Summary
Steroid hormones control important developmental processes and are linked to many diseases. To systematically identify genes and pathways required for steroid production, we performed a Drosophila genome-wide in vivo RNAi screen and identified 1,906 genes with potential roles in steroidogenesis and developmental timing. Here, we use our screen as a resource to identify mechanisms regulating intracellular levels of cholesterol, a substrate for steroidogenesis. We identify a conserved fatty acid elongase that underlies a mechanism which adjusts cholesterol trafficking and steroidogenesis with nutrition and developmental programs. Additionally, we demonstrate the existence of an autophagosomal cholesterol mobilization mechanism and show that activation of this system rescues Niemann Pick type C1-deficiency that causes a disorder characterized by cholesterol accumulation. These cholesterol trafficking mechanisms are regulated by TOR and feedback signaling that couples steroidogenesis with growth and ensure proper maturation timing. These results reveal genes regulating steroidogenesis during development that likely modulate disease mechanisms.
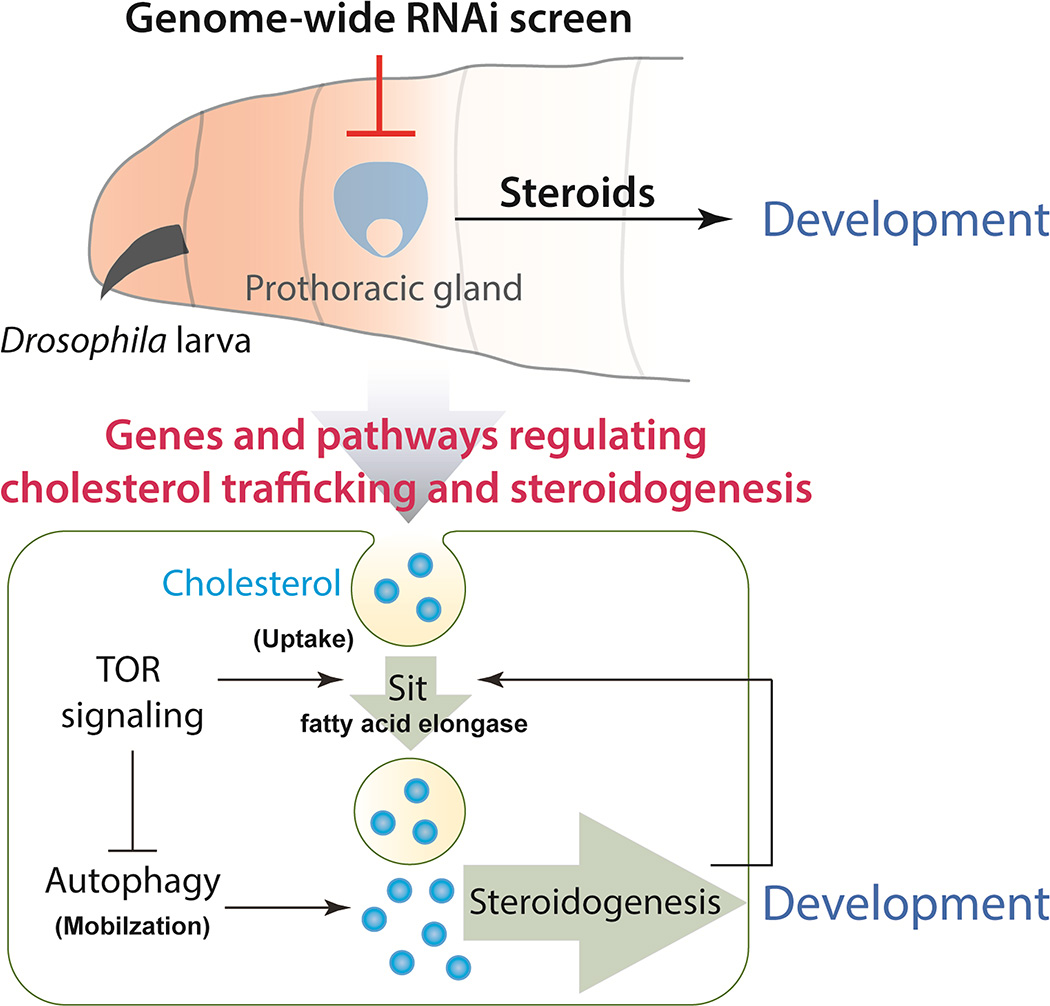
Graphical Abstract
Introduction
Steroid hormone signaling controls important biological functions during development and underlies pathologies of many disorders (Rewitz et al., 2013; Risbridger et al., 2010). During postembryonic development the production and release of steroids from endocrine cells are critical for developmental transitions including growth termination and maturation (Colombani et al., 2015; Garelli et al., 2015; Vallejo et al., 2015). Cells responsible for the production of steroids share similar mechanisms by which cholesterol or other sterol precursors are converted into hormones that are released into circulation to regulate a myriad of physiological and developmental processes. Similar to mammals, the production of steroids in model organisms such as the fruit fly Drosophila melanogaster is mediated by cytochrome P450 enzymes that convert cholesterol or other suitable sterols to active steroids (Carvalho et al., 2010; Lavrynenko et al., 2015; Rewitz et al., 2013). The production of steroids is regulated by multiple signals including insulin, TOR, Ras/ERK, TGFβ and feedback control. In Drosophila, the prothoracic gland (PG) produces the steroid hormone ecdysone from cholesterol and integrates different signals to adjust hormone production in response to nutrient conditions and development (Yamanaka et al., 2013). This ensures that growth is coordinated with activation of steroid production that triggers maturation or metamorphosis to form an adult with a characteristic body size.
Many of the signaling pathways that regulate cholesterol trafficking and steroid production are evolutionarily conserved, and dysregulation of these pathways is associated with disease. In humans, cholesterol is derived from two sources – dietary uptake and de novo synthesis – with uptake being the primary source of intracellular cholesterol supply in both endocrine cells and many tumors (Miller and Auchus, 2011; Yue et al., 2014). Intracellular trafficking of LDL-derived cholesterol from receptor-mediated endocytosis accounts for ~80% of the cholesterol in mammalian cells. Similarly, Drosophila acquires cholesterol by an LDL-related mechanism, since it lacks the ability to synthesize sterols (Carvalho et al., 2010; Huang et al., 2008). Intracellular trafficking of LDL-derived cholesterol provides a substrate for steroid synthesis. Genetic studies have shown that trafficking involves Niemann Pick type C1 (NPC1) disease associated genes that promote mobilization of LDL-derived cholesterol from late endosomes (Schwend et al., 2011). Mutations that disrupt NPC1 genes cause fatal lipid storage disorders, characterized by accumulation of cholesterol and other lipids in late endosomes/lysosomes, for which there is no cure. Loss of Npc1a, the Drosophila NPC1 homolog, causes insufficient cholesterol delivery to support ecdysone production (Huang et al., 2005). In steroid-related cancers such as prostate cancer, loss of the tumor suppressor PTEN and the subsequent upregulation of the PI3K/AKT/TOR pathway increases cholesterol uptake leading to an accumulation of cholesterol that drives cancer progression (Yue et al., 2014). Furthermore, studies in mice and Drosophila have linked metabolic disorders and disruption of the conserved insulin-like system to altered steroid signaling and delayed onset of maturation and reproduction (Colombani et al., 2005; Daftary and Gore, 2005; Tennessen and Thummel, 2011).
Our current understanding of steroidogenesis is largely based on cell-culture models, which have limitations since cell lines are unlikely to fully recapitulate physiological processes that occur in endocrine cells in vivo. Therefore, identifying such mechanisms is key to a better understanding of developmental processes and the mechanisms that underlie steroid-related disease. Here, we present a genome-wide in vivo RNAi screen in Drosophila to systematically uncover genes important for steroidogenic tissue function. We identify stuck in traffic (sit), a homolog of a fatty acid elongase linked to prostate cancer, and show that it is important for cholesterol trafficking in steroidogenic cells. Knockdown of sit results in accumulation of cholesterol-rich lipid droplets, likely due to impaired sphingolipid synthesis, which blocks cholesterol delivery and reduces steroid production. Additionally, our data identify an autophagic cholesterol trafficking system and we show that inhibition of autophagy leads to accumulation of cholesterol-rich lipid droplets in the PG. We further provide evidence that TOR signaling and steroid feedback coordinate cholesterol uptake and trafficking in PG cells. Our characterization of genes and mechanisms regulating cholesterol levels in endocrine cells provides insight into how steroidogenesis is controlled in a developmental context during the juvenile-adult transition, and molecular clues concerning mechanisms underlying certain cancers and lipid storage disorders.
