knuckle mutated recombinant protein gave a reading of 3.9 ppb (Figure 3). This indicates that the zinc-knuckle motif of hSlu7 binds a zinc atom at this region and that this interaction can be disrupted through four amino acid mutations. Further calculations showed that this figure corresponds with the stoichiometric ratio of one zinc atom per one hSlu7 protein (see Supplementary Material for calculations).
Figure 3.
Zinc-knuckle motif of hSlu7 binds zinc. The amount of zinc released from recombinant wild-type hSlu7 (hSlu7-WT) and a zinc-knuckle mutant version of the same protein (hSlu7-mut) was measured using an ICP-AES. The figures are an average of three measurements. H2O, the metal-free water we used for our samples; buffer only, a mock sample with no protein; and hSlu7-WT, purified recombinant wild-type hSlu7; hSlu7-mut, purified recombinant zinc-knuckle mutant hSlu7 (SSGS mutant; see text). SD is in ppb.
Multiple Strong NLS Signals Combine to Direct hSlu7 to the Nucleus
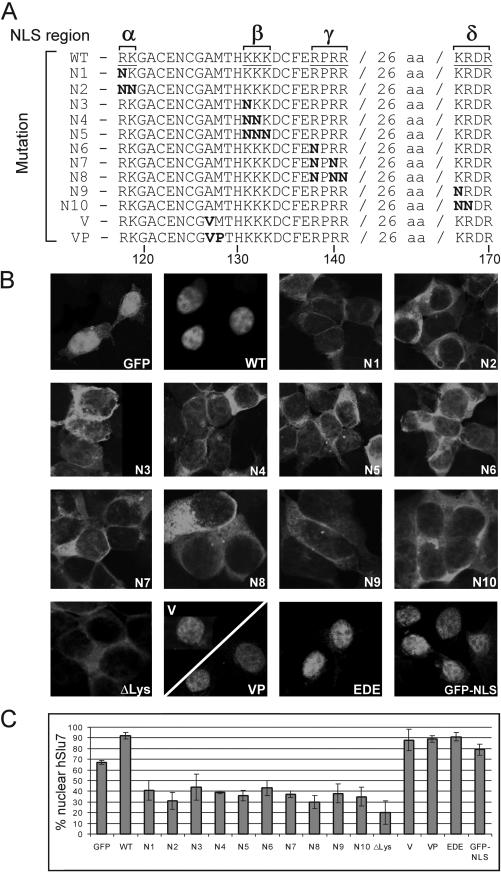
We performed a systematic mutagenesis study at the potential bipartite NLS consensus amino acids, and in the additional conserved sequences downstream of the motif, to determine whether these sequences affect cellular localization. Specifically, the NLS was divided into four regions (see Figure 4A, denoted α, β, γ, and δ), where α and β are the software-identified NLS, and γ and δ are the conserved basic residues downstream of it. Lysine (K) and arginine (R) were mutated to a noncharged amino acid, asparagine (N), to eliminate the positive charge required for NLS function, concomitant with minimizing the disruption to the protein structure (also see Robbins et al., 1991; Ha et al., 2002). Mutated hSlu7 was cloned downstream of the GFP reporter gene; plasmids were transfected into 293T cells; and after 48 h, before fixation, live cells were observed for their hSlu7-GFP mutant localization to confirm that the fixation process did not alter the cellular localization (our unpublished data).
Figure 4.
An extended bipartite nuclear localization signal is required for hSlu7's nuclear import. A systematic mutagenesis study at the evolutionary conserved amino acids of the potential NLS was performed. (A) The NLS was divided into four regions (denoted α, β, γ, and δ from left to right), and mutations were introduced cumulatively in each region from one to two or three amino acids. The conserved basic amino acids lysine (K) and arginine (R) were mutated to the uncharged asparagines (N). Mutated hSlu7 sequences were cloned down-stream of the GFP reporter protein. Numbers at the bottom indicate amino acid position. (B) Wild-type and mutated plasmids were transiently transfected and expressed in 293T cells. The cellular localization was observed after fixation. In other constructs, AM amino acids were mutated to VP (A, alanine; M, methionine; V, valine; P, proline; see A for sequence); the EDE amino acids at position 238–240 (located in the following semiconserved sequence KDHNSEDEDEDKYADDIDM; also see DISCUSSION) were mutated to AAA (E, glutamate; D, aspardic acid; and A, alanine), and the lysine-rich region was deleted (ΔLys; amino acids 498–507). (C) Quantification of the nuclear concentration of the various hSlu7 mutants.
