Abstract
To address questions about algal virus persistence (i.e., continued existence) in the environment, rates of decay of infectivity for two viruses that infect Chlorella-like algae, ATCV-1 and CVM-1, and a virus that infects the prymnesiophyte Chrysochromulina parva, CpV-BQ1, were estimated from in situ incubations in a temperate, seasonally frozen pond. A series of experiments were conducted to estimate rates of decay of infectivity in all four seasons with incubations lasting 21 days in spring, summer and autumn, and 126 days in winter. Decay rates observed across this study were relatively low compared with previous estimates obtained for other algal viruses, and ranged from 0.012 to 11% h−1. Overall, the virus CpV-BQ1 decayed most rapidly whereas ATCV-1 decayed most slowly, but for all viruses the highest decay rates were observed during the summer and the lowest were observed during the winter. Furthermore, the winter incubations revealed the ability of each virus to overwinter under ice as ATCV-1, CVM-1 and CpV-BQ1 retained up to 48%, 19% and 9% of their infectivity after 126 days, respectively. The observed resilience of algal viruses in a seasonally frozen freshwater pond provides a mechanism that can support the maintenance of viral seed banks in nature. However, the high rates of decay observed in the summer demonstrate that virus survival and therefore environmental persistence can be subject to seasonal bottlenecks.
Introduction
Since the revelation that viruses are the numerically dominant component of aquatic environments, the burgeoning field of viral ecology has begun to illuminate the roles that viruses play in these ecosystems (Bergh et al., 1989; Wommack and Colwell, 2000; Brussaard, 2004; Suttle, 2007; Short, 2012). Their high abundance and obligately parasitic lifestyle allow viruses to exert top-down control of cellular organism populations, which is illustrated most dramatically through the implication that viruses are involved in the termination of some algal blooms (e.g., Bratbak et al., 1993; Tarutani et al., 2000; Wilson et al., 2002; Brussaard et al., 2005: Gobler et al., 2007; Tomaru et al., 2007). More subtly, viruses contribute to the mortality of bacteria, phytoplankton and higher trophic levels of the aquatic food web (Proctor and Fuhrman, 1990; Suttle, 1994; Baudoux et al., 2006). Viral lysis of algae (Haaber and Middelboe, 2009) and bacteria (Middelboe et al., 2003) can cause the release of particulate and dissolved organic matter, and dissolved organic matter can also be leaked from algal cells currently infected with viruses (Shiek et al., 2014). Liberated particulate organic matter and dissolved organic matter due to viral lysis or leakage from infected cells can be utilized by bacteria (Bratbak et al., 1998; Middelboe et al., 2003; Haaber and Middelboe, 2009; Sheik et al., 2014) or by primary producers (Shelford et al., 2012). Together, these observations demonstrate that viruses can have direct effects on ecosystems via viral lysis of host cells and altered population dynamics, and indirect effects such as enhanced nutrient recycling (Fuhrman, 1999). Further, by altering the flow of nutrients, viruses can even stimulate primary production (Weinbauer et al., 2011; Staniewski and Short, 2014).
Historically, aquatic virus ecology has focused on marine environments, but high viral abundances have been observed in both the water column and the sediments of freshwater systems (Maranger and Bird, 1996; Filippini and Middelboe, 2007). The importance of viruses as agents of freshwater phytoplankton mortality has also been established through a number of modified dilution experiments (Gobler et al., 2008; Tijdens et al., 2008; Staniewski et al., 2012), and algal virus diversity surveys of various lakes and rivers have been conducted (e.g., Short and Short, 2008; Clasen and Suttle, 2009; Gimenes et al., 2012; Zhong and Jacquet, 2014). Seasonal studies of algal virus abundance in lakes (e.g., Short and Short, 2009; Short et al., 2011a; Hewson et al., 2012; Rozon and Short, 2013) have revealed distinct seasonality with patterns of ‘boom or bust' oscillations, or constant abundance depending on the particular virus examined. Similar abundance patterns have been observed for virus-like particles and phages in rivers (e.g., Mathias et al., 1995; Farnell-Jackson and Ward, 2003) and lakes (Hofer and Sommaruga, 2001; Bettarel et al., 2004; Hewson et al., 2012; Zhong et al., 2015). Additionally, observations of thousands of viral genomes in metagenomic studies of aquatic environments (Breitbart et al., 2002), and of persistent viruses that exist at low but detectable abundances throughout much of the year (Waterbury and Valois, 1993; Short and Short, 2009; Short et al., 2011a; Zhong et al., 2013), have provided evidence for an environmental ‘seed bank.' In the context of aquatic viruses, the concept of a seed bank is borrowed from terrestrial plant ecology and implies that an inactive pool of viruses persist in the environment waiting for appropriate conditions for ‘germination' or replication (Short et al., 2011a). In turn, this idea is based on the ‘Bank model' hypothesis of Breitbart and Rohwer (2005). Their metagenomics study demonstrated that only a few virus genomes are highly abundant, and most are rare and part of a ‘bank' fraction maintained at low abundances resisting destruction until their hosts reach abundances high enough to promote their replication.
In contrast to observations of ‘seed bank' viruses, experimentally derived decay rates of aquatic viruses are variable, but can be high, ranging from 0.13% to 54% of particles every hour (% h−1; Heldal and Bratbak, 1991; Cottrell and Suttle, 1995; Noble and Fuhrman, 1997; Garza and Suttle, 1998; Hewson et al., 2012; Frada et al., 2014). To date, there are few environmental decay rate estimates for algal viruses. For marine algal viruses, decay rates of MpV-SP1 that infects the prasinophyte Micromonas pusilla (Cottrell and Suttle, 1995) and the virus EhV that infects the coccolithophore Emiliania huxleyi (Frada et al., 2014) have been determined via loss of infectivity and were also variable, but suggested relatively high turnover (2–3% and 28–30% h−1 for EhV and MpV-SP1, respectively); a decay rate of 2% h−1 is equal to a half-life of only 34 h. Interestingly, decay rates of freshwater algal viruses most closely related to Acanthocystis turfacea Chlorella virus 1 (ATCV-1) were estimated by tracking loss of viral DNA in experimental incubations of water samples from a lake in New York, and were much lower than previous reports of algal virus decay with an estimated half-life of 22 days (the decay constant, λ=0.13% h−1; Hewson et al., 2012).
Since virus decay rates are potentially high, the seasonality of algal hosts of many viruses presents an obstacle to the continued production, and thus the persistence (i.e., continued existence), of virus particles throughout the year. It is well established that many freshwater algae species drop below detectable levels throughout much of the year (e.g., Munawar and Munawar; 1986; Reynolds, 2006). Because estimated host abundance thresholds for virus transmission range from 103 to 104 host cells ml−1 for both cyanophages (e.g., Wiggins and Alexander, 1985; Suttle and Chan, 1994) and algal viruses (e.g., Cottrell and Suttle, 1995; Jacquet et al., 2002), it is clear that both viral infection and production are dependent upon host availability. Given the apparently contradictory observations of viral ‘seed banks' and seemingly high decay rates for the few virus taxa that have been studied, it is vital to further explore viruses' ability to persist in the environment when their hosts are absent.
The purpose of our study was three-fold: (i) to gain information about the environmental persistence of algal viruses in freshwater, (ii) to test if virus decay rates, and hence their ability to survive outside of host cells, varies seasonally, and (iii) to determine if algal viruses survive during the winter months in a temperate freshwater habitat. With these goals in mind, decay rates of three strains of freshwater algal viruses including two chloroviruses, ATCV-1 and Chlorella virus Marburg-1 (CVM-1), and a newly isolated virus (CpV-BQ1; Mirza et al., 2015) which infects the prymnesiophyte Chrysochromulina parva were estimated from seasonal in situ incubations. These particular viruses were used in this study because their hosts can be grown in the laboratory and, therefore, titers of infectious viruses can be estimated. Furthermore, close relatives of each of these viruses are known in local freshwaters; CpV-BQ1 was isolated from Ontario waters (Mirza et al., 2015) and DNA polymerase gene sequences closely related to ATCV-1, CVM-1 and CpV-BQ1 have been amplified from the study site (Short et al., 2011b).
