Abstract
The amoeba Dictyostelium is a simple genetic system for analyzing substrate adhesion, motility and phagocytosis. A new adhesion-defective mutant named phg2 was isolated in this system, and PHG2 encodes a novel serine/threonine kinase with a ras-binding domain. We compared the phenotype of phg2 null cells to other previously isolated adhesion mutants to evaluate the specific role of each gene product. Phg1, Phg2, myosin VII, and talin all play similar roles in cellular adhesion. Like myosin VII and talin, Phg2 also is involved in the organization of the actin cytoskeleton. In addition, phg2 mutant cells have defects in the organization of the actin cytoskeleton at the cell-substrate interface, and in cell motility. Because these last two defects are not seen in phg1, myoVII, or talin mutants, this suggests a specific role for Phg2 in the control of local actin polymerization/depolymerization. This study establishes a functional hierarchy in the roles of Phg1, Phg2, myosinVII, and talin in cellular adhesion, actin cytoskeleton organization, and motility.
INTRODUCTION
Cell-substrate adhesion and cell motility are essential in many crucial biological processes such as development, wound healing, lymphocyte migration, metastasis, and phagocytosis. These complex cellular events involve cell surface receptors, signaling molecules, the actin cytoskeleton, and many associated proteins. In one of the best-studied examples, cell surface integrins bind to the extracellular matrix and connect it with the actin cytoskeleton and a complex network of cytosolic proteins controlling cell spreading, migration, proliferation, and survival (for review, see Schwartz, 2001). In mammalian cells, adhesion receptors, signaling molecules, and the actin cytoskeleton are concentrated in discrete structures known as focal adhesions at sites of contact with the substrate. To date, >50 proteins have been identified in focal adhesions, most of which contain multiple domains for interaction with other proteins, and the structure of focal adhesions seems increasingly complex (Zamir and Geiger, 2001). Thus, although numerous protein–protein interactions have been mapped, the precise functional role and relative importance of each of these proteins is often difficult to establish. Whereas the crucial role of proteins such as focal adhesion kinase (FAK) in adhesion and motility has been established by analyzing the phenotypes of knockout mice (Ilic et al., 1995), few gene products have been analyzed in this manner.
The amoeba Dictyostelium discoideum is a widely used model for studying a number of fundamental eucaryotic functions such as cellular adhesion, phagocytosis, cytokinesis, and cell motility. It has been suggested that during the course of evolution, essential adhesion and motility mechanisms first arose in primitive cells such as amoebae, and these strategies were then conserved in most cell types throughout further evolution of metazoan organisms (Friedl et al., 2001). Indeed, the genome sequencing project identified a high proportion of genes common to Dictyostelium and vertebrates (Eichinger and Noegel, 2003). Furthermore, because this amoeba is haploid and easily amenable to genetic analysis, an integrated view of these various cellular functions can be obtained. Based on the analysis of specific knockout mutants, several orthologues of mammalian adhesion proteins were shown to play essential roles in cell-substrate adhesion in Dictyostelium (Bracco et al., 2000). For example, the actin-binding protein talin is necessary for phagocytosis and adhesion in Dictyostelium (Niewohner et al., 1997). Dictyostelium myosin VII null mutants also have defects in both cell-cell and cell-substrate adhesion (Tuxworth et al., 2001). Homologues of VASP and small GTP-binding proteins of the ras family also have been identified in Dictyostelium, and mutant phenotypes are indicative of roles in adhesion and regulation of the actin cytoskeleton (Han et al., 2002; Lim et al., 2002). In addition, two membrane proteins, Phg1 (Cornillon et al., 2000) and SadA (Fey et al., 2002), are thought to function as cell surface receptors or as regulators of adhesion in Dictyostelium.
In this study, we identified a novel kinase protein, named Phg2, and examined its function by characterizing the phenotype of phg2 null cells. We also generated null mutants for several other adhesion molecules (Phg1, talin, and myosin-VII) in the same genetic background and compared their phenotype with phg2. Whereas these proteins share a common role in cellular adhesion and phagocytosis, Phg2 plays a unique role in the modeling of actin-rich focal sites and in cell motility.
MATERIALS AND METHODS
Cell Culture and Mutagenesis
D. discoideum cells were routinely grown at 21°C in HL5 medium (Cornillon et al., 1998) and subcultured twice a week to maintain a density of no >106 cells/ml.
All mutant strains used in this study were derived directly from the subclone DH1–10 (Cornillon et al., 2000) of the D. discoideum wild-type strain DH1 (Caterina et al., 1994). The phg1 mutant was described previously (Cornillon et al., 2000) The myoVII and talin knockout mutants were derived from DH1–10 by using the knockout plasmids described previously (Niewohner et al., 1997; Titus, 1999). The phg2 mutant strain was isolated following an identical procedure to that used to isolate the phg1 mutant (Cornillon et al., 2000). Briefly cells were mutagenized by restriction enzyme-mediated integration (REMI) of the pUCBsrΔBamHI plasmid and transfected cells selected in the presence of Blasticidin S hydrochloride. A fluorescence-activated cell sorter was used to select mutant cells that phagocytosed less fluorescent latex beads than wild-type cells in HL5 medium. Genomic DNA was extracted for each mutant clone, and the inserted plasmid recovered with the genomic flanking regions at the insertion site which were then sequenced. The rescued plasmid was then used to knockout the PHG2 gene by homologous recombination in a DH1–10 wild-type strain, and these phg2 knockout cells used for further characterization.