Figure 4B shows the cellular localization of each hSlu7-GFP mutant protein (as outlined in Figure 4A), and Figure 4C quantitatively summarizes the average percentage of hSlu7 protein in the nucleus for each of the mutants (see MATERIALS AND METHODS). Plasmids expressing GFP only localized mainly to the nucleus with some cytoplasmic staining (Figure 4, B and C, GFP), whereas wild-type hSlu7 is exclusively localized to the nucleus (Figure 4, WT). hSlu7-GFP mutants, even with one-point mutation in the α or β NLS regions, alter the nuclear staining of hSlu7, leading to cytoplasmic accumulation, suggesting that the bipartite NLS is active (Figure 4, N1–N5). It also is noticeable that the downstream conserved basic residues (regions γ and δ) serve as functional NLS, because independent mutations at these regions cause hSlu7 to accumulate in the cytoplasm (Figure 4, N6–N10). The lysine-rich deleted hSlu7 (deletion of amino acids 498–507) shows a remarkable change in accumulation of the protein with only 20% average nuclear presence (Figure 4, B and C, ΔLys). Additional mutations between the α and β NLS regions, without disrupting the putative zinc-knuckle conserved amino acids, were introduced and shown to have no effect on the cellular distribution of hSlu7 (Figure 4, V and VP; and EDE, at amino acid position 238–240). We note that the spacer region of the NLS indeed was shown to tolerate point mutations without affecting localization (Robbins et al., 1991; Cokol et al., 2000; Romanelli and Morandi, 2002), which strengthens the functional specificity of the four identified NLS regions. Interestingly, fusing the NLS region only to the GFP protein increased its nuclear localization by 12% (Figure 4, B and C, GFP-NLS), even though GFP is a small enough protein to freely diffuse in and out of the nucleus (Xiao et al., 2003). This same NLS region, fused to a larger cytoplasmic protein, increased its nuclear localization by 80% (see Supplementary Material). These results indicate that hSlu7 possesses at least four functional NLS regions that target the protein to the nucleus and that the lysine-rich domain, at the C terminal of the protein, also plays a role in its nuclear localization.
The Zinc-Knuckle Motif of hSlu7 Controls Its Cellular Localization
Numerous proteins containing zinc-finger or -knuckle motifs are associated with nuclear localization (Yu et al., 1998; Pandya and Townes, 2002), localization to subnuclear speckles (Dye and Patton, 2001), and nuclear import (Braem et al., 2002). None of them, however, overlap with a bipartite NLS region. Given that the NLS region of hSlu7 does harbor a zinc-knuckle motif, we wanted to determine whether these domains cross talk or function independently.
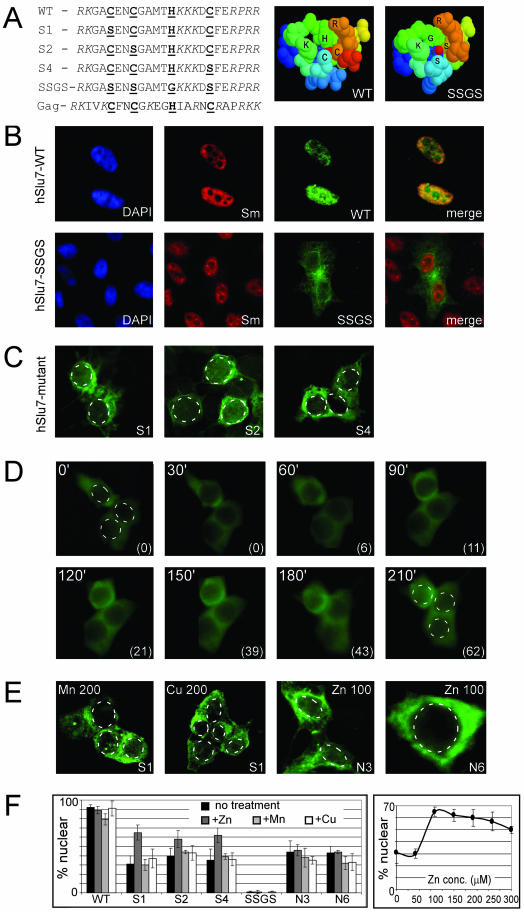
The conserved basic amino acids of the zinc-knuckle, cysteine (C) and histidine (H), were mutated to serine (S) and glycine (G), respectively, to eliminate the ability to bind the zinc atom (see Figure 4A for mutated sequences). We assume that we did not affect the overlapping NLS, because only the basic amino acids (R/K) control the NLS's function (also see Robbins et al., 1991; Cokol et al., 2000; Romanelli and Morandi, 2002). Before mutational studies, and to predict the mutational affect on the three-dimensional structure of hSlu7's zinc-knuckle domain, we performed the following procedure. A profile was built, based on the conserved zinc-knuckle and bipartite NLS domain of hSlu7 (see MATERIALS AND METHODS). The profile was used to identify proteins with similar sequences in the TrEMBL database. Several hundred proteins possess either motif separately. However, none possess both motifs in an overlapping manner as in hSlu7, and only the viral Gag polyprotein contains a zinc-knuckle motif with some basic amino acids on either side similarly to hSlu7 (Figure 5A, Gag). This protein happens to have a solved three-dimensional structure, and, based on its coordinates, Swiss-PDB viewer was used to build the three-dimensional structure of hSlu7's zinc-knuckle domain. This model was mutated to observe the effect on the zinc-knuckle domain as well. Comparison of the wild-type (WT) model to the SSGS mutant revealed that the mutant lost the tight duct around the zinc atom (Figure 5A). The S1, S2, and S4 mutants did not cause a detectable change in the three-dimensional conformation (see Supplementary Material). This modeling software reports possible clash among amino acids after virtual mutagenesis is performed. No such clash was detected in these structures, increasing the fidelity of the predicted model.
