Abstract
The distribution of major clades of Prochlorococcus tracks light, temperature and other environmental variables; yet, the drivers of genomic diversity within these ecotypes and the net effect on biodiversity of the larger community are poorly understood. We examined high light (HL) adapted Prochlorococcus communities across spatial and temporal environmental gradients in the Pacific Ocean to determine the ecological drivers of population structure and diversity across taxonomic ranks. We show that the Prochlorococcus community has the highest diversity at low latitudes, but seasonality driven by temperature, day length and nutrients adds complexity. At finer taxonomic resolution, some ‘sub-ecotype' clades have unique, cohesive responses to environmental variables and distinct biogeographies, suggesting that presently defined ecotypes can be further partitioned into ecologically meaningful units. Intriguingly, biogeographies of the HL-I sub-ecotypes are driven by unique combinations of environmental traits, rather than through trait hierarchy, while the HL-II sub-ecotypes appear ecologically similar, thus demonstrating differences among these dominant HL ecotypes. Examining biodiversity across taxonomic ranks reveals high-resolution dynamics of Prochlorococcus evolution and ecology that are masked at phylogenetically coarse resolution. Spatial and seasonal trends of Prochlorococcus communities suggest that the future ocean may be comprised of different populations, with implications for ecosystem structure and function.
Introduction
The numerical dominance of Prochlorococcus spp. in open oligotrophic oceans (Partensky et al., 1999; Flombaum et al., 2013) is driven in part by the exceptional genomic diversity of this genus of cyanobacteria (Scanlan et al., 2009; Partensky and Garczarek, 2010; Biller et al., 2015). Although its populations can coexist within a water column (Scanlan and West 2002; Zinser et al., 2007), across ocean basins the distributions of the major clades of Prochlorococcus are structured by a combination of environmental variables, notably temperature and light (Bouman et al., 2006; Johnson et al., 2006). The importance of these and other drivers is supported by physiological studies that have demonstrated differences among its clades in optimal light intensity (Moore and Chisholm, 1999), temperature (Moore et al., 1995; Johnson et al., 2006), light harvesting (Hess et al., 2001), virus specificities (Sullivan et al., 2003) and nutrient assimilation (Moore et al., 2002, 2005) among other properties. Hence, the major phylogenetic clades of Prochlorococcus are often referred to as ‘ecotypes' (sensu Cohan, 2002) because they partition the ocean based on their niche envelope.
Genomic (Rocap et al., 2003; Malmstrom et al., 2013) and metagenomic (Rusch et al., 2007; Martiny et al., 2009a; Dupont et al., 2014) evidence provides additional mechanistic support for ecological diversification among the phylogenetic clades and further shows distinct patterns of gene gain and loss (Kettler et al., 2007) associated with the environment. In particular, it has been hypothesized that environmental variables have a hierarchical influence on genomic diversification across taxonomic ranks (that is, the levels of the Prochlorococcus phylogenetic tree ranging from the basal clades to the leaves) and thus drive phylogenetic structure. Specifically, light and temperature drove ancient niche differentiation at higher taxonomic levels, nutrient acquisition drove mid-level differentiation and phage/grazer resistance drove more recent fine-scale genotype selection at the ‘leaves' of the Prochlorococcus phylogenetic tree (Coleman and Chisholm, 2007). For example, phosphorus or nitrate acquisition genes have been shown to be polyphyletic in Prochlorococcus (Martiny et al., 2006; Berube et al., 2014), whereas photosynthesis and light harvesting genes show more basal patterns (Scanlan et al., 2009). However, genomic signatures for other important environmental variables such as temperature have yet to be identified (Kettler et al., 2007; Scanlan et al., 2009), even though there are clear physiological and biogeographic differences among ecotypes (Johnson et al., 2006). Nevertheless, if environmental selection drives genomic diversification in a hierarchical manner we may expect to identify monophyletic clades that have variable environmental response traits across taxonomic levels (Violle et al., 2011) when Prochlorococcus community diversity is assessed at high resolution.
These broad ecogenomic patterns appear even more complex, in light of recent studies that show Prochlorococcus clades can be composed of hundreds of sub-populations that apparently diverged a few million years ago and possibly represent ancient niche partitioning (Kashtan et al., 2014; Johnson and Martiny, 2015). This ‘micro-diversity' is a dominant feature of broader bacterial communities (Acinas et al., 2004), but how micro-diversity maps onto a hierarchical genomic diversification framework or its impact on microbial community structure and function remain unclear (Koeppel et al., 2008) in spite of some evidence of its importance for ecological specialization (Hunt et al., 2008; Yung et al., 2014). Further, while the environmental variables that define the niche space of major clades of Prochlorococcus are increasingly well established, it is unknown how these environmental variables influence α- and β-diversity and its biogeography at higher taxonomic resolutions. Models suggest that ocean warming may lead to shifts in both the distribution and composition of the Prochlorococcus community (Thomas et al., 2012) through multiple mechanisms (Flombaum et al., 2013). Thus, mapping environmental traits onto appropriate taxonomic ranks will help to refine predictions of these present and future patterns and the mechanisms that drive them.
Here, we use intra- and inter-seasonal variability in the North Pacific Ocean over multiple spatial and temporal scales to examine how the environment affects fine-scale Prochlorococcus community α- and β-diversity across taxonomic ranks. We show that the relative dominance of major HL ecotypes of Prochlorococcus is largely driven by changes in temperature, but that seasonal shifts in community composition reflect additional changes in their sub-ecotype component populations. HL-I sub-ecotype populations are highly variable and structured along environmental gradients, whereas HL-II sub-ecotype populations are more seasonally stable. These results have implications for understanding the mechanisms driving present-day distributions and for predicting the response of Prochlorococcus communities to climate change.
Materials and methods
Field sampling
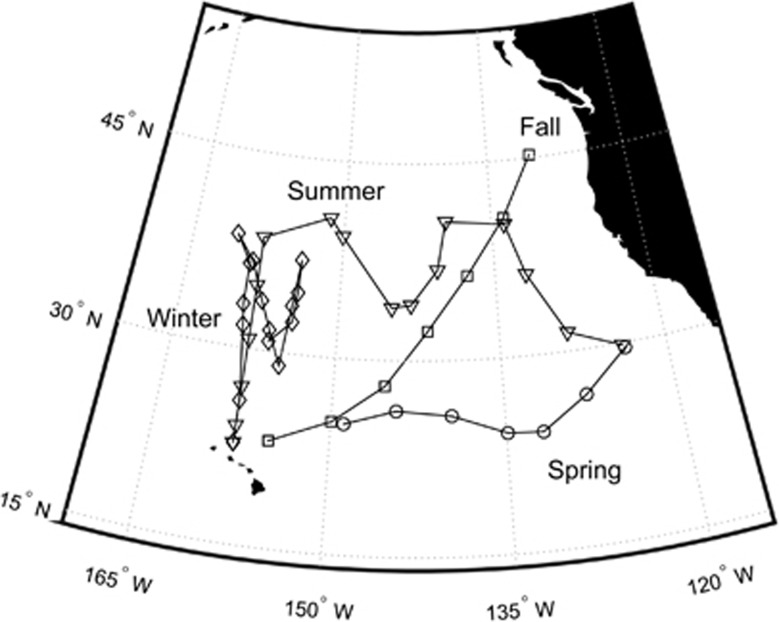
Samples from 56 stations were collected aboard four oceanographic cruises (Phytoplankton of Warming Ocean Waters—POWOW0: 27 October–12 November 2011; POWOW2: 10 January–8 February 2013; POWOW1: 29 February–11 March 2012; POWOW3: 1–28 July 2013) in the North Pacific Ocean (Figure 1) roughly corresponding to the four seasons (fall, winter, spring and summer, respectively). Water for POWOW0 was collected from the vessel's on-board flow-through seawater system (intake depth ~7 m) at 0700 h local time. For the other cruises, water was collected using a Niskin rosette system (depth ~10 m) and sampled between ~0200 and 0500 h local time.