Materials and methods
In situ incubations to estimate virus decay
To assess the seasonality of algal virus decay in a freshwater environment, decay rates of ATCV-1, CVM-1 and CpV-BQ1 were estimated via experiments conducted throughout the year in a storm water management pond on the University of Toronto Mississauga campus. Seasonal decay rates were estimated by incubating 500 ml natural water samples with known concentrations of infectious viruses in situ during incubation experiments initiated within a few days of the spring and fall equinoxes and the summer solstice, and within a month of the winter solstice on 23 May 2013, 21 June 2013, 3 October 2013 and 2 December 2013. The infectious titers in incubation bottles were determined before and after incubation to provide estimates of loss of infectivity (i.e., decay). Additionally, to compare biotic and abiotic components of decay, each incubation experiment involved two treatments; infectious viruses were incubated with either unfiltered water or water filtered to remove microorganisms larger than viruses.
At the beginning of each decay incubation, ~15 l water samples collected from the University of Toronto Mississauga pond were passed through a 210- μm pore-size Nitex mesh to remove large particulates and floating debris before being split for the two different treatments. The so-called unfiltered water (i.e., whole water) treatment used filtrate from the Nitex mesh as medium for in situ virus incubations while the other treatment used water filtered through a 142 mm dia., 0.45- μm pore-size HVLP membrane filter (EMD Millipore, Etobicoke, Canada). For each experiment and treatment (whole water and filtered water), triplicate 500 ml polycarbonate bottles (VWR International, Mississauga, Canada) were filled with the appropriate natural water to which final concentrations of 1.40–4.31 × 106 infectious viruses ml−1 for ATCV-1, 7.02 × 105 to 2.96 × 106 for CVM-1 and 2.20–3.53 × 104 for CpV-BQ1 were added. CpV-BQ1 had not been isolated at the time of the spring incubation and thus was not used in the May 2013 experiment. PC bottles, although ultraviolet (UV) opaque, were used for this study because of their durability and because they are not known to have deleterious effects on algal viruses. Given the lengthy incubations that were conducted, using incubations bottles that could maintain sample integrity by withstanding potential disturbances from local fauna as well as a wide range of environmental conditions was deemed essential. For the spring, summer and autumn decay experiments triplicate bottles for each treatment were destructively sampled after incubating in situ for 1, 4, 7 and 21 days. Winter incubations were sampled on days 1, 4 and 7, and again after 126 days to test the survivability of algal viruses when frozen in the pond over the winter months. During incubations, the bottles were secured in an arbitrary order to a PVC frame tethered to float unshaded at the surface of the University of Toronto Mississauga pond. Ice thickness was monitored for the first 7 days of the winter experiment.
Water temperatures were measured at each time point with a digital thermometer (VWR International). Upon destructive sampling of triplicate incubation bottles at each time point and for each treatment, 100 ml of H2O from each bottle was sequentially filtered through a 47 mm dia., GC50 glass fiber filter (0.5 μm nominal rating, Advantec AMD Manufacturing Inc., Mississauga, Canada) followed by a 47 mm dia., 0.45 μm pore-size HVLP membrane filter (EMD Millipore). The infectious titers of each virus in the resultant 0.45 μm filtrates were determined and used to calculate environmental decay rates for each sample. Infectious titers were estimated within 2 weeks after destructive sampling at each time point; an earlier experiment demonstrated that 4 °C storage of a 0.45 μm filtrated lysate of ATCV-1 retained 100% of its infectivity even after 86 days (data not shown).
Cell culture conditions and estimating virus titers
Cell cultures of Micractinium conductrix strain Pbi (formerly Chlorella strain Pbi) were grown in FES medium (Reisser et al., 1986), while cultures of Chlorella heliozoae strain SAG 3.83 were grown in modified Bold's basal medium (Van Etten et al., 1983), and Chrysochromulina parva were grown in DY-V medium (Anderson et al., 2005). Viral lysates were generated for ATCV-1, CVM-1 and CpV-BQ1 by inoculating 1 ml of infectious viruses (i.e., 0.45- μm-filtered viral lysates) into 150 ml cultures of the appropriate host. After the cell cultures cleared, the resulting viral lysates were filtered with a 47- mm dia. 0.45- μm pore-size HVLP membrane filter (Merck Millipore, Billerica, MA, USA). Filtered viral lysates were stored at 4 °C until utilization in environmental decay experiments. Infectious titers of the viral lysates were estimated using plaque assays for ATCV-1 and CVM-1, and a most probable number assay (MPN) for CpV-BQ1. Infectious titers remaining in each bottle after in situ incubation were determined using the same methods.
Plaque assays for ATCV-1 used the host C. heliozoae grown on modified Bold's basal medium—agar medium (Van Etten et al., 1983), whereas plaque assays for CVM-1 were carried out using the same protocol, but with M. conductrix grown on FES agar medium. Plaque assays were performed in triplicate for every sample. At the time of this study C. parva had not been successfully cultivated on solid medium, so MPN assays were used to titer CpV-BQ1. MPNs were carried out in 96-well microtiter plates with 100 μl of C. parva cells (107 cells ml−1) and 100 μl of the 0.45 μm-filtered samples from each incubation bottle diluted serially from 100 to 10−10; each column of eight wells in the plates were replicates of a single dilution level. A column of wells with 100 μl of cells and 100 μl of DY-V was used as a control in each plate. MPNs were calculated as described in Jarvis et al. (2010).
Decay rate calculations and statistical analyses
Decay rates were calculated as previously described (Noble and Fuhrman, 1997). Briefly, linear regressions were calculated for natural log transformed infectious titers plotted against time, as decay of infectivity follows the exponential model N(t)=N0e−λt, where N0 is the infectious titer at time zero, N(t) is the infectious titer at time t and λ is the decay constant. The slope of the regressions represent the decay constants (units are h−1), the reciprocal of the decay constants are turnover times and decay rates expressed as percentage infectivity lost per hour were calculated by multiplying λ by 100 and half-lives were calculated by dividing ln(2) by λ. In the summer incubation, time points 7 and 21 days were excluded from the CpV-BQ1 calculations in both treatments due to the absence of detectable infectious CpV-BQ1 viruses. Statistical comparisons using summer CpV-BQ1 data also exclude these time points.
Linear regression analyses and analysis of covariance (ANCOVA) statistical tests were conducted using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA). To be consistent with the decay rate calculations, statistical tests were calculated using natural log transformed data. A significance level of 0.05 was used for linear regression. To determine if decay rates of the three viruses were significantly different, ANCOVA was used to compare the slopes of linear regressions of different viruses incubated during the same season and in the same treatments. Similarly, to determine if the two treatments (whole-water incubation versus incubation in 0.45 μm filtrate) had a significant effect on viral decay rates, ANCOVA was used to compare slopes of two treatments for the same viruses within the same season, while tests for the effect of seasons on viral decay rates were based on comparisons of the same viruses in the same treatment incubated during different times of the year. To account for the multiple pairwise comparisons, Bonferroni corrections were applied to adjust the significance levels for seasonal (α=0.00167), treatment (α=0.0045) and virus-to-virus (α=0.0025) comparisons.
Results
Environmental parameters
The average number of daylight hours over the course of the spring, summer, autumn and winter experiments were 15.25, 15.36, 11.05 and 10.22 h, respectively, with cumulative daylight hours and irradiation greatest during the summer and lowest in winter (Table 1). The average water temperature over the five time points was 21.85, 27.08, 17.36 and 3.18 °C during the spring, summer, autumn and the first four time points of winter, respectively. The temperature at the final time point of the winter experiment after the pond thawed was 5.9 °C. During the winter incubation, ice covered the pond for all 126 days with a thickness of at least 1.5 cm and snowpack up to 25 cm according to data from Environment Canada for nearby Toronto International Airport (approx. 14 km from the study site). During this winter incubation, snow began to accumulate on the seventh day of the incubation and stayed until the 109th day of the 126 day incubation. The daily average snow cover thickness was 5, 6, 15 and 5 cm, during the months of December, January, February and March, respectively.