Figure 5.
The zinc-knuckle domain controls nucleocytoplasmic shuttling of hSlu7. (A) Sequences: Zinc-knuckle hSlu7 mutants, mild (one amino acid change; S1, S2, and S4) and severe (four amino acid change; SSGS), were cloned downstream of the GFP reporter gene. Zinc-knuckle conserved amino acids are bold and underlined, and NLS motifs are italicized. The Gag sequence represents the zinc-knuckle motif (bold and underlined) of the viral Gag polyprotein sequence with adjacent basic amino acids (italicized). Right, computerized predictive three-dimensional models of the zinc-knuckle and NLS domain of wild-type (WT) and the SSGS mutant (SSGS). The zinc atom is represented by the red dot at the center. (B) Forty-eight hours posttransfection, 293T cells were fixed, and the cellular localization of hSlu7 and Sm proteins was determined by immunofluorescence. Wild-type hSlu7 colocalizes with Sm proteins in the nucleus (hSlu7-WT), whereas the zinc-knuckle SSGS mutants accumulates in the cytoplasm (hSlu7-SSGS). (C) Mild hSlu7 zinc-knuckle mutants (S1, S2 and S4) also gather in the cytoplasm. The nuclear borders are marked by a broken line. See panel F for quantitation. (D) 293T cells were transfected with the hSlu7 S1 mutant plasmid. After 48 h, zinc at 100 μM was added to the cellular medium, and the live cells were immediately taken for fluorescence time-course image analysis. Time in minutes is indicated at the top left corner of each image, and the percentage of hSlu7 inside the nucleus is marked in brackets at the bottom right-hand corner. The nuclear borders are marked by a broken line in the first and last images. (E) Similar to D, except that 293T cells were transfected either with the hSlu7 S1 mutant plasmid or with N3 and N6 hSlu7 NLS mutants (see Figure 3A for sequence). After 48 h, either zinc (Zn) at 100 μM or manganese (Mn) or copper (Cu) at 200 μM was added to the cellular medium. Cells were fixed and visualized as described above. Numbers indicate concentration in micromolar. The nuclear borders are marked by a broken line. (F) Left, quantitation of the results described above. Transfected plasmid types are indicated on the x-axis (see A and Figure 4A for sequence). Bars, from left to right, in decreasing shading intensity represent; no treatment (black), addition of zinc (100 μM; dark gray), manganese (100 μM; light gray), or copper (100 μM; white). Right, dose-dependent entrance of hSlu7 S1 mutant into the nucleus measured at 210 min after addition of zinc.
We then examined the effect of zinc-knuckle mutations on hSlu7 cellular localization. The zinc-knuckle-mutated hSlu7 protein (SSGS; see Figure 5A) localizes to the cytoplasm. In the same cells, the Sm proteins localize to the nucleus, and there is no overlapping yellow color (Figure 5B). Even the mild mutations of hSlu7's zinc-knuckle (S1, S2 and S4; Figure 5A) accumulate in the cytoplasm (Figure 5C and quantitated in 5F). This indicates that the zinc-knuckle motif, which is embedded within the NLS, is also required for the nuclear localization of hSlu7.
To discover whether the zinc-knuckle motif determines the cellular localization through the disruption of zinc binding to the zinc-knuckle, we tried to reverse the effect by adding zinc into the medium of the growing cells. Indeed, zinc added at ≥100 μM to the cellular medium partially restored nuclear staining of hSlu7 to S1, S2, and S4 mutants, but not to the SSGS mutant. Figure 5D shows real-time images of the hSlu7 S1 mutant protein partially entering the nucleus following addition of 100 μM zinc into the cellular medium. This effect was specific to zinc and not to other divalent metal ions, even at 200 μM concentration (Figure 5, E and F; our unpublished data). We believe that the addition of zinc did not permeabilize the nuclear membrane, thus allowing free diffusion of hSlu7 into the nucleus, because mutant hSlu7's cytoplasmic concentration was not altered by this treatment. Imbalances in intracellular zinc pools lead to improper regulation of cell death and proliferation. However, the concentrations affecting mutant hSlu7 were below the reported concentration that induces apoptosis (Watjen et al., 2002). This implies that the binding of zinc to the zinc-knuckle was involved in the localization of the protein to the nucleus, either directly or indirectly, by exerting some effect on the NLS, possibly via a structural change. To test whether the addition of zinc triggered selective gene expression (O'Halloran, 1993) and thus affected cellular localization, we incubated the transfected cells with zinc and a transcription inhibitor (actinomycin-D). This process did not alter the cellular localization of hSlu7, suggesting that the localization is independent of inhibitor of transcription (our unpublished data).