Figure 1.
Station locations in the North Pacific Ocean (fall: POWOW0/27 October–12 November 2011; winter: POWOW2/10 January–8 February 2013; spring: POWOW1/29 February–11 March 2012; summer: POWOW3/1–28 July 2013).
Environmental variables
Hydrographic data were collected from depth using a CTD system or a ship DAS system. Nutrients (NH4, NO2, NO3, PO4 and SiO4) and pH were measured as previously described (Holmes et al., 1999; Johnson et al., 2013) using certified reference materials. Prochlorococcus cellular abundances were quantified on live samples using a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) as previously described (Johnson et al., 2010). Seasonal trends in surface temperature for selected stations were estimated using remotely sensed microwave-infrared sea surface temperatures (Remote Sensing Systems—ftp://data.remss.com/SST/daily_v04.0/).
Prochlorococcus DNA extraction and quantitative PCR
Community genomic DNA was extracted as described previously (Zinser et al., 2006), using a protocol optimized for Prochlorococcus quantification. The abundance of eHL-I and eHL-II was calculated using quantitative PCR as previously described (Zinser et al., 2006), with eHL-I- and eHL-II-specific primers targeting the 23S rRNA gene (Ahlgren et al., 2006; Lin et al., 2013) and synthesized plasmids as standards. Synthesized plasmids were linearized and quantified as previously described (Zinser et al., 2006). The 23S plasmid inserts corresponded to fragments of genomes representative of the target ecotype and had the following sequences: eHL-I—GAAGCTGCGG ATAAATTTAT TTATGGTAGG GGAGCGTTCT ATGTTGGGTG AAGCGTTAGC GTAAGCGGAC GTGGACGGCATAGAAGTGAGAATGT CGGCTTGAGTAGCGAAAACATGGGTGAGAATCCCATGCCCCGAAACCCTAAGGGTT CCTCCGGCAGGCTCGTCCGCGGAGGGTTAGTCTGGACCTAAGGCGA GGCCGAAAGGCG TAGTCGATGGATAACAGGTCAATATTCCTGTACCAGTTATGTTTTGGGAAGGGGGA and eHL-II—TCGGGGAGTTGGAAGCACACTTTGATCCGGGAATTTCCGAATGGGGCAACCCCAT GTACGGCCAACTGAA TATATAGG.
Internal transcribed spacer amplicon libraries
The internal transcribed spacer (ITS) region was PCR amplified using Prochlorococcus-specific primers tRNA_789-F (5′-TGTCAGCGGTTCGAGTCCG-3′) (designed in this study) and 23S-R (5′-TCATCGCCTCTGTGTGCC-3′) (Martiny et al., 2009b), with barcodes and regions specific for 454 sequencing. DNA was added to 20 μl reactions (1 × buffer, 200 μm dNTPs, 0.3 mg ml−1 BSA, 50 nm primers, 0.0125 units Jumpstart Taq; Sigma-Aldrich, St Louis, MO, USA) and amplified as follows: 95 °C for 10 min, followed by 30 cycles of 95 °C 15 s, 58 °C 45 s, and 72 °C 60 s and a final extension step at 72 °C for 10 min. A 1:10 dilution of the PCR product was reconditioned (Thompson et al., 2002) for three additional cycles. Products (with different barcodes) were pooled and sequenced (454 GS FLX+ Titanium, Roche, Basel, Switzerland) resulting in 739 437 reads. The mean bp length of the sequencing output was 338 bp, with a maximum of 841 bp. Sequences for this project are deposited in NCBI under SRP065205.
ITS sequences quality filtering and operational taxonomic unit grouping
The sequencing output was demultiplexed and quality filtered using QIIME (Caporaso et al., 2010) as follows: length truncated to 250 bp, sequences with a mean quality score of less than Q20 in a running window of 16 bp and/or homopolymers longer than 8 bp were removed from the analysis. The USEARCH algorithm (Edgar, 2013) was used to dereplicate sequences, remove singletons and de-novo chimeras and cluster operational taxonomic units (OTUs) using centroid-based clustering. A total of 408 294 sequence reads were grouped into OTUs based on 97% sequence similarity. In all, 14 OTUs (559 sequences) identified (BLASTN) as not Prochlorococcus by their highest BLASTN hit against the refseq_genomic database (NCBI) were removed from the analysis. A total of 1748 OTUs (407 735 sequences) remained for subsequent analyses.
Phylogenetic tree construction
Representative sequences from each OTU were manually-aligned to a reference full-length ITS alignment (Rocap et al., 2002) of 20 strains of Prochlorococcus and 2 strains of Synechococcus (WH8102 and CC9605). Tree building was performed in RaxML version 8.0.20 (Stamatakis, 2014) using a maximum likelihood search of 1000 non-parametric bootstraps with algorithmic shortcuts (‘Rapid Bootstrapping') and a GTR model with multi-gamma distributed rate variation among sites. Finally, OTUs were identified based on their phylogenetically closest reference sequences and their highest BLASTN hit against a Prochlorococcus-only database that included full genomes from cultured isolates and other uncultured clades (Huang et al., 2012). OTUs that were most closely related to low light clades were removed from subsequent analyses (571 OTUs). Phylogenetic trees are deposited in TreeBase (TB2:S18421).
Library rarefaction
The appropriate sequencing depth to accurately assess the patterns in Prochlorococcus α- and β-diversity was determined (Lundin et al., 2012). Briefly, OTU sequence abundance was rarefied (5 × replicated) to a range of sampling depths. Bacterial β-diversity (Bray-Curtis dissimilarity) and α-diversity metrics (OTU richness and Shannon's Diversity Index) were calculated for each rarefaction depth and replicate. For β-diversity, the Mantel-Test Coefficient of Variation (r2M) was used to determine how similar the rarefied data sets were to the full data set. For the α-diversity metrics, P-values were calculated using a Welch's T-Test and used to determine whether the rarefied data sets were significantly different from the diversity trends of the full data set. At a rarefaction depth of 5000, trends in both α- and β-diversity metrics were not significantly different from those calculated for the full data set (Supplementary Figure S1). Additionally, the sequence coverage of each station was visually inspected (Caporaso et al., 2010); and all curves that had greater than 5000 sequences appeared to be approaching asymptotic saturation (Supplementary Figure S2). The full OTU table was re-sampled 100 times at a sequencing depth of 5000; and the median value for each OTU was used for further analysis. Good's Coverage was ⩾0.94 for all samples.
Culturing
Axenic Prochlorococcus strains MIT9312 (VOL4) of HL-II and MED4 (VOL7) of HL-I were grown in modified AMP1 basal medium (replacing 1 mm TAPS for HEPES and adding 2 mm NaHCO3) at various salinities and adding ‘ProZ' nutrients consisting of 40 μm NaH2PO4, 800 μm NH4Cl, 0.15 μm H3BO3 and Pro99 metals (Moore et al., 2007; Morris and Zinser, 2013). Cultures were grown at ~80 μmol quanta m−2 sec−1 in 14:10 light:dark incubators at 21 °C. Growth was monitored using a calibrated 10-AU fluorometer (Turner Designs, Sunnyvale, CA, USA; Lin et al., 2013).
Statistical analysis
All diversity calculations and statistical analyses were performed in R (R software, v. 3.1.2, Vienna, Austria). The ‘vegan' package (Oksanen et al., 2015) was used to calculate Shannon's Diversity Index, and the best Euclidean combination of environmental variables for describing Bray-Curtis dissimilarities (via ‘bioenv'). OTU abundances were square-root transformed and submitted to a Wisconsin double standardization before Bray-Curtis dissimilarities were calculated. The ‘ecodist' package (Goslee and Urban, 2007) was used to examine Prochlorococcus community composition through a non-metric multidimensional scaling (NMDS) ordination of Bray-Curtis dissimilarity. Environmental variables and the rarefied sequence abundances of the Prochlorococcus clades were fitted to the NMDS ordination using the ‘vf' function in ‘ecodist'.