Table 1. Environmental parameters for seasonal decay experiments.
| Season | Sampling times (h) | Water temperature (°C) | Cummulative daylight hours | Ice thickness (cm) |
|---|---|---|---|---|
| Spring | 0 | 20.6 | 0 | — |
| 24 | 15.3 | 15.03 | — | |
| 96 | 20.7 | 60.31 | — | |
| 168 | 27.1 | 105.78 | — | |
| 504 | 25.6 | 320.05 | — | |
| Summer | 0 | 25.1 | 0 | — |
| 24 | 26.7 | 15.45 | — | |
| 96 | 29.4 | 61.78 | — | |
| 168 | 27.1 | 108.07 | — | |
| 504 | 27.1 | 322.63 | — | |
| Fall | 0 | 21.2 | 0 | — |
| 24 | 18.5 | 11.07 | — | |
| 96 | 18.1 | 45.45 | — | |
| 168 | 18.3 | 79.38 | — | |
| 504 | 10.8 | 232.13 | — | |
| Wintera | 0 | 3.5 | 0 | 3 |
| 24 | 4.0 | 10.12 | 4.5 | |
| 96 | 2.8 | 37.37 | 1.5 | |
| 168 | 2.4 | 64.37 | 5.5 | |
| 3072 | 5.9 | 1288.53 | 0 |
Cummulative daylight hours do not necessarily reflect the exposure of viruses to daylight during this incubation due to persistent ice and snow cover.
Environmental decay
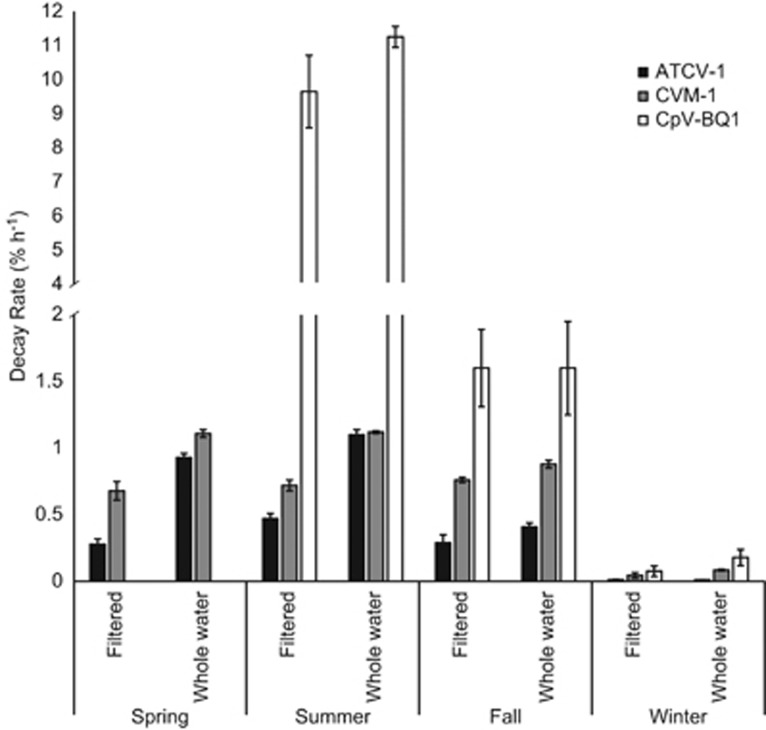
All three algal viruses experienced lowest decay rates in the winter filtered-water treatment and the highest in the summer whole-water treatment (Figure 1). ATCV-1 infectivity decayed at rates ranging from 0.012 to 1.10% h−1 with the lowest rates in the winter and the highest in the summer (Figure 1). Half-lives for ATCV-1 ranged from 2.6 to 240 days. CVM-1 decay rates ranged from 0.047 to 1.2% h−1 with half-lives from 2.4 to 61 days, and for CpV-BQ1, decay rates ranged from 0.077 to 11% h−1, with half-lives from 0.26 to 38 days.
Figure 1.
Seasonal decay rates of algal viruses. Each data point is the average of three calculated decay rates from triplicate incubations for each time point and treatment. Error bars represent standard deviation. Note the split Y-axis with different scaling above and below the split.
Statistical comparisons of decay rates
Complete statistical comparisons of slopes from the regressions of the natural logarithm of virus abundance versus time (i.e., decay rates) for each virus during every season and each treatment are compiled in Table 2 and Supplementary Tables S1–S3. Except for the regression slope for ATCV-1 during the winter decay incubation in the whole-water treatment, the regression slopes for all decay incubations were significantly non-zero, with P-values <0.05 (Table 2). Pairwise comparisons of the regression slopes were conducted to detect differences in decays rates between seasons, between filtered- or whole-water treatments, and between the different viruses themselves, and significant differences were observed among all three sets of comparisons.
Table 2. Linear regression analysis of decay curves.
| Season | Virus | Treatment | Slope | 95% confidence interval | Slope signficantly non-zero? | F | DFn, DFd | P-value |
|---|---|---|---|---|---|---|---|---|
| Spring | ATCV-1 | Filtered | −0.0028±0.00044 | −0.0038 to −0.0019 | Yes | 41.03 | 1, 13 | <0.0001 |
| Whole water | −0.0093±0.00043 | −0.0102 to −0.0084 | Yes | 474.4 | 1, 13 | <0.0001 | ||
| CVM-1 | Filtered | −0.0068±0.00034 | −0.0075 to −0.0061 | Yes | 410.4 | 1, 13 | <0.0001 | |
| Whole water | −0.011±0.00041 | −0.012 to −0.0102 | Yes | 733.5 | 1, 13 | <0.0001 | ||
| Summer | ATCV-1 | Filtered | −0.0047±0.00056 | −0.0059 to −0.0035 | Yes | 70.8 | 1, 13 | <0.0001 |
| Whole water | −0.011±0.00019 | −0.011 to −0.0105 | Yes | 3144 | 1, 13 | <0.0001 | ||
| CVM-1 | Filtered | −0.0072±0.00063 | −0.0086 to −0.0059 | Yes | 132.3 | 1, 13 | <0.0001 | |
| Whole water | −0.012±0.00062 | −0.013 to −0.0103 | Yes | 351.5 | 1, 13 | <0.0001 | ||
| CpV-BQ1 | Filtered | −0.096±0.0042 | −0.11 to −0.087 | Yes | 530.5 | 1, 7 | <0.0001 | |
| Whole water | −0.11±0.0041 | −0.12 to −0.102 | Yes | 761.8 | 1, 7 | <0.0001 | ||
| Autumn | ATCV-1 | Filtered | −0.0030±0.00022 | −0.0034 to −0.0025 | Yes | 178 | 1, 13 | <0.0001 |
| Whole water | −0.0040±0.00028 | −0.0047 to −0.0034 | Yes | 206.9 | 1, 13 | <0.0001 | ||
| CVM-1 | Filtered | −0.0076±0.00024 | −0.0081 to −0.00702 | Yes | 956.3 | 1, 13 | <0.0001 | |