Nuclear Export Is Mediated through the CRM1 Pathway
LMB is a specific inhibitor of CRM-1-mediated nuclear export (CRM1; Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Kudo et al., 1999), a process that has been shown to be important for the function of several proteins, including transcription factors (Komeili and O'Shea, 2000). The presence of several putative NES (Figure 1) on hSlu7 suggested that, in addition to being transported into the nucleus, hSlu7 may be exported out of the nucleus. To test this hypothesis, cells were transfected with hSlu7, wild-type, zinc-knuckle mutants and NLS mutants, and then subjected to LMB at various concentrations for 3 h. After fixation and staining, the cellular localization of the various proteins was observed. We found that all hSlu7 zinc-knuckle-containing mutations, including the SSGS mutant, localized to the nucleus after treatment of cells with LMB, even at a concentration as low as 2–5 ng/ml (Figure 6A; our unpublished data). However, a similar low dose LMB causes the NLS mutations to remain in the cytoplasm (see Supplementary Material; our unpublished data). This indicates that the NLS of hSlu7, but not the zinc-knuckle domain, is a bona fide motif directing the protein into the nucleus.
Figure 6.
A functional zinc-knuckle is required to lock hSlu7 in the nucleus. (A) LMB, an inhibitor of the CRM1 NES pathway, maintains zinc-knuckle mutated hSlu7 in the nucleus. Forty-eight hours after plasmid transfection and 3 h after addition of LMB (5 ng/ml), 293T cells were fixed and stained with anti-hSlu7. Transfection was carried out with wild-type hSlu7 (WT) and SSGS mutant hSlu7 (SSGS). (B) Zinc-knuckle mutants are probably under a constant nucleocytoplasmic shuttling balance. 293T cells were transfected with the SSGS mutant hSlu7 plasmid. After 48 h, LMB (5 ng/ml) was added, and the slide containing the live cells was immediately taken for time-course image analysis. Top left image is a transmission image of the cells, outlined in white. The other images are fluorescent images of the hSlu7 mutant taken over time (in minutes; top left corner of each image) and the percentage of hSlu7 inside the nucleus (in brackets, bottom right corner of each image).
In light of the findings, we had two opposing lines of results. The first is that SSGS mutants are exclusively cytoplasmic, and the second is that LMB, a CRM1 nuclear export inhibitor, traps the mutant protein in the nucleus. If hSlu7-SSGS cytoplasmic staining is caused by its inability to enter the nucleus due to its defective sequences, we could not explain how, after LMB treatment, the SSGS mutant is trapped within the nucleus. To test whether the zinc-knuckle–mutated proteins can enter the nucleus and to ask whether the SSGS mutant is still under nucleocytoplasmic shuttling that favors the cytoplasm, we performed a time-course analysis of live cells treated with LMB and transfected with the SSGS mutant hSlu7 plasmid. Without LMB incubation, the hSlu7 mutant showed exclusive cytoplasmic localization (Figure 6B, marked as 0 time). After 60 min of LMB addition, the mutant protein began accumulating in the nucleus, reaching its maximum level after 180 min (Figure 6B, 180). This suggests that hSlu7 shuttles between the nucleus and cytoplasm by using its zinc-knuckle in addition to its NLS. A functional zinc-knuckle is not required for hSlu7's entrance into the nucleus but rather to maintain hSlu7 inside it, preventing its shuttle back to the cytoplasm via the CRM1 pathway. Thus, the zinc-knuckle motif of hSlu7 determines the cellular localization of the protein through a nucleocytoplasmic sensitive shuttling balance.
The Functional Role of the hSlu7 Zinc-Knuckle in mRNA Splicing
To test whether the cellular localization of hSlu7 has a functional significance in mRNA splicing, we used two mini-genes containing the genomic sequence of adenosine deaminase gene ADAR2 from exon 7–9, in which exon 8 is an alternatively spliced Alu exon (Lev-Maor et al., 2003). We have shown that increasing the nuclear concentration of wild-type hSlu7 in vivo selects a proximal 3′ ss AG only when the distance between the proximal and distal 3′ ss exceeds 10 nucleotides (ADAR2 + 10 mini-gene; see lower part of Figure 7B for the mini-gene sequences at the 3′ ss of the Alu exon). We reasoned that if the hSlu7 zinc-knuckle mutant is localized to the cytoplasm, it will exert no effect on 3′ ss selection compared with wild-type hSlu7. Thus, we cotransfected 293T cells with plasmids containing either wild-type hSlu7 or zinc-knuckle SSGS mutant and the ADAR2 mini-genes. Forty-eight hours posttransfection, cells were observed to confirm equal transfection amounts (Figure 7A, top) and expression levels (Figure 7A, bottom) of the hSlu7 proteins, cytoplasmic RNA was collected, and the splicing pattern was examined by RT-PCR by using specific primers, after separation in agarose gel (Figure 7B). An increased amount of wild-type hSlu7 (hSlu7-WT; Figure 7B) does not change 3′ ss selection of the alternatively spliced Alu exon, but brings about increased selection of the ADAR2 + 10 internal exon (12%) and a shift in the 3′ ss selection from the distal to the proximal site. Interestingly, an increased quantity of mutant zinc-knuckle hSlu7 (hSlu7-SSGS, which was shown to localize to the cytoplasm in Figure 5B) does not cause selection of the Alu exon. This indicates that mutant hSlu7, which localizes to the cytoplasm, does not affect the 3′ ss selection and raises the possibility that nuclear concentrations of hSlu7, controlled by its binding to zinc, might be responsible for 3′ ss regulation. The splicing pattern of adenovirus E1A gene shows no effect after transfection of hSlu7 (our unpublished data). This was used as a control in order to ensure that the hSlu7 effect is specific to 3′ ss exon selection, because this gene uses three alternative 5′ ss spliced to a single 3′ ss.