To assess environmental trends of the sub-ecotype clades, the abundance-weighted mean latitude, mean day length, mean temperature and median phosphate for each OTU were calculated. Non-rarefied OTU abundances were used in this specific analysis to avoid losing data on the tail ends of the OTU distributions and thus avoid biasing the environmental mean for each OTU. Median values were used for nutrients. Non-parametric two-sample t-tests with 9999 Monte Carlo permutations were used to compare the sub-ecotype clades (Supplementary Table S1). Additionally, Abouheif's test of phylogenetic proximity was performed to determine whether environmental trait clustering across taxonomic ranks was significantly different from a random model using ‘abouheif.moran' in ‘adephylo' (Supplementary Table S2) (Abouheif, 1999; Pavoine et al., 2008; Jombart et al., 2010). Finally, the relative influence of environmental variables on rarefied sequence abundance across taxonomic ranks was assessed by a variance partitioning analysis using ‘calc.relimp' in the ‘relaimpo' package. To determine which environmental variables were significantly correlated with the rarefied OTU abundance, data matrix redundant or highly correlated (R2>0.90) variables were removed and a stepwise ordination significance test was performed using ‘ordistep' in ‘vegan'. The correlations between the environmental variables identified by ‘ordistep' and the OTU abundance matrix for each sub-ecotype clade were then confirmed to be significant using a permutational multivariate analysis of variance test (‘adonis' in ‘vegan') (Supplementary Table S3). The significant environmental metrics were then used in the variance partitioning analysis.
Results and discussion
Phylogenetic placement and OTU classification
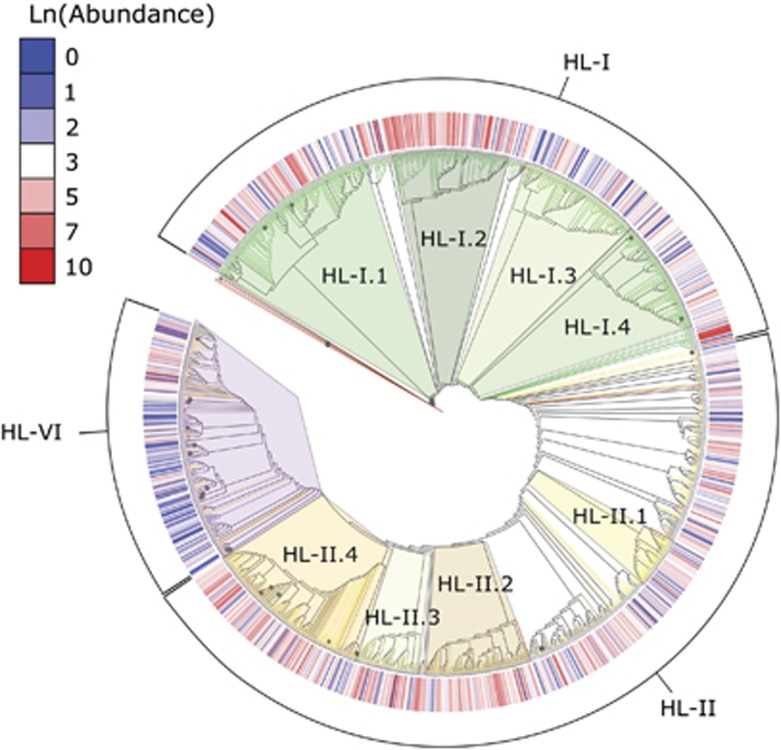
Across all stations and seasons, a total of 1177 Prochlorococcus HL OTUs were identified. Based on Prochlorococcus database BLAST results and the phylogenetic tree structure, these OTUs grouped into three major clades (458 OTUs—HL-I; 537 OTUs—HL-II and 182 OTUs—HL-VI) (Figure 2). Because of the short amplicon length of the ITS sequences (250 bp), the phylogenetic tree had short branch length and low bootstrap support (Supplementary Figure S3). However, representative sequences that identified as eHL-I or eHL-II via BLAST grouped with their respective cultured reference sequences. Additionally, a small portion of the OTU representative sequences were identified as eHL-VI (Huang et al., 2012) and were grouped into a single monophyletic clade at the tip of the tree. In contrast to eHL-I and eHL-II, eHL-VI was more recently discovered, and while it has no cultured representatives, it has been hypothesized to be associated with high-nutrient, low-chlorophyll conditions (Huang et al., 2012). Similar to past phylogenies, eHL-VI is closely related to eHL-II and more distantly related to eHL-I (Figure 2). These major clades could be further sub-divided into other clades based on their branching patterns, abundance and relationship with environmental variables.
Figure 2.
Maximum likelihood cladistic phylogenetic tree of ITS fragments from representative OTUs. Branches with >80% bootstrap support are marked with filled circles. Reference ITS sequences include isolates MED4, MIT9515, AS9601, MIT9215, MIT9301, MIT9312, MIT9211, NATL1A, NATL2A and SS120, and are indicated by red branches. Green, yellow and purple branch color as well as the external brackets indicate ecotype assignment. Internal shading indicates the sub-ecotype clades (green: eHL-I.1–4 and yellow: eHL-II.1–4) and eHL-VI (purple). Unshaded clades were included in ecotype analyses but were not examined in sub-ecotype clade analyses. The external heat map is the natural log of the sum across all stations of the rarefied sequence abundance for each OTU.
Four clades were chosen from within eHL-I and eHL-II for further analysis. These sub-ecotype clades were chosen based on clade size (OTUs and number of sequences) and phylogenetic distribution. The four largest HL-I clades had similar numbers of branches, or OTUs (138 OTUs—HL-I.1; 87 OTUs—HL-I.2; 92 OTUs—HL-I.3 and 86 OTUs—HL-I.4). The HL-II clades, on the other hand, were relatively uneven. Therefore, the two largest clades (77 OTUs—HL-II.2 and 127 OTUs—HL-II.4) and two additional clades from other portions of the phylogenetic tree (38 OTUs—HL-II.1 and 48 OTUs—HL-II.3) were selected. The sub-clades analyzed represent 89% of the OTUs and 74% of the total sequences for HL-I. For HL-II, they are 55% and 73%, respectively.
Significant environmental drivers of HL Prochlorococcus populations
The full suite of environmental variables from all cruise transects was narrowed down to those variables that were significantly correlated with community composition. The variables assessed included latitude, temperature, salinity, day length, phosphate, silicate, nitrate+nitrite, ammonia, oxygen, density, pH, mixed layer depth, chlorophyll and total Prochlorococcus concentration. Of these variables, nitrate+nitrite, ammonia, pH, mixed layer depth and Prochlorococcus concentration were found to be not significant (P⩾0.19) and removed from subsequent analyses. The nitrogen species were likely found to be non-significant due to their low concentration in marine surface waters. Mixed layer depth was likely found to be non-significant since this analysis only examines the HL populations of Prochlorococcus. Mixed layer depth has been found to be important for Prochlorococcus community composition in the past because it influences the relative proportions of HL and LL ecotypes present in the community as a whole (Bouman et al., 2006). However, mixed layer depth may be less important for the HL community alone. Similarly, a recent study found that mixed layer depth was not significantly correlated with the eHL-II to eHL-I ratio.