| Whole water | −0.0088±0.00047 | −0.0098 to −0.0078 | Yes | 348.3 | 1, 13 | <0.0001 | ||
| CpV-BQ1 | Filtered | −0.016±0.0029 | −0.023 to −0.0101 | Yes | 31.8 | 1, 13 | <0.0001 | |
| Whole water | −0.016±0.0035 | −0.024 to −0.0083 | Yes | 20.6 | 1, 13 | 0.00061 | ||
| Winter | ATCV-1 | Filtered | −0.00014±4.8e−005 | −0.00025 to −3.7e−005 | Yes | 8.5 | 1, 13 | 0.012 |
| Whole water | −0.00012±6.8e−005 | −0.00027 to 2.7e−005 | No | 3.1 | 1, 13 | 0.10 | ||
| CVM-1 | Filtered | −0.00047±6.1e−005 | −0.00059 to −0.00034 | Yes | 61.04 | 1, 13 | <0.0001 | |
| Whole water | −0.00086±6.5e−005 | −0.001001 to −0.00072 | Yes | 178.4 | 1, 13 | <0.0001 | ||
| CpV-BQ1 | Filtered | −0.00077±0.00013 | −0.0011 to −0.00048 | Yes | 34.1 | 1, 13 | <0.0001 | |
| Whole water | −0.0018±0.00017 | −0.0021 to −0.0014 | Yes | 103.8 | 1, 13 | <0.0001 |
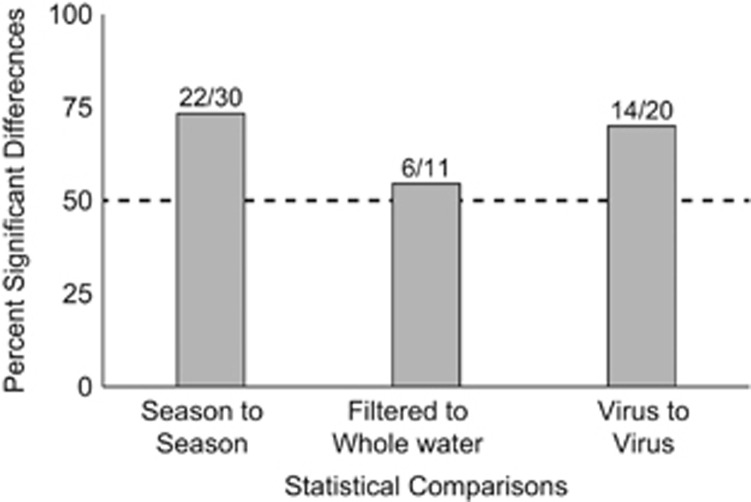
As noted above, for each of the three algal viruses studied slopes were most negative in the summer and least negative in the winter; that is, decay rates were highest in the summer and lowest in the winter (Table 2). In general, regression slopes in different seasons were significantly different from each other (P-value <0.00167; Supplementary Table S1) but there were some exceptions. For both ATCV-1 and CVM-1, certain comparisons of spring and summer, spring and autumn, and summer and autumn slopes were not significantly different. On the other hand, for CpV-BQ1 all seasonal comparisons produced significant differences. Overall, seasonal comparisons were statistically significant for 73% of all slopes compared (Figure 2).
Figure 2.
Percentage of statistically significant differences in the comparisons between seasons, filtration treatment or viruses. Values were derived from the season to season, whole water to filtered water and virus-to-virus comparisons using the statistical tests from Supplementary Tables S1–S3, respectively. The numbers above each bar show the number of significantly different comparisons and the total number of comparisons.
During all seasons and for every virus, regression slopes were more negative in the whole-water treatment compared with filtered water. The regression slopes of the two treatments were significantly different (P-value <0.0045; Supplementary Table S2) except for viruses in the autumn incubation experiment, as well as ACTV-1 in the winter, and CpV-BQ1 in the summer. For both treatments in every seasonal incubation experiment, ATCV-1 decayed most slowly (least negative regression slopes), while CpV-BQ1 decayed most rapidly (most negative slope). Treatment effects were significant in 55% of the statistical comparisons (Figure 2). The differences of the decay rates between the three algal viruses were generally statistically significant, with P-values <0.0025 (Supplementary Table S3). Notable exceptions include comparisons of the regression slopes of CVM-1 and ATCV-1 in the summer and spring whole-water treatments, and the spring filtered-water treatment, and CVM-1 and CpV-BQ1 comparisons during the autumn and in the winter filtered-water treatment. Overall, virus-to-virus comparisons were statistically significant for 70% of the time (Figure 2).
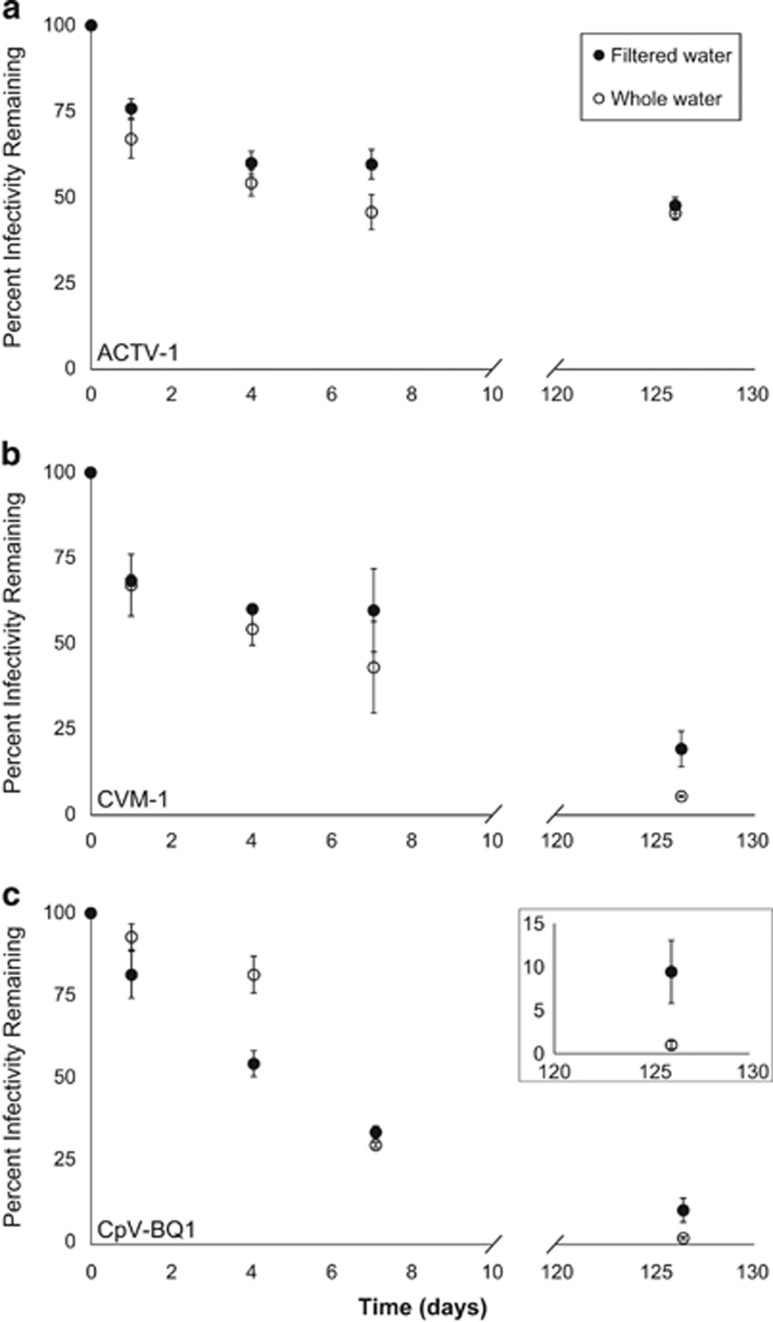
After 126 days in a frozen freshwater pond, infectious viruses were detected for all three of the algal viruses and in both treatments. ATCV-1 retained 47.82% of its original infectivity in the filtered-water treatment and 45.58% in the whole-water treatment (Figure 3a), while CVM-1 retained 18.82% in filtered water and 5% in whole water (Figure 3b), and CpV-BQ1 retained 9.22% in filtered water and 0.79% in whole water (Figure 3c). Statistical comparisons demonstrated that there was no difference in decay rates for the filtered-water versus whole-water treatments for ATCV-1 in the winter, but rates for these treatments were significantly different in the winter for CVM-1 and CpV-BQ1 (Supplementary Table S2).
Figure 3.