Results
A genome-wide in vivo RNAi screen for genes involved in steroidogenesis in Drosophila
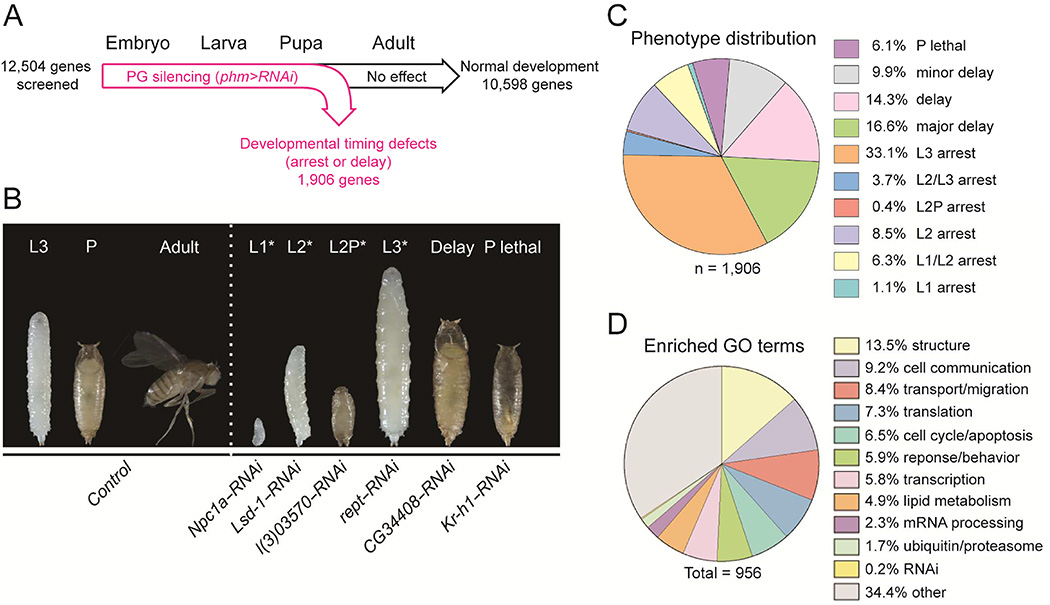
To systematically identify genes required for endocrine steroidogenic cell function, we performed a genome-wide in vivo RNAi screen in the steroid-producing PG of Drosophila. For this purpose, we used the Drosophila RNAi library (Dietzl et al., 2007) to reduce expression of 12,504 individual genes [~90% of the protein coding genes (Matthews et al., 2015)] specifically in the PG cells. The phm-Gal4 (phm>) driver (Ono et al., 2006) was crossed to UAS-controlled transgenic RNAi lines to direct tissue-specific silencing in the PG (Figure 1A). Ecdysone is required for developmental transitions between larval stages, the onset (pupariation) and completion of metamorphosis. Therefore, impaired production and release of this steroid from the PG causes a gradient of phenotypes ranging from simple delayed development and overgrowth to a more severe developmental arrest at different larval instar stages (Danielsen et al., 2014; Enya et al., 2014; Layalle et al., 2008; Rewitz et al., 2009). Based on developmental defects (Table S1) that range from arrest in the first (L1), second (L2) and third (L3) larval instar to developmental delay [~1 day (minor delay); ~2 days (delay); ~3 days (major delay)], we identified 1,906 (15.2%) candidate genes, of which 1,289 have human homologs. Additionally, we screened for premature entry into metamorphosis after the second larval instar (L2 prepupa: L2P) and for lethality during the pupal stage (P lethal) (Figure 1B and 1C). The arrest in the different developmental stages likely reflects a failure to produce an ecdysone pulse required to trigger entry into the next stage. Gene hits associated with arrest in L1 or L2, the strongest phenotypes, include genes directly involved in the ecdysone biosynthetic pathway (shroud, phantom, disembodied, shadow and Cyp6t3). The screen also identified genes associated with cholesterol trafficking (Npc1a, GstE14/Nobo and snmp1), genes in major signaling pathways such as insulin/TOR (InR, Akt1, raptor, Tif-1A), PTTH/Torso/Ras (torso, Ras85D) and TGFβ (put and tkv), and transcription factors (vvl, kni, mld, ouib, br, E75B, EcR and USP) that are known to regulate ecdysone production in the PG (Caceres et al., 2011; Colombani et al., 2005; Danielsen et al., 2014; Gibbens et al., 2011; Huang et al., 2005; Komura-Kawa et al., 2015; Koyama et al., 2014; Layalle et al., 2008; Mirth et al., 2005; Moeller et al., 2013; Niwa et al., 2010; Niwa and Niwa, 2014; Ou et al., 2011; Rewitz et al., 2009; Talamillo et al., 2013; Zhou et al., 2004). This shows that our screen was successful in identifying genes with known steroidogenic roles, and an additional ~1,800 genes that have not been linked to either steroidogenesis, steroidogenic cell function or general gland viability; indeed, many of these genes have no identified function.
Figure 1. Genome-wide in vivo screen for genes involved in steroid hormone production in Drosophila.
(A) Scheme of the screen design depicting the procedure for prothoracic gland (PG) specific RNAi mediated gene silencing. Virgin females of the PG-specific phm> driver line were crossed to a library of transgenic UAS-RNAi males to specifically reduce expression of genes in the PG. In total 12,504 RNAi lines, each targeting individual genes, were used. (B) Results from the screen reveal 1,906 candidate genes causing developmental defects including arrest in L1 (L1*), arrest in L2 (L2*), pupariation of L2 larvae (L2P*), arrest in L3 (L3*), developmental delays (delay) and pupal lethality (P lethal), indicating that the genes are important for steroidogenic tissue function. (C) Diagram showing the distribution of the phenotypic categories. (D) Gene ontology (GO) analysis of the gene set showing the top enriched functional categories. Genes were grouped into common functional categories based on GO terms from both Drosophila genes and their human orthologs. Numbers indicate total number of GO terms. See also Figure S1, Table S1 and S2.
In order to identify biological processes important for steroidogenic cell activity, we performed functional Gene Ontology (GO) term enrichment analysis of the gene hits identified in our screen. We found significant enrichment for multiple cellular processes, such as structure-related processes, cell communication, transport/migration, translation and cell cycle/apoptosis (Figure 1D, S1 and Table S2). Intriguingly, our analysis also revealed significant enrichment of gene functions related to lipid metabolism. Furthermore, many of the most highly expressed genes in the Drosophila ring gland, an endocrine organ largely comprised of the PG cells, are related to lipid metabolism (Ou et al., 2016), suggesting a specific role of these genes in steroidogenesis. These include genes involved in uptake and transport of lipids, and regulation of lipid synthesis and modification (Npc1a, S2P, Hmgcr, Hmgs, GstE14/Nobo, cueball, Fatp and CG5278). Strikingly, reduced expression of these genes in the PG, except for Npc1a and cueball, results in similar developmental delay phenotypes (Table S1). Lipids are components of cell membranes and control important cellular processes (Wymann and Schneiter, 2008), yet their roles in regulating steroidogenesis are largely unknown. We therefore further investigated CG5278/stuck in traffic (sit), which encodes an uncharacterized fatty acid elongase homolog.
Loss of the fatty acid elongase homolog sit causes accumulation of cholesterol and impairs steroidogenic activity
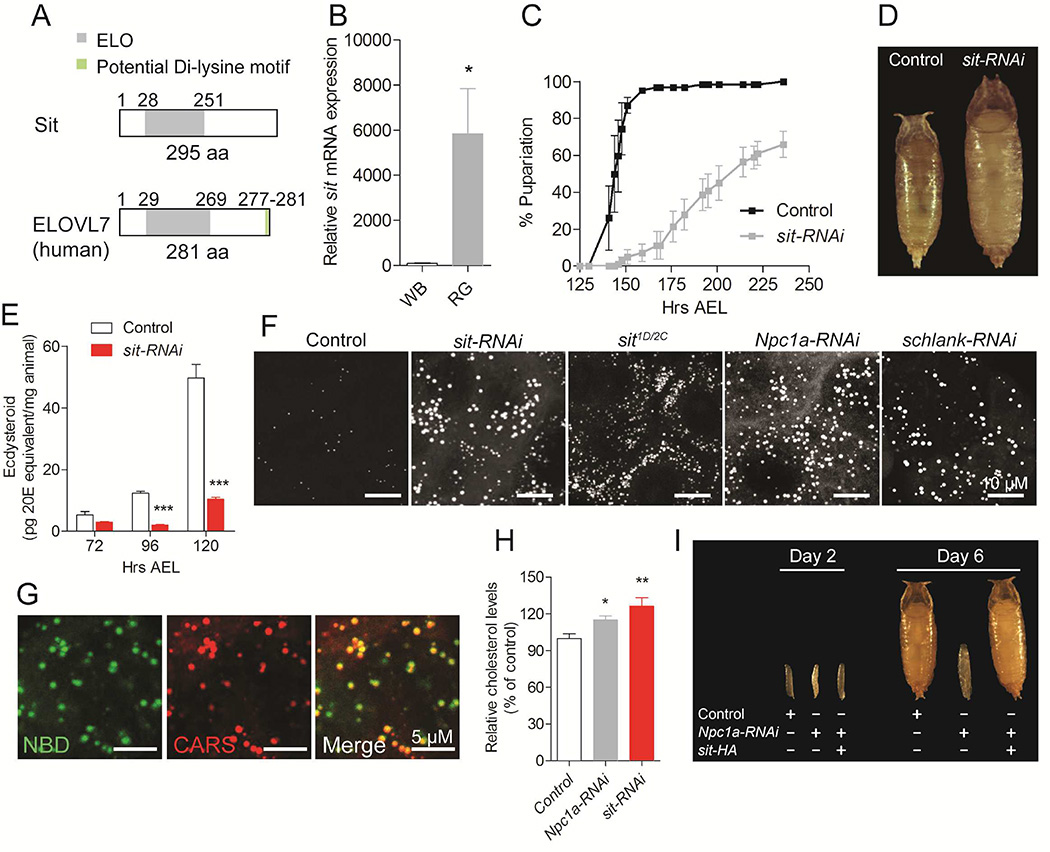
sit encodes a homolog of the human fatty acid elongases ELOVL1/7 (Figure 2A and S2A), which are specifically linked to breast and prostate cancer (Hilvo et al., 2011; Tamura et al., 2009; Tolkach et al., 2015). ELOVL enzymes are involved in the rate-limiting step in the elongation of very long chain fatty acids. Expression of sit is high in the Drosophila ring gland compared to other tissues, suggesting a specific role in steroidogenesis (Figure 2B and S2B). Silencing of sit in the PG resulted in a developmental delay, incomplete pupariation and overgrowth (Figure 2C, 2D and S2C), consistent with a failure in the production and/or release of ecdysone from the PG that triggers pupariation (Colombani et al., 2012; Rewitz et al., 2009). To confirm that sit is required for steroidogenic activity, we measured ecdysone levels in animals with reduced expression of sit in the PG. Indeed, reduced expression of sit in the PG caused low levels of ecdysone during the L3 stage (Figure 2E). We next generated two mutant lines carrying a deletion of the sit coding sequence (Figure S2D and S2E). Animals homozygous for either of two different sit deletions (sitD1 and sitC2) or a heteroallelic combination (sitD1/C2) showed developmental lethality with the majority of animals dying in larval stages (Figure S2F).