Relative ecotype abundance
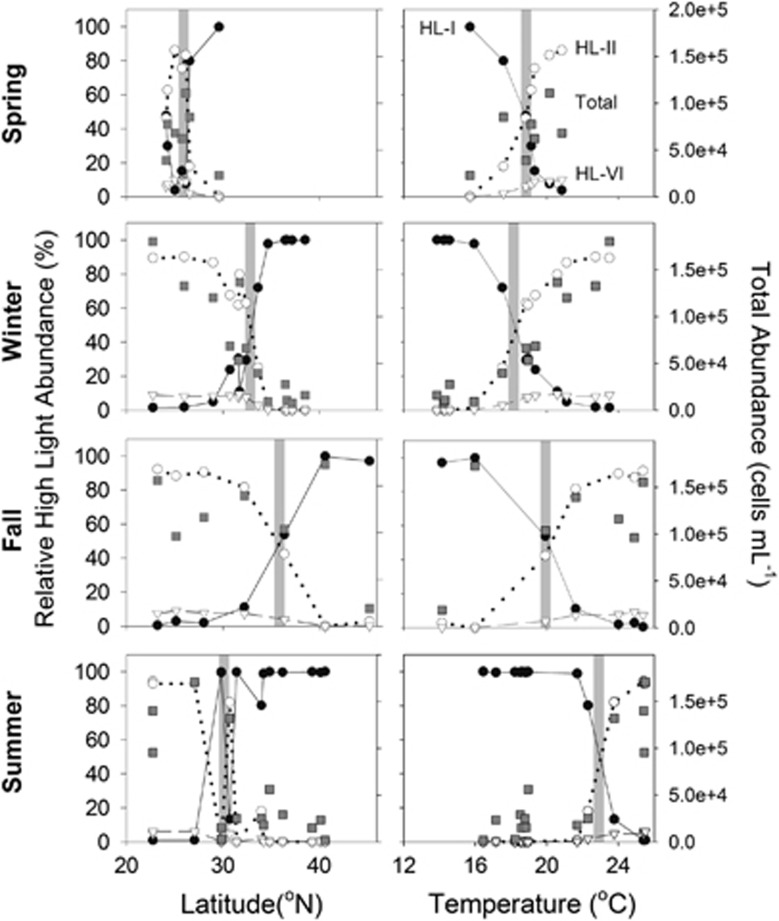
As in the Atlantic Ocean, Pacific Ocean Prochlorococcus communities assessed with amplicon libraries were dominated by eHL-II at lower latitudes and warmer temperatures and eHL-I at higher latitudes and colder temperatures (Johnson et al., 2006) (Figure 3). eHL-VI was also most abundant at low latitudes, but was about an order of magnitude less abundant than eHL-II. Quantitative abundances (cells per ml) available from quantitative PCR and the relative amplicon abundances were highly correlated (r2=0.97) (Supplementary Figure S4), suggesting that amplicon libraries can be used as quantitative estimates of Prochlorococcus spp. population abundance.
Figure 3.
Total abundance (flow cytometry) and relative ecotype abundance (rarefied OTU abundance) of Prochlorococcus across latitudinal and temperature gradients. The shaded boxes are crossover regions where the ratio of HL-I to HL-II+HL-VI is ~1:1.
Although the overall trends were conserved between seasons, there are notable differences between seasons in the location and temperature of transition between dominance by eHL-II and eHL-I (Figure 3). For example, the transition point in the summer occurred at ~31°N (~23 °C), shifted north to 36.3°N (19.9 °C) in the fall, and returned to 33.6°N (17.5 °C) in the winter, with a total range corresponding to ~450 km. The spring transect did not cover a large latitudinal range, but the transition between eHL-I and eHL-II along longitudinal gradients occurred at ~19 °C. Although we hypothesized that the transition point would shift to a higher latitude in the summer, the transition point was unexpectedly observed at a lower latitude during the summer and higher latitude in the fall. In addition to changes in the relative ecotype frequency, at higher latitudes (above 32°N) Prochlorococcus community abundance (cell ml−1) increased in the fall and was lowest in the summer. A latitudinal transect along an ~18 °C isotherm at the eastern edge of our summer transect was dominated by eHL-I in the surface samples along the entire transect leg (Supplementary Figure S5), showing that temperature has a dominant, but not exclusive role in structuring ecotype frequency. These patterns are generally consistent with observations from other ocean basins of both spatial (latitudinal and longitudinal) and seasonal transitions in Prochlorococcus ecotypes (Bouman et al., 2006; Rusch et al., 2007, 2010; Malmstrom et al., 2013).
Whereas the spring and fall transitions between eHL-I and eHL-II dominance occurred at the expected temperature of ~19 °C, which is the temperature wherein eHL-I and eHL-II cultures have equal growth rates under laboratory conditions (Johnson et al., 2006), the winter and summer transitions were significantly different (17.5 °C and 23 °C, respectively). To determine whether this difference represented a lagged population response to temperature change, sea surface temperatures were examined at the winter transition point where eHL-II was unexpectedly high (17.5 °C) and two stations in the summer where eHL-II had unexpectedly low abundance (21.3 °C and 21.7 °C). At the winter crossover station, there was a medium to high rate of temperature decrease before the winter sampling period (Supplementary Figure S6). Similarly, both of the summer stations experienced either high or increasing rates of temperature increase directly before the summer sampling period (Supplementary Figure S6). A trait-based model based on laboratory growth rates as a function of temperature (Johnson et al., 2006) and forced using daily remote surface seawater temperature observations also predicts that these ‘anomalies' from expectations are consistent with lagged population responses. Finally, strong correlations between major ecotypes and salinity, which are not consistent with the direct effects of salinity on growth rate (Supplementary Figure S7), are instead likely because of salinity acting as an indicator of the boundary between the oligotrophic gyre and higher latitudes. At these three diagnostic stations salinity generally displayed stochastic fluctuations around a mean value, as opposed to temperature, which showed stable, seasonal fluctuations (Supplementary Figure S8). These data, along with the evidence presented here that Prochlorococcus populations shift to higher latitudes in the fall, suggest that there is a time lag in the response of, Prochlorococcus ecotypes to changing temperature, as has been seen in other microbial communities (Gilbert et al., 2012; Chow et al., 2013) and in Prochlorococcus communities in other ocean basins (Malmstrom et al., 2010). This evidence also suggests that temperature is the dominant environmental variable poising broad scale ecotype distributions, though this relationship differs at the sub-ecotype level (Supplementary Figure S9).
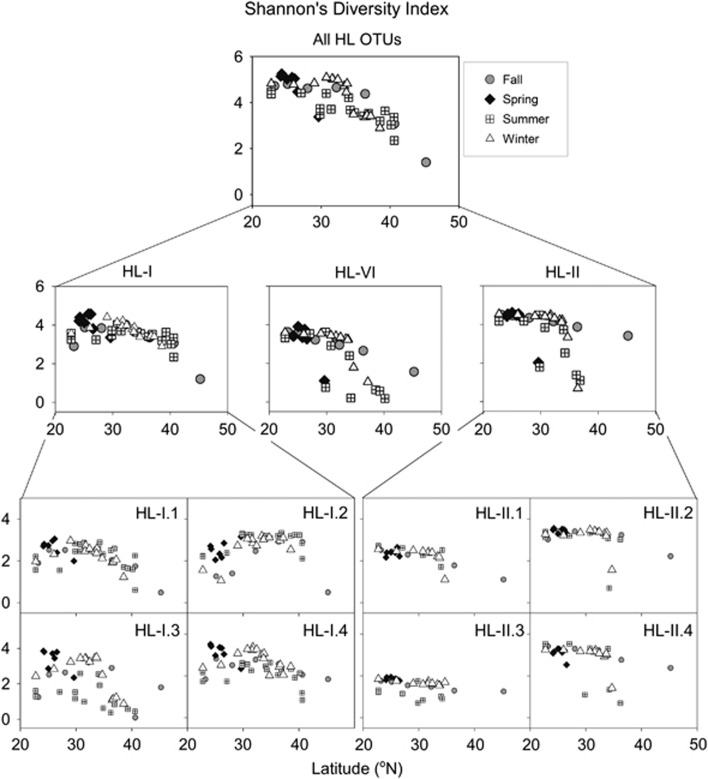
Latitudinal and seasonal trends in α-diversity
In addition to latitudinal and seasonal transitions in relative ecotype frequencies, the Prochlorococcus community also showed latitudinal trends in α-diversity as quantified by Shannon's Diversity Index (Figure 4). Similar to patterns of microbial diversity for whole communities across ocean basins (Pommier et al., 2007; Fuhrman et al., 2008; Ladau et al., 2013; Swan et al., 2013), the Prochlorococcus HL community had the highest diversity at low latitudes, with remarkably similar spatial patterns between seasons. These trends were also apparent within ecotypes with some additional seasonal and spatial complexity: eHL-II and eHL-VI had higher diversity at lower latitudes, but with precipitous decreases at latitudes above ~35°N in the winter and summer. At low latitudes eHL-I had lower diversity than eHL-II, but decreased in diversity more slowly at higher latitudes resulting in higher overall diversity for eHL-I than eHL-II between ~36 and 42°N. These patterns may be driven in part by abundance as OTU richness for both eHL-I and eHL-II increased in a logarithmic manner with increasing ecotype-specific abundance (Supplementary Figure S10). However, the same relationship was not observed between abundance and Shannon's Diversity Index (data not shown).