Overwintering of algal viruses in a seasonally frozen freshwater pond. Mean percent infectivity remaining in triplicate bottles incubated through the winter months was plotted against time in days for the viruses ACTV-1 (a), CVM-1 (b) and CpV-BQ1 (c). Filled circles are data points from the filtered-water treatment while open circles are data points from the whole-water treatment. Error bars represent standard deviation. The inset figure in panel (c) shows a close-up view of the percent infectivity remaining for CpV-BQ1 at the final time point.
Discussion
Decay of aquatic viruses
Viruses can be destroyed, inactivated or removed from aquatic environments by exposure to sunlight (both UV and photosynthetically active radiation (PAR)), extreme temperatures, heat-labile organic matter such as nucleases, consumption by heterotrophic nanoflagelletes, attachment to non-host cells, and adsorption to detritus and subsequent sinking (Wommack and Colwell, 2000). The role of other microbes in virus destruction has also been demonstrated in several studies showing increased survival of viruses when other microbes were inactivated through antibiotics, autoclaving or filtration, and many microorganisms are known to produce enzymes with antiviral properties (Gerba, 2005). These observations, together with the knowledge of threshold abundances for virus production and succession in phytoplankton assemblages, have led to questions about the survival and environmental persistence of aquatic viruses. In general, the results of our study corroborate past work on aquatic virus decay since rates were variable among different viruses, and the highest decay rates were observed during summer incubations, which received the greatest irradiation, and whole-water incubations, which included microbes <210 μm in size. Most significantly, the results of our study also demonstrate the ability of freshwater algal viruses to overwinter and remain viable after freezing, supporting the hypothesis that some viruses, even in the absence of ongoing production, can form a persistent seed bank in aquatic environments.
Using cultivation-based techniques to estimate numbers of infectious viruses, environmental decay rates of the algal viruses ATCV-1, CpV-BQ1 and CVM-1 were estimated from in situ incubations. The ranges of decay rates estimated in this study for ATCV-1 (0.012–1.10% h−1), CVM-1 (0.047–1.12% h−1) and CpV-BQ1 (0.077–11.26% h−1) were relatively low compared with decay rates determined using a similar experimental approach, but for bacteriophages incubated in direct sunlight using water from Santa Monica Bay, USA (4.1–11% h−1; Noble and Fuhrman, 1997). Given the inclusion of seasonal estimates of decay for the study reported here, and structural differences between different types of viruses, and the fact that the Santa Monica study included UV exposure, it is not surprising that decay rates estimated for bacteriophages and algal viruses are not directly comparable. However, even when compared with other algal viruses, ATCV-1 and CVM-1 decay rates were relatively low compared with rates reported in other studies (Cottrell and Suttle, 1995; Hewson et al., 2012; Frada et al., 2014). On the other hand, the decay rates estimated for CpV-BQ1 were the highest of the three algal viruses used in this study and, at least for the summer experiments, were within the range of previously reported values for algal viruses. That the rates estimated in this study were generally low compared with previous studies of algal viruses is likely due to the different experimental approaches that were used, and more importantly because previous studies estimated decay in only a single season. It is important to note that the decay rates estimated in this study likely represent underestimates, especially for the summer incubations, due to the use of polycarbonate bottles that are essentially UV opaque. UV radiation, and UV-B in particular, is an important factor for bacteriophage decay, and decay rates estimated in the absence of UV-B can be much lower (e.g., 20%) than the values estimated from incubations in full sunlight (Suttle and Chen, 1992). However, in some cases, PAR can be responsible for more viral decay than UV radiation (e.g., Wommack et al., 1996; Baudoux et al., 2012). Since the PC bottles used in this study are PAR transparent, it is certain that the algal viruses in this study were subjected to some photochemical or sunlight-mediated decay, but relative contributions of UV and PAR to algal virus decay cannot be resolved. Additionally, as is the case with any microcosm study conducted using closed incubation bottles, attachment to detritus and sinking is not a mechanism of decay that can be estimated in this study. Therefore, true environmental decay rates are likely higher than the estimates presented in this, or indeed any, study of viral decay.
The decay rate of 0.13% h−1 for the ATCV-1-like environmental virus reported in Hewson et al. (2012) was, as the authors acknowledged, not an estimate of decay of infectivity but rather an estimate of decay of genomic DNA based on quantitative PCR. Infectivity decays more rapidly than virus particles or genomes because virion damage can compromise attachment to host cells, or other critical steps such as cell entry and unpackaging despite the fact that the virus particle and even genome can remain intact (Suttle and Chen, 1992; Wommack et al., 1996; Noble and Fuhrman, 1997). MpV-SP1 infectivity was estimated to decay at a rate of 28–30% h−1 during incubations carried out in March and April in unattenuated sunlight using water from the Gulf of Mexico (Cottrell and Suttle, 1995). Overall, decay rates determined during our study were often low compared with the few literature values available for algal viruses, but the rates we observed during the spring, summer and autumn were similar to estimates determined for E. huxleyi viruses in the North Atlantic (i.e., 2–3% h−1; Frada et al., 2014). Furthermore, as expected, the lowest rates estimated during this study came from the winter incubation, and for ATCV-1 were 10 times lower than the lowest estimate previously reported for any algal virus. However, if the algal viruses in this study followed the pattern observed for marine bacteriophage where the decay rate in the absence of UV-B is only 20% of the decay rate in the presence of UV-B (Suttle and Chen, 1992), the decay estimates obtained in this study can be normalized to account for UV-B-mediated decay by multiplying the rates by a factor of 5. This yields decay estimates ranging from 0.06 to 5.5% h−1 for ACTV-1, 0.235 to 5.6% h−1 for CVM-1 and 0.385 to 56.3% h−1 for CpV-BQ1. As noted above, this normalization may be conservative for the summer incubations and exaggerated for the winter incubations, but the highest corrected decay rates observed in this study actually exceeds the decay estimate obtained from the Gulf of Mexico for the algal virus MpV-SP1 (Cottrell and Suttle, 1995).
Seasonality and variability in rates of decay
Although other more general studies of aquatic virus decay have noted the lowest rates in the winter (Thomas et al., 2011), the winter incubation experiment with the freshwater algal viruses ATCV-1, CVM-1 and CpV-BQ1 produced the lowest estimated decay rates observed for any algal viruses in nature. It is likely that these unprecedented low rates of decay were due to the fact that the freshwater pond was frozen over during the winter incubation, which would dramatically reduce exposure to sunlight, especially when ice is covered by snow (Perovich et al., 1993; Bertilsson et al., 2013). As expected, for all three algal viruses decay rates were highest in the summer and intermediate for the spring and autumn. Similarly, decay rates of cyanophage in the Gulf of Mexico, Texas (Garza and Suttle, 1998) and Lake Donghu, China (Cheng et al., 2007) were also shown to be highest during the summer. Seasonality in virus decay is likely due to seasonal fluctuations in temperature and sunlight (both UV and PAR), which have both been implicated in virus inactivation (Lo et al., 1976; Suttle and Chen, 1992; Garza and Suttle, 1998; Baudoux et al., 2012). Sunlight exposure can cause photochemical damage of viruses directly, while increased temperatures may act indirectly through antiviral increased microbial and enzymatic activity (e.g., Yates et al., 1985; Gersberg et al., 1987; Garza and Suttle, 1998).
Exposure to UV radiation and PAR can deactivate viruses (Cottrell and Suttle, 1995; Furuta et al., 1997; Jacquet and Bratbak, 2003; Baudoux et al., 2012), yet some algal viruses encode genetic machinery to repair light-induced DNA damage. Many, but not all, genomes of chloroviruses, including ATCV-1 and CVM-1, contain homologs of a UV repair gene, denV, which encodes a UV-specific DNA glycosylase-pyrimidine lyase (Fitzgerald et al., 2007; Jeanniard et al., 2013) known to be functional in the strain PBCV-1 (Furuta et al., 1997). Moreover, other nucleic acid metabolism genes encoded by algal viruses that could aid in environmental persistence include DNA ligase, DNA polymerase δ, proliferating cell nuclear antigen, as well as genes involved in base incision repair and nucleotide incision repair (Dunigan et al., 2006; Redrejo-Rodriguez and Salas, 2014). The absence of denV homologs in some chlorovirus strains such as KS1B, and other DNA repair genes in other algal virus genomes, demonstrates that these capabilities are not universally present among algal viruses (Jeanniard et al., 2013). Furthermore, the genome of EhV-86, a coccolithovirus, contains a pyrimidine dimer-specific glycosylase, while the genome of EsV-1, a phaeovirus, does not (Dunigan et al., 2006). While photo-induced pyrimidine dimers were likely not a major source of DNA damage in this study, the variability of denV is illustrative of the variability in the genetic potential of specific algal virus strains to repair DNA damage.