We wanted to test whether mutant zinc-knuckle hSlu7 is active in mRNA splicing, therefore we depleted HeLa nuclear extract of hSlu7, added back rhSlu7 and mutant rhSlu7 (SSGS; also see Figure 2Ai) and then tested their ability to restore splicing in vitro. Nondepleted nuclear extract shows accumulation of second step splicing products as compared with first step splicing products (75 and 25%, respectively, out of the total amount of splicing products; Figure 7D, lane 1, and E, NE). However, nuclear extract depleted of hSlu7 shows accumulation of first step splicing products (58% of total products; Figure 7D, lane 2, and E, Control). As reported by Chua and Reed (1999a), a total inhibition of the second step of splicing by using in vitro depletion could not be attained. Addition of both recombinant wild-type and mutant hSlu7 to the depleted extract increase the amount of step two splicing that occurs to almost the same level as the nondepleted extract (33 and 37%, respectively; Figure 7D, compare lanes 1–3 and 4; E, compare NE to rhSlu7 and rhSlu7 mut). As a control, a GST protein was added to the hSlu7-depelted nuclear extract, showing no effect on the accumulation of first step splicing products (62%; Figure 7D, compare lanes 2 and 5; E, compare control and GST). Thus, we conclude that hSlu7 with a nonfunctional zinc-knuckle motif is active in mRNA splicing in vitro [similarly to the conserved RNA recognition motif 3 of U2AF(65), which is essential in vivo but dispensable for activity in vitro; Banerjee et al., 2004], although in the cell it localizes to the cytoplasm.
DISCUSSION
We identified three domains of the hSlu7 protein that have distinct roles in its subcellular localization—a NLS, a zinc-knuckle, and a lysine-rich region—and also nuclear export activity that was identified through the use of a specific inhibitor (LMB). The zinc-knuckle and NLS regions overlap in a unique organization that determines hSlu7-subcellular localization. We also showed that hSlu7 nucleocytoplasmic shuttling is dependent on hSlu7 binding to zinc. Namely, the entrance from the cytoplasm to the nucleus is independent of a functional hSlu7 zinc-knuckle domain, but to maintain the protein inside the nucleus and to prevent its shuttling back to the cytoplasm (via the CRM1 pathway), a functional zinc-knuckle domain is required.
One method to regulate a protein's function in vivo is to control the level of transcription or translation of alternatively spliced regulatory proteins. This type of regulation was shown for SF2/ASF splicing protein (Hanamura et al., 1998). In addition, the long half-lives of many splicing factors might argue against the involvement of this type of mechanism in the short-term regulation of these protein levels. A more subtle regulation controls the subcellular localization when a given protein is shuttled out of the nucleus where splicing takes place, thus reducing the nuclear concentration of that protein and affecting splicing or alternative-splicing profiles of genes (such as for polypyrimidine tract-binding protein, Xie et al., 2003; or hnRNP A1; van der Houven van Oordt et al., 2000). Such finer regulation also was observed for hSlu7, in which the nucleocytoplasmic shuttling is dependent on binding to zinc, which controls its cytoplasmic accumulation.
Intracellular trafficking of metal ions within cells in a chaperone-like manner is regulated by a group of proteins termed metallochaperones. In prokaryotes, copper, for example, was shown to stimulate a transcription activator at a sensitivity that is less than one atom per cell (Changela et al., 2003). Although no eukaryotic chaperones for metals other than copper have been identified to date, it is likely that analogous cofactor trafficking pathways exist for other proteins and metals (for review, see O'Halloran and Culotta, 2000; Rosenzweig, 2002; Burdette and Lippard, 2003), possibly regulating hSlu7 binding to zinc.
We showed that hSlu7's cellular localization is controlled by three motifs: NLS, zinc-knuckle, and a lysine-rich region. As mentioned above, the NLS and zinc-knuckle motifs overlap. In a survey of all proteins in the TrEMBL database, we found no protein with a similar motif, and only the viral Gag polyprotein possesses some overlapping amino acids (Figure 5A, Gag; its basic amino acids were associated with nuclear localization and membrane binding; Zhou et al., 1994; Parent et al., 1996; Scarlata and Carter, 2003; also see Yu et al., 1998 for some similarity to motifs in the orphan nuclear receptor TR2 protein). The unique functional zinc-knuckle domain embedded within the NLS is required to “lock” hSlu7 inside the nucleus. We assume that zinc binding to the zinc-knuckle induces a conformational change which is required for this nuclear retention. When the zinc atom is removed, presumably as occurs in response to changing physiological conditions (unpublished data), hSlu7 is partially translocated from the nucleus to the cytoplasm. This might be a fine-tuned regulation that controls the nuclear concentration of hSlu7.
