Abstract
A variety of proteins involved in gene expression have been localized within mammalian cell nuclei in a speckled distribution that predominantly corresponds to interchromatin granule clusters (IGCs). We have applied a mass spectrometry strategy to identify the protein composition of this nuclear organelle purified from mouse liver nuclei. Using this approach, we have identified 146 proteins, many of which had already been shown to be localized to IGCs, or their functions are common to other already identified IGC proteins. In addition, we identified 32 proteins for which only sequence information is available and thus these represent novel IGC protein candidates. We find that 54% of the identified IGC proteins have known functions in pre-mRNA splicing. In combination with proteins involved in other steps of pre-mRNA processing, 81% of the identified IGC proteins are associated with RNA metabolism. In addition, proteins involved in transcription, as well as several other cellular functions, have been identified in the IGC fraction. However, the predominance of pre-mRNA processing factors supports the proposed role of IGCs as assembly, modification, and/or storage sites for proteins involved in pre-mRNA processing.
INTRODUCTION
Interphase mammalian nuclei are compartmentalized into a large number of structures or organelles that are likely to contribute to the fidelity and efficiency of the many functions that occur within this compartment, including transcription, pre-mRNA processing, DNA replication, DNA repair/recombination, assembly of ribosomal subunits, and nucleocytoplasmic protein/ribonucleoprotein (RNP) trafficking (for a review, see Spector, 1993; Lamond and Earnshaw, 1998; Misteli, 2000). Although some nuclear functions can be reproduced in in vitro systems (i.e., transcription and pre-mRNA splicing), these systems may be less efficient than their in vivo counterparts (Corden and Patturajan, 1997). Therefore, in vivo spatial and temporal coordination may have a significant influence on gene expression and other nuclear processes. Among those nuclear organelles thus far identified in normal and cancer cells (for a review, see Spector, 2001) are interchromatin granule clusters (IGCs), perichromatin fibrils, nucleoli, paraspeckles, perinucleolar compartment, Cajal bodies, gemini of Cajal bodies, and promyelocytic leukemia nuclear bodies. Several of these organelles have been shown to have a relationship to various disease states, including cancer and spinal muscular atrophy (Spector et al., 1992; Matera, 1999; Huang, 2000). Recently, several nuclear structures, including the nuclear pore complex (Rout et al., 2000; Cronshaw et al., 2002), nuclear envelope (Schirmer et al., 2003), and nucleoli (Andersen et al., 2002; Scherl et al., 2002) have been isolated, and their protein composition was characterized by mass spectrometry analysis. In addition, in vitro-assembled spliceosomes, the U1 small nuclear ribonucleoprotein particle (snRNP), and the U4/U6.U5 tri-snRNP have been analyzed using this approach (Neubauer et al., 1997, 1998; Gottschalk et al., 1999; Rappsilber et al., 2002; Zhou et al., 2002). Analysis of the yeast nuclear pore complex (NPC) identified 174 proteins in total of which 40 were found to be associated with the NPC in the form of nucleoporins (29 proteins) or transport factors (11 proteins) (Rout et al., 2000). In the case of the NPC from rat liver nuclei, 94 proteins in total were identified, 29 of which were classified as nucleoporins and 18 were classified as NPC-associated proteins (Cronshaw et al., 2002). By using a subtractive proteomics approach to analyze a mouse nuclear envelope fraction, 13 known nuclear envelope integral proteins were identified as well as 67 uncharacterized open reading frames with predicted membrane spanning regions (Schirmer et al., 2003). Proteomic analysis of human nucleoli has identified 271 (Andersen et al., 2002) to ∼350 (Scherl et al., 2002) proteins, 30% of which are encoded by novel human genes (Andersen et al., 2002). Analysis of in vitro assembled spliceosomes has identified 145 (Zhou et al., 2002) or 311 proteins (Rappsilber et al., 2002).
One of the most intensely studied nuclear substructures, the IGCs, are thought to play a role in efficiently coupling transcription and pre-mRNA splicing in nuclei (for a review, see Lamond and Spector, 2003). IGCs measure ∼1.0–1.5 μm along their widest length and are composed of clusters of 20- to 25-nm granules that often seem to be connected by short fibers (for a review, see Fakan and Puvion, 1980). The IGCs were initially shown to contain a subset of pre-mRNA splicing factors by immunofluorescence and immunoelectron microscopy (for a review, see Spector, 1993). More recent studies have shown that the IGCs are enriched in a number of pre-mRNA splicing factors and the large subunit of RNA polymerase II (Bregman et al., 1995; Mortillaro et al., 1996), however, transcription and pre-mRNA splicing do not generally seem to take place within these nuclear regions (Cmarko et al., 1999; Misteli and Spector, 1999). Instead, splicing factor assembly, modification and/or storage are thought to occur within these nuclear compartments (for a review, see Misteli and Spector, 1998; Lamond and Spector, 2003). IGCs are dynamic nuclear structures from which splicing factors have been shown to be recruited to sites of active transcription in living cells (Misteli et al., 1997; Janicki et al., 2004). Studies using fluorescence recovery after photobleaching have shown that there is a continuous flux of proteins between the IGCs and the nucleoplasm (Kruhlak et al., 2000; Phair and Misteli, 2000). However, it is unclear whether the IGC proteins move as monomers, small complexes, or as a large complex such as individual 20- to 25-nm granules to sites of transcription. In addition, the specific composition of individual interchromatin granules remains to be determined.
We have previously established a protocol to biochemically isolate IGCs from mouse liver nuclei (Mintz et al., 1999) and in our initial characterization of this fraction by mass spectrometry, we identified 33 protein constituents of IGCs. Here, we have extended these studies to saturation and have identified 146 IGC proteins as well as 32 novel protein candidates. We have characterized the 146 proteins based upon their motifs and localization. Our analysis has identified 31 RS domain-containing proteins as well as proteins involved in other aspects of mRNA metabolism. Interestingly, we have found a significant overlap (63%) between our analysis and the recently reported analyses of the protein composition of spliceosomes (Neubauer et al., 1998; Rappsilber et al., 2002; Zhou et al., 2002). Our findings support a proposed role of IGCs in the assembly, modification, and/or storage of proteins involved in pre-mRNA processing.
MATERIALS AND METHODS
IGC Purification and Mass Spectrometry Analysis
Approximately 3 mg of IGCs was purified from 120 5- to 6-wk-old female Swiss Webster mice (27–30 g) according to a procedure described previously (Mintz et al., 1999). The purified IGC fraction was directly dissolved in 2 M urea-phosphate-buffered saline-0.1 mM EDTA, allowing us to recover IGC proteins with high efficiency, rather than our previous approach, whereby we resuspended proteins in TM5 (0.25 M sucrose, 10 mM Tris-HCl, pH 7.4, 5 mM MgCl2). In addition, in the present study we started with 6 times the number of mice relative to our previous report, which yielded ∼10 times more IGC proteins based on measurement of protein concentrations by mass spectrometry analysis. One-third of the dissolved IGC proteins were biotinylated at Cys residues with the chemical cross-linker Biotin-HPDP followed by trypsin digestion, whereas the remaining two-thirds of the IGC proteins were directly digested with trypsin. Cys-containing peptides were selected through avidin chromatography to reduce the complexity of the peptide mixture, thus increasing the chances of detecting low abundant peptides with Cys residues that are normally masked by abundant peptides (Spahr et al., 2000). The selected Cys-containing peptides, as well as a mixture of trypsin-digested peptides without Cys selection, were analyzed by liquid chromatography and tandem mass spectrometry (MS/MS). Fragment ion spectra were batch searched against nonredundant protein sequences in databases. Resulting peptide matches were manually evaluated and confirmed. Motif analysis of each identified protein was performed using SMART (http://smart.embl-heidelberg.de/) (Schultz et al., 1998; Letunic et al., 2002). Database for Tables 1, 2, 3, 4 is available at http://spectorlab.cshl.edu.
Table 1.