Figure 2. CG5278/stuck in traffic (sit) is a conserved fatty acid elongase homolog important for steroid production that affects cholesterol levels.
(A) Sit encodes a protein composed of 295 amino acids (aa) containing a conserved ELO domain similar to the human ELOVL7 fatty acid elongase protein. (B) Expression of sit is high in the ring gland (RG) compared to whole body (WB) 96 hours after egg laying (AEL). Knock down of sit expression specifically in the PG cells results in (C) delayed and impaired pupariation and (D) an increased pupal size. (E) RNAi knock down of sit in PG cells reduces ecdysone levels in the larvae during the L3 stage. (F) Lipid droplets detected by CARS microscopy in PG cells of L3 larvae with PG-specific RNAi silencing and in sit1D/2C mutants. (G) Co-localization of lipid droplets detected by CARS microscopy with NBD-cholesterol in the PG of animals with PG-specific silencing of sit. (H) L3 larvae with ubiquitous RNAi mediated silencing of Npc1a or sit contain higher levels of cholesterol. The RNAi effect was conditionally induced in L2 larvae 96 hours AEL by shifting larvae from 18°C to 29°C and assayed two days later. (I) Overexpression of an HA-tagged sit (sit-HA) in the PG rescues the developmental arrest phenotype caused by knock down of Npc1a. Day numbers refer to day AEL. Detailed description of genotypes is given in Supplemental Experimental Procedures. 20E; 20-hydroxyecdysone. Error bars indicate SEM (*P<0.05; **P<0.01; ***P<0.001). See also Figure S2 and S3.
Given that sit encodes a fatty acid elongase homolog, we next sought to detect abnormalities in lipid metabolism using labeling-free Coherent Anti-stokes Raman Scattering (CARS) microscopy that detects lipids (Nan et al., 2003). Surprisingly, we observed a significant accumulation of lipid droplets in sit-deficient PG cells (Figure 2F, S2G and S2H). By contrast, we found no obvious change in fat body lipid droplets of the sit mutants (Figure S2I), consistent with sit expression being PG-specific, indicating that its primary role is related to endocrine steroidogenic cell function. Steroidogenic cells have a high demand for cholesterol as it is a precursor for steroid synthesis, and intracellular cholesterol typically accumulates in lipid droplets (Miller and Bose, 2011). To examine whether the lipid droplets in the PG detected by CARS contain cholesterol, we used an ex vivo uptake assay with a fluorescent cholesterol analog, NBD-cholesterol (Gimpl, 2010), which showed that lipid droplets detected by CARS in PG cells are cholesterol-rich (Figure 2G). Dysfunction of NPC1 genes is known to cause accumulation of free unesterified cholesterol (Karten et al., 2009). Consistent with this, we observed a strong accumulation of lipid droplets in Npc1a-deficient PG cells, similar to the effects caused by loss of sit (Figure 2F and S2G). Together this suggests that loss-of-sit function is associated with intracellular accumulation of cholesterol-rich droplets, similar to what occurs in Npc1a-deficient cells. Next, we conditionally induced global RNAi-mediated knockdown of sit and Npc1a during the L2 stage to avoid the early developmental arrest caused by early reduction in the expression of these genes, and then measured cholesterol and cholesterol ester levels in the L3 larvae. Global reduction of sit or Npc1a indeed resulted in accumulation of free unesterified cholesterol (Figure 2H and S2J).
To investigate whether loss-of-sit function results in a block of cholesterol delivery for the steroidogenic pathway, we examined the dependence on dietary cholesterol of the PG-specific sit-RNAi phenotype. Previously, it has been shown that the cholesterol limitation that causes the ecdysone deficiency of Npc1a and GstE14/Nobo-deficient PG cells can be rescued by increasing dietary cholesterol (Danielsen et al., 2014; Enya et al., 2014). We confirmed that increasing dietary cholesterol rescues the Npc1a loss of function phenotype (Figure S3A). Increasing dietary cholesterol concentrations completely rescued the developmental delay caused by reduced sit expression in the PG cells, indicating that cholesterol is limiting in these cells (Figure S3B). An ex vivo uptake assay confirmed that PG cells lacking sit accumulate NBD-cholesterol (Figure S3C). Taken together, these data suggest that loss of sit causes reduction of intracellular cholesterol transport. Since loss of Npc1a and sit produce similar phenotypes suggestive of altered cholesterol transport, we asked if overexpression of sit is sufficient to rescue the cholesterol trafficking defect in Npc1a-deficient cells. Knockdown of Npc1a in the PG causes L1 arrest. Remarkably, we found that overexpression of sit rescues the phenotype caused by lack of Npc1a in the PG (Figure 2I) suggesting that these genes act in an interconnected cholesterol trafficking pathway and we therefore named this gene stuck in traffic (sit).
Reduction of sit phenocopies reduction in sphingolipid levels
We next considered the mechanism that could contribute to the cholesterol-rich droplet accumulation in sit-deficient PG cells. Abnormalities in sphingolipid metabolism is a common feature of many lysosomal storage diseases, including NPC1 disease, and interaction between sphingolipids and cholesterol likely contributes to the pathogenesis (Vanier, 2015). Fatty acid elongases, like Sit, provide the long chain fatty acid substrate for synthesis of the ceramide precursors for sphingolipids, which are essential structural components of cell membranes (Sassa and Kihara, 2014). The accumulation of cholesterol in sit-deficient cells might therefore be linked to aberrant sphingolipid synthesis. To investigate this possibility, we depleted sphingolipids in the PG by silencing schlank, a gene encoding the single ceramide synthase in Drosophila required for sphingolipid synthesis (Bauer et al., 2009). In our screen, PG-specific loss of schlank was found to cause a phenotype similar to the knockdown of sit, supporting a functional relation between sit and schlank (Table S1). We also observed accumulation of lipid droplets and NBD-cholesterol in schlank-deficient PG cells (Figure 2F and S3C and S3D). This suggests that sphingolipid depletion causes a developmental phenotype and accumulation of cholesterol similar to the loss of sit, making it possible that Sit-dependent sphingolipid synthesis plays a major role in regulating trafficking of cholesterol. To investigate whether knockdown of sit results in alterations of ceramide levels, we performed an analysis of ceramides using LC-MS. Importantly, this confirmed that knockdown of sit causes a reduction in specific ceramide species (Figure S3E), consistent with the notion that its function is required for production of some sphingolipids.
Cholesterol is trafficked through the endosomal-lysosomal pathway. To further examine the effect of sit on cholesterol trafficking, we used a ubiquitously expressed GFP-LAMP marker (Pulipparacharuvil et al., 2005). GFP-LAMP is trafficked through the endosomal-lysosomal pathway where GFP is degraded, but the GFP signal accumulates when this pathway is blocked. Silencing of sit led to a strong accumulation of GFP-LAMP in the PG, indicating a trafficking defect in sit-deficient cells (Figure S3F). To further investigate the effect of sit knockdown on the endosomal pathway, we expressed Rab7-GFP, a late endosomal marker in combination with sit-RNAi. Loss of sit caused enlargement of Rab7 vesicles in the PG compared to the control, consistent with a defect in endosomal pathway (Figure S3G and S3H). We also found oversized Rab7 vesicles and accumulation of GFP-LAMP in schlank deficient PG cells, further supporting the idea that the cholesterol trafficking defect in cells lacking sit is linked to aberrant sphingolipid synthesis (Figure S3F–S3H).