Even though UV-B and much of UV-A were attenuated in this study, the facts that ATCV-1 and CVM-1 encode pyrimidine dimer-specific glycosylases as well as other DNA repair genes and had 10-fold lower decay rates than CpV-BQ1 in summer months suggest that CpV-BQ1 might not encode similar DNA repair machinery. Genome sequence information from CpV-BQ1 could resolve this hypothesis and generate other interesting questions about the genetic basis for the environmental stability of aquatic viruses. Furthermore, the results presented here reveal both intra- and inter-genus variability as the chloroviruses ATCV-1 and CVM-1 and the newly isolated, putative prymnesiovirus CpV-BQ1 all decayed at different rates. Differential rates of decay could drive differences in virus–host dynamics among different algal viruses supporting the notion that individual virus–host pairs are ecologically unique (Rozon and Short, 2013). It is worth noting that decay rates of CVM-1 were estimated via MPNs versus plaque assays to compare these different approaches (data not shown), but were not significantly different, suggesting that differences in decay rates between the chloroviruses and prymnesiovirus were not due to experimental methods alone.
Differences in decay rates between the filtered- and whole-water treatments implicate the antiviral effect of microbes as the filtered-water treatment produced decay rates that were often lower than the whole-water treatment. An active microbial community is known to accelerate rates of viral decay through adsorption to non-host cells, consumption of viruses by nanoflagellates, or the activity of extracellular nucleases and other enzymes (Gerba, 2005). This effect was most significant in the spring and summer incubations for ATCV-1 and CVM-1 when temperatures, and presumably microbial activity, were highest. On the other hand, at least one study has demonstrated that particulate material that was presumably removed during our filtration could actually reduce algal virus decay rates. The viruses ATCV-1 and CVM-1 can dynamically attach and detach to host cells and host cell debris, and remain infectious, and this process has been implicated in increased virus survival (Agarkova et al., 2014). As such, depending on the particular samples there may have been particulate material filtered out of the whole water that could have enhanced virus decay or enhanced virus survival. These contrasting effects of different particulate materials on virus decay may explain why only a little more than half of the filtered-water treatments yielded decay rates that were significantly different than rates from the corresponding whole-water treatment. The highest estimated decay rates observed in this study coincided with the maxima for water temperature, sunlight and daylight hours, providing further evidence that temperature and sunlight are major factors in the inactivation of virus particles in aquatic environments and drove the seasonality observed in the decay of ATCV-1, CVM-1 and CpV-BQ1.
Algal virus overwintering
This study presents the first experimental evidence that algal viruses can persist in ice-covered, freshwater environments. For the winter incubation, samples bottles were initially placed underneath the existing thin ice cover and remained in the water column, unfrozen, for the first 7 days of the experiment. After 7 days, the ice was too thick to break and the bottles were left in situ for the rest of the season. During this time, the exact date that the samples froze is unknown, but before the ice became covered in snow the bottles were clearly stuck in the thick ice cover and the liquid in the bottles was visibly frozen. Following their winter-long incubation, samples were recovered after the ice on the pond had thawed and every incubation bottle contained infectious viruses. Furthermore, in a lab study conducted to simulate freezing in the environment, ATCV-1, CVM-1 and CpV-BQ1 retained approximately 92%, 85% and 89% of their infectivity after they were chilled in ice water for 3 h, stored at −20 °C for 15 h and were subsequently thawed in ice water (data not shown). Thus it is apparent that some algal viruses can survive overwinter in the ice of seasonally frozen ponds and lakes. The survival of ATCV-1 and CVM-1 after freezing is perhaps surprising as PBCV-1, a close relative, can be inactivated by freezing (Van Etten et al., 1991). Although we have provided the first direct evidence that some algal viruses can survive in frozen environments, other viruses have been previously shown to tolerate similar conditions. For example, human enteric viruses persisted for several months when incubated in situ in dialysis bags filled with autoclaved marine water, and were most stable during winter months (Lo et al., 1976). Also, viruses have been shown to exist in high abundances in frozen Antarctic lakes (Foreman et al., 2011), viral genomes have been detected in ~700-year-old frozen caribou feces (Ng et al., 2014), and even more incredible, a putatively 30 000-year-old giant virus of amoebas has been recovered from Siberian permafrost (Legendre et al., 2014). Hence, it is plausible, even likely, that reduced decay rates experienced during winter months may constitute a major mechanism for algal virus survival and the establishment of viral seed banks in many aquatic environments.
Conclusions
Over the year, algal virus decay rates were highly variable, but the winter decay rates observed here are among the lowest reported for aquatic viruses. The winter half-life of the least resilient virus examined, CpV-BQ1, was 38 days, long enough for a substantial fraction of the virus population to survive as a seed bank for the subsequent ice-free growing season. The observation that decay rates were greatly reduced in the winter and viruses maintained infectivity after freezing provides direct evidence that algal viruses can persist in the environment for many months. However, during the summer when decay rates peaked the half-life of even the most resilient virus, ATCV-1, was only 2 days, suggesting that even this population would be rapidly destroyed; hypothetically, starting with 105 viruses ml−1 only 6 viruses ml−1 would remain after 30 days. This suggests that the summer represents a seasonal bottleneck for virus survival, and ongoing virus production or some other means of escaping destruction such as mixing into deeper waters is necessary to maintain virus populations through these months; the decay experiments described here were conducted at the surface of the pond where sunlight was maximal, and it is known that algal virus decay rates attenuate rapidly with depth (Cottrell and Suttle, 1995). Although virus survival when experiencing high decay rates necessitates constant production, environmental refugia or non-lytic infections, viruses that withstand the summer in temperate aquatic environments should be able to overwinter until the subsequent growing season of their hosts. Therefore, the results of this study clearly demonstrate the importance of seasonality in the environmental persistence of algal viruses.
In future studies, quantitative PCR methods that were established to monitor the abundance of diverse, uncultivated phycodnaviruses (Short and Short, 2009; Short et al., 2011a) could be combined with cultivation approaches to determine relationships between decay of infectivity and decay of viral nucleic acids. In turn, these relationships could be used to infer decay rates for viruses that have not yet been cultivated, which constitutes the vast majority of environmental viruses. Knowing true, functional rates of decay for a range of algal viruses is essential to establish boundaries related to their resilience and environmental persistence. Constraining estimates of virus environmental persistence is a vital step towards realistic models of virus–host dynamics in aquatic environments.