Identified IGC proteins
| Protein Description | Accession Code | Chromosomal Locus | RNA Binding Motif | RS | Speckle Localization | Reference | Other Domains and Motif(s)a | Low Complexity Region |
|---|---|---|---|---|---|---|---|---|
| Pre-mRNA splicing | ||||||||
| 30 kDa splicing factor | AAC64086 | 10q23 | TUDOR, coiled coil | Yes | ||||
| 45 kDa splicing factor | AAC64085 | 10p15.1 | RRM | Coiled coil, G_patch | Yes | |||
| cdc5-related protein (KIAA0432) | BAA24862 | 6p21 | Yes (IF) | Burns CG., 1999 | SANT domains, coiled coil | Yes | ||
| DEAD/H box polypeptide 15 | O43143 | 4p15.3 | HELICc, HA2 | Yes | ||||
| Formin binding protein (PRP40 homolog) | AAD39463 | 2q24.1 | Signal peptide, WW and FF domain repeats | Yes | ||||
| Heterogeneous ribonucleoprotein A0 | AAA65094 | 5q31 | 2 RRMs | Yes | ||||
| hnRNP A2/B1 | P22626 | 2 RRMs | Yes | |||||
| hnRNP A3 | P51991 | 10q11.21 | 2 RRMs | Yes | ||||
| hnRNP C | A26885 | RRM | Coiled coil | Yes | ||||
| hnRNP C like | A44192 | 2 RRMs | Yes | |||||
| hnRNP C1/C2 | AAD03717 | 14q11.2 | RRM | Coiled coil | Yes | |||
| hnRNP D | BAA09522 | 4q21.1–q21.2 | 2 RRMs | Yes | ||||
| hnRNP E1 | CAA55016 | 2p13–p12 | 3 KHs | |||||
| hnRNP E2 | CAA55015 | 3 KHs | ||||||
| hnRNP F/H | P52597 | 10q11.21–q11.22 | 3 RRMs | Yes (IF) | Matunis et al., 1994 | Yes | ||
| hnRNP H′ | P55795 | Xq22 | 3 RRMs | Yes | ||||
| hnRNP I | P26599 | 4 RRMs | Signal peptide | Yes | ||||
| hnRNP K | Q07244 | 9q21.32–q21.33 | 3 KHs | Yes | ||||
| hnRNP K like/sub2.3 | CAA82631 | 2 KHs | ||||||
| hnRNP L | P14866 | 19q13.2 | 3 RRMs | Yes | ||||
| hnRNP M | P52272 | 7q11 | 3 RRMs | Yes | ||||
| hnRNP U (SAF A) | Q00839 | 1q44 | SAP, SPRY, coiled coil | Yes | ||||
| hnRNP A/B related protein | Q99020 | 5q35.3 | 2 RRMs | Yes | ||||
| hnRNPA1 | P09651 | 2 RRMs | Yes | |||||
| hnRNP G | P38159 | Xq26 | RRM | Yes | ||||
| Homolog of C. elegans smu-1 | NP_060695 | 9p12 | LisH, CTLH, 7 WD | |||||
| 40 repeats | ||||||||
| KH type splicing regulatory factor | AAB53222 | 19p13.3 | 4 KHs | Yes | ||||
| nhp2/rs6 family protein | P55770 | Yes | ||||||
| Nuclear matrix protein 55 | AAC51852 | Xq13.1 | 2 RRMs | Coiled coil | Yes | |||
| Nuclear RNA-binding protein 54-kD | Q15233 | Xq13.1 | 2 RRMs | Coiled coil, | Yes | |||
| Plenty-of-prolines-101 | AAC17422 | 1p36.11 | Yes (FP) | Mintz et al., 1999 | PWI | Yes | ||
| PTB associated splicing factor | P23246 | 1p34.3 | 2 RRMs | Coiled coil | Yes | |||
| RNPS1 | AAC39791 | 16p13.3 | RRM | Yes | Yes (IF) | Mayeda et al., 1999 | Yes | |
| SAP 114/SF3a | Q15459 | 22q12.2 | 2 SWAP, UBQ | Yes | ||||
| SAP 130/SF3b (KIAA0017) | NP_036558 | 16q21–22 | Yes (FP) | Mintz et al., 1999 | Yes | |||
| SAP 14/SF3b (pre-mRNA branch site protein p14) | AAK94041 | 2pter-p25.1 | RRM | Yes | ||||
| SAP 145/SF3b 150 | Q13435 | 11q13.1 | SAP, coiled coil | Yes | ||||
| SAP 155/SF3b | AAC97189 | 2q33 | Yes (FP) | Schmidt-Zachmann et al., 1998 | Coiled coil | Yes | ||
| SAP 49/SF3b | Q15427 | 1q12-q21 | 2 RRMs | Yes | ||||
| SAP 61/SF3a | A55749 | Coiled coil, 1 Znf_C2H2 | ||||||
| SAP 62/SF3a66 | Q62203 | 19p13.3-p13.2 | 1 ZnF_U1, 1 ZnF_C2H2 | Yes | ||||
| SF3b14b/PHD-finger 5a | NP_116147 | 22q13.2 | ||||||
| Siah binding protein 1 | AAB41656 | 8q24.2-qtel | 3 RRMs | Yes | ||||
| SnRNP Sm B/B′ | P27048 | Sm | Yes | |||||
| SnRNP Sm D1 | P13641 | 18q11.2 | Sm | Yes | ||||
| SnRNP Sm D2 | P43330 | 19q13.2 | Sm | |||||
| SnRNP Sm E | P08578 | 1q32 | Sm | Yes | ||||
| SnRNP Sm F | NP_003086 | 12q23.1 | Sm | Yes | ||||
| SnRNP Sm G | Q15357 | 2p13.3 | Sm | |||||
| SnRNP Sm D3 | P43331 | 22q11.23 | Sm | Yes | ||||
| SnRNP U1A | S42114 | 2 RRMs | Yes | |||||
| Splicing factor 9G8 | A57198 | 2p22-21 | RRM | Yes | Yes (IF) | Caceres et al., 1998 | 1 ZnF_C2HC | Yes |
| Splicing factor HCC1 | AAA16347 | Xp11.3 | 3 RRMs | Yes | Yes (IF) | Imai et al., 1993 | Yes | |
| Splicing factor hPRP17 | AAC39730 | 6q22.1 | 7 WD 40 repeats | Yes | ||||
| Splicing factor SC35 | Q01130 | 17q25.3 | RRM | Yes | Yes (IF) | Fu et al., 1992 | Yes | |
| Splicing factor SF1 | AAC29484 | KH | ||||||
| Splicing factor SF2/ASF | S26404 | 17q21.3-q22 | 2 RRMs | Yes | Yes (IF) | Caceres et al., 1997 | Yes | |
| Splicing factor SF3b10 | NP_112577 | 6q24.1 | ||||||
| Splicing factor SRp20 | P23152 | 6p21 | RRM | Yes | Yes (IF) | Caceres et al., 1997 | Yes | |
| Splicing factor SRp30 | Q13242 | 15q24-25 | 2 RRMs | Yes | Yes (IF) | Zahler et al., 1992 | Yes | |
| Splicing factor SRp40 | Q13243 | 14q23-24 | 2 RRMs | Yes | Yes (IF) | Zahler et al., 1992 | Yes | |
| Splicing factor SRp55 | AAA93072 | 6 20q12-q13.1 | 2 RRMs | Yes | Yes (IF) | Zahler et al., 1992 | Yes | |
| Splicing factor SRp75 | Q08170 | 1p35.2 | 2 RRMs | Yes | Yes | |||
| Splicing factor YT521-B (KIAA1966) | NP_588611 | 4q13.3 | Coiled coil | Yes | ||||
| TLS-associated serinearginine protein | NP_006616 | 1p36.11 | RRM | Yes | Yes | |||
| Tra-2 beta homolog | AAC28242 | 3q28 | RRM | Yes | Yes (IF) | Beil et al., 1997 | Yes | |
| U1 small ribonucleoprotein 1 | AAF19255 | 14q24 | RRM | Yes | PWI, coiled coil, RD/E dipeptide repeats | Yes | ||
| U1 snRNP 70 | P08621 | 19q13.3 | RRM | Yes | Coiled coil, RD/E dipeptide repeats | Yes | ||
| U1 snRNP C | P09234 | 6p21.31 | 1 ZnF_U1 | Yes | ||||
| U2 snRNP-A′ | P09661 | LRRcap | Yes | |||||
| U2AF35 | Q01081 | 15q12-13 | RRM | Yes | 2 ZnF_C3H1 | Yes | ||
| U2AF65 | P26368 | 19q13.4 | 3 RRMs | Yes | Yes | |||
| U4/U6-associated RNA splicing factor (PRP3) | AAC09069 | 1q21.1 | PWI | Yes | ||||
| U5 snRNP 200kD protein (KIAA0788) | O75643 | 2q11.2 | 2DEXDc, 2HELICc, SEC63 | Yes | ||||
| U5 snRNP 220kD protein | NP_006436 | 17p13.3 | JAB_MPN | Yes | ||||
| U5 snRNP 40 kDa protein (38 kDa splicing factor) | AAC69625 | 1p35.1 | 7 WD 40 repeats | Yes | ||||
| U5 snRNP 116 kDa protein (KIAA0031) | AAC53299 | 17q21 | Yes (IF) | Fabrizio et al., 1997 | 1 ZnF_NFX | Yes | ||
| U5 snRNP-associated 102 kDa protein | AAF66128 | 20q13.33 | Coiled coil, 13 HAT repeats | Yes | ||||
| RNA-associated proteins | ||||||||
| ATP dependent RNA helicase A | Q08211 | 1q25 | 2 DSRMs | DEXDc, HELICc, HA2 | Yes | |||
| DAM1 (breast carcinoma amplified sequence 2) | BAA34863 | 1p13.3-21 | Coiled coil | Yes | ||||
| DEAD/H box polypeptide 3 | O00571 | Xp11.3-p11.23 | Yes | HELICc | Yes | |||
| DEAD/H box RNA helicase p68 | Q61656 | 17q21 | HELICc | Yes | ||||
| DEAD/H box RNA helicase p72 | Q92841 | HELICc | Yes | |||||
| Double-stranded RNA binding nuclear protein, DRBP76 | CAC01405 | 19p13.2 | 2 DSRMs | DZF | Yes | |||
| E1B-55 kDa associated protein | CAA07548 | 19q13.31 | SAP, SPRY | Yes | ||||
| Elav-like 1 | P70372 | 3 RRMs | ||||||
| Interleukin enhancer binding factor 3 | AAC71052 | 19p13.2 | 2 DSRMs | DZF | Yes | |||
| Matrin 3 | P43244 | 2 RRMs | 1 ZnF_U1, 1 ZnF_C2H2 | Yes | ||||
| Nuclear cap binding protein 20 kDa (CBP20) | P52298 | 3q29 | RRM | Yes | ||||
| Nuclear cap binding protein 80 kd | Q09161 | 9q34.1 | MIF4G, coiled coil | Yes | ||||
| Nuclear protein NP220 | BAA11748 | 2p13.2-p13.1 | 2 RRMs | Yes | Yes (IF) | Inagaki et al., 1996 | 2 ZnF_C2H2, 2 ZnF_U1, scattered 9-meric repeats | Yes |
| Nuclear RNA helicase BAT1 | Q13838 | 6p21.3 | DEXDc, HELICc | |||||
| Pleiotropic regulator 1 | AAD24799 | 7q22 | 7 WD 40 repeats | |||||