Figure 4.
Latitudinal trends of Prochlorococcus α-diversity as calculated by Shannon's Diversity Index for subsets of OTUs grouped at the whole HL community, ecotype and sub-ecotype taxonomic levels.
Partitioning these diversity trends into the sub-ecotype clades reveals more intricate relationships across latitudes. Although the HL-II sub-ecotype clades generally showed similar patterns to the ecotype as a whole (with the exception of HL-II.3 in summer), each of the HL-I sub-ecotype clades had a unique seasonal and spatial pattern of diversity across latitudes (Figure 4). These results demonstrate that while the spatial patterns of the Prochlorococcus community α-diversity are similar between seasons, the components of this diversity can be dynamic across seasons, suggesting niche complementarity among its members.
Latitudinal and seasonal trends in β-diversity
To gain insight into how and why the composition of the Prochlorococcus HL community and its components shifted across seasons and stations, we performed NMDS to help detect relationships. This NMDS analysis demonstrates that the summer stations are clearly distinct from the other seasons, whereas the remaining seasons are generally more interspersed (Figure 5a). This suggests that Prochlorococcus communities in the summer have significant shifts in composition when compared with other seasons. Key environmental variables quantified for all four seasons and fitted to the ordination space show that day length is the dominant driver distinguishing summer from other seasons. The latitudinal distribution in community composition is in part related to gradients in temperature, but other variables such as silicate, phosphate and salinity may also be related to the spatial trends. To confirm that these metrics were the best Euclidean combination of environmental variables that explained the greatest variability in Bray-Curtis dissimilarities, we performed a BIO-ENV test (Clarke and Ainsworth, 1993) on the full suite of variables that were significantly correlated with community composition for the spring, summer and winter cruises (only limited environmental data are available for the fall cruise). As before, latitude, temperature, salinity and day length were the most explanatory metrics and were highly correlated with Bray-Curtis dissimilarity (r=0.81). The importance of this suite of environmental variables is consistent with other latitudinal oceanographic studies, and these variables are likely representative of a range of abiotic processes affecting broader microbial community composition (Johnson et al., 2006; Fuhrman et al., 2008; Sjostedt et al., 2014).
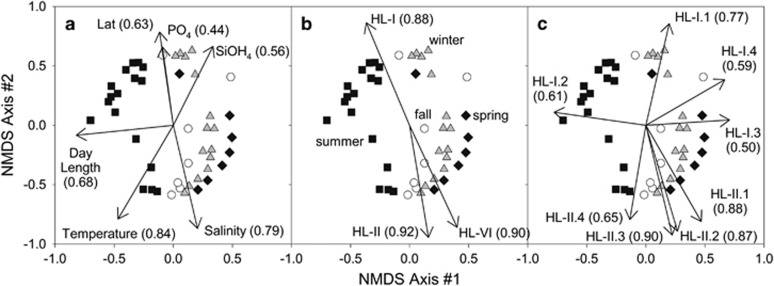
Figure 5.
NMDS of Bray-Curtis dissimilarities. Ordination of all HL OTUs with (a) environmental variables, (b) rarefied ecotype sequence abundance and and (c) rarefied sub-ecotype clade sequence abundance fitted to the distribution of sampling sites in the ordination space. The correlation of each variable to the distribution of sampling sites is represented in parentheses. Symbols correspond to the summer (squares), spring (diamonds), fall (circles) and winter (triangles) stations.
We extended this analysis to examine how ecotype and sub-ecotype abundance influenced the β-diversity of community diversity (Figures 5b and c). Relative ecotype abundance (eHL-I, eHL-II and eHL-VI) aligns similarly with latitude and salinity (that is, lagged temperature), confirming the importance of broad scale oceanographic environmental variables in structuring major clades/ecotypes of Prochlorococcus. However, sub-ecotype clades are more dispersed in the NMDS analysis, suggesting different niche spaces for these groups. In particular, while the sub-ecotype clades of HL-II generally align with the composite eHL-II clade directionality in NMDS space (Figures 5b vs c), the HL-I sub-ecotype clades are more divergent. For example, HL-I.2 and HL-I.3 are aligned in opposite directions along NMDS axis 1, while HL-I.1 is orthogonal to these clades and aligns closely with NMDS axis 2. These data suggest that structuring of major clades (that is, ecotypes) can follow (and are driven by) large-scale oceanographic variability, but that finer scale changes in community diversity may be driven by additional processes or unmeasured environmental properties.
Environmental drivers of sub-ecotype clade abundance
The correlations between Bray-Curtis distance matrices of OTU abundance for each sub-ecotype clade and environmental variables were tested using a permutational multivariate analysis of variance (Oksanen et al., 2015). A ‘Type III' adjustment was applied due to the correlation between environmental metrics. In this statistical technique, the effect of each variable is evaluated only after both the effects and the interactive effects of all other variables have been accounted for in a sum of squares model. Thus, this is a highly conservative approach that only assesses the variability that is uniquely attributable to each environmental metric. The result of the test shows that latitude, temperature, salinity, day length, silicate, phosphate, oxygen and chlorophyll are all significantly correlated with at least one sub-ecotype clade (Supplementary Table S3). However, a much higher proportion of the environmental variables are significantly correlated with the HL-I sub-ecotype clades than the HL-II sub-ecotype clades. As the Type III adjustment eliminates interactive effects, it is possible that the interaction between multiple environmental variables is important for driving HL-II community composition. Thus, to fully understand the environmental associations of the sub-ecotype clades, further analysis was needed.
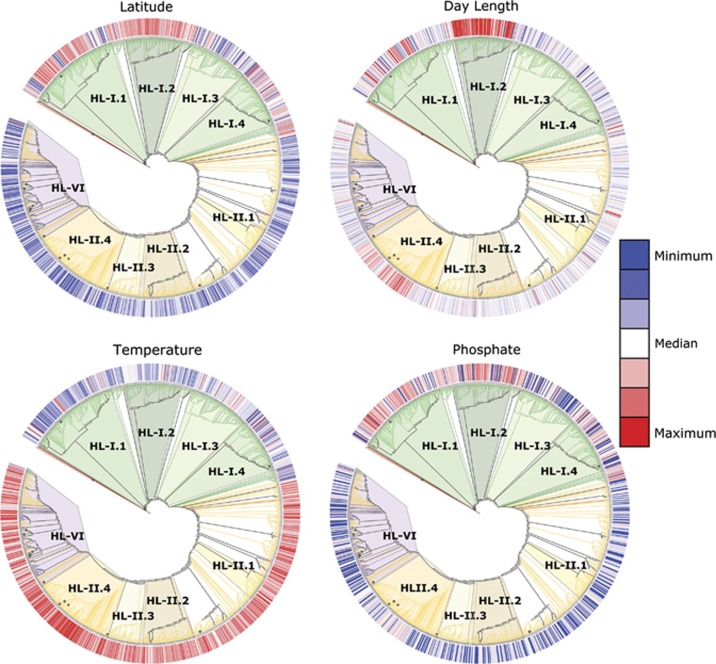
To explore the environmental drivers and niche space of sub-ecotype clades, we examined the phylogenetic association of key variables (Figure 6). As expected, eHL-I is generally associated with low temperatures and high latitudes, whereas eHL-II is, with higher temperatures and low latitudes. However, additional complexity exists among the sub-ecotype clades. For example, HL-I.2 is enriched in high latitudes relative to HL-I.3, and these patterns are apparent with temperature as well. HL-I.2 shows a clear enrichment in longer day lengths (that is, summer samples) relative to other HL-I clades. The HL-I clades also show general cohesion for specific phosphate environments. These sub-ecotype clade associations with specific environments for HL-I do not appear to be as clearly defined for HL-II. Although HL-II.4 is enriched in longer day lengths, higher temperature and high phosphate, the remaining sub-ecotype clades are not clearly delineated among these key variables.