Acknowledgments
Special thanks to Dr James Van Etten and colleagues at University of Nebraska, Lincoln for providing cell cultures of M. conductrix and C. heliozoae as well as isolates of ATCV-1 and CVM-1. We are also grateful to Cindy Short and Samia Mirza for their support in maintaining cell culture lines. This research was supported in part by the Canadian Foundation for Innovation Leaders Opportunity Fund and NSERC Discovery grants awarded to SMS.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Agarkova I, Hertel B, Zhang X, Lane L, Tchourbanov A, Dunigan DD et al. (2014). Dynamic attachment of Chlorovirus PBCV-1 to Chlorella variabilis. Virol 466: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RA, Berges JA, Harrison PJ, Watanabe MM. (2005) Appendix A—Recipes for freshwater and seawater media. In: Andersen RA (ed), Algal Culturing Techniques. Academic Press/Elsevier: New York, pp 429–538. [Google Scholar]
- Baudoux AC, Hendrix RW, Lander GC, Bailly X, Podell S, Paillard C et al. (2012). Genomic and functional analysis of Vibrio phage SIO-2 reveals novel insights into ecology and evolution of marine siphoviruses. Environ Microbiol 14: 2071–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudoux AC, Noordeloos AAM, Veldhuis MJW, Brussaard CPD. (2006). Virally induced mortality of Phaeocystis globosa during two spring blooms in temperate coastal waters. Aquat Microb Ecol 44: 207–217. [Google Scholar]
- Bergh O, Borsheim KY, Bratbak G, Heldal M. (1989). High abundance of viruses found in aquatic environments. Nature 340: 467–468. [DOI] [PubMed] [Google Scholar]
- Bertilsson S, Burgin A, Carey CC, Fey SB, Grossart H, Grubisic LM et al. (2013). The under-ice microbiome of seasonally frozen lakes. Limnol Oceanogr 58: 1998–2012. [Google Scholar]
- Bettarel Y, Sime-Ngando T, Amblard C, Dolan J. (2004). Viral activity in two contrasting lake ecosystems. Appl Environ Microbiol 70: 2941–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratbak G, Egge JK, Heldal M. (1993). Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser 93: 39–48. [Google Scholar]
- Bratbak G, Jacobsen A, Heldal M. (1998). Viral lysis of Phaeocystis pouchetii and bacterial secondary production. Aquat Microb Ecol 16: 11–16. [Google Scholar]
- Breitbart M, Rohwer F. (2005). Here a virus, there a virus, everywhere the same virus? Trends Microbiol 13: 278–284. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D et al. (2002). Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA 99: 14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard CPD. (2004). Viral control of phytoplankton populations—a review. J Euk Microbiol 51: 125–138. [DOI] [PubMed] [Google Scholar]
- Brussaard CPD, Mari X, Van Bleijswijk JDL, Veldhuis MJW. (2005). A mesocosm study of Phaeocystis globosa (Prymnesiophyceae) population dynamics—II. Significance for the microbial community. Harmful Algae 4: 875–893. [Google Scholar]
- Cheng K, Zhao Y, Du X, Zhang Y, Lan S, Shi Z. (2007). Solar radiation-driven decay of cyanophage infectivity, and photoreactivation of the cyanophage by host cyanobacteria. Aquat Microb Ecol 48: 13–18. [Google Scholar]
- Clasen JL, Suttle CA. (2009). Identifying freshwater Phycodnaviridae and their potential phytoplankton hosts using DNA pol sequence fragments and a genetic distance analysis. Appl Environ Microbiol 75: 991–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell MT, Suttle CA. (1995). Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol Oceanogr 40: 730–739. [Google Scholar]
- Dunigan DD, Fitzgerald LA, Van Etten JL. (2006). Phycodnaviruses: a peek at genetic diversity. Virus Res 117: 119–132. [DOI] [PubMed] [Google Scholar]
- Farnell-Jackson EA, Ward AK. (2003). Seasonal patterns of viruses, bacteria and dissolved organic carbon in a riverine wetland. Freshwat Biol 48: 841–851. [Google Scholar]
- Filippini M, Middelboe M. (2007). Viral abundance and genome size distribution in the sediment and water column of marine and freshwater ecosystems. FEMS Microbiol Ecol 60: 397–410. [DOI] [PubMed] [Google Scholar]
- Fitzgerald LA, Graves MV, Li X, Hartigan J, Pfitzner AJP, Hoffart E et al. (2007). Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic Chlorella strain of the heliozoon Acanthocystis turfacea. Virol 362: 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman CM, Dieser M, Greenwood M, Cory RM, Laybourn-Parry J, Lisle JT et al. (2011). When a habitat freezes solid: microorganisms over-winter within the ice column of a coastal Antarctic lake. FEMS Microbiol Ecol 76: 401–412. [DOI] [PubMed] [Google Scholar]
- Frada MJ, Schatz D, Farstey V, Ossolinski JE, Sabanay H, Ben-Dor S et al. (2014). Zooplankton may serve as transmission vectors for viruses infecting algal blooms in the ocean. Curr Biol 24: 2592–2597. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399: 541–548. [DOI] [PubMed] [Google Scholar]
- Furuta M, Schrader JO, Schrader HS, Kokjohn TA, Nyaga S, McCullough AK et al. (1997). Chlorella virus PBCV-1 encodes a homolog of the bacteriophage T4 UV damage repair gene denV. Appl Environ Microbiol 63: 1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza DR, Suttle CA. (1998). The effect of cyanophages on the mortality of Synechococcus spp. and selection for UV resistant viral communities. Microb Ecol 36: 281–292. [DOI] [PubMed] [Google Scholar]
- Gerba CP. (2005) Survival of viruses in the marine environment. In: Belken S, Colwell RR (ed), Oceans and Health: Pathogens in the Marine Environment. Springer: New York, pp 133–142. [Google Scholar]
- Gersberg RM, Lyon SR, Brenner R, Elkins BV. (1987). Fate of viruses in artificial wetlands. Appl Environ Microbiol 53: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenes MV, Zanotto PMA, Suttle CA, da Cunha HB, Mehnert DU. (2012). Phylodynamics and movement of Phycodnaviruses among aquatic environments. ISME J 6: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobler CJ, Anderson OR, Gastrich MD, Wilhelm SW. (2007). Ecological aspects of viral infection and lysis in the harmful brown tide alga Aureococcus anophagefferens. Aquat Microb Ecol 47: 25–36. [Google Scholar]
- Gobler CJ, Davis TW, Deonarine SN, Saxton MA, Lavrentyev PJ, Jochem FJ et al. (2008). Grazing and virus-induced mortality of microbial populations before and during the onset of annual hypoxia in Lake Erie. Aquat Microb Ecol 51: 117–128. [Google Scholar]
- Haaber J, Middelboe M. (2009). Viral lysis of Phaeocystis pouchetii: Implications for algal population dynamics and heterotrophic C, N and P cycling. ISME J 3: 430–441. [DOI] [PubMed] [Google Scholar]
- Heldal M, Bratbak G. (1991). Production and decay of viruses in aquatic environments. Mar Ecol Prog Ser 72: 205–212. [Google Scholar]
- Hewson I, Barbosa JG, Brown JM, Donelan RP, Eaglesham JB, Eggleston EM et al. (2012). Temporal dynamics and decay of putatively allochthonous and autochthonous viral genotypes in contrasting freshwater lakes. Appl Environ Microbiol 78: 6583–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer JS, Sommaruga R. (2001). Seasonal dynamics of viruses in an alpine lake: importance of filamentous forms. Aquat Microb Ecol 26: 1–11. [Google Scholar]
- Jacquet S, Bratbak G. (2003). Effects of ultraviolet radiation on marine virus-phytoplankton interactions. FEMS Microbiol Ecol 44: 279–289. [DOI] [PubMed] [Google Scholar]
- Jacquet S, Heldal M, Iglesias-Rodriguez D, Larsen A, Wilson W, Bratbak G. (2002). Flow cytometric analysis of an Emiliana huxleyi bloom terminated by viral infection. Aquat Microb Ecol 27: 111–124. [Google Scholar]
- Jarvis B, Wilrich C, Wilrich PT. (2010). Reconsideration of the derivation of Most Probable Numbers, their standard deviations, confidence bounds and rarity values. J Appl Microbiol 109: 1660–1667. [DOI] [PubMed] [Google Scholar]