| Poly(A) binding protein II | AAC39596 | 14q11.2-q13 | RRM | Yes (IF) | Bregman et al., 1995 | Coiled coil | Yes | |
| Ribonucleoprotein L | BAA24237 | 19q13.2 | RRM | |||||
| RNA binding motif protein 14 | NP_063922 | 11q13.1 | 2 RRMs | Yes | ||||
| RNA binding motif protein 5 | AAH02957 | 3p21.3 | 2 RRMs | 1 ZnF_RBZ | Yes | |||
| RNA binding motif protein EWS | Q01844 | RRM | 1 ZnF_RBZ | Yes | ||||
| RNA binding protein FUS/TLS | P35637 | 16p11.2 | RRM | 1 ZnF_RBZ | Yes | |||
| RNA binding protein HuR | AAB41913 | 19p13.2 | 3 RRMs | |||||
| RNA binding protein Raly/Merc | A47318 | 20q11.21-q11.23 | RRM | Yes | ||||
| RNA helicase (KIAA0801) | NP_055644 | 5q31.2 | Yes | Yes (FP) | This study | DEXDc, HELICc, coiled coil | Yes | |
| Rnpc2 | AAH04000 | 3 RRMs | Yes | |||||
| Son protein (KIAA1019) | P18583 | 21q22.11 | DSRM | Yes | Yes (FP) | This study | 11 mer repeats, 16 tandem decameric repeats, 12 tandem heptameric repeats, 15 heptameric repeats, 3 tandem 11 mer repeats, 13 heptameric repeats, G_patch, coiled coil | |
| SR140: U2-associated SR140 protein (KIAA0332) | BAA20790 | 3q23 | RRM | Yes | Yes(IF) | Will et al., 2002 | SWAP, coiled coil, RPR, 5 octamer repeats | Yes |
| SYT interacting protein (RNA binding motif protein 14) | NP_006319 | 11q13.1 | 2 RRMs | Brett et al., 1997 | Yes | |||
| Zinc finger RNA binding protein, ZFR (KIAA1086) | AAC25762 | 5p13.3 | 3 ZnF_U1, 3 ZnF_C2H2, DZF | Yes | ||||
| Cleavage and polyadenylation | ||||||||
| CPSF 100 kDa subunit | AAB66830 | 14q31.1 | Coiled coil | Yes | ||||
| CPSF 160 kDa subunit | Q10569 | Yes | ||||||
| CPSF 30 kDa subunit | AAC53567 | 5 ZnF_C3H1 | Yes | |||||
| CPSF 73 kDa subunit | AAB70268 | 2p25.2 | ||||||
| CSTF 64 kDa | P33240 | Xq22.1 | RRM | Yes | ||||
| Pre-mRNA cleavage factor Im | NP_008938 | 12q13.2 | RRM | Yes | RD/E dipeptide repeats | Yes | ||
| RNA polymerase II subunits | ||||||||
| RNA polymerase II 16 kDa subunit | O15514 | 2q21 | RPOL4c | |||||
| RNA polymerase II 19 kDa subunit | P52433 | 11q13.1 | S1 (Ribosomal protein S1-like RNA binding domain) | |||||
| RNA polymerase II 23 kDa subunit | P19388 | 19p13.3 | Yes | |||||
| RNA polymerase II 140 kDa subunit | P30876 | 4q12 | Yes | |||||
| RNA polymerase II Largest subunit | P24928 | 17p12-13 | Yes (IF) | Bregman et al., 1995 | RPOLA_N, coiled coil, C-terminal 7 residue repeats | Yes | ||
| Transcription | ||||||||
| POZ domain protein FBI-1 | NP_056982 | 19p13.3 | Yes (IF) | Pendergrast et al., 2002 | BTB, 4 ZnF_C2H2 | Yes | ||
| POZ/zinc finger transcription factor, ODA-8 | NP_062752 | 3q13.2 | BTB, 5 ZnF_C2H2 | Yes | ||||
| Skip | Q13573 | 14q24.3 | Coiled coil | Yes | ||||
| Tho2 | AAM28436 | Xq25-q26.3 | Coiled coil | Yes | ||||
| RNApolymerase II holoenzyme component SRB7 | Q13503 | 12p12.1 | ||||||
| mRNA export, NMD | ||||||||
| Aly | AAD09608 | 17q25.3 | RRM | Yes (IF) | Zhou, et al., 2000 | Yes | ||
| Mago-nashi homolog | NP_002361 | 1p34-p33 | Yes (IF) | Kataoka et al., 2001 | ||||
| Rae1/mRNP41 | P78406 | 20q13.31 | 4 WD 40 repeats | Yes | ||||
| RNA binding motif protein 8 (Y14) | AAD21089 | 14q22-23 | RRM | Yes | Yes (IF) | Kataoka et al., 2000 | Yes | |
| Apoptosis | ||||||||
| Acinus/SAP152 (KIAA0670) | NP_055792 | 14q11.2 | RRM | Yes | Yes (FP) | This study | SAPdomain, coiled coil, RD/E dipeptide repeats | Yes |
| Bcl-2-associated transcription factor, Btf (KIAA0164) | AAH34300 | 6q22-23 | Yes | Yes (FP) | This study | Yes | ||
| Others | ||||||||
| actin | P02571 | 17q25 | Yes (IF) | Spector, unpublished data | ||||
| APOBEC-1 stimulating protein | CAB94754 | 10q21.1 | 3 RRMs | Yes | ||||
| CAF1/p48 | Q09028 | 1p34.3 | Yes (FP) | Saitoh, N. unpublished data | 6 WD 40 repeats | |||
| Cell division cycle 2-like 1, Clk | NP_277025 | 1p36 | Yes (FP) | Sacco-Bubulya et al., 2002 | Coiled coil | Yes | ||
| eIF4A III (KIAA0111) | P38919 | 17q25.3 | Yes (FP) | Sacco-Bubulya, P. unpublished data | DEXDc, HELICc | Yes | ||
| Galectin | O08573 | Yes (IF) | GLECT | |||||
| Glutathione transferase | S-P08011 | Yes (IF) | Bennett et al., 1986 | MAPEG | ||||
| Hsp 70/Hsc 70 | NP_005338 | 9q33-q34.1 | Yes (IF) | Maheswaran et al., 1998 | Signal peptide | |||
| Nuclear matrix protein NMP200 | CAB51857 | 11q12.2 | U box, 7 WD 40 repeats | Yes | ||||
| Pinin | NP_002678 | 14q13.3 | Yes (IF) | Brandner et al., 1997 | Coiled coil | Yes | ||
| Protein phosphatase 1, regulatory subunit 10/FB19 protein | JE0291 | 6p21.3 | RRM | TFS2N, 1 ZnF_C3H1 | Yes | |||
| Rod1 | BAA75465 | 5q22 | 4 RRMs | Yes | ||||
| SAF B | AAC29479 | 19p13.2-13.3 | RRM | Yes (FP) | Nayler et al., 1998 | Coiled coil | Yes | |
| SCAF10 | JC5314 | Yes | Yes (IF) | Mortillaro et al., 1998 | Pro_isomerase | Yes | ||
| SCAF6/DAN16 | AAN77183 | 19p13.1 | Yes | SWAP, RPR, Trp containing repeat region, G_patch, | Yes | |||
| SRm300 (KIAA0324) | AAF21439 | 16p13.3 | Yes | Yes | ||||
| Wilms' tumour 1-associating protein, WTAP (KIAA0105) | NP_004897 | 6q25-q27 | Yes (IF) | Little et al., 2000 | Coiled coil | Yes |
IF, Immunofluorescence; FP, fluorescent protein.
Database for motif and domain searches: SMART (http://smart.embl-heidelberg.de), (Schultz et al., 1998; Letunic et al., 2002). Only those proteins containing SR dipeptides were manually searched for RD/E dipeptide repeats and other repetitive amino acid sequences
Table 2.
Potential IGC proteins
| Protein Description | Accession Code | RNA Binding Motif | RS | Other Domains and Motif(s)a | Low Complexity Region |
|---|---|---|---|---|---|
| A-kinase anchor protein 8K | Q63014 | 1 ZnF_C2H2 | Yes | ||
| Aladin (Adracalin). | P58742 | 4 WD 40 repeats | Yes | ||
| Aquarius (KIAA0560) | AAB50008 | Yes | |||
| Ash2 | AAC13564 | SPRY | Yes | ||
| Ataxin-1 | P54254 | AXH | Yes | ||
| BAF53A | AAC94992 | Actin | |||
| BAF57 | AAC04509 | HMG, coiled coil | Yes | ||
| BMAL1(HLH/PAS protein) | O00327 | Yes | |||
| C/EBPa | P53566 | BRLZ | Yes | ||
| C/EBPb | P28033 | BRLZ | Yes | ||
| Calsyntenin 1 (KIAA0911) | NP_075538 | Signal peptide, cadherin repeats, transmembrane, coiled coil | Yes | ||
| CyP-60 (cyclophilin-like protein) | S64705 | Ubox, pro_isomerase | Yes | ||
| Dna J protein homolog 2 | P31689 | Dna J, DnaJ CXXCXGXG, DnaJ C | Yes | ||
| dpy-30-like protein | NP_115963 | Dpy-30 | |||
| Early lymphoid activation protein | I56219 | ||||
| eIF4AI | P04765 | DEXDc, HELICc | |||
| FB19 protein | JE0291 | TFS2N, 1 ZnF_C3H1 | Yes | ||
| G10 protein | AAC14190 | ||||
| GC-rich sequence DNA-binding factor candidate | AAD34617 | Yes | |||
| General transcription factor IIIC, polypeptide 2 | NP_001512 | 4 WD 40 repeats | Yes | ||
| Hepatocyte nuclear factor 1 alpha | P15257 | HOX | Yes | ||
| Hepatocyte nuclear factor 4 alpha | P41235 | 1 ZnF_C4, HOLI | Yes | ||
| Homeobox protein zhx-1 | JC4863 | 2 ZnF_C2H2, 5 HOX | Yes | ||
| Interleukin enhancer binding factor 2 | NP_080650 | DZF | Yes | ||
| IRA1 | AAG44738 | LisH, 8 WD 40 repeats | Yes | ||
| LIM-domain protein LMP-1 | AAD13197 | PDZ, 3 LIM | Yes | ||
| Lupus La protein | P32067 | RRM | LA | Yes | |
| Mader/NAB | S31927 | Yes | |||
| MAX-like bHLHZIP protein, transcription factor-like 4 | NP_037515 | HLH | |||
| mRNA associated protein MRNP 41 (RAE1 homolog) | P78406 | 4 WD 40 repeats | Yes | ||
| mRNA export factor TAP | Q99JX7 | RRM | LRR, LRRcap, NTF2, TAPC | Yes | |