We also believe that the shuttling kinetics is unlikely to be caused by protein behavior that may have been altered by GFP tagging or overexpression from transient transfection. The wild-type hSlu7-GFP fusion protein localizes to similar sub-nuclear compartments as the endogenous counterparts (Figure 2B; also see Figure 5B, WT; although no speckles are visible in the GFP-tagged protein and it is also more diffused within the nucleus) and tagged proteins were shown to be active in mRNA splicing in vitro (Figure 2 and 7; although it is possible that the GST and GFP hindrance to the protein is different). In addition, cells expressing the lowest detectable levels of transfected hSlu7 showed cellular staining similar to that of high level transfected hSlu7-expressing cells. Thus, the differences in cellular localizations of the mutant hSlu7 are likely to reflect the endogenous hSlu7 behavior of proteins as well.
An intriguing question raised is what happens to the spliceosome when hSlu7 is shuttled out of the nucleus and thus hSlu7 concentrations are reduced. The cells that possess lower nuclear levels of hSlu7 probably form an hSlu7-lacking spliceosome. Many comprehensive spliceosomal proteomic studies were performed to identify components involved in the splicing reaction (for review, see Jurica and Moore, 2003). Although each individual experiment is limited by its purification method and to the stage of spliceosome assembly, only five of these studies, out of more than a dozen, identified hSlu7/Slu7 as an integral component of the spliceosome (Jurica and Moore, 2003). Chua and Reed (1999a,b) observed, in vitro, a novel functional spliceosome lacking hSlu7. They showed that this complex contained all snRNPs and indicated that, in the absence of hSlu7, the 5′ exon is held loosely within the spliceosome. A possible redundancy in function of hSlu7 with another protein may explain how the spliceosome can function in vivo in the absence of hSlu7. The most probable candidate is hPrp18 (another second-step splicing protein known to interact with Slu7 in yeast), because, in yeast, these proteins have shown some redundant function in vitro (Zhang and Schwer, 1997).
It is possible that a protein that binds the zinc-knuckle domain of hSlu7 somehow facilitates the use of the NLS or occludes the use of the putative NES. The most probable candidate, again, is hPrp18 because yeast Prp18 was shown to interact with Slu7 (Zhang and Schwer, 1997; James et al., 2002). However, immunoprecipitation by using anti-hSlu7 antibodies on total cell extract did not detect any binding of hPrp18 to wild-type or mutated hSlu7 (even at mild conditions used; our unpublished data). The other possible interaction domain (amino acids EDE, at position 238–240, which is in the same region that abrogated Slu7 binding to Prp18 in yeast; James et al., 2002), had no effect on hSlu7 cellular localization (Figure 4, B and C). We cannot exclude that other proteins bind to hSlu7.
Before this study, comprehensive research was performed on hSlu7 to elucidate its function during the second step and its 3′ ss selection mechanism, in vitro (Chua and Reed, 1999a,b). Here, we have elucidated the domains affecting the cellular localization of hSlu7 in vivo, in particular its zinc-dependence maintenance in the nucleus which presents a new feature of regulation, generally, and a specific regulation of the hSlu7 splicing factor.
Acknowledgments
We thank Joel A. Hirsch and Yarden Opatowsky for guidance through recombinant hSlu7 purification techniques and Gabriel Rosenblum and Vasili Rosen for ICP assistance. We thank the Ast group members and the teams working at the laboratories of Yossi Shiloh, Karen B. Avraham, and Shimon Efrat for helpful discussions. We appreciate the work carried out by the rotation students in our laboratory: Aviv Shaul, Kfir Molakandov, Nurit Asia, and Vicky Meltser. We are grateful for Leeat Anker's comments on the manuscript. This work was supported by a grant from the Israel Science Foundation and, in part, by a grant from the Israel Cancer Association, MOP India-Israel, FD Hope, and the chief scientist of Israel Health Ministry to G.A.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-02-0152. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-02-0152.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Ast, G., and Weiner, A.M. (1986). A U1/U4/U5 snRNP complex induced by a 2′-O-methyl-oligoribonucleotide complementary to U5 snRNA. Science. 272, 881-884. [DOI] [PubMed] [Google Scholar]
- Banerjee, H., Rahn, A., Gawande, B., Guth, S., Valcarcel, J., and Singh, R. (2004). The conserved RNA recognition motif 3 of U2 snRNA auxiliary factor (U2AF 65) in essential in vivo but dispensable for activity in vitro. RNA 10, 240-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas, T. (1993). Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. 3, 193-227. [PubMed] [Google Scholar]
- Braem, C.V., Kas, K., Meyen, E., Debiec-Rychter, M., Van De Ven, W.J., and Voz, M.L. (2002). Identification of a karyopherin alpha 2 recognition site in PLAG1, which functions as a nuclear localization signal. J. Biol. Chem. 277, 19673-19678. [DOI] [PubMed] [Google Scholar]
- Brow, D.A. (2002). Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36, 333-360. [DOI] [PubMed] [Google Scholar]
- Brown, V., Krynetski, E., Krynetskaia, N., Grieger, D., Mukatira, S., Murti, K., Slaughter, C., Park, H.W., and Evans, W. (2003). A novel CRM1-mediated nuclear export signal governs nuclear accumulation of GAPDH following genotoxic stress. J. Biol. Chem. 279, 5984-5992. [DOI] [PubMed] [Google Scholar]
- Burdette, S.C., and Lippard, S.J. (2003). Meeting of the minds: metalloneurochemistry. Proc. Natl. Acad. Sci. USA 100, 3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. (1999). Splicing of precursors to mRNA by the spliceosome. In: The RNA World, ed. R.F. Gesteland and J.F. Atkins, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 525-560.