Cholesterol uptake and trafficking are correlated with TOR and ecdysone signaling
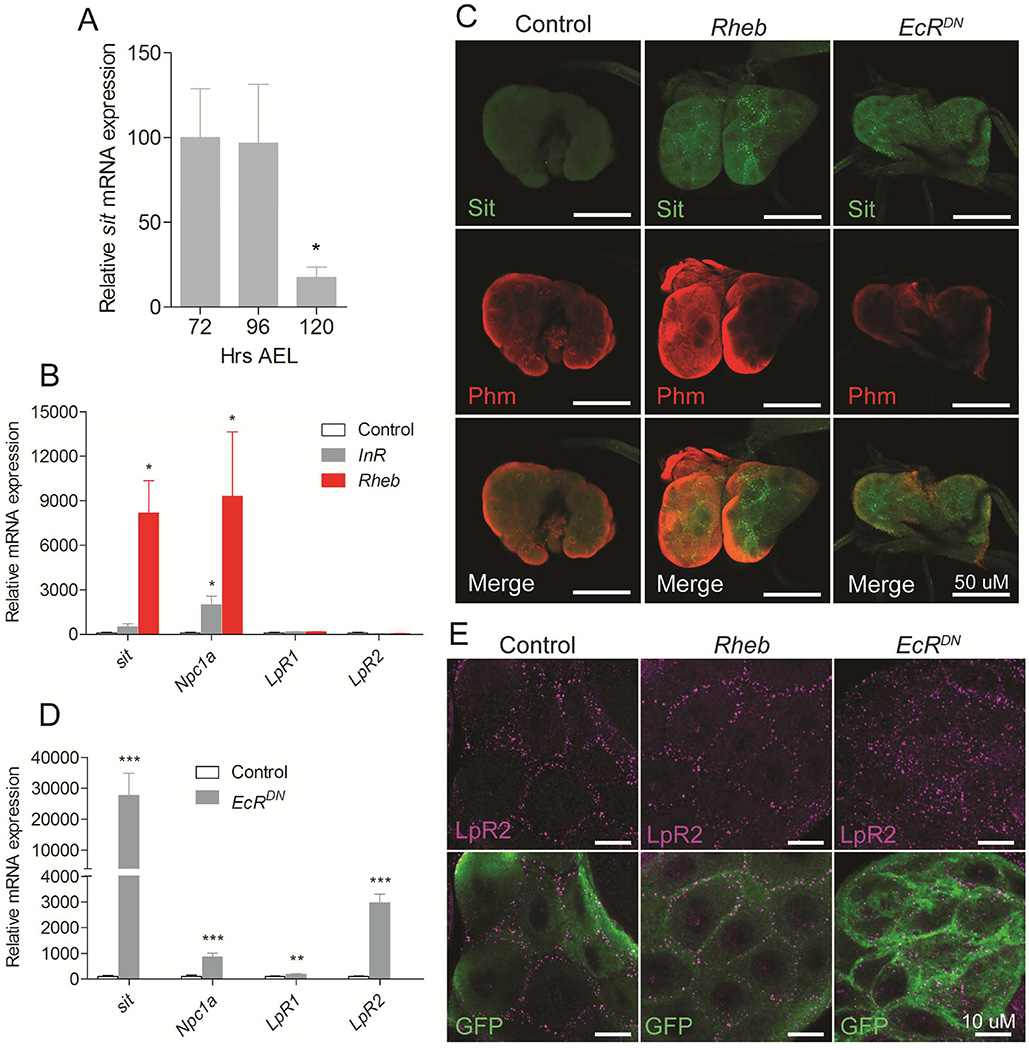
Since steroids are synthesized preferentially from cholesterol, we hypothesized that change in nutritional and developmental cues that control the steroidogenic activity of the PG regulate cholesterol uptake and transport in these endocrine cells to coordinate cholesterol availability with steroidogenic activity. To test this, we determined the expression of sit in dissected ring glands at different time points during the L3 stage. We found a dramatic downregulation of sit mRNA levels during the non-feeding wandering stage 120 hours after egg laying (AEL) (Figure 3A). We, therefore, asked whether insulin and TOR, the two major nutrient-sensing and growth-promoting signaling pathways that affect ecdysone biosynthesis in the PG (Colombani et al., 2005; Layalle et al., 2008), regulate sit expression. Activating the TOR pathway in the PG by overexpression of Rheb upregulated sit mRNA and protein levels (Figure 3B and 3C). In contrast, activation of insulin signaling by overexpression of the insulin receptor (InR) in the PG had little effect on sit expression, suggesting that sit is regulated preferentially by the TOR pathway.
Figure 3. TOR and ecdysone signaling affect cholesterol transport mechanisms in steroid producing tissue.
(A) Expression of sit in the ring gland decreases from the L3 feeding stages (72 and 96 hours AEL) to the late L3 wandering stage (120 hours AEL). (B) Effect on genes involved in cholesterol trafficking in ring glands with activated TOR signaling by overexpression of Rheb or activated insulin signaling by overexpression of the insulin receptor (InR) in the PG. (C) Immunolocalization of Sit using a CRISPR/Cas9 generated knock in of a Venus tag on the endogenous genomic sit locus (Sit-Venus). Detection of the ecdysone biosynthetic enzyme Phantom (Phm) using anti-Phm (red) and Sit-Venus using anti-GFP shows that Sit protein levels (green) increases in the PG cells of the ring glands when Rheb or EcRDN are overexpressed. (D) Effect on genes involved in cholesterol trafficking in ring glands with inhibition of ecdysone feedback regulation by overexpression of a dominant negative form of EcR (EcRDN) in the PG. (E) LpR2 is localized at the cell membrane in both control PG cells and upon Rheb overexpression, while expression EcRDN results in increased and scattered LpR2 distribution throughout cytosol. Detailed description of genotypes is given in Supplemental Experimental Procedures. Error bars indicate SEM (*P<0.05; **P<0.01; ***P<0.001).
The decline in sit coincides with the elevated ecdysone signaling during the non-feeding wandering stage that triggers pupariation. Previously, we reported that ecdysone production in the PG is regulated by feedback through the ecdysone receptor (EcR) in the PG during this stage (Moeller et al., 2013). We therefore asked whether EcR acts as a negative regulator of sit and is involved in inhibition of cholesterol delivery for steroidogenesis in the PG during the non-feeding stage. Consistent with this idea, we found a dramatic increase in sit mRNA levels when EcR signaling was blocked in the PG by expression of a dominant negative form of EcR (EcRDN) (Figure 3C and 3D). Together, these observations suggest that TOR signaling upregulates sit expression during feeding stages in response to nutrient intake, while EcR is a negative regulator of sit, responsible for its downregulation during the non-feeding wandering stage.
Given the strong influence of TOR and EcR on Sit, we investigated whether TOR and ecdysone signaling coordinate cholesterol uptake and transport in the PG by analyzing the expression of genes involved in cholesterol uptake and trafficking. Interestingly, our data show that TOR and also insulin signaling stimulate Npc1a expression in the ring gland, while having no effects on LpR1 and LpR2, which encode LDL-like receptors important for uptake of neutral lipids such as cholesterol (Parra-Peralbo and Culi, 2011) (Figure 3B). In contrast, we found that EcR is a strong negative regulator of Npc1a, as well as LpR1 and LpR2 (Figure 3D). We next investigated the effects of TOR and EcR signaling on LpR2 protein levels in the PG. While LpR2 localize mostly to the plasma membrane in control PG cells, cells with reduced EcR signaling display increased LpR2 staining and also in the cytoplasm (Figure 3E). Together these results suggest that TOR and EcR signaling regulate cholesterol trafficking in opposite directions.
Cholesterol accumulation is driven by TOR and inhibited by ecdysone signaling
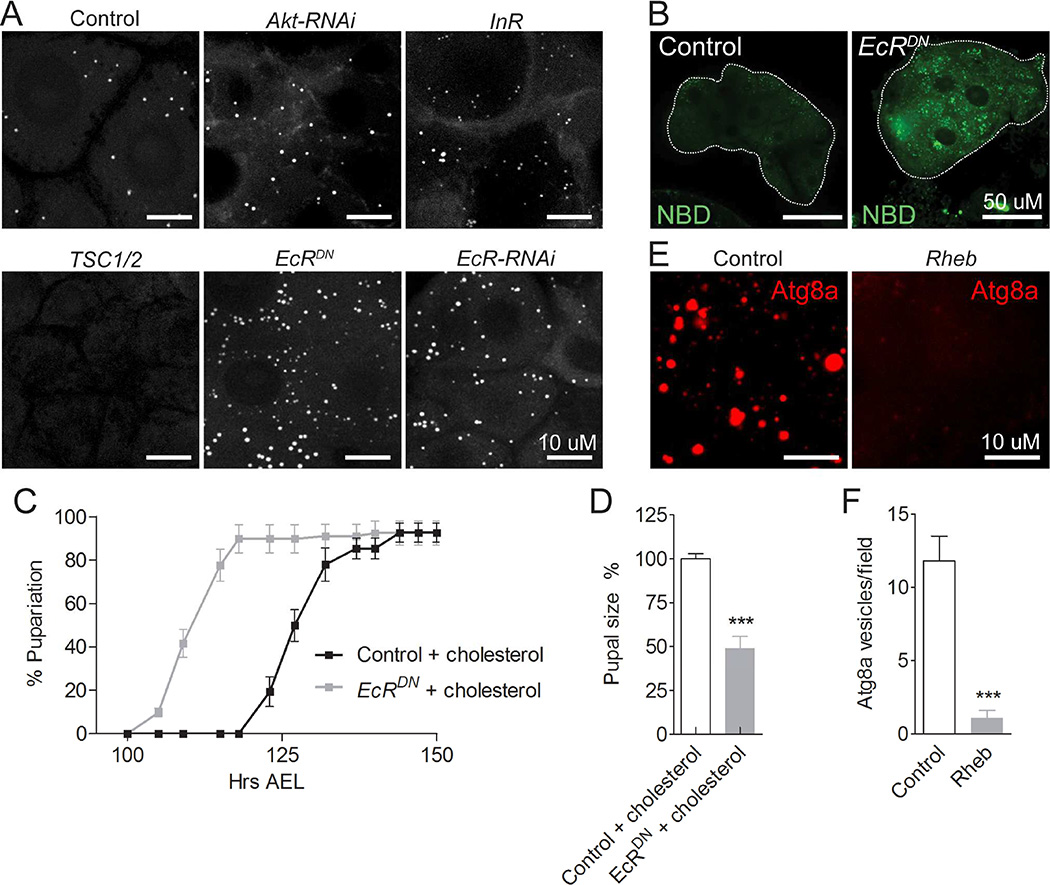
Consistent with our data indicating that TOR promotes cholesterol uptake and trafficking, we found that suppressing TOR activity through expression of TSC1 and TSC2 (TSC1/2) (Layalle et al., 2008) in the PG leads to a complete lack of lipid droplets (Figure 4A and S4A). In contrast, we found that inhibition of ecdysone signaling, either by expression of EcRDN or by silencing using EcR-RNAi, caused a strong increase in the number of lipid droplets in the PG, indicating that EcR suppresses cholesterol uptake and trafficking (Figure 4A and S4B). To further determine whether loss-of-EcR function is sufficient to drive cholesterol influx, we analyzed ex vivo uptake of NBD-cholesterol and found accumulation of NBD-cholesterol in the PG when EcR signaling was repressed (Figure 4B). We therefore rationalized that loss of EcR would enhance uptake and delivery of cholesterol and consequently increase ecdysone production in the PG under conditions with high cholesterol. As expected, we found that increasing dietary cholesterol concentrations led to a strong (~15 hours) acceleration of pupariation in animals expressing EcRDN in the PG compared to the control under these conditions (Figure 4C). Animals with reduced EcR signaling in the PG pupariate prematurely ~110 hours AEL on a high cholesterol diet, which shortens the larval growth period and causes ~50% reduction in pupal size (Figure 4D). These data suggest that inhibition of EcR signaling enhances the ability of the PG cells to take up and deliver cholesterol, which provides excess substrate that increases steroidogenesis and causes premature pupariation. Altogether, these results suggest an essential role for TOR and steroid signaling in the coordination of cellular cholesterol accumulation and mobilization whereby TOR activity promotes cholesterol uptake and trafficking, while EcR activity suppresses it in the PG.