| Ngfi-A binding protein 1 | NP_032693 | NCD1, NCD2, Nab1 | Yes | ||
| Nuclear Factor I-X | P09414 | DWA | Yes | ||
| Nuclear protein ZAP3 | Q9R0I7 | Coiled coil | Yes | ||
| Nuclear receptor coactivator 5 | NP_066018 | HGTP anticodon, coiled coil | Yes | ||
| Nuclear VCP-like protein NVLp.1 | AAB70460 | AAA | Yes | ||
| NuMA | A42184 | Coiled coil | Yes | ||
| p150TSP (KIAA0155) | BAA09925 | 9 TPR, coiled coil | Yes | ||
| PCAF-associated factor 400, PAF400 | AAD04629 | FAT, PI3Kc, FATc | Yes | ||
| Peptidylprolyl isomerase (cyclophilin)-like 1 | NP_057143 | ||||
| Polymyositis/Scleroderma autoantigen 1, PM/SCL-75 | Q9JH17 | RNase_PH, RNase_PH_C | Yes | ||
| Predicted osteoblast protein | BAA13251 | Signal peptide | |||
| Prox1 | Q92786 | ||||
| RAD50 | AAC52894 | Rad50_zn_hook, coiled coil | Yes | ||
| RuvB like DNA helicase | NP_035434 | AAA | |||
| SEC13-related protein | NP_109598 | 6 WD 40 repeats | |||
| Symplekin, Huntingtin interacting protein I | XP_017129 | Yes | |||
| SYT interacting protein SIP | AAC64058 | 2 RRMs | Yes | ||
| TAFII30 protein | CAB59510 | Signal peptide | Yes | ||
| TAR DNA binding protein | NP_663531 | 2 RRMs | Yes | ||
| TAR DNA-binding protein-43 | I38977 | 2 RRMs | Yes | ||
| Thyroid hormone receptor-associated protein 100 kDa·· (KIAA0130) | NP_035999 | Yes | |||
| Thyroid hormone receptor-associated protein 150 kDa | AAD22034 | Yes | Yes | ||
| Transcription elongation factor B (SIII) polypeptide 2, elongin B | NP_112391 | UBQ | |||
| Transcription factor NF-AT 45 | A54857 | DZF | |||
| Transcription factor-like protein 4 | JC5333 | HLH | Yes | ||
| Transcription intermediary factor 1-beta, TIF1-beta | Q62318 | Signal peptide, 2 RING, 2 BBOX, BBC, PHD, BROMO | Yes | ||
| Transcriptional co-activator CRSP77 | XP_048386 | ||||
| Transcriptional intermediary factor 2 | CAA66263 | HLH, PAS, PAC | Yes | ||
| Transducin (beta) like 1 protein | CAA73319 | Yes | |||
| Trf-proximal protein | NP_064432 | ||||
| Tuftelin-interacting protein 33 | NP_061253 | G_patch | Yes | ||
| Tumor protein D52 | P55327 | TPD52 | |||
| WD repeat domain 5 protein | NP_060058 | 7 WD40 repeats | Yes | ||
| WD repeat protein BIG-3 | AAL27006 | 7 WD40 repeats | Yes | ||
| XPA-binding protein 2, XAB2 (KIAA1177) | BAB15807 | 11 HAT | Yes | ||
| XPE UV-damaged DNA binding protein | CAA05770 | Yes | |||
| ZAN75 | BAA31522 | 2 ZnF_C2H2 | Yes | ||
| Zinc finger DNA binding protein 99 ZBP-99 | AAD21084 | 4 ZnF_C2H2 | Yes | ||
| Zinc finger protein | CAB70967 | 4 ZnF_C2H2 | Yes |
Database for motif and domain searches: SMART (http://smart.embl-heidelberg.de), (Schultz et al., 1998; Letunic et al., 2002). Only those proteins containing SR dipeptides were manually searched for RD/E dipeptide repeats and for other repetitive amino acid sequences
Table 3.
Unexpected proteins
| Protein Description | Accession Code |
|---|---|
| 14-3-3 protein | P31946 |
| 40s ribosomal protein s4, X isoform | P12750 |
| 40s ribosomal protein S10 | P46783 |
| 40s ribosomal protein S14 | P13471 |
| 40s ribosomal protein S16 | P17008 |
| 40s ribosomal protein s2 (s4) (llrep3 protein) | P15880 |
| 40s ribosomal protein S28 | P25112 |
| 40s ribosomal protein s3a. 12/1998 | P49241 |
| 40s ribosomal protein s5. 7/1999 | P46782 |
| 40s ribosomal protein s6 (phosphoprotein np33) | P10660 |
| 40s ribosomal protein S7 | P06584 |
| 40s ribosomal protein s8 | P09058 |
| 40s ribosomal protein S9 | P29314 |
| 60s acidic ribosomal protein p0 | P05388 |
| 60s acidic ribosomal protein p1 | P47955 |
| 60s ribosomal protein L12 | P30050 |
| 60s ribosomal protein L13 | P41123 |
| 60s ribosomal protein L14 | P50914 |
| 60s ribosomal protein L15 | P39030 |
| 60s ribosomal protein L19 | P14118 |
| 60s ribosomal protein L23 | P23131 |
| 60s ribosomal protein L24 | P38663 |
| 60s ribosomal protein L27a. | P46776 |
| 60s ribosomal protein L31 | P12947 |
| 60s ribosomal protein L35 | P42766 |
| 60s ribosomal protein L4 | P36578 |
| 60s ribosomal protein L7a | P11518 |
| 60s ribosomal protein L8 | P25120 |
| Acetyl-CoA carboxylase | Q13085 |
| aryl sulfotransferase | P52840 |
| Clathrin heavy chain 1 (CLH-17) | Q00610 |
| Coilin p80 | P38432 |
| CRM1 | BAA23415 |
| Cytochrome c oxidase polypeptide VIB | P56391 |
| Cytochrome p450 | Q64458 |
| DNA polymerase e | Q07864 |
| DNA ligase I | P37913 |
| DNA repair protein XRCC4 | NP_071801 |
| Endo/exonuclease Mre 11 | AAB04955 |
| Enhancer of rudimentary homolog | Q14259 |
| Exosome complex exonuclease RRP45/PMSCL1 | Q06265 |
| Fibrillarin | P22087 |
| Fibrinogen, alpha polypeptide | XP_130931 |
| Glucocorticoid receptor | P06537 |
| Glucokinase regulatory protein | Q07071 |
| Histone deacetylase (HD1) | Q13547 |
| Histone H1 | P15864 |
| Histone H2a | P02262 |
| Histone H2b | P02278 |
| Histone H3 | P06351 |
| Histone H4 | P02304 |
| Host cell factor C1 HCF | P51610 |
| HP1 | P45973 |
| Immunoglobulin Heavy Chain Binding Protein | P11021 |
| Importin alpha | P52294 |
| Importin beta | Q14974 |
| Integral membrane glycoprotein gp210 | P11654 |
| Lamin A | P02545 |
| Lamin B1 | P14733 |
| Lamin B2 | P21619 |
| Lamin B3 | P48680 |
| Lamin C | P02545 |
| Lamina-associated polypeptide 2 LAP2 | P42166 |
| Metalloproteinase inhibitor 1 precursor | P01033 |
| Methyl-CpG binding domain-containing protein MBD3 | AAC68877 |
| Mi2 chromodomain helicase-dna-binding protein 4 | Q14839 |
| Microfibrillar-associated protein 1 | P55081 |
| Mitotic phosphoprotein 44 | AAL86380 |
| MTA1-like protein (KIAA1266) | BAAC36562 |
| myb-binding protein p160 | AAC39954 |
| Myosin light chanin alkali, non-muscle isoform | P16475 |
| Nuclear pore complex protein Nup84 | AAB52419 |
| Nuclear pore complex protein Nup153 | P49791 |
| Nuclear pore complex protein nup155 | O75694 |
| Nuclear pore complex protein Nup50 | AAC53278 |
| Nuclear receptor corepressor N-CoR | S60254 |
| Nucleolar phosphoprotein p130 | I38073 |
| Nucleolar protein family A, member 1 | NP_080854 |
| Nucleolar protein NAP57/CBF5 | O60832 |
| Nucleolar protein NOP10 | NP_061118 |
| Nucleolar protein NOP5/NOP58 | AAD27610 |
| Nucleolar protein NOP56 | O00567 |
| Nucleoporin Nup75 | NP_079120 |
| Nucleoporin Nup84 | AAB52419 |
| Nuclear Pore Complex Protein NUP155 | O75694 |
| O-linked GlcNAc transferase | AAB63466 |
| PCAF associated factor 400 | AF110377 |
| PML | AAA97601 |
| Protein disulfide isomerase A3 precursor, ER-60 | P30101 |
| RAD50 homolog | NP_033038 |
| Ran GAP1 | P46061 |
| Ran GTPase | NP_033417 |
| RanBP2 (Nup 358) | P49792 |
| Recombination signal binding protein | AAA16254 |
| RelA-associated inhibitor | XP_030918 |
| REST corepressor (KIAA0071) | NP_055971 |
| Ribosomal protein S30 | AAD1774 |
| S164/presenilin | AAC97961 |
| SAP18 (sin3 associated polypeptide p18) | AAD41090 |
| Sin3 | AAB01610 |
| SWI/SNF BAF155 | AAC50693 |
| SWI/SNF related, BAF170, Rsc8 | NP_003066 |
| SWI/SNF related, member 5 | BAA25173 |
| TPR protein | S33124 |
| Transcription repressor p66 (KIAA1150) | AAL39081 |
| Tryptophan 2,3-dioxygenase | P48776 |
| Tubulin b | P07437 |
| Ubiquinol cytochrome C reductase complex protein 2 | Q9DB77 |
| Ubiquitin-conjugating enzyme E2L 3 | NP_003338 |
| Ubiquitin-like protein SMT3A | P55854 |
| UDP-glucuronosyl transferase | P09875 |
| Vimentin | P08670 |
Table 4.