- Caceres, J.F., Misteli, T., Screaton, G.R., Spector, D.L., and Krainer, A.R. (1997). Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138, 225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraburtty, K. (1999). Functional interaction of yeast elongation factor 3 with yeast ribosomes. Int. J. Biochem. Cell Biol. 31, 163-173. [DOI] [PubMed] [Google Scholar]
- Changela, A., Chen, K., Xue, Y., Holschen, J., Outten, C.E., O'Halloran, T.V., and Mondragon, A. (2003). Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383-1387. [DOI] [PubMed] [Google Scholar]
- Chua, K., and Reed, R. (1999a). Human step II splicing factor hSlu7 functions in restructuring the spliceosome between the catalytic steps of splicing. Genes Dev. 13, 841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, K., and Reed, R. (1999b). The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature 402, 207-210. [DOI] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1, 411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S., and Maitra, U. (2001). Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog. Nucleic Acid Res. Mol. Biol. 70, 207-231. [DOI] [PubMed] [Google Scholar]
- Dye, B.T., and Patton, J.G. (2001). An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to sub-nuclear speckles. Exp. Cell Res. 263, 131-144. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., and Aebi, U. (2003). The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell. Biol. 4, 757-766. [DOI] [PubMed] [Google Scholar]
- Frank, D., and Guthrie, C. (1992). An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 6, 2112-2124. [DOI] [PubMed] [Google Scholar]
- Frank, D., Patterson, B., and Guthrie, C. (1992). Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol. Cell. Biol. 12, 5197-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier, M., Moulinier, L., and Giege, R. (2000). A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J. 19, 2371-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M., and Nishida, E. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308-311. [DOI] [PubMed] [Google Scholar]
- Ha, S., Park, S., Yun, C.H., and Choi, Y. (2002). Characterization of nuclear localization signal in mouse ING1 homolog protein. Biochem. Biophys. Res. Commun. 293, 163-166. [DOI] [PubMed] [Google Scholar]
- Hanamura, A., Caceres, J.F., Mayeda, A., Franza, B.R., Jr., and Krainer, A.R. (1998). Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4, 430-444. [PMC free article] [PubMed] [Google Scholar]
- Harris, E.D. (2002). Cellular transporters for zinc. Nutr. Rev. 60, 121-124. [DOI] [PubMed] [Google Scholar]
- Hastings, M.L., and Krainer, A.R. (2001). Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13, 302-309. [DOI] [PubMed] [Google Scholar]
- Henderson, B.R., and Eleftheriou, A. (2000). A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256, 213-224. [DOI] [PubMed] [Google Scholar]
- James, S.A., Turner, W., and Schwer, B. (2002). How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8, 1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, K.B., Dredge, B.K., Stefani, G., Zhong, R., Buckanovich, R.J., Okano, H.J., Yang, Y.Y., and Darnell, R.B. (2000). Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359-371. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Horowitz, D.S., and Xu, R.M. (2000). Crystal structure of the functional domain of the splicing factor Prp18. Proc. Natl. Acad. Sci. USA 97, 3022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.H., Frank, D.N., and Guthrie, C. (1995). Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl. Acad. Sci. USA 92, 9687-9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S., and Moore, M.J. (2003). Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12, 5-14. [DOI] [PubMed] [Google Scholar]
- Kitagawa, H., Hotta, Y., Fujiki, K., and Kanai, A. (1996). Cloning and high expression of rabbit FKBP25 in cornea. Jpn. J. Ophthalmol. 40, 133-141. [PubMed] [Google Scholar]
- Komeili, A., and O'Shea, E.K. (2000). Nuclear transport and transcription. Curr. Opin. Cell Biol. 12, 355-360. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M., and Sharp, P.A. (1986). Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46, 845-855. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Matsumori, N., Taoka, H., Fujiwara, D., Schreiner, E.P., Wolff, B., Yoshida, M., and Horinouchi, S. (1999). Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96, 9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena, M.J., Chalmers, K.J., Llamazares, S., Lamond, A.I., and Valcarcel, J. (2002). Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109, 285-296. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and Spector, D.L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4, 605-612. [DOI] [PubMed] [Google Scholar]
- Leary, D.J., Terns, M.P., and Huang, S. (2003). Components of U3 snoRNA containing complexes shuttle between nuclei and the cytoplasm and differentially localize in nucleoli: implications for assembly and function. Mol. Biol. Cell 15, 281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Maor, G., Sorek, R., Shomron, N., and Ast, G. (2003). The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300, 1288-1291. [DOI] [PubMed] [Google Scholar]