Figure 4. Opposing effects of TOR and EcR on cholesterol and lipid accumulation.
(A) CARS images of lipid droplets in PG cells with inhibition of TOR (expression of TSC1/2) or ecdysone feedback (expression of EcRDN or EcR-RNAi), or activation (expression of InR) or inhibition (Akt-RNAi) of insulin signaling. To allow development to L3, overexpression of TSC1/2 was induced 96 hours AEL by shifting L2 larvae from 18°C to 29°C and PG cells were imaged two days later. For all other genotypes larvae were reared at 25°C and the PG was assayed 120 hours AEL. (B) Ex vivo incubation assay reveals that inhibition of ecdysone feedback by expression of EcRDN leads to excessive NBD-cholesterol accumulation in PG cells. Developmental timing of pupariation (C) and pupal size (D) of animals overexpressing EcRDN in the PG compared to control animals on a high cholesterol diet (+ cholesterol). (E, F) Effect of TOR on autophagy (Atg8a punctae) in the PG. Detailed description of genotypes is given in Supplemental Experimental Procedures. Error bars indicate SEM (**P<0.01; ***P<0.001). See also Figure S4.
Cholesterol trafficking involves autophagy and depends on nutrient availability
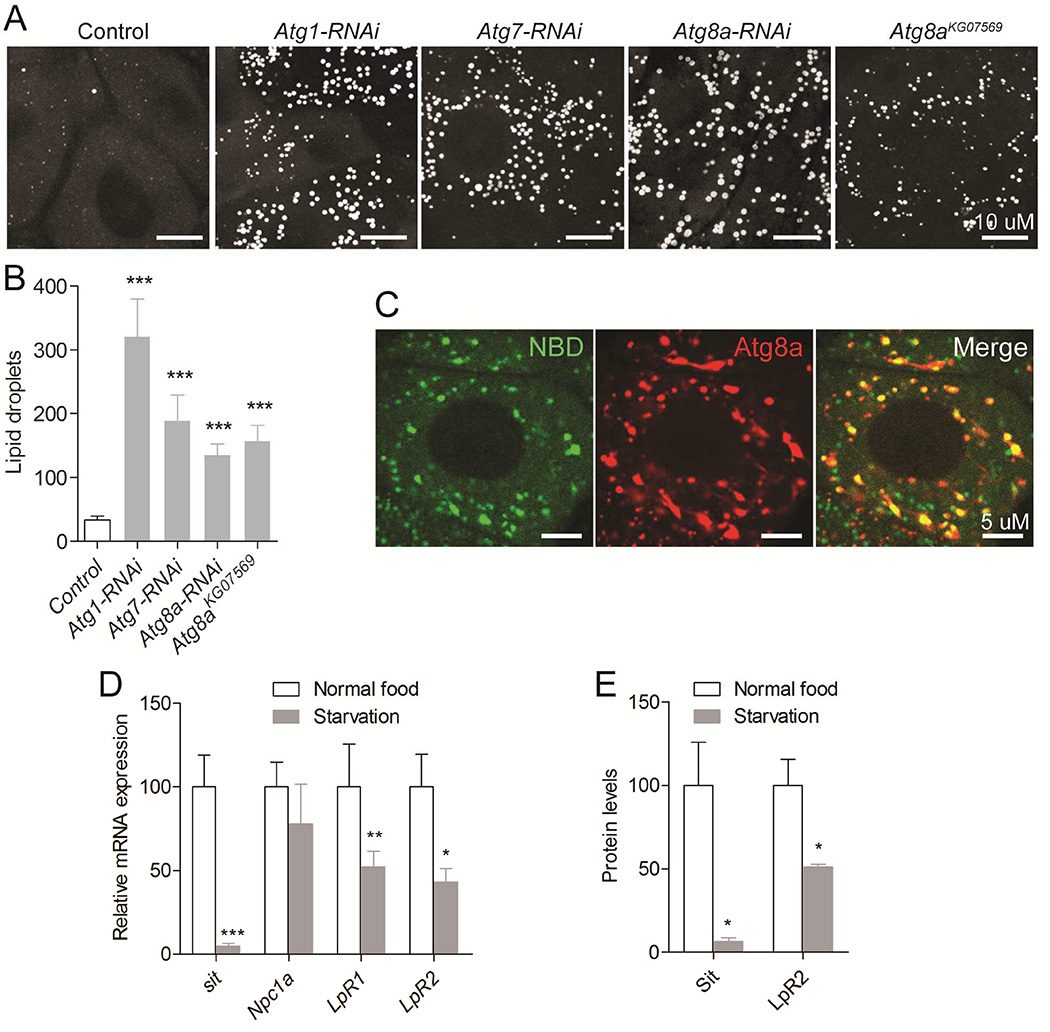
Our data show that inhibition of TOR in L2 results in almost complete lack of lipid droplets in the PG of L3 larvae, which indicates that inhibition of TOR may also promote a process that degrades cholesterol-rich lipid droplets. TOR is a negative regulator of a conserved process known as macroautophagy (hereafter referred to as autophagy), which is an intracellular degradation pathway for cytoplasmic components. Defective autophagy has been associated with NPC1 disease (Sarkar et al., 2013), indicating that autophagy could play a specific role in regulating cholesterol trafficking. First, we asked whether TOR affects autophagy in the PG by analyzing mCherry-positive puncta in larvae expressing UAS-mCherry-Atg8a, a tagged Atg8a protein that labels autophagic vesicles (Chang and Neufeld, 2009). Activation of TOR by overexpression of Rheb decreased Atg8a puncta in the PG, confirming that TOR is a repressor of autophagy (Figure 4E and 4F). To test whether autophagy plays a role in regulating cholesterol trafficking in the PG, we analyzed lipid droplet numbers in Atg8a mutant PG cells as well as in those where essential autophagy genes Atg1, Atg7 and Atg8a were knocked down. Inhibition of autophagy was sufficient to cause massive accumulation of lipid droplets in the PG (Figure 5A and 5B). This suggests that autophagy plays a specific role in the breakdown of cholesterol-rich droplets in the PG. To test directly whether autophagic vesicles sequester cholesterol-rich droplets, we incubated larval PG cells expressing UAS-mCherry-Atg8a with NBD-cholesterol. Co-localization shows that Atg8a vesicles sequester NBD-cholesterol (Figure 5C), suggesting that autophagy contributes to the mobilization of cholesterol from lipid droplets in the PG.
Figure 5. Inhibition of autophagy leads to lipid accumulation, which is coupled to nutrition.
(A) Lipid droplets accumulate upon RNAi mediated depletion of Atg1, Atg7, Atg8a in the PG and in the PG of Atg8aKG07569 mutants, quantified in (B). (C) Ex vivo incubation assay shows that NBD-cholesterol (green) co-localize with mCherry-Atg8 positive vesicles (red) in PG cells. mRNA levels (D) and protein levels (E) of genes involved in cholesterol uptake and trafficking in L3 larvae fed on normal food versus L3 larvae starved for 10 hours. Detailed description of genotypes is given in Supplemental Experimental Procedures. Error bars indicate SEM (*P<0.05; **P<0.01; ***P<0.001).
Because autophagy is a cellular response to starvation, we further investigated whether cholesterol uptake and metabolism is adjusted according to nutrient intake in steroidogenic cells. We examined whether genes involved in cholesterol uptake and trafficking are regulated in response to starvation. Expression of sit was reduced after starvation (Figure 5D). Furthermore, starvation also decreased expression of LpR1 and LpR2. When we examined Sit and LpR2 protein levels by Western blotting, we found that both proteins were also reduced in response to starvation (Figure 5E). Taken together, these results indicate that the endocrine cells of the PG coordinate cholesterol uptake, transport and mobilization in response to nutritional cues to adjust ecdysone production to environmental conditions.