Novel IGC protein candidates
| Protein Description | Accession Code | RNA Binding Motif | RS | Other Domains and Motif(s)a | Low Complexity Regions | Notes |
|---|---|---|---|---|---|---|
| DNA segment, Chr 6, Wayne State University 176 | NP_613053 | Transmembrane | ||||
| Epidermal Langerhans cell protein LCP1 | NP_075923 | HMG box | Yes | |||
| GC-rich sequence DNA-binding factor candidate | NP_037461 | Coiled coil | Yes | |||
| Hypothetical protein FLJ10637 | NP_060634 | Coiled coil | ||||
| Hypothetical protein FLJ11305 | BAA91611 | Yes | Similar (89% identity) to unnamed protein product (AK001302) | |||
| Hypothetical protein MGC28864 | AAH17152 | Coiled coil, HGTP anticodon domain | Yes | |||
| KIAA0052 protein | AAH28604.2 | DEXDc, HELICc, coiled coil | Yes | Homologous to “putative helicase”, “RNA helicase Mtr4” | ||
| KIAA0460 protein | T00074 | Yes | ||||
| KIAA0663 protein | T00368 | 3 ZnF_C3H1 | Yes | Slightly similar to “lacunin”, large multidomain extracellular matrix Zinc finger protein, CPSF (clipper/cleavage and polyadenylation stimulation factor) | ||
| KIAA1160 protein | BAA86474 | Coiled coil | ||||
| mKIAA1125 protein | BAC41468 | PHD, BROMO, PWWP, 1 ZnF_NFX, coiled-coil | Yes | |||
| Putative 40-2-3 protein | AAH28253 | Yes | ||||
| RIKEN cDNA 1110015K06 | AAH10333 | Yesa | ||||
| RIKEN cDNA 1700016A15 | XP_127067 | 1 ZnF_NFXa | Yes | Similar (97% identity) to nuclear protein UKp68 | ||
| Wdr18 protein | AAH32968.1 | 4 WD40 repeats | Yes | Similar (74% identity) to hypothetical protein R32184_1 | ||
| RIKEN cDNA 2410002M20 | NP_766285 | PRP38 family | Yes | Weakly similar to splicing factor, arginin/serine-rich 2 | ||
| RIKEN cDNA 2410008G02 (KIAA0095) | AAH23140 | NIC | Yes | Related to NIC96 | ||
| RIKEN cDNA 2500003M10 | NP_075704 | Yes | ||||
| RIKEN cDNA 2610015J01 | NP_081625 | RRM | Yes | |||
| RIKEN cDNA 2610034N24 | NP_081532 | HEAT PBS | Yes | |||
| RIKEN cDNA 2610511G16 | NP_080477 | Yes | SAP, coiled coil | Yes | ||
| RIKEN cDNA 2610528A15 (KIAA0052) | NP_082427 | DEXDc, HELICc, coiled coil | Yes | |||
| RIKEN cDNA 2810013E07 | NP_835213 | TPR | Yes | Similar (91% identity) to hypothetical protein FLJ20530 | ||
| RIKEN cDNA 5730555F13/modulator of estrogen induced transcription | NP_079966 | Coiled coil | Yes | |||
| RIKEN cDNA 9330151F09 gene | NP_666265 | Yes | Similar (60% identity) to thyroid hormone receptor-associated protein, 150 kDa subunit | |||
| Similar to a C.elegans protein encoded in cosmid K12D12(Z49069) (KIAA0225 protein) | BAA13214 | Yes | ||||
| Similar to alcohol dehydrogenase PAN1B-like protein | XP_223159 | Yes | Short-chain alcohol dehydrogenase | |||
| Similar to CG11943 gene product | AAH45524 | Yes | ||||
| Similar to hypothetical protein | XP_290525 | SAP, SPRY | Yes | Similar (92% identity) to nuclear calmodulin-binding protein | ||
| Similar to KIAA0138 gene product | XP_128733 | RRM | SAP, coiled coil | Yes | Similar (75% identity) to scaffold attachment factor B | |
| Similar to thyroid hormone receptor-associated protein, 150 kDa | XP_233523 | Yes | Yes | |||
| Unnamed protein product | BAA96656 | LisH, CTLH, 6 WD40 repeats | Homologous to “brain-enriched WD-repeat protein” |
Database for motif and domain searches: SMART (http://smart.embl-heidelberg.de) (Schultz et al., 1998; Letunic et al., 2002). Only those proteins containing SR dipeptides were manually searched for RD/E dipeptide repeats and other repetitive amino acid sequences
Transient Transfection of Cells and Immunofluorescence Microscopy
Four cDNA clones that correspond to newly identified IGC proteins (KIAA0164, 0670, 0801, and 1019) were kindly provided by Dr. Nagase (Kazusa DNA Research Institute, Chiba, Japan). The clones were fused in frame, to enhanced yellow fluorescent protein at their N termini by using the pEYFP-C expression vector (BD Biosciences Clontech, Palo Alto, CA). A431 cells were transfected with the resultant constructs using FuGENE6 transfection reagent (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions and incubated for 16 h. Cells were processed for immunofluorescence as described previously (Spector et al., 1998). Antibody to SC35 (Fu and Maniatis, 1990) was used at 1:1000 dilution to label IGCs, followed by Texas Red-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). Images were acquired on an Axioplan 2i fluorescence microscope (Carl Zeiss, Thornwood, NY)with a plan-APO 100×/1.4 numerical aperture objective lens using Openlab Software (Improvision, Lexington, MA) and an Orca charge-coupled device camera (Hammamatsu, Middlesex, NJ).
RESULTS
The IGC Proteome
We have previously reported on the development of a biochemical strategy to purify and characterize IGCs from mouse liver nuclei. Using this approach combined with mass spectrometry analysis, we identified 33 known proteins (Mintz et al., 1999) and expressed sequence tags (ESTs) encoding at most 16 proteins after searching a nonredundant protein database or dbEST (National Center for Biotechnology Information and DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank) with the uninterpreted MS/MS spectra. We have now extended this study by scaling up our purification and optimizing the sample preparation (see MATERIALS AND METHODS) to identify a larger complement of IGC proteins. The IGC fraction was digested with trypsin and subjected to liquid chromatography electrospray ionization MS/MS followed by uninterpreted fragment ion searching of nonredundant and expressed sequence tag databases (dbEST) in a data-dependent manner. Our analysis will identify proteins that are enriched in IGCs and therefore localize in a speckled pattern by immunofluorescence microscopy (i.e., snRNPs and serine-arginine proteins), as well as other proteins that may be equally distributed throughout the nucleoplasm, including the IGCs and diffuse nuclear pools (i.e., hnRNP A and C). We performed five rounds of the analysis and reached saturation as we repeatedly obtained the same set of peptide sequences. As a result, 2214 peptide sequences were obtained, which correspond to 360 proteins. We categorized the proteins based upon their known function, motifs, and/or localization: identified IGC proteins (41%), potential IGC proteins (19%), novel IGC protein candidates (9%), and unexpected IGC proteins (31%) (Tables 1, 2, 3, 4).
Identified IGC Proteins
The group of identified IGC proteins (Table 1, 146 proteins) contains the most frequently detected proteins and is composed of previously identified IGC proteins, as well as proteins whose functions are similar to well-characterized IGC proteins. Because many of the proteins that have been localized to IGCs contain RNA binding motifs and RS domains that are stretches of dipeptide repeats of arginine (R) and serine (S) (Birney et al., 1993), we systematically surveyed all of the detected proteins with regard to these motifs. Nineteen percent of the identified IGC proteins contain an RS domain, and 50% contain one to four RNA binding motifs (Table 1). The presence of an RS domain and/or basic region has been reported to act as a speckle localization signal for some pre-mRNA splicing factors as well as a protein interaction domain (for a review, see Fu, 1995; Graveley, 2000). In addition, each of the identified proteins was characterized with regard to the presence of other motifs and its localization to nuclear speckles. Twenty-seven percent of the identified IGC proteins have previously been reported to localize in nuclear speckles. We did not detect any sequence motifs common to all identified IGC proteins. Two frequently detected motifs in this group are the DEAD box helicase motif (Linder et al., 1989; Luking et al., 1998) and an RNA binding motif (Birney et al., 1993). The absence of a specific localization signal, aside from the RS domain contained within a subset of proteins, may reflect a more transient interaction of many proteins with nuclear speckles or may indicate that these proteins are targeted to and/or associate with nuclear speckles through other RS-domain–containing interaction partners.
A profile of this protein group (Figure 1) indicates that 54% of the identified IGC proteins have a role in pre-mRNA splicing, 20% of the proteins are classified as RNA-associated proteins, and 7% have roles in other aspects of pre-mRNA processing, such as 3′ RNA processing, mRNA export, and nonsense-mediated decay (see DISCUSSION). Together, 81% of the IGC proteins likely participate in pre-mRNA/mRNA metabolism.
Figure 1.
Profile of the Identified IGC proteins. One hundred forty-six identified IGC proteins are categorized based upon their proposed functions; 81% of the proteins are involved in activities related to RNA metabolism.
IGCs have been proposed to be important for the coupling of RNA polymerase II transcription and pre-mRNA splicing, because numerous proteins are recruited from nuclear speckles to sites of transcription (for a review, see Lamond and Spector, 2003). Six percent of the identified IGC proteins are involved in transcription (Table 1). Several subunits of RNA polymerase II, including the largest subunit, which has previously been localized to nuclear speckles (Bregman et al., 1995; Mortillaro et al., 1996), and several transcription factors have been identified in this fraction. Most general transcription factors were diffusely distributed throughout the nucleoplasm and were not identified in the IGC fraction. However, the proportion of transcription factors may be underrepresented, because we have categorized many proteins as potential IGC proteins (Table 2) due to the lack of information on their specific subnuclear localization. As expected, we did not detect RNA polymerases I or III in the IGC fraction.
Interestingly, several proteins were identified that have previously been characterized as having structural roles in cells. These proteins include actin (Nakayasu and Ueda, 1984), matrin 3 (Belgrader et al., 1991; Nakayasu and Berezney, 1991), lamin A/C (Jagatheesan et al., 1999), and pinin (Ouyang and Sugrue, 1996; Brandner et al., 1997; Ouyang et al., 1997). Although all of these proteins have been localized to IGCs, they do not form an underlying protein scaffold for attachment of IGCs (Sacco-Bubulya and Spector, 2002). Instead, they may be integral components of individual interchromatin granules and their role(s) is yet to be determined.