Figure 6.
Phylogenetic association of latitude, day length, temperature and phosphate. Heat-map values (red=high, white=median, blue=low) represent the abundance-weighted mean (or median for phosphate) environmental value across all observations of a given OTU for the full sample set. Branches of cladistic phylogenetic tree with >80% bootstrap support are marked with filled circles.
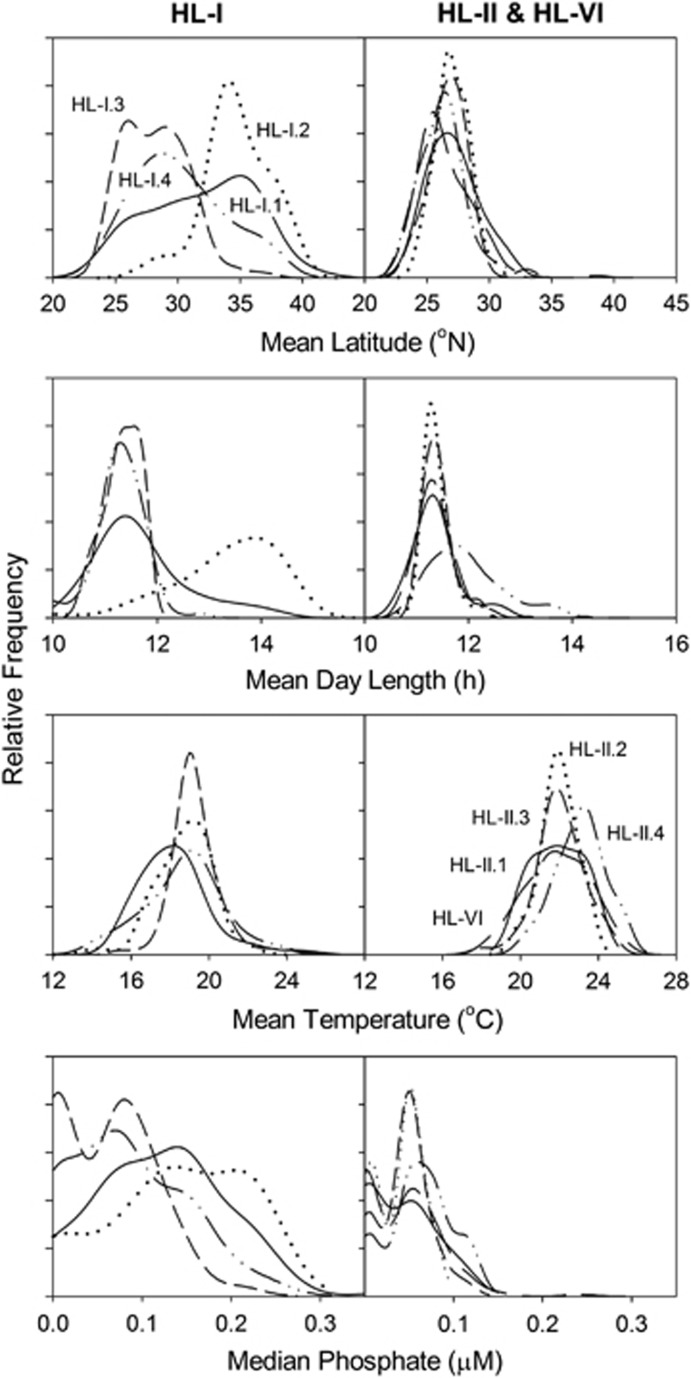
Frequency distributions of the abundance-weighted mean (or median for phosphate) environmental values for OTUs in each sub-ecotype clade also demonstrate that, with the exception of day length and temperature for HL-II.4, the HL-II clades are remarkably consistent in their environmental distributions (Figure 7). Conversely, HL-I clades have different distributions for each of the variables. The distribution of mean values for each sub-ecotype clade was compared with pairwise non-parametric two-sample t-tests, which showed that the HL-I sub-ecotype clades had a higher proportion of significant differences in the distribution of their environmental means than the HL-II clades (P<0.05) (Supplementary Table S1). Other environmental variables including silicate, oxygen and chlorophyll show similar patterns between HL-I and HL-II (Supplementary Figure S11). This analysis reveals that many of the HL-I sub-ecotype clades appear to have cohesive (within clades), yet distinct (across clades) environmental associates (and presumably niche envelopes), whereas HL-II clades appear to have more similar environmental niche space.
Figure 7.
Histogram density curves of abundance-weighted mean and median environmental variables for OTUs within each sub-ecotype clade for the full sample set.
To ensure that these differences in clade-specific environmental traits were significantly different from a random model, we performed Abouheif's test of phylogenetic proximity. This test was performed at the full tree, ecotype and sub-ecotype levels by separating the tree into sub-trees (Abouheif, 1999; Pavoine et al., 2008; Pavoine and Ricotta, 2013). P-values from this test show that phylogenetic signal, or partitioning, can be detected in environmental traits across all taxonomic ranks (Supplementary Table S2). This suggests that both the ecotypes and some of the sub-ecotype clades can be partitioned into ecologically distinct groups. However, this test does not indicate what evolutionary or environmental forces, such as selection, drift, convergence and conservatism, are driving this phylogenetic partitioning of OTU traits. Therefore, further analysis was needed to discover the major drivers of clade-specific abundance.