Inhibition of TOR and induction of autophagy provides rescue of a Drosophila model of NPC1 disease
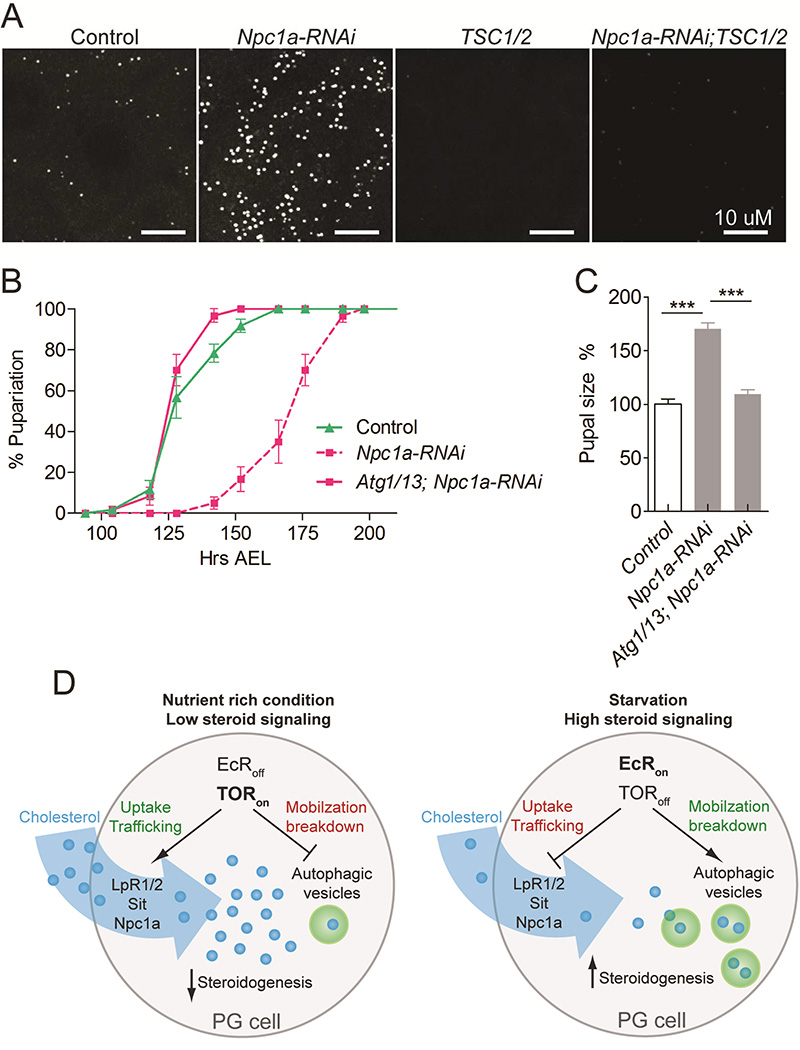
To assess whether TOR inhibition is a potential rational approach to target the deleterious accumulation of cholesterol underlying the pathogenic effects of Npc1a deficiency, we expressed TSC1/2 in Npc1a-deficient PG cells using the P0206-Gal4 (P0206>) lines that drives weaker expression in the PG (Layalle et al., 2008) compared to phm>. The dramatic lipid accumulation in Npc1a-deficient PG cells was suppressed by TCS1/2 overexpression that inhibits TOR (Figure 6A and S4C). Furthermore, we asked whether activation of autophagic degradation would be a means to rescue the impaired cholesterol metabolism associated with loss of NPC1 activity. Remarkably, we found that ectopic expression of Atg1 and Atg13 (Atg1/13), which is sufficient to induce autophagy (Scott et al., 2007), rescues the phenotype associated with loss of Npc1a in the PG (Figure 6B and 6C). Thus, our data suggest that the inhibition of TOR signaling and the induction of autophagy may form the basis for future strategies aimed at treating NPC1 disease.
Figure 6. Rescue of Npc1a-deficiency phenotypes by inhibition of TOR and activation of autophagy.
(A) Inhibition of TOR in PG cells by overexpression of TSC1/2 suppresses accumulation of lipid droplet due to Npc1a-RNAi. (B) Developmental timing of pupariation and (C) pupal size. Stimulation of autophagy by overexpression of Atg1/13 in the PG rescues the Npc1a loss-of-function phenotype, indicating that increased autophagic clearance is sufficient to overcome the block in cholesterol trafficking in Npc1a deficient cells. (D) A model for TOR and EcR mediated regulation of cholesterol trafficking mechanisms in the PG cells. Detailed description of genotypes is given in Supplemental Experimental Procedures. Error bars indicate SEM (***P<0.001). See also Figure S4.
Discussion
Here, we report a genome-wide in vivo RNAi screen in a Drosophila model, which allows systematic dissection of the genes and pathways that regulate the production of steroids in endocrine cells during development. Importantly, some of the genes that we identified as important for steroidogenesis had no known function until now, but have human homologs that have been associated with diseases where steroid signaling and cholesterol transport are dysregulated. Our data thus have the potential to uncover genes that play important roles in regulating steroidogenesis during development, which also have general relevance for diseases including some of the most common cancers and NPC1 disease. This is highlighted by our discovery and characterization of sit, an uncharacterized gene (CG5278) encoding a fatty acid elongase homolog. Our data show that sit is involved in a mechanism that controls cellular uptake and trafficking of cholesterol in the PG to produce the steroid pulse that triggers maturation in Drosophila. Elevated expression of ELOVL7, a human homolog of sit, is associated with steroidogenic tissues and prostate cancer progression (Tamura et al., 2009), yet the molecular basis for this relationship remains unclear. Prostate cancer cells acquire the ability to enhance cholesterol uptake, potentially making the cancers more aggressive, but the underlying molecular basis is poorly understood (Peck and Schulze, 2014; Yue et al., 2014). Our data suggest that Sit may play a role in this process.
The exact mechanism by which cholesterol exits endosomes after uptake and moves to other organelles is largely unknown. However, the sterol-sensing NPC proteins have been demonstrated to play a crucial role in this process and are required for trafficking of cholesterol (Huang et al., 2008; Vanier, 2015). We observed that loss of sit in the PG resulted in lipid droplet accumulation that mimicked loss-of-Npc1a function. Furthermore, the loss of sit was accompanied by accumulation of LAMP-GFP and enlarged endosomal vesicles indicating that Sit is required for vesicle trafficking in the endosomal-lysosomal pathway. Studies in yeast support our finding that very long chain fatty acids are required for proper late endosome trafficking (Obara et al., 2013), but leave open the question as to how silencing of sit leads to endosomal vesicle trafficking defects and cholesterol accumulation. Our observation that knockdown of sit affects ceramide levels and is phenocopied by silencing of the ceramide synthase schlank, together with the fact that most long chain fatty acids are found as constituents of sphingolipids (Sassa and Kihara, 2014), suggests that membrane sphingolipid composition is important for endosomal trafficking and movement of cholesterol between organelles, perhaps through alterations of membrane fusion dynamics. Consistent with this view, ceramide stimulates NPC-mediated cholesterol transfer (Abdul-Hammed et al., 2010) and we find that reduced Npc1a function is rescued by overexpression of sit, which suggests a close relationship between Npc1a and sit in cholesterol trafficking. Given that sit is highly expressed in PG cells, which have a high demand for cholesterol, we propose that this fatty acid elongase homolog is required for trafficking of cholesterol, and thereby provides a molecular context for understanding the association between the dysregulation of its human homolog and certain cancers.
We have previously shown that EcR-mediated feedback control of ecdysone biosynthesis is critical for pupal development in Drosophila (Moeller et al., 2013). Our data show that expression of sit is repressed by EcR, which reduces cellular uptake of cholesterol in the PG. Why does EcR mediate a negative feedback that blocks cholesterol uptake? The most likely explanation is that blocking cholesterol accumulation is required as part of an efficient negative feedback circuit in coordination with downregulation of the ecdysone biosynthetic pathway to generate the temporal steroid pulse that drives developmental progression. Under this view, intracellular cholesterol homeostasis is under tight feedback regulation to control steroid production. Alterations in such feedback mechanisms may cause reprogramming of cholesterol metabolism that allows cells to evade cellular cholesterol homeostatic control in certain cancers. In mammals, cholesterol levels are regulated by liver X receptor (LXR), an ortholog of EcR that protects cells from cholesterol overload (King-Jones and Thummel, 2005; Zhao and Dahlman-Wright, 2010). Our studies suggest that EcR-deficiency strongly enhances cholesterol influx, which indicates that EcR is required for homeostatic control to prevent cholesterol overload similar to LXR.
Our data suggest that uptake and trafficking of cholesterol require low EcR signaling in the presence of TOR activity, a condition that occurs during the feeding stage. Previous work has shown that EcR and TOR influence ecdysone biosynthesis in the PG (Layalle et al., 2008; Moeller et al., 2013). Thus, our results suggest that these signaling pathways adjust uptake and trafficking of cholesterol with dietary intake and developmental cues, thereby coordinating substrate delivery with activity of the ecdysone biosynthetic pathway. According to this view, TOR promotes both gland growth and ensures cholesterol uptake during the feeding stage, while EcR represses it during the non-feeding wandering stage. Interestingly, induction of ELOVL7 is mediated by mTOR activity (Purdy et al., 2015), which suggests that the TOR pathway has a conserved role in regulating ELOVL genes and synthesis of long chain fatty acids that promote uptake of cholesterol. Altogether, our findings indicate that TOR and EcR are key regulatory mediators of distinct programs that couple intracellular cholesterol homeostasis with steroidogenic activity, nutrition and developmental progression.
Recent evidence indicates a link between vesicle trafficking defects in lysosomal storage diseases and abnormalities in autophagy that may contribute to the accumulation of intracellular cholesterol (Walkley and Vanier, 2009). We demonstrate the existence of an autophagosomal mechanism through which cholesterol is trafficked. We find that autophagic Atg8a vesicles sequester cholesterol-rich lipid droplets, indicating a critical function of autophagy in cholesterol metabolism. Our study illustrates that TOR and EcR function as a regulatory switch that adjusts cholesterol uptake and trafficking to nutrient intake and steroid levels (Figure 6D). Based on our observations, we propose that TOR and EcR together regulate cholesterol uptake/trafficking and mobilization/breakdown, the two main processes that determine intracellular cholesterol levels. Our study suggests that these signaling pathways regulate uptake and trafficking of cholesterol through the endosomal pathway. On the other hand, cholesterol-rich lipid droplets are sequestered by autophagosomes and degraded in a process regulated by TOR to control intracellular cholesterol mobilization in response to availability of nutrients and cholesterol.
Defective autophagy is associated with many neurodegenerative diseases, including NPC1 disease that is characterized by excessive cholesterol accumulation (Nixon, 2013; Sarkar et al., 2013). Because cholesterol accumulation is underlying the pathology of the disease, we asked whether we could rescue cholesterol accumulation and restore dysfunction in NPC1-deficient cells. Interestingly, inhibition of TOR suppresses cholesterol accumulation caused by Npc1a-defiency in the PG cells. A recent paper has provided initial evidence of a link between NPC1-deficiency and impaired autophagy (Sarkar et al., 2013). Our findings indeed suggest that stimulating autophagy rescues the phenotype associated with Npc1a loss, raising the possibility of targeting TOR and autophagy as a strategy for treatment of NPC1 disease.