In addition, our analysis identified several proteins that were recently shown by others to have roles in pre-mRNA splicing or to be localized to nuclear speckles. These include acinus (Boucher et al., 2001; Schwerk et al., 2003), eIF4Aiii (Li et al., 1999; Holzmann et al., 2000), RNA binding motif protein 8 (Y14) (Kataoka et al., 2001), and the RNA export protein Aly (Zhou et al., 2000). Surprisingly, our analysis did not reveal some proteins that have previously been reported to localize to nuclear speckles, for example, casein kinase II and protein phosphatase 1 (Trinkle-Mulcahy et al., 2001). Protein phosphatase 1 has only one trypsin cleavage site, so it would likely be underrepresented in our peptide identification by mass spectrometry. Other proteins that were not identified associate with IGCs with low affinity and therefore may dissociate during the purification procedure. Alternatively, association of proteins such as kinases and phosphatases may be more sensitive to changes in phosphorylation state during IGCs purification.
Potential and Unexpected IGC Proteins
We found 70 proteins whose nuclear localizations, for the most part, have not been characterized, although these proteins have been studied at the biochemical and/or molecular levels (Table 2). We categorized this group of proteins as potential IGC proteins. Many of these potential IGC proteins have roles in transcription, such as DNA cis-element binding factors (i.e., transcription factor NF-AT45, nuclear factor I-X, and C/EBPs), components of a chromatin remodeling complex (BAF53A and BAF57), and transcription mediators (transcriptional coactivator CRSP77, thyroid hormone receptor-associated proteins, and transcriptional intermediary factors). Seven percent of the potential IGC proteins are possible molecular chaperones because they contain either a cyclophilin type peptidyl-prolyl cis-trans-isomerase motif or AAA ATPase family motif. Four percent are DNA repair proteins, and the remaining proteins have varied functions or they have not been studied at the molecular level. Although the subnuclear distribution of each protein remains to be determined, the identification of these proteins in the IGC fraction suggests that IGCs may be major sites for coupling transcription and pre-mRNA processing, thus promoting efficient gene expression. Furthermore, some of the molecular chaperone proteins included in this category may be responsible for the formation/maintenance of the structure of IGCs.
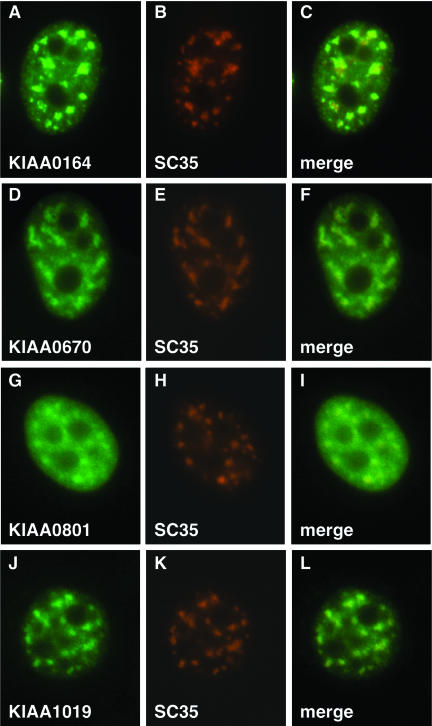
To determine whether these proteins are bona fide IGC constituents, we made cDNA fusion constructs to tag them with yellow fluorescent protein and expressed them in A431 cells. Four representative cDNAs shown in Figure 2 all encoded for proteins that localize to IGCs (KIAA0670, KIAA0801, and KIAA1019) or their periphery (KIAA0164), further confirming the specificity of our preparation. Because we now have evidence that they are bona fide IGC proteins, we have included these four proteins in Table 1.
Figure 2.
In vivo localization of several novel IGC proteins. The cDNAs for several novel IGC protein candidates (KIAA0164 = Btf, KIAA0670 = acinus, KIAA0801 = RNA helicase, KIAA1019 = son protein) were fused to yellow fluorescent protein and expressed in A431 cells. The cells were fixed and labeled with an antibody to the pre-mRNA splicing factor SC35 (Fu and Maniatis, 1990), which localizes in IGCs.
In our previous study, we showed that the IGC fraction was highly purified and free of detectable contaminants, such as other nuclear structures. When examined using transmission electron microscopy, the final fraction was significantly homogeneous, containing granules measuring 20–25 nm in diameter that were immunolabeled with anti-SC35 antibody, a marker protein for IGCs (Mintz etal., 1999). In addition, immunoblot analysis showed that a subset of known IGC proteins are highly enriched in the IGC fraction, whereas minimal contamination of protein components of other nuclear structures, such as the nuclear envelope, promyelocytic leukemia bodies, or Cajal bodies were detected in the IGC fraction (Mintz et al., 1999). Nonetheless, by mass spectrometry we did detect numerous proteins, which have previously been characterized as components of other cellular structures, and therefore we have classified them as unexpected proteins (Table 3). Because these proteins are relatively abundant and mass spectrometry is a highly sensitive technique, it is likely that they are protein contaminants in our preparation.
Novel IGC Protein Candidates
In addition, and most interestingly, we found 32 proteins for which no available biological information is available, except for sequence information (Table 4). Each of these proteins was analyzed for known motifs. Four proteins have various similarities to other proteins involved in RNA metabolism. These examples include a protein with a RNA helicase C-terminal domain (KIAA0052), a protein slightly similar to cleavage and polyadenylation stimulation factor (KIAA0663), a putative splicing factor (RIKEN cDNA 2410002M20), and a protein with similarity to SAF-B (similar to KIAA0138 gene product), which is known to be in IGCs. Thus, these proteins are highly likely to be IGC components. Two other proteins contain an SAP motif, one also with a poly-A binding domain (RIKEN cDNA 2610511G16) and the other with SPRY and Ffh domains (similar to hypothetical protein). The SAP motif is named after SAF-A/B, acinus and PIAS (Aravind and Koonin, 2000). SAF-B and acinus are localized in the IGCs (Table 1 and Figure 2), and PIAS has been shown to be associated with RNA helicase II/ATP-dependent RNA helicase (Valdez et al., 1997). The SAP motif is defined as a sequence homologous to the N-terminal DNA binding region of SAF-A and has been found in several other nuclear proteins (Aravind and Koonin, 2000). Proteins with a SAP domain often contain an additional motif that is involved in the assembly of RNA-processing complexes (Aravind and Koonin, 2000). Therefore, it has been proposed that such proteins are associated both with chromatin and RNA. Additionally, they may function to deliver the RNA processing machinery to the site of transcription (Aravind and Koonin, 2000), which overlaps with a proposed function of IGCs.
RS Domain-containing Proteins
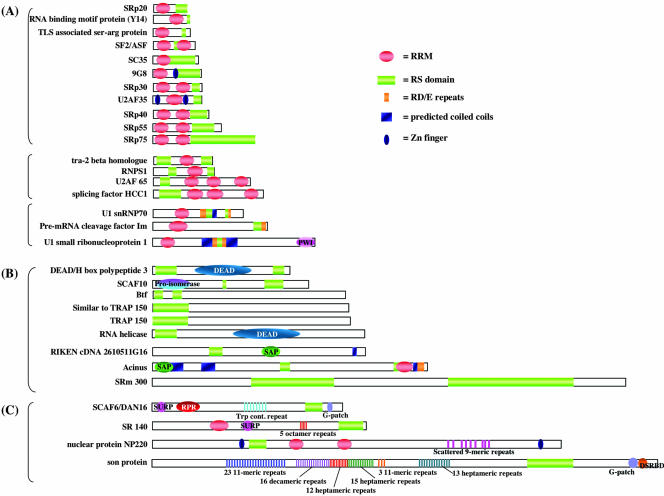
In the IGC proteomic analysis, we detected 31 proteins with RS dipeptide motifs, including two novel IGC candidates (Tables 1, 2, and 4). Of these, 17 proteins have actually been shown to localize to IGCs by either immunofluorescence analysis or expression of the fluorescently tagged proteins in cells (Table 1). By comparing these proteins, based upon the organization of their other motifs relative to the RS domains, we sorted them into three major groups (Figure 3). The first group (Figure 3A) represents proteins with an RS motif and one to three RNA recognition motifs (RRMs). This group can be further divided into three subgroups. Proteins in the first subgroup, from SRp20 to SRp75, are small proteins with N-terminal RRMs and a C-terminal RS motif. Among this group are members of the SR family of pre-mRNA splicing factors (SRp20, SF2/ASF, SC35, 9G8, SRp30, SRp40, SRp55, and SRp75). Proteins in the second sub-group, from the tra-2 beta homologue to splicing factor HCC1, are also small splicing factors, but they have an N-terminal RS motif and a C-terminal RRM(s). Proteins in the third subgroup are related to the first two subgroups because they have N-terminal RRM and C-terminal or middle region RS motifs; however, they are larger proteins and their RS motifs are continuous to RD or RE dipeptides, which could provide them with additional functional properties (see DISCUSSION).
Figure 3.
RS domain-containing proteins detected in the IGCs. Thirty-one proteins with RS motifs were detected in the IGC fraction and were categorized into three subgroups. Proteins in the first group (A) are of relatively low molecular mass, contain one or more RRMs, and many are founding members of the SR protein family. Proteins in the second group (B) are of larger molecular mass, and most do not contain an RRM but do contain additional motifs. Proteins in the third group (C) are also of higher molecular mass and contain repetitive sequences.
Proteins in the second group (Figure 3B) are medium-to-large proteins, ranging from 663 to 2297 amino acids. All (except for acinus) do not have a recognizable RRM motif, and they are characterized by the presence of compositionally biased regions. Among them, Btf and a protein called “similar to TRAP150” have significant sequence similarities to TRAP150 (60 and 33% sequence identity, respectively). TRAP150 has been shown to be a transcriptional mediator component (Johnson et al., 2002). Proteins categorized in this second group contain additional domains, such as a cyclophilin type peptidyl-prolyl cis-trans-isomerase (proisomerase) domain, a SAP domain, and a DEAD box helicase motif, thus they may have additional interactions and/or functions. Indeed, SRm 300 is a splicing coactivator (Blencowe et al., 2000), and acinus is involved in chromatin condensation in the late stage of apoptosis (Sahara et al., 1999) as well as in pre-mRNA processing (Schwerk et al., 2003). Btf also was reported to be involved in apoptosis (Kasof et al., 1999).