- Jeanniard A, Dunigan DD, Gurnon JR, Agarkova IV, Kang M, Vitek J et al. (2013). Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics 14: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A et al. (2014). Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci USA 111: 4274–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S, Gilbert J, Hetrick F. (1976). Stability of human enteroviruses in estuarine and marine Waters. Appl Environ Microbiol 32: 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranger R, Bird DE. (1996). High concentrations of viruses in the sediments of Lac Gilbert, Quebec. Microb Ecol 31: 141–151. [DOI] [PubMed] [Google Scholar]
- Mathias CB, Kirschner AKT, Velimirov B. (1995). Seasonal-variations of virus abundance and viral control of the bacterial production in a backwater system of the Danube River. Appl Environ Microbiol 61: 3734–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelboe M, Riemann L, Steward GF, Hansen V, Nybroe O. (2003). Virus-induced transfer of organic carbon between marine bacteria in a model community. Aquat Microb Ecol 33: 1–10. [Google Scholar]
- Mirza SF, Staniewski MA, Short CM, Long AM, Chaban YV, Short SM. (2015). Isolation and characterization of a virus infecting the freshwater algae Chrysochromulina parva. Virology 486: 105–115. [DOI] [PubMed] [Google Scholar]
- Munawar M, Munawar IF. (1986). The seasonality of phytoplankton in the North American Great Lakes, a comparative synthesis. Hydrobiologia 138: 85–115. [Google Scholar]
- Ng TFF, Chen L, Zhou Y, Shapiro B, Stiller M, Heintzman PD et al. (2014). Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc Natl Acad Sci USA 111: 16842–16847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble RT, Fuhrman JA. (1997). Virus decay and its causes in coastal waters. Appl Environ Microbiol 63: 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perovich DK, Cota GF, Maykut GA, Grenfell TC. (1993). Bio-optical observations of first-year arctic sea ice. Geophys Res Lett 20: 1059–1062. [Google Scholar]
- Proctor LM, Fuhrman JA. (1990). Viral mortality of marine-bacteria and cyanobacteria. Nature 343: 60–62. [Google Scholar]
- Redrejo-Rodriguez M, Salas ML. (2014). Repair of base damage and genome maintenance in the nucleo-cytoplasmic large DNA viruses. Virus Res 179: 12–25. [DOI] [PubMed] [Google Scholar]
- Reisser W, Becker B, Klein T. (1986). Studies on ultrastructure and host range of a Chlorella attacking virus. Protoplasma 135: 162–165. [Google Scholar]
- Reynolds CS. (2006) The Ecology of Phytoplankton. Cambridge University Press: Cambridge. [Google Scholar]
- Rozon RM, Short SM. (2013). Complex seasonality observed amongst diverse phytoplankton viruses in the Bay of Quinte, an embayment of Lake Ontario. Freshwat Biol 58: 2648–2663. [Google Scholar]
- Sheik AR, Brussaard CPD, Lavik G, Lam P, Musat N, Krupke A et al. (2014). Responses of the coastal bacterial community to viral infection of the algae Phaeocystis globosa. ISME J 8: 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelford EJ, Middelboe M, Moller EF, Suttle CA. (2012). Virus-driven nitrogen cycling enhances phytoplankton growth. Aquat Microb Ecol 66: 41–46. [Google Scholar]
- Short CM, Rusanova O, Short SM. (2011. a). Quantification of virus genes provides evidence for seed-bank populations of phycodnaviruses in Lake Ontario, Canada. ISME J 5: 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM, Rusanova O, Staniewski MA. (2011. b). Novel phycodnavirus genes amplified from Canadian freshwater environments. Aquat Microb Ecol 63: 61–67. [Google Scholar]
- Short SM, Short CM. (2008). Diversity of algal viruses in various North American freshwater environments. Aquat Microb Ecol 51: 13–21. [Google Scholar]
- Short SM, Short CM. (2009). Quantitative PCR reveals transient and persistent algal viruses in Lake Ontario, Canada. Environ Microbiol 11: 2639–2648. [DOI] [PubMed] [Google Scholar]
- Short SM. (2012). The ecology of viruses that infect eukaryotic algae. Environ Microbiol 14: 2253–2271. [DOI] [PubMed] [Google Scholar]
- Staniewski MA, Short CM, Short SM. (2012). Contrasting community versus population-based estimates of grazing and virus-induced mortality of phytoplankton. Microb Ecol 64: 25–38. [DOI] [PubMed] [Google Scholar]
- Staniewski MA, Short SM. (2014). Potential viral stimulation of primary production observed during experimental determinations of phytoplankton mortality. Aquat Microb Ecol 71: 239–256. [Google Scholar]
- Suttle CA, Chan AM. (1994). Dynamics and distribution of cyanophages and their effect on marine Synechococcus Spp. Appl Environ Microbiol 60: 3167–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA, Chen F. (1992). Mechanisms and rates of decay of marine viruses in seawater. Appl Environ Microbiol 58: 3721–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. (1994). The significance of viruses to mortality in aquatic microbial communities. Microb Ecol 28: 237–243. [DOI] [PubMed] [Google Scholar]
- Suttle CA. (2007). Marine viruses - major players in the global ecosystem. Nat Rev Microbiol 5: 801–812. [DOI] [PubMed] [Google Scholar]
- Tarutani K, Nagasaki K, Yamaguchi M. (2000). Viral impacts on total abundance and clonal composition of the harmful bloom-forming phytoplankton Heterosigma akashiwo. Appl Environ Microbiol 66: 4916–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Berdjeb L, Sime-Ngando T, Jacquet S. (2011). Viral abundance, production, decay rates and life strategies (lysogeny versus lysis) in Lake Bourget (France). Environ Microbiol 13: 616–630. [DOI] [PubMed] [Google Scholar]
- Tijdens M, van de Waal DB, Slovackova H, Hoogveld HL, Gons HJ. (2008). Estimates of bacterial and phytoplankton mortality caused by viral lysis and microzooplankton grazing in a shallow eutrophic lake. Freshwat Biol 53: 1126–1141. [Google Scholar]
- Tomaru Y, Hata N, Masuda T, Tsuji M, Igata K, Masuda Y et al. (2007). Ecological dynamics of the bivalve-killing dinoflagellate Heterocapsa circularisquama and its infectious viruses in different locations of western Japan. Environ Microbiol 9: 1376–1383. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Burbank DE, Kuczmarski D, Meints RH. (1983). Virus-infection of culturable Chlorella-like algae and development of a plaque-assay. Science 219: 994–996. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Lane LC, Meints RH. (1991). Viruses and viruslike particles of eukaryotic algae. Microbiol Rev 55: 586–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterbury JB, Valois FW. (1993). Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol 59: 3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG, Bonilla-Findji O, Chan AM, Dolan JR, Short SM, Simek K et al. (2011). Synechococcus growth in the ocean may depend on the lysis of heterotrophic bacteria. J Plankton Res 33: 1465–1476. [Google Scholar]
- Wiggins BA, Alexander M. (1985). Minimum bacterial density for bacteriophage replication—implications for significance of bacteriophages in natural ecosystems. Appl Environ Microbiol 49: 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WH, Tarran GA, Schroeder D, Cox M, Malin G. (2002). Isolation of viruses responsible for the demise of an Emiliania huxleyi bloom in the English Channel. J Mar Biol Ass UK 82: 369–377. [Google Scholar]
- Wommack KE, Colwell RR. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64: 69–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Hill RT, Muller TA, Colwell RR. (1996). Effects of sunlight on bacteriophage viability and structure. Appl Environ Microbiol 62: 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates MV, Gerba CP, Kelley LM. (1985). Virus persistence in groundwater. Appl Environ Microbiol 49: 778–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Berdjeb L, Jacquet S. (2013). Temporal dynamics and structure of picocyanobacteria and cyanomyoviruses in two large and deep peri-alpine lakes. FEMS Microbiol Ecol 86: 312–326. [DOI] [PubMed] [Google Scholar]
- Zhong X, Guidoni B, Jacas L, Jacquet S. (2015). Structure and diversity of ssDNA Microviridae viruses in two peri-alpine lakes (Annecy and Bourget, France). Res Microbiol 166: 644–654. [DOI] [PubMed] [Google Scholar]
- Zhong X, Jacquet S. (2014). Contrasting diversity of phycodnavirus signature genes in two large and deep western European lakes. Environ Microbiol 16: 759–773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.