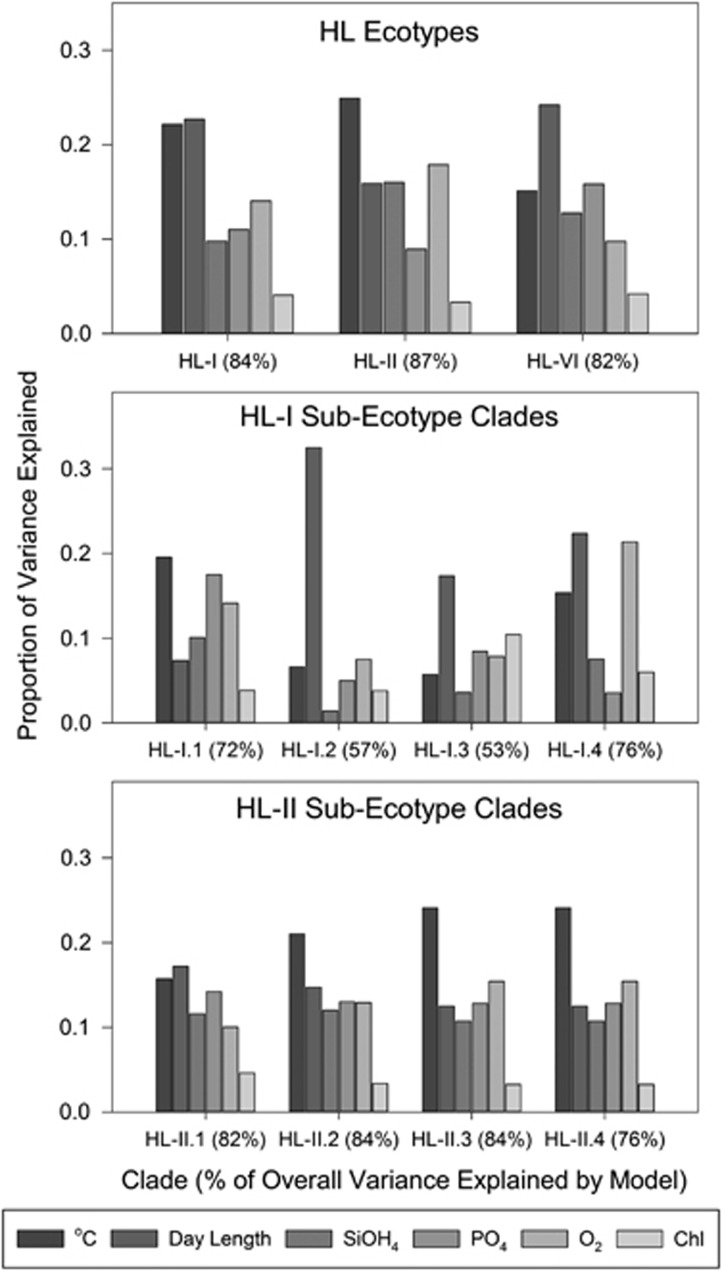
Finally, to determine relative importance of the identified environmental variables across taxonomic ranks we partitioned variance (Figure 8) focusing on environmental variables that were significantly correlated with community composition (P<0.05), including temperature, day length, silicate, phosphate, oxygen and chlorophyll. Latitude and salinity were removed from this analysis as they are not believed to be the proximal drivers of Prochlorococcus abundance, merely correlated with large-scale oceanographic trends. Temperature and day length (season) explained the greatest proportion of variability for the eHL-I, eHL-II and eHL-VI abundance. Similarly, at the sub-ecotype level, these two variables were generally the most explanatory. Other variables including oxygen and phosphate were important for some sub-ecotype clades, but not others. More broadly, the HL-II clades have the same general ranking in their partitioning of variance across environmental variables. This is in contrast to the HL-I clades that have dramatically different associations. For example, day length is most explanatory for HL-I.2 whereas for HL-I.1 day length is less explanatory. Variance partitioning along with phylogenetic association of environmental variables (Figures 6 and 7), α-diversity (Figure 4) and β-diversity (Figure 5) suggests that HL-I clades have coherent and unique niche space, whereas HL-II clades are generally less differentiated and do not have significant correlations with the measured environmental variables (Supplementary Table S3). Additional variables not measured here such as grazer pressure or viral susceptibility may distinguish HL-II clades into more refined, ecologically distinct groups. Regardless of niche differences between the clades of eHL-I and eHL-II, overall it appears that different combinations of variables (or their associated response curves) may be responsible for defining the niche space of sub-ecotype clades rather than a strict trait hierarchy per se (Coleman and Chisholm, 2007). Varying combinations of traits (through flexible genomic content) could promote the diversification and maintenance of numerous distinct Prochlorococcus genomes each with its own niche space (Kashtan et al., 2014).
Figure 8.
Percent of variance in rarefied sequence abundance partitioned by environmental variables across taxonomic ranks.
Niche differentiation of Prochlorococcus sub-ecotype clades
Fundamentally different ecological processes may drive the distribution of HL-I and HL-II diversity and community composition. The sub-ecotype clades appear to partition the ocean both spatially and temporally, relying on adaptations that affect their overall niche space. Whereas HL-I sub-ecotype clades are found in a more variable environment and show highly diverse responses to environmental changes, the HL-II sub-ecotype clades are more similar in their environmental niches. The extent of environmental differences between sub-ecotypes within clades suggests that the HL-I sub-ecotype clades experience a greater degree of bottom-up control of their spatial and temporal distribution. This finding is in concordance with studies of both terrestrial and coastal marine microbial communities that show that environmental variables, such as temperature, explain differences in composition better than neutral processes on regional spatial scales and interannual temporal scales (Martiny et al., 2011; Hatosy et al., 2013). However, HL-II sub-ecotype clades responded to environmental variability with a much smaller degree of differentiation than the Hl-I clades did, suggesting that in the more stable oligotrophic environments other unmeasured abiotic and biotic variables (such as interactions with other microbes, phage resistance or grazing), as well as neutral variability, may be more important for driving niche partitioning in these clades.
Multiple modeling studies have suggested that there is a geographic and ecological separation between the seasonally stable oligotrophic ocean and the more seasonally variable high latitudes (Barton et al., 2010; Hellweger et al., 2014). Hellweger et al. (2014) used a global ocean circulation model to show that differences between microbial communities at the HOT station and the Gulf of Alaska could arise through neutral genetic mutation alone. Similarly, Barton et al. (2010) used a global ocean ecosystem model combined with an idealized resource competition framework to determine that random dispersal and variability of the environment were the greatest controls on phytoplankton diversity. In the Barton et al. model, seasonal variability in the high latitudes resulted in competitive exclusion of all but a few phytoplankton species, whereas seasonal stability at lower latitudes allowed the coexistence of physiologically distinct, but equally fit, phytoplankton. Results from this study support this division between seasonally variable, bottom-up driven high-latitude microbial communities and seasonally stable mid-to-low latitude microbial communities.
Conclusions
Prochlorococcus communities undergo large-scale shifts in composition across both inter- and intra-seasonal environmental gradients. Broadly, like other microbial communities (for example, Fuhrman et al., 2008) the Prochlorococcus community is more abundant and more diverse at lower latitudes, but sub-ecotype clades show differing responses to environmental gradients adding complexity to broad community patterns. Indeed, changes in sub-ecotype clades appear to drive differences in community composition across seasons. The responses of these sub-ecotype clades appear to be driven by multiple environmental variables (temperature, day length and nutrients) that define their niche space, rather than by a hierarchy of traits. Finally, this study provides evidence for the geographic separation between bottom-up driven seasonally variable Prochlorococcus populations in temperate northerly latitudes and seasonally stable subtropical Prochlorococcus populations that are likely driven by both bottom-up and top-down forces, which may in part explain differences in variance partitioning among the HL-I and HL-II sub-clades. In the future ocean, the distribution and variability of key environmental variables identified here are predicted to change and available niche space models predict substantial changes for Prochlorococcus community distributions (Flombaum et al., 2013). These changes in community abundance will translate into seasonal and spatial shifts in community composition; the impact of these compositional shifts on ecological interactions or biogeochemical processes such as carbon fixation and nutrient cycling remains unknown.