The third group (Figure 3C) also represents proteins of medium-to-large (917–2427 amino acid length) size with interesting repetitive sequences. Especially notable is son protein, which contains six types of repetitive sequences that cover approximately one-third of its sequence. The functions of these proteins are not well characterized; however, NP220 was reported to be a DNA and nuclear matrix binding protein (Inagaki et al., 1996), and SR140 is associated with U2 snRNP (Will et al., 2002).
DISCUSSION
We have performed an in-depth analysis of the protein composition of IGCs derived from mouse liver nuclei. As expected, we detected numerous proteins involved in pre-mRNA processing. In addition, we detected transcription factors, RNA polymerase II subunits, and proteins with unexpected roles in apoptosis and DNA repair. We also identified numerous novel IGC protein candidates.
IGCs and Spliceosomes
Extensive evidence has suggested that the nucleus is compartmentalized with respect to gene expression (for a review, see Spector, 2003). IGCs are enriched in pre-mRNA splicing factors, yet these nuclear regions are not sites of splicing or transcription. Rather, they are sites of splicing factor assembly/modification and/or storage (for a review, see Lamond and Spector, 2003) from which factors are recruited to nearby sites of active transcription. The C-terminal domain of the large subunit of RNA polymerase II and phosphorylation of the RS domain of SR splicing factors play a major role in supplying these factors to the site of active transcription (Misteli et al., 1998; Misteli and Spector, 1999). However, it has not been determined whether different splicing factors are targeted to a site of transcription individually, or as subcomplexes as needed for different stages of pre-mRNA processing. The latter is a possibility, because individual interchromatin granules are of a consistent size with ribosomes and are therefore large enough to contain such subcomplexes of proteins. When we made a comparison of protein components of the spliceosome (Zhou et al., 2002) versus IGC components, we found significant (63%), but not total overlap, between these two structures, although each complex was initially purified from an entirely distinct nuclear fraction.
Because there is considerable overlap of IGC components (modification/assembly and/or storage sites) with spliceosome components (functional sites), there is a possibility that interchromatin granules move from the IGCs to the site of active transcription, rather than each protein moving individually. It has been shown that fluorescently tagged splicing factors are highly mobile in living cells, but they move slowly enough to suggest that the proteins move in a complex, rather than as a monomer (Kruhlak et al., 2000). By time-lapse microscope analysis, it was shown that “spheres” seem to bud off of the surface of nuclear speckles when cells are actively transcribing (Eils et al., 2000). It remains to be determined whether these spheres correspond to an individual granule or clusters of IGC granules.
Apoptosis and Other Functions
In addition to proteins functioning in pre-mRNA splicing and transcription, we detected proteins that are involved in other nuclear functions. For example, acinus (KIAA0670) has been reported to be involved in a late step of an apoptotic pathway (Sahara et al., 1999). An in vitro system using permeabilized cells and apoptotic cell lysates revealed that acinus is activated by caspase 3 cleavage, and it induces apoptotic chromatin condensation in the absence of DNA fragmentation (Sahara et al., 1999). It was also shown that acinus is important for apoptotic chromatin condensation in vivo by using antisense RNA (Sahara et al., 1999). Recently, a complex called ASAP, containing RNPS1 (splicing factor), acinus and SAP18 (Sin3-associated protein; a component of a histone deacetylase complex), was isolated and the complex was shown to promote both pre-mRNA splicing and apoptosis, suggesting a possible link among apoptosis, splicing, and chromatin modification (Schwerk et al., 2003). Interestingly, acinus contains an RS domain (Boucher et al., 2001) that accounts for its localization to IGCs (Figure 2).
A second protein implicated in apoptosis, Btf (KIAA0164), was identified as a protein associated with the adenovirus oncoprotein E1B 19K as well as Bcl-2 family members. Btf has a transcriptional repression activity and its sustained overexpression induces apoptosis and suppresses transformation by E1A and E1B-19K or mutant p53 (Kasof et al., 1999). Although we have found that acinus colocalized within IGCs, Btf is localized at the periphery of IGCs (Figure 2).
As potential IGC proteins, we detected DNA repair proteins such as XPE UV-damaged DNA binding protein and XPA-binding protein 2 (Table 2). It is also interesting that we detected several types of “chaperone” proteins such as Hsp70, Dna J protein homolog, or RuvB like DNA helicase. In the developing kidney, Hsp70 is colocalized with Wilms tumor suppressor WT-1 in a speckled nuclear distribution pattern (Maheswaran et al., 1998). In the plant Brassica napus, it was shown that Hsp70 becomes associated with RNP structures in the interchromatin region and the nucleolus upon stress treatment to induce embryogenesis of microspores (Segui-Simarro et al., 2003). Although the localization of Hsp70 in IGCs remains to be confirmed, it would be interesting to analyze the changes in protein components in IGCs throughout the stages of development, oncogenesis, or environmental changes.
Recently, it has been suggested that transcription and translation are coupled. A small amount of translation, which might be important for quality control of gene products, has been reported to take place in the nucleus before export of mRNAs to the cytoplasm where the majority of translation occurs (Iborra et al., 2001). Thus far, we have detected two isoforms of eukaryotic initiation factor 4A, eIF4Ai and iii, in our proteomics analysis of IGCs. We and others also have found that fluorescently tagged eIF4Aiii is localized to IGCs (Holzmann et al., 2000). It has been shown that eIF4Ai, ii, and iii all confer RNA-dependent RNA helicase and ATP-dependent RNA helicase activities. However, they seem to function differently because eIF4Ai and ii facilitate translation, but eIF4Aiii inhibits translation in a reticulocyte lysate (Li et al., 1999). Recently, eIF4Aiii has been shown to be involved in nonsense-mediated decay (NMD) (Ferraiuolo et al., 2004). NMD is an RNA surveillance mechanism that serves to degrade mRNAs containing premature translation termination codons (for a review, see Maniatis and Reed, 2002; Wilkinson and Shyu, 2002; Singh and Lykke-Andersen, 2003). In our IGC fraction, we identified numerous members of the exon-exon junction complex that contains factors that are required for both mRNA export and NMD [Aly, RNPS1, RNA binding motif protein 8 (Y14), and mago-nashi homolog (MAGOH)]. This finding raises the possibility that proteins involved in these processes may be recruited from IGCs to transcription sites.
Motif Analysis
As expected, we detected many proteins with RNA binding motifs, RS motifs, and RNA helicase motifs, including ATP binding DEAD box helicases. However, thus far we have not detected a single sequence motif that is common among all IGC proteins. Therefore, aside from the RS domain, which serves to target certain proteins to IGCs, many other IGC-associated proteins may assemble into these structures by specific protein–protein and/or protein–RNA interactions rather than by a single targeting signal. Interestingly, 82% of the identified IGC proteins contain low complexity regions, such as a long stretch of a single type of amino acid, which could be involved in interactions with RNA or other proteins.
Because the RS motif seems to be unique among IGC proteins, we focused on a more in depth analysis of proteins containing an RS domain. We found that this group of proteins can be divided into several subgroups (Figure 3). In addition to the typical small RS domain-containing proteins that contain one or more RRMs, among which are members of the SR family of pre-mRNA splicing factors, there are larger RS domain-containing proteins containing additional domains and/or regions containing short repeats. It is plausible to imagine that these repeats are likely to perform a scaffolding function, as is found for certain HEAT repeat-containing proteins (Neuwald and Hirano, 2000). Also interesting are four proteins, U1 snRNP70, pre-mRNA cleavage factor Im, U1 small ribonucleoprotein 1, and acinus, that have degenerated RS domains in which the RS repeat itself contains, or is continuous with, RD/E dipeptides. RE repeats were previously found in the splicing factor YT521-B and were shown to be important for localization to the YT body, a subnuclear structure that is similar to but distinct from nuclear speckles (Nayler et al., 2000). The RD/E dipeptide motif is reminiscent of a phosphorylated RS domain, because the serine residue in RS is replaced with a negatively charged aspartic acid or glutamic acid. Interestingly, YT521-B was shown to localize to transcriptionally active sites and was suggested to play a role in grouping genes into higher order structures (Nayler et al., 2000). Thus, proteins with both RS and RD/E motifs may bridge sites of active transcription with IGCs.
In summary, we have characterized the proteome of IGCs purified from mouse liver nuclei. Although the protein identification supports a role of these nuclear domains in events relating to pre-mRNA processing, a significant number of new proteins have been identified, as well as interesting domains of known proteins. These will provide the impetus for future studies aimed at deciphering the organization and additional function(s) associated with this nuclear organelle.
Acknowledgments
This work was supported by grant 42694 to D.L.S. from the National Institute of General Medical Sciences/National Institutes of Health. N.S. was funded by a postdoctoral fellowship from the American Cancer Society (PF-00-008-01-CSM) and by the Breast Cancer Research Program from the U.S. Army Medical Research and Material Command (BC990019).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0253. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0253.