Experimental procedures
Drosophila stocks and transgenes
Drosophila larvae were raised on standard cornmeal food (Nutri-Fly, Bloomington Formulation) under a 12:12 hour light:dark cycle, 50–70% humidity and 25°C unless otherwise stated. Transgenic RNAi fly lines for the genome-wide in vivo RNAi screen were obtained from Vienna Drosophila RNAi Center (VDRC). The RNAi screen was performed using the KK collection (phiC31 inserted at a specific AttP site). In cases where it was not possible to obtain KK lines, GD lines (randomly integrated) were used instead as to get the best coverage of the genome (Dietzl et al., 2007). Additional stocks used in the study include: tub-Gal80ts, UAS-CD8-GFP (UAS-GFP), UAS-Rab7-GFP, UAS-EcRDN from the Bloomington Drosophila Stock Center (BDSC); tub-GFP-LAMP (Akbar et al., 2009); UAS-TSC1, UAS-TSC2 (UAS-TSC1/2) (Tapon et al., 2001); UAS-Rheb (Patel et al., 2003); UAS-InR29.4 (UAS-InR) (Mirth et al., 2005); Act-Gal4;Gal80ts a generous gift from Stephen Cohen; P0206-Gal4 (Colombani et al., 2005); Atg8aKG0769 a generous gift from Thomas Neufeld; phm-Gal4 (Ono et al., 2006); w1118 VDRC #60000; UAS-Npc1a-RNAi #42782 VDRC; UAS-Atg1-RNAi #34340 Transgenic RNAi project (TRIP) (Ni et al., 2011), UAS-Atg7-RNAi #27707 TRIP, UAS-Atg8a-RNAi #26732 TRIP. The sit deletion mutants were generated with two guide RNAs (5’-GTGGGAGCAAGAGTCCAACG-3’ and 5’-GCTGCAGCAGGAGAAGCAGA-3’) designed to eliminate the entire coding sequence of CG5278 except the last few nucleotides by using the CRISPR/Cas9 system (Kondo and Ueda, 2013). Oligonucleotides containing these CRISPR target sites were cloned into pBFv-U6.2B vector as described previously. The resulting vector was injected into y2 cho2 v1; attP40(nos-Cas9)/CyO embryos by BestGene Inc (Chino Hills, CA) and the progeny were screened for deletions by genomic PCR using following primers: CG5278-forward, 5’-TCGCAAAACTCGTTCGTTGCGACG-3’ and CG5278-reverse, 5’-TGGCTCTCAGT-GGGTGTTTCTACC-3’ and sequenced. To generate a UAS-sit-HA line, first the sit (CG5278) coding sequence together with a 3×HA tag in the C-terminal end, before the stop codon, was obtained from GeneArt Strings DNA Fragments (LifeTechnologies) and cloned into the pUAST vector at the EcoRI site using In-Fusion cloning (Clontech). The cloned pUAS-sit-HA plasmid was sequenced and injected by BestGene into embryos for generation of transgenic UAS-sit-HA animals. To generate a transgene carrying a sit-venus, the CRISPR/Cas9 system was used to knock-in a venus tag on the endogenous sit genomic locus. The venus was placed immediately downstream of the final coding codon if sit by homologous recombination, to generate a locus encoding a sit with a C-terminal venus. Details of the targeting vector will be described elsewhere.
Genome-wide in vivo RNAi screen
Six virgins of phm-Gal4 (phm>) 3–4 days after eclosion were crossed with four transgenic UAS-RNAi males from the VDRC collection and allowed to lay eggs on standard food in 25 mm vials for 24 hours. Phenotypes of F1 offspring were scored at day 11–13 for developmental delay or arrest. Two replicates were collected from each RNAi cross. Each batch of 100 genetic crosses included an isogenic control phm>+ (phm> crossed to w1118; the genetic background for the RNAi library from VDRC).
Imaging of lipid droplets using Coherent Anti-Stokes Raman Scattering (CARS) microscopy
Coherent Anti-Stokes Raman Scattering (CARS) imaging was performed using a Leica TCS SP8 system with a CARS laser, picoEmerald (OPO, > 600 mW @ 780 nm to 940 nm, pulse width 5 to 6 ps, 80 MHz; Pump, > 750 mW @ 1,064 nm, pulse length 7 ps, 80 MHz) and the LAS AF/X software. The lasers were adapted to the symmetrical C-H stretch range by tuning the pump beam to 816.4 nm while keeping the Stokes beam constant at 1,064.6 nm. The output of both lasers was set to 1.3 W and the scan speed to 400 Hz. Only signal from the epi-CARS (E-CARS) and epi-SHG (E-SHG) detectors were collected. Images were processed using ImageJ (NIH), and quantification of lipid droplets were done with CellProfiler (Jones et al., 2008) and manual counting.
Statistical Analysis
Error bars indicate standard error of the mean (SEM) and statistical difference between datasets were calculated using two-tailed Student’s t-test.
Supplementary Material
In brief.
Steroid hormones play important roles in development and disease. Danielsen et al. show that TOR and steroid feedback signaling regulate cholesterol substrate levels for steroid production through processes involving the fatty acid elongase Sit and autophagy. This reveals mechanisms regulating steroidogenesis during development with implications for certain diseases, including cancers.
Highlights.
A resource for identifying regulators of steroidogenesis and developmental timing
The FA elongase Sit is involved in cholesterol trafficking and steroidogenesis
Intracellular cholesterol is sequestered by autophagosomes to mobilize cholesterol
Cholesterol is regulated by TOR and feedback signaling in steroidogenic cells
Acknowledgments
We thank the people that contributed reagents. This work was supported by the Danish Council for Independent Research, Natural Sciences grant 11-105446 and by the Novo Nordisk Foundation grant 10929 to KFR. M.B.O, K.A.O and M.J.H were supported by NIH grant GM093301 to M.B.O. N.Y was supported by NIH grants K99/R00 HD073239 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contribution
E.T.D., M.E.M., N.Y., K.K., M.B.O. and K.F.R. conceived the research. E.T.D., M.E.M., N.Y., Q.O., C.S., R.Z., B.P., K.T., J.Z., S.K., C.H.N., E.H., N.J.F., M.J.H., K.A.O., K.K., M.B.O. and K.F.R. performed the experiments. E.T.D., M.E.M., N.Y., Q.O., C.S., C.H.N., E.H., N.J.F., K.K., M.B.O. and K.F.R. analyzed the data. E.T.D., M.E.M., N.Y., N.J.F., K.K., M.B.O. and K.F.R. wrote the manuscript.
References
- Abdul-Hammed M, Breiden B, Adebayo MA, Babalola JO, Schwarzmann G, Sandhoff K. Role of endosomal membrane lipids and NPC2 in cholesterol transfer and membrane fusion. Journal of lipid research. 2010;51:1747–1760. doi: 10.1194/jlr.M003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar MA, Ray S, Kramer H. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol Biol Cell. 2009;20:1705–1714. doi: 10.1091/mbc.E08-03-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R, Voelzmann A, Breiden B, Schepers U, Farwanah H, Hahn I, Eckardt F, Sandhoff K, Hoch M. Schlank, a member of the ceramide synthase family controls growth and body fat in Drosophila. EMBO J. 2009;28:3706–3716. doi: 10.1038/emboj.2009.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres L, Necakov AS, Schwartz C, Kimber S, Roberts IJ, Krause HM. Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Genes Dev. 2011;25:1476–1485. doi: 10.1101/gad.2064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M, Schwudke D, Sampaio JL, Palm W, Riezman I, Dey G, Gupta GD, Mayor S, Riezman H, Shevchenko A, et al. Survival strategies of a sterol auxotroph. Development. 2010;137:3675–3685. doi: 10.1242/dev.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Andersen DS, Boulan L, Boone E, Romero N, Virolle V, Texada M, Leopold P. Drosophila Lgr3 Couples Organ Growth with Maturation and Ensures Developmental Stability. Curr Biol. 2015;25:2723–2729. doi: 10.1016/j.cub.2015.09.020. [DOI] [PubMed] [Google Scholar]
- Colombani J, Andersen DS, Leopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336:582–585. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carre C, Noselli S, Leopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- Daftary SS, Gore AC. IGF-1 in the brain as a regulator of reproductive neuroendocrine function. Experimental biology and medicine. 2005;230:292–306. doi: 10.1177/153537020523000503. [DOI] [PubMed] [Google Scholar]
- Danielsen ET, Moeller ME, Dorry E, Komura-Kawa T, Fujimoto Y, Troelsen JT, Herder R, O'Connor MB, Niwa R, Rewitz KF. Transcriptional Control of Steroid Biosynthesis Genes in the Drosophila Prothoracic Gland by Ventral Veins Lacking and Knirps. PLoS Genet. 2014;10:e1004343. doi: 10.1371/journal.pgen.1004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Enya S, Ameku T, Igarashi F, Iga M, Kataoka H, Shinoda T, Niwa R. A Halloween gene noppera-bo encodes a glutathione S-transferase essential for ecdysteroid biosynthesis via regulating the behaviour of cholesterol in Drosophila. Sci Rep. 2014;4:6586. doi: 10.1038/srep06586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garelli A, Heredia F, Casimiro AP, Macedo A, Nunes C, Garcez M, Dias AR, Volonte YA, Uhlmann T, Caparros E, et al. Dilp8 requires the neuronal relaxin receptor Lgr3 to couple growth to developmental timing. Nature communications. 2015;6:8732. doi: 10.1038/ncomms9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbens YY, Warren JT, Gilbert LI, O'Connor MB. Neuroendocrine regulation of Drosophila metamorphosis requires TGFbeta/Activin signaling. Development. 2011;138:2693–2703. doi: 10.1242/dev.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpl G. Cholesterol-protein interaction: methods and cholesterol reporter molecules. Sub-cellular biochemistry. 2010;51:1–45. doi: 10.1007/978-90-481-8622-8_1. [DOI] [PubMed] [Google Scholar]
- Hilvo M, Denkert C, Lehtinen L, Muller B, Brockmoller S, Seppanen-Laakso T, Budczies J, Bucher E, Yetukuri L, Castillo S, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71:3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- Huang X, Suyama K, Buchanan J, Zhu AJ, Scott MP. A Drosophila model of the Niemann- Pick type C lysosome storage disease: dnpc1a is required for molting and sterol homeostasis. Development. 2005;132:5115–5124. doi: 10.1242/dev.02079. [DOI] [PubMed] [Google Scholar]