- Liu, X., Jiang, Q., Mansfield, S.G., Puttaraju, M., Zhang, Y., Zhou, W., Cohn, J.A., Garcia-Blanco, M.A., Mitchell, L.G., and Engelhardt, J.F. (2002). Partial correction of endogenous DeltaF508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat. Biotechnol. 20, 47-52.11753361 [Google Scholar]
- Lopato, S., Gattoni, R., Fabini, G., Stevenin, J., and Barta, A. (1999). A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol. Biol. 39, 761-773. [DOI] [PubMed] [Google Scholar]
- Mahe, D., Fischer, N., Decimo, D., and Fuchs, J.P. (2000). Spatiotemporal regulation of hnRNP M and 2H9 gene expression during mouse embryonic development. Biochim. Biophys. Acta 1492, 414-424. [DOI] [PubMed] [Google Scholar]
- Meshorer, E., Erb, C., Gazit, R., Pavlovsky, L., Kaufer, D., Friedman, A., Glick, D., Ben-Arie, N., and Soreq, H. (2002). Alternative splicing and neuritic mRNA translocation under long-term neuronal hypersensitivity. Science 295, 508-512. [DOI] [PubMed] [Google Scholar]
- Mintz, P.J., and Spector, D.L. (2000). Compartmentalization of RNA processing factors within nuclear speckles. J. Struct. Biol. 129, 241-251. [DOI] [PubMed] [Google Scholar]
- Modrek, B., and Lee, C. (2002). A genomic view of alternative splicing. Nat. Genet. 30, 13-19.11753382 [Google Scholar]
- O'Halloran, T.V. (1993). Transition metals in control of gene expression. Science 261, 715-725. [DOI] [PubMed] [Google Scholar]
- O'Halloran, T.V., and Culotta, V.C. (2000). Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 275, 25057-25060. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari, B., Bachelerie, F., and Dargemont, C. (1997). Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278, 141-144. [DOI] [PubMed] [Google Scholar]
- Pandya, K., and Townes, T.M. (2002). Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J. Biol. Chem. 277, 16304-16312. [DOI] [PubMed] [Google Scholar]
- Parent, L.J., Wilson, C.B., Resh, M.D., and Wills, J.W. (1996). Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J. Virol. 70, 1016-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, J., Dilworth, S.M., Laskey, R.A., and Dingwall, C. (1991). Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64, 615-623. [DOI] [PubMed] [Google Scholar]
- Romanelli, M.G., and Morandi, C. (2002). Importin alpha binds to an unusual bipartite nuclear localization signal in the heterogeneous ribonucleoprotein type I. Eur. J. Biochem. 269, 2727-2734. [DOI] [PubMed] [Google Scholar]
- Rosenzweig, A.C. (2002). Metallochaperones: bind and deliver. Chem. Biol. 9, 673-677. [DOI] [PubMed] [Google Scholar]
- Scarlata, S., and Carter, C. (2003). Role of HIV-1 Gag domains in viral assembly. Biochim. Biophys. Acta 1614, 62-72. [DOI] [PubMed] [Google Scholar]
- Shomron, N., and Ast, G. (2003). Boric acid reversibly inhibits the second step of pre-mRNA splicing. FEBS Lett. 552, 219-224. [DOI] [PubMed] [Google Scholar]
- Shomron, N., Malca, H., Vig, I., and Ast, G. (2002). Reversible inhibition of the second step of splicing suggests a possible role of zinc in the second step of splicing. Nucleic Acids Res. 30, 4127-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.W., and Valcarcel, J. (2000). Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25, 381-388. [DOI] [PubMed] [Google Scholar]
- Stoilov, P., Meshorer, E., Gencheva, M., Glick, D., Soreq, H., and Stamm, S. (2002). Defects in pre-mRNA processing as causes of and predisposition to diseases. DNA Cell Biol. 21, 803-818. [DOI] [PubMed] [Google Scholar]
- Stros, M. (2001). Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry 40, 4769-4779. [DOI] [PubMed] [Google Scholar]
- Umen, J.G., and Guthrie, C. (1995). The second catalytic step of pre-mRNA splicing. RNA 1, 869-885. [PMC free article] [PubMed] [Google Scholar]
- van der Houven van Oordt, W., Diaz-Meco, M.T., Lozano, J., Krainer, A.R., Moscat, J., and Caceres, J.F. (2000). The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 149, 307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., and Manley, J.L. (1995). Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA 1, 335-346. [PMC free article] [PubMed] [Google Scholar]
- Watjen, W., Haase, H., Biagioli, M., and Beyersmann, D. (2002). Induction of apoptosis in mammalian cells by cadmium and zinc. Environ Health Perspect. 110, 865-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, K.M., and Crothers, D.M. (1992). RNA binding assays for Tat-derived peptides: implications for specificity. Biochemistry 31, 10281-10287. [DOI] [PubMed] [Google Scholar]
- Xiao, Z., Latek, R., and Lodish, H.F. (2003). An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene 22, 1057-1069. [DOI] [PubMed] [Google Scholar]
- Xie, J., Lee, J.A., Kress, T.L., Mowry, K.L., and Black, D.L. (2003). Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 100, 8776-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z., Lee, C.H., Chinpaisal, C., and Wei, L.N. (1998). A constitutive nuclear localization signal from the second zinc-finger of orphan nuclear receptor TR2. J. Endocrinol. 159, 53-60. [DOI] [PubMed] [Google Scholar]
- Zhang, X., and Schwer, B. (1997). Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 25, 2146-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., Parent, L.J., Wills, J.W., and Resh, M.D. (1994). Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68, 2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]