References
- Andersen, J.S., Lyon, C.E., Fox, A.H., Leung, A.K., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1-11. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (2000). SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25, 112-114. [DOI] [PubMed] [Google Scholar]
- Belgrader, P., Dey, R., and Berezney, R. (1991). Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J. Biol. Chem. 266, 9893-9899. [PubMed] [Google Scholar]
- Birney, E., Kumar, S., and Krainer, A.R. (1993). Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21, 5803-5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J., Bauren, G., Eldridge, A.G., Issner, R., Nickerson, J.A., Rosonina, E., and Sharp, P.A. (2000). The SRm160/300 splicing coactivator subunits. RNA 6, 111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher, L., Ouzounis, C.A., Enright, A.J., and Blencowe, B.J. (2001). A genome-wide survey of RS domain proteins. RNA 7, 1693-1701. [PMC free article] [PubMed] [Google Scholar]
- Brandner, J.M., Reidenbach, S., and Franke, W.W. (1997). Evidence that “pinin”, reportedly a differentiation-specific desmosomal protein, is actually a widespread nuclear protein. Differentiation 62, 119-127. [DOI] [PubMed] [Google Scholar]
- Bregman, D.B., Du, L., van der Zee, S., and Warren, S.L. (1995). Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129, 287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko, D., Verschure, P.J., Martin, T.E., Dahmus, M.E., Krause, S., Fu, X.D., van Driel, R., and Fakan, S. (1999). Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell 10, 211-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden, J.L., and Patturajan, M. (1997). A CTD function linking transcription to splicing. Trends Biochem. Sci. 22, 413-416. [DOI] [PubMed] [Google Scholar]
- Cronshaw, J.M., Krutchinsky, A.N., Zhang, W., Chait, B.T., and Matunis, M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eils, R., Gerlich, D., Tvarusko, W., Spector, D.L., and Misteli, T. (2000). Quantitative imaging of pre-mRNA splicing factors in living cells. Mol. Biol. Cell 11, 413-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan, S., and Puvion, E. (1980). The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int. Rev. Cytol. 65, 255-299. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo, M.A., Lee, C.S., Ler, L.W., Hsu, J.L., Costa-Mattioli, M., Luo, M.J., Reed, R., and Sonenberg, N. (2004). A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl. Acad. Sci. USA 101, 4118-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.D. (1995). The superfamily of arginine/serine-rich splicing factors. RNA 1, 663-680. [PMC free article] [PubMed] [Google Scholar]
- Fu, X.D., and Maniatis, T. (1990). Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343, 437-441. [DOI] [PubMed] [Google Scholar]
- Gottschalk, A., Neubauer, G., Banroques, J., Mann, M., Luhrmann, R., and Fabrizio, P. (1999). Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J 18, 4535-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. (2000). Sorting out the complexity of SR protein functions. RNA 6, 1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann, K., Gerner, C., Poltl, A., Schafer, R., Obrist, P., Ensinger, C., Grimm, R., and Sauermann, G. (2000). A human common nuclear matrix protein homologous to eukaryotic translation initiation factor 4A. Biochem. Biophys. Res. Commun. 267, 339-344. [DOI] [PubMed] [Google Scholar]
- Huang, S. (2000). Review: perinucleolar structures. J. Struct. Biol. 129, 233-240. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., Jackson, D.A., and Cook, P.R. (2001). Coupled transcription and translation within nuclei of mammalian cells. Science 293, 1139-1142. [DOI] [PubMed] [Google Scholar]
- Inagaki, H., Matsushima, Y., Nakamura, K., Ohshima, M., Kadowaki, T., and Kitagawa, Y. (1996). A large DNA-binding nuclear protein with RNA recognition motif and serine/arginine-rich domain. J. Biol. Chem. 271, 12525-12531. [DOI] [PubMed] [Google Scholar]
- Jagatheesan, G., Thanumalayan, S., Muralikrishna, B., Rangaraj, N., Karande, A.A., and Parnaik, V.K. (1999). Colocalization of intranuclear lamin foci with RNA splicing factors. J. Cell Sci. 112, 4651-4661. [DOI] [PubMed] [Google Scholar]
- Janicki, S.M., et al. (2004). From silencing to gene expression: real-time analysis in single cells. Cell 116, 683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K.M., Wang, J., Smallwood, A., Arayata, C., and Carey, M. (2002). TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16, 1852-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasof, G.M., Goyal, L., and White, E. (1999). Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol. Cell. Biol. 19, 4390-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, N., Diem, M.D., Kim, V.N., Yong, J., and Dreyfuss, G. (2001). Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 20, 6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak, M.J., Lever, M.A., Fischle, W., Verdin, E., Bazett-Jones, D.P., and Hendzel, M.J. (2000). Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J. Cell Biol. 150, 41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I., and Earnshaw, W.C. (1998). Structure and function in the nucleus. Science 280, 547-553. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and Spector, D.L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4, 605-612. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P., and Bork, P. (2002). Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30, 242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Imataka, H., Morino, S., Rogers, G.W., Jr., Richter-Cook, N.J., Merrick, W.C., and Sonenberg, N. (1999). Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 19, 7336-7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, P., Lasko, P.F., Ashburner, M., Leroy, P., Nielsen, P.J., Nishi, K., Schnier, J., and Slonimski, P.P. (1989). Birth of the D-E-A-D box. Nature 337, 121-122. [DOI] [PubMed] [Google Scholar]
- Luking, A., Stahl, U., and Schmidt, U. (1998). The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33, 259-296. [DOI] [PubMed] [Google Scholar]
- Maheswaran, S., et al. (1998). Inhibition of cellular proliferation by the Wilms tumor suppressor WT1 requires association with the inducible chaperone Hsp70. Genes Dev. 12, 1108-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., and Reed, R. (2002). An extensive network of coupling among gene expression machines. Nature 416, 499-506. [DOI] [PubMed] [Google Scholar]
- Matera, A.G. (1999). RNA splicing: more clues from spinal muscular atrophy. Curr. Biol. 9, R140-R142. [DOI] [PubMed] [Google Scholar]
- Mintz, P.J., Patterson, S.D., Neuwald, A.F., Spahr, C.S., and Spector, D.L. (1999). Purification and biochemical characterization of interchromatin granule clusters. EMBO J 18, 4308-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. (2000). Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J. Cell Sci. 113, 1841-1849. [DOI] [PubMed] [Google Scholar]
- Misteli, T., Caceres, J.F., Clement, J.Q., Krainer, A.R., Wilkinson, M.F., and Spector, D.L. (1998). Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143, 297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., Caceres, J.F., and Spector, D.L. (1997). The dynamics of a pre-mRNA splicing factor in living cells. Nature 387, 523-527. [DOI] [PubMed] [Google Scholar]
- Misteli, T., and Spector, D.L. (1998). The cellular organization of gene expression. Curr. Opin. Cell Biol. 10, 323-331. [DOI] [PubMed] [Google Scholar]
- Misteli, T., and Spector, D.L. (1999). RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3, 697-705. [DOI] [PubMed] [Google Scholar]
- Mortillaro, M.J., Blencowe, B.J., Wei, X., Nakayasu, H., Du, L., Warren, S.L., Sharp, P.A., and Berezney, R. (1996). A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA 93, 8253-8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu, H., and Berezney, R. (1991). Nuclear matrins: identification of the major nuclear matrix proteins. Proc. Natl. Acad. Sci. USA 88, 10312-10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu, H., and Ueda, K. (1984). Small nuclear RNA-protein complex anchors on the actin filaments in bovine lymphocyte nuclear matrix. Cell Struct. Funct. 9, 317-325. [DOI] [PubMed] [Google Scholar]
- Nayler, O., Hartmann, A.M., and Stamm, S. (2000). The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J. Cell Biol. 150, 949-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer, G., Gottschalk, A., Fabrizio, P., Seraphin, B., Luhrmann, R., and Mann, M. (1997). Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. USA 94, 385-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer, G., King, A., Rappsilber, J., Calvio, C., Watson, M., Ajuh, P., Sleeman, J., Lamond, A., and Mann, M. (1998). Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20, 46-50. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F., and Hirano, T. (2000). HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 10, 1445-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, P., and Sugrue, S.P. (1996). Characterization of pinin, a novel protein associated with the desmosome-intermediate filament complex. J. Cell Biol. 135, 1027-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, P., Zhen, Y.Y., and Sugrue, S.P. (1997). Cloning and analysis of cDNA encoding murine pinin. Gene 197, 115-120. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., and Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604-609. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B.T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco-Bubulya, P., and Spector, D.L. (2002). Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156, 425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara, S., Aoto, M., Eguchi, Y., Imamoto, N., Yoneda, Y., and Tsujimoto, Y. (1999). Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature 401, 168-173. [DOI] [PubMed] [Google Scholar]
- Scherl, A., Coute, Y., Deon, C., Calle, A., Kindbeiter, K., Sanchez, J.C., Greco, A., Hochstrasser, D., and Diaz, J.J. (2002). Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13, 4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, E.C., Florens, L., Guan, T., Yates, J.R., 3rd, and Gerace, L. (2003). Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380-1382. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95, 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk, C., Prasad, J., Degenhardt, K., Erdjument-Bromage, H., White, E., Tempst, P., Kidd, V.J., Manley, J.L., Lahti, J.M., and Reinberg, D. (2003). ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 23, 2981-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui-Simarro, J.M., Testillano, P.S., and Risueno, M.C. (2003). Hsp70 and Hsp90 change their expression and subcellular localization after microspore embryogenesis induction in Brassica napus L. J. Struct. Biol. 142, 379-391. [DOI] [PubMed] [Google Scholar]
- Singh, G., and Lykke-Andersen, J. (2003). New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 28, 464-466. [DOI] [PubMed] [Google Scholar]
- Spahr, C.S., Susin, S.A., Bures, E.J., Robinson, J.H., Davis, M.T., McGinley, M.D., Kroemer, G., and Patterson, S.D. (2000). Simplification of complex peptide mixtures for proteomic analysis: reversible biotinylation of cysteinyl peptides. Electrophoresis 21, 1635-1650. [DOI] [PubMed] [Google Scholar]
- Spector, D.L. (1993). Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9, 265-315. [DOI] [PubMed] [Google Scholar]
- Spector, D.L. (2001). Nuclear domains. J. Cell Sci. 114, 2891-2893. [DOI] [PubMed] [Google Scholar]
- Spector, D.L. (2003). The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72, 573-608. [DOI] [PubMed] [Google Scholar]
- Spector, D.L., Goldman, R.D., and Leinwand, L.A. (1998). Cells: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Spector, D.L., Lark, G., and Huang, S. (1992). Differences in snRNP localization between transformed and nontransformed cells. Mol. Biol. Cell 3, 555-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy, L., Sleeman, J.E., and Lamond, A.I. (2001). Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 114, 4219-4228. [DOI] [PubMed] [Google Scholar]
- Valdez, B.C., Henning, D., Perlaky, L., Busch, R.K., and Busch, H. (1997). Cloning and characterization of Gu/RH-II binding protein. Biochem. Biophys. Res. Commun. 234, 335-340. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.F., and Shyu, A.B. (2002). RNA surveillance by nuclear scanning? Nat. Cell Biol. 4, E144-E147. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Urlaub, H., Achsel, T., Gentzel, M., Wilm, M., and Luhrmann, R. (2002). Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21, 4978-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. (2002). Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182-185. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Luo, M.J., Straesser, K., Katahira, J., Hurt, E., and Reed, R. (2000). The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407, 401-405. [DOI] [PubMed] [Google Scholar]