- Huang X, Warren JT, Gilbert LI. New players in the regulation of ecdysone biosynthesis. J Genet Genomics. 2008;35:1–10. doi: 10.1016/S1673-8527(08)60001-6. [DOI] [PubMed] [Google Scholar]
- Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC bioinformatics. 2008;9:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karten B, Peake KB, Vance JE. Mechanisms and consequences of impaired lipid trafficking in Niemann-Pick type C1-deficient mammalian cells. Biochimica et biophysica acta. 2009;1791:659–670. doi: 10.1016/j.bbalip.2009.01.025. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors--a perspective from Drosophila. Nat Rev Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Komura-Kawa T, Hirota K, Shimada-Niwa Y, Yamauchi R, Shimell M, Shinoda T, Fukamizu A, O'Connor MB, Niwa R. The Drosophila Zinc Finger Transcription Factor Ouija Board Controls Ecdysteroid Biosynthesis through Specific Regulation of spookier. PLoS Genet. 2015;11:e1005712. doi: 10.1371/journal.pgen.1005712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Rodrigues MA, Athanasiadis A, Shingleton AW, Mirth CK. Nutritional control of body size through FoxO-Ultraspiracle mediated ecdysone biosynthesis. eLife. 2014;3 doi: 10.7554/eLife.03091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrynenko O, Rodenfels J, Carvalho M, Dye NA, Lafont R, Eaton S, Shevchenko A. The ecdysteroidome of Drosophila: influence of diet and development. Development. 2015;142:3758–3768. doi: 10.1242/dev.124982. [DOI] [PubMed] [Google Scholar]
- Layalle S, Arquier N, Leopold P. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev Cell. 2008;15:568–577. doi: 10.1016/j.devcel.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Matthews BB, Dos Santos G, Crosby MA, Emmert DB, St Pierre SE, Gramates LS, Zhou P, Schroeder AJ, Falls K, Strelets V, et al. Gene Model Annotations for Drosophila melanogaster: Impact of High-Throughput Data. G3. 2015;5:1721–1736. doi: 10.1534/g3.115.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocrine reviews. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL, Bose HS. Early steps in steroidogenesis: intracellular cholesterol trafficking. J Lipid Res. 2011;52:2111–2135. doi: 10.1194/jlr.R016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C, Truman JW, Riddiford LM. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr Biol. 2005;15:1796–1807. doi: 10.1016/j.cub.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Moeller ME, Danielsen ET, Herder R, O'Connor MB, Rewitz KF. Dynamic feedback circuits function as a switch for shaping a maturation-inducing steroid pulse in Drosophila. Development. 2013;140:4730–4739. doi: 10.1242/dev.099739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Cheng JX, Xie XS. Vibrational imaging of lipid droplets in live fibroblast cells with coherent anti-Stokes Raman scattering microscopy. Journal of lipid research. 2003;44:2202–2208. doi: 10.1194/jlr.D300022-JLR200. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nature methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Namiki T, Ito K, Shimada-Niwa Y, Kiuchi M, Kawaoka S, Kayukawa T, Banno Y, Fujimoto Y, Shigenobu S, et al. Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the 'Black Box' of the ecdysteroid biosynthesis pathway. Development. 2010;137:1991–1999. doi: 10.1242/dev.045641. [DOI] [PubMed] [Google Scholar]
- Niwa R, Niwa YS. Enzymes for ecdysteroid biosynthesis: their biological functions in insects and beyond. Biosci Biotechnol Biochem. 2014;78:1283–1292. doi: 10.1080/09168451.2014.942250. [DOI] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nature medicine. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Obara K, Kojima R, Kihara A. Effects on vesicular transport pathways at the late endosome in cells with limited very long-chain fatty acids. J Lipid Res. 2013;54:831–842. doi: 10.1194/jlr.M034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono H, Rewitz KF, Shinoda T, Itoyama K, Petryk A, Rybczynski R, Jarcho M, Warren JT, Marques G, Shimell MJ, et al. Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev Biol. 2006;298:555–570. doi: 10.1016/j.ydbio.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Ou Q, Magico A, King-Jones K. Nuclear Receptor DHR4 Controls the Timing of Steroid Hormone Pulses During Drosophila Development. PLoS Biol. 2011;9:e1001160. doi: 10.1371/journal.pbio.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Q, Zeng J, Yamanaka N, Brakken-Thal CM, O'Connor MB, King-Jones K. The insect prothoracic gland as a model for steroid hormone biosynthesis and regulation. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.05.053. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Peralbo E, Culi J. Drosophila lipophorin receptors mediate the uptake of neutral lipids in oocytes and imaginal disc cells by an endocytosis-independent mechanism. PLoS Genet. 2011;7:e1001297. doi: 10.1371/journal.pgen.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Thapar N, Guo L, Martinez M, Maris J, Gau CL, Lengyel JA, Tamanoi F. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. Journal of cell science. 2003;116:3601–3610. doi: 10.1242/jcs.00661. [DOI] [PubMed] [Google Scholar]
- Peck B, Schulze A. Cholesteryl esters: fueling the fury of prostate cancer. Cell metabolism. 2014;19:350–352. doi: 10.1016/j.cmet.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Akbar MA, Ray S, Sevrioukov EA, Haberman AS, Rohrer J, Kramer H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci. 2005;118:3663–3673. doi: 10.1242/jcs.02502. [DOI] [PubMed] [Google Scholar]
- Purdy JG, Shenk T, Rabinowitz JD. Fatty acid elongase 7 catalyzes lipidome remodeling essential for human cytomegalovirus replication. Cell reports. 2015;10:1375–1385. doi: 10.1016/j.celrep.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, Gilbert LI, O'Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326:1403–1405. doi: 10.1126/science.1176450. [DOI] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, O'Connor MB. Developmental checkpoints and feedback circuits time insect maturation. Curr Top Dev Biol. 2013;103:1–33. doi: 10.1016/B978-0-12-385979-2.00001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbridger GP, Davis ID, Birrell SN, Tilley WD. Breast and prostate cancer: more similar than different. Nat Rev Cancer. 2010;10:205–212. doi: 10.1038/nrc2795. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Carroll B, Buganim Y, Maetzel D, Ng AH, Cassady JP, Cohen MA, Chakraborty S, Wang H, Spooner E, et al. Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell reports. 2013;5:1302–1315. doi: 10.1016/j.celrep.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa T, Kihara A. Metabolism of very long-chain Fatty acids: genes and pathophysiology. Biomolecules & therapeutics. 2014;22:83–92. doi: 10.4062/biomolther.2014.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwend T, Loucks EJ, Snyder D, Ahlgren SC. Requirement of Npc1 and availability of cholesterol for early embryonic cell movements in zebrafish. J Lipid Res. 2011;52:1328–1344. doi: 10.1194/jlr.M012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamillo A, Herboso L, Pirone L, Perez C, Gonzalez M, Sanchez J, Mayor U, Lopitz-Otsoa F, Rodriguez MS, Sutherland JD, et al. Scavenger receptors mediate the role of SUMO and Ftz-f1 in Drosophila steroidogenesis. PLoS Genet. 2013;9:e1003473. doi: 10.1371/journal.pgen.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Makino A, Hullin-Matsuda F, Kobayashi T, Furihata M, Chung S, Ashida S, Miki T, Fujioka T, Shuin T, et al. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009;69:8133–8140. doi: 10.1158/0008-5472.CAN-09-0775. [DOI] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Thummel CS. Coordinating growth and maturation - insights from Drosophila. Curr Biol. 2011;21:R750–R757. doi: 10.1016/j.cub.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkach Y, Merseburger A, Herrmann T, Kuczyk M, Serth J, Imkamp F. Signatures of Adverse Pathological Features, Androgen Insensitivity and Metastatic Potential in Prostate Cancer. Anticancer research. 2015;35:5443–5451. [PubMed] [Google Scholar]
- Vallejo DM, Juarez-Carreno S, Bolivar J, Morante J, Dominguez M. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science. 2015;350:aac6767. doi: 10.1126/science.aac6767. [DOI] [PubMed] [Google Scholar]
- Vanier MT. Complex lipid trafficking in Niemann-Pick disease type C. J Inherit Metab Dis. 2015;38:187–199. doi: 10.1007/s10545-014-9794-4. [DOI] [PubMed] [Google Scholar]
- Walkley SU, Vanier MT. Secondary lipid accumulation in lysosomal disease. Biochimica et biophysica acta. 2009;1793:726–736. doi: 10.1016/j.bbamcr.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymann MP, Schneiter R. Lipid signalling in disease. Nature reviews Molecular cell biology. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- Yamanaka N, Rewitz KF, O'Connor MB. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol. 2013;58:497–516. doi: 10.1146/annurev-ento-120811-153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B, Cheng L, Masterson TA, Liu X, Ratliff TL, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Dahlman-Wright K. Liver X receptor in cholesterol metabolism. J Endocrinol. 2010;204:233–240. doi: 10.1677/JOE-09-0271. [DOI] [PubMed] [Google Scholar]
- Zhou X, Zhou B, Truman JW, Riddiford LM. Overexpression of broad: a new insight into its role in the Drosophila prothoracic gland cells. J Exp Biol. 2004;207:1151–1161. doi: 10.1242/jeb.00855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.