Acknowledgments
We acknowledge the Armbrust team (UW) for collecting samples on TN271 (POWOW0), other members of the science teams aboard the three POWOW cruises, and BCO-DMO for assistance with data deposition and quality control. This work was financially supported by NSF DBI09-59630, OCE10-31064 and OCE10-30518 (USA) and the Alexander von Humboldt Foundation (Germany).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abouheif E. (1999). A method for testing the assumption of phylogenetic independence in comparative data. Evol Ecol Res 1: 895–909. [Google Scholar]
- Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. (2004). Divergence and redundancy of 16S rRNA sequences in genomes with multiple rRNA operons. J Bacteriol 186: 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren NA, Rocap G, Chisholm SW. (2006). Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol 8: 441–454. [DOI] [PubMed] [Google Scholar]
- Barton AD, Dutkiewicz S, Flierl G, Bragg J, Follows MJ. (2010). Patterns of diversity in marine phytoplankton. Science 327: 1509–1511. [DOI] [PubMed] [Google Scholar]
- Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH et al. (2014). Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J 9: 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Berube PM, Lindell D, Chisholm SW. (2015). Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol 13: 13–27. [DOI] [PubMed] [Google Scholar]
- Bouman HA, Ulloa O, Scanlan DJ, Zwirglmaier K, Li WKW, Platt T et al. (2006). Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science 312: 918–921. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CE, Sachdeva R, Cram JA, Steele JA, Needham DM, Patel et al. (2013). Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J 7: 2259–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Ainsworth M. (1993). A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser 92: 205–219. [Google Scholar]
- Cohan FM. (2002). What are bacterial species? Annu Rev Microbiol 56: 457–487. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. (2007). Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol 15: 398–407. [DOI] [PubMed] [Google Scholar]
- Dupont CL, McCrow JP, Valas R, Moustafa A, Walworth N, Goodenough U et al. (2014). Genomes and gene expression across light and productivity gradients in eastern subtropical Pacific microbial communities. ISME J 9: 1076–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Flombaum P, Gallegos JL, Gordillo RA, Rincon J, Zabala LL, Jiao N et al. (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. PNAS 110: 9824–9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. PNAS 105: 7774–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrueck L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslee SC, Urban DL. (2007). The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22: 1–19. [Google Scholar]
- Hatosy SM, Martiny JBH, Sachdeva R, Steele J, Fuhrman JA, Martiny AC. (2013). Beta diversity of marine bacteria depends on temporal scale. Ecology 94: 1898–1904. [DOI] [PubMed] [Google Scholar]
- Hellweger FL, van Sebille E, Fredrick ND. (2014). Biogeographic patterns in ocean microbes emerge in a neutral agent-based model. Science 345: 1346–1349. [DOI] [PubMed] [Google Scholar]
- Hess WR, Rocap G, Ting CS, Larimer F, Stilwagen S, Lamerdin J et al. (2001). The photosynthetic apparatus of Prochlorococcus: Insights through comparative genomics. Photosynth Res 70: 53–71. [DOI] [PubMed] [Google Scholar]
- Holmes RM, Aminot A, Kerouel R, Hooker BA, Peterson BJ. (1999). A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci 56: 1801–1808. [Google Scholar]
- Huang S, Wilhelm SW, Harvey HR, Taylor K, Jiao N, Chen F. (2012). Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J 6: 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320: 1081–1085. [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740. [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Shyam R, Ritchie AE, Mioni C, Lance VP, Murray JW et al. (2010). The effect of iron- and light-limitation on phytoplankton communities of deep chlorophyll maxima of the western Pacific Ocean. J Mar Res 68: 283–308. [Google Scholar]
- Johnson ZI, Wheeler BJ, Blinebry SK, Carlson CM, Ward CS, Hunt DE. (2013). Dramatic variability of the carbonate system at a temperate coastal ocean site (Beaufort, North Carolina, USA) is regulated by physical and biogeochemical processes on multiple timescales. PLoS One 8: e85117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Martiny AC. (2015). Techniques for quantifying phytoplankton biodiversity. Annu Rev Mar Sci 7: 299–324; null. [DOI] [PubMed] [Google Scholar]
- Jombart T, Balloux F, Dray S. (2010). adephylo: new tools for investigating the phylogenetic signal in biological traits. Bioinformatics 26: 1907–1909. [DOI] [PubMed] [Google Scholar]
- Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe et al. (2014). Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344: 416–420. [DOI] [PubMed] [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S et al. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3: 2515–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner A, Ward DM et al. (2008). Identifying the fundamental units of bacterial diversity: A paradigm shift to incorporate ecology into bacterial systematics. PNAS 105: 2504–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladau J, Sharpton TJ, Finucane MM, Jospin G, Kembel SW, O'Dwyer J et al. (2013). Global marine bacterial diversity peaks at high latitudes in winter. ISME J 7: 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Gazsi K, Lance VP, Larkin A, Chandler J, Zinser ER et al. (2013). In situ activity of a dominant Prochlorococcus ecotype (eHL-II) from rRNA content and cell size. Environ Microbiol 15: 2736–2747. [DOI] [PubMed] [Google Scholar]
- Lundin D, Severin I, Logue JB, Ostman O, Andersson AF, Lindstrom ES. (2012). Which sequencing depth is sufficient to describe patterns in bacterial alpha- and beta-diversity? Environ Microbiol Rep 4: 367–372. [DOI] [PubMed] [Google Scholar]
- Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, Zinser ER et al. (2010). Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J 4: 1252–1264. [DOI] [PubMed] [Google Scholar]
- Malmstrom RR, Rodrigue S, Huang KH, Kelly L, Kern SE, Thompson et al. (2013). Ecology of uncultured Prochlorococcus clades revealed through single-cell genomics and biogeographic analysis. ISME J 7: 184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Coleman ML, Chisholm SW. (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. PNAS 103: 12552–12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Kathuria S, Berube PM. (2009. a). Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. PNAS 106: 10787–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Tai APK, Veneziano D, Primeau F, Chisholm SW. (2009. b). Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol 11: 823–832. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC. (2011). Drivers of bacterial beta-diversity depend on spatial scale. PNAS 108: 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LR, Goericke R, Chisholm SW. (1995). Comparative physiology of Synechococcus and Prochlorococcus—Influence of light and temperature on growth, pigments, fluorescence and adsorptive properties. Mar Ecol Prog Ser 116: 259–275. [Google Scholar]
- Moore LR, Chisholm SW. (1999). Photophysiology of the marine cyanobacterium Prochlorococcus: Ecotypic differences among cultured isolates. Limnol Oceanogr 44: 628–638. [Google Scholar]
- Moore LR, Post AF, Rocap G, Chisholm SW. (2002). Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr 47: 989–996. [Google Scholar]
- Moore LR, Ostrowski M, Scanlan DJ, Feren K, Sweetsir T. (2005). Ecotypic variation in phosphorus acquisition mechanisms within marine picocyanobacteria. Aquat Microb Ecol 39: 257–269. [Google Scholar]
- Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D et al. (2007). Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Methods 5: 353–362. [Google Scholar]
- Morris JJ, Zinser ER. (2013). Continuous hydrogen peroxide production by organic buffers in phytoplankton culture media. J Phycol 49: 1223–1228. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara R et al. (2015), Package ‘vegan'. Community ecology package, version 2.2-1.
- Partensky F, Hess WR, Vaulot D. (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63: 106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partensky F, Garczarek L. (2010). Prochlorococcus: advantages and limits of minimalism. Annu Rev Mar Sci 2: 305–331. [DOI] [PubMed] [Google Scholar]
- Pavoine S, Ollier S, Pontier D, Chessel D. (2008). Testing for phylogenetic signal in phenotypic traits: New matrices of phylogenetic proximities. Theor Popul Biol 73: 79–91. [DOI] [PubMed] [Google Scholar]
- Pavoine S, Ricotta C. (2013). Testing for phylogenetic signal in biological traits: the ubiquity of cross-product statistics. Evolution 67: 828–840. [DOI] [PubMed] [Google Scholar]
- Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880. [DOI] [PubMed] [Google Scholar]
- Rocap G, Distel DL, Waterbury JB, Chisholm SW. (2002). Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68: 1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA et al. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. (2010). Characterization of Prochlorococcus clades from iron-depleted oceanic regions. PNAS 107: 16184–16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan DJ, West NJ. (2002). Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol 40: 1–12. [DOI] [PubMed] [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostedt J, Martiny JBH, Munk P, Riemann L. (2014). Abundance of broad bacterial taxa in the Sargasso Sea explained by environmental conditions but not water mass. Appl Environ Microbiol 80: 2786–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Waterbury JB, Chisholm SW. (2003). Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424: 1047–1051. [DOI] [PubMed] [Google Scholar]
- Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, Gonzalez JM et al. (2013). Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. PNAS 110: 11463–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MK, Kremer CT, Klausmeier CA, Litchman E. (2012). A global pattern of thermal adaptation in marine phytoplankton. Science 338: 1085–1088. [DOI] [PubMed] [Google Scholar]
- Thompson JR, Marcelino LA, Polz MF. (2002). Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR'. Nucleic Acids Res 30: 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violle C, Nemergut DR, Pu Z, Jiang L. (2011). Phylogenetic limiting similarity and competitive exclusion. Ecol Lett 14: 782–787. [DOI] [PubMed] [Google Scholar]
- Yung C-M, Vereen MK, Herbert A, Davis KM, Yang J, Kantorowska et al. (2014). Thermally adaptive tradeoffs in closely-related marine bacterial strains. Environ Microbiol 17: 2421–2429. [DOI] [PubMed] [Google Scholar]
- Zinser ER, Coe A, Johnson ZI, Martiny AC, Fuller NJ, Scanlan DJ et al. (2006). Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol 72: 723–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, Chisholm SW. (2007). Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr 52: 2205–2220. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.