To determine the full coding sequence of the PHG2 gene, two cDNA fragments and one reverse transcription-polymerase chain reaction (PCR) product were sequenced. A Dictyostelium Lambda ZapII (a kind gift from Dr. Gräf, München, Germany) was screened using a probe corresponding to a 3′ region of the PHG2 gene. The probe was obtained by PCR amplification of genomic DNA with the primers: 5′-TAGAAGTAAAGGATACATGGG-3′ and 5′-TTCAACGACTTCTGTCATCC-3′. It was labeled with dATP32 by random priming (Nonaprimer kit I; Appligene, Oncor, Graffenstaden, France). Two cDNAs were isolated and sequenced and found to represent partial cDNAs (nucleotides 340-2852 and 1021-Stop in the coding sequence, respectively). A cDNA pool was prepared by reverse transcription of vegetative Dictyostelium mRNAs (GeneRacerT; Invitrogen, Carlsbad, CA) and the 5′ coding region of PHG2 (nucleotides -30–380) was amplified by PCR with appropriate oligonucleotides and sequenced. The assembled coding sequence of the PHG2 gene was found to be almost identical to the predicted open reading frame in the Dictyostelium genome (DD186362C-CU). This sequence was deposited in European Molecular Biology Laboratory database (accession no. AJ585374).
Northern blot analysis was performed as described previously (Cornillon et al., 1998), by using the PHG2 labeled probe described above.
Phagocytosis and Fluid Phase Uptake
Phagocytosis and fluid phase uptake were determined by shaking the cells for 20 min at 21°C in HL5 medium with either 1-μm-diameter Fluoresbrite YG carboxylate microspheres (Polysciences, Warrington, PA) or with fluorescein isothiocyanate-dextran (Molecular Probes, Eugene, OR) and measuring the internalized fluorescence in a fluorescence-activated cell sorter as described previously (Cornillon et al., 2002).
Cellular Adhesion and Motility
To evaluate the adhesion of cells to their substrate, the radial flow detachment assay was performed according to Decave et al. (2002a) and Decave et al. (2002b). Briefly, cells were adhered to a glass plate in HL5 for 10 min. A flat stainless disk pierced in its center was placed above at a defined distance, and HL5 medium flowed at a constant rate through its central orifice. After 7min, the disk was removed and the radius at which 50% of the cells were detached (r50%) determined by microscopic examination. The stress at this distance to the center was σ50% = 3Dη/πe2r50%, where D is the flow rate, e the distance between the plate and the disk, and η the fluid viscosity. Alternatively, when indicated the adhesion and the detachment assay were done in phosphate buffer (PB: 2 mM Na2HPO4, 14.7 mM KH2PO4, pH 6.5). Critical stresses were expressed relative to the parental cell line DH1–10.
To determine random velocity of cells, cells were allowed to settle onto a plastic substrate (cell culture six-well plates; Falcon, Cowley, United Kingdom) for 30 min in PB. Their movement was then recorded at 2-min intervals with a Zeiss Axiovert 100 microscope coupled to a Hamamatsu Orca camera (OpenLab3 software). Cells engaging in prolonged cell-cell contact were not taken into consideration, because this modified their velocity. The position of each cell was determined every 4min, and its average velocity over 1 h was calculated. For each cell type, at least 30 cells from at least three independent experiments were analyzed, and the average and SEM were calculated.
Shear-stress induced motility was determined in a lateral flow chamber according to Decave et al. (2003). Briefly, cells were introduced in the flow chamber and adhered during 2 min. PB supplemented with 1 mM CaCl2 was then applied at a low shear stress to wash out unbound cells. A flow corresponding to a 2-Pa shear stress, inferior to the stress required to detach the cells (at least 3 Pa), was then applied. Cell positions were recorded for 15 min (30 images), and the trajectories reconstructed using a semiautomatic procedure. Cells entering into contact with others or detaching during the time course of the experiment were discarded from the analysis. The speed (V) and the directional speed (Vx) were obtained by averaging >40 cells and 10 min (400 data points), the instantaneous cell speed or the projection of the instantaneous cell speed on the axis corresponding to the direction of the flow. The speed calculated from cell movements recorded at zero shear stress gives V0, the random explorative cell speed.
Microscopy
To express a fusion protein of Phg2 with the green fluorescent protein (Phg2-GFP), the full coding sequence of PHG2 preceded by the coding sequence of GFP was introduced in the pDXA-3C expression vector (Manstein et al., 1995). This vector was then introduced in phg2 mutant cells and clones expressing Phg2-GFP were isolated.
To visualize filamentous actin, cells adhered on a glass coverslip for 2 or 10 min in phosphate buffer were fixed in PB containing 4% paraformaldehyde for 30 min. This fixation was sufficient to permeabilize the cells. The actin cytoskeleton was labeled by incubating the cells for 1 h in phosphate-buffered saline (PBS) containing 0.2% bovine serum albumin and 1 μg/ml tetramethylrhodamine B isothiocyanate (TRITC)-labeled phalloidin. The coverslips were then washed twice, mounted and observed by laser scanning confocal microscopy (Zeiss LSM 510). When both TRITC-labeled phalloidin and Phg2-GFP were imaged, appropriate controls (cells expressing no GFP, cells not stained with TRITC-phalloidin) were used to check that no leakage occurred between the red and green fluorescence channels (our unpublished data). Unless otherwise specified, the images presented corresponded to optical sections at the site of contact between the cells and the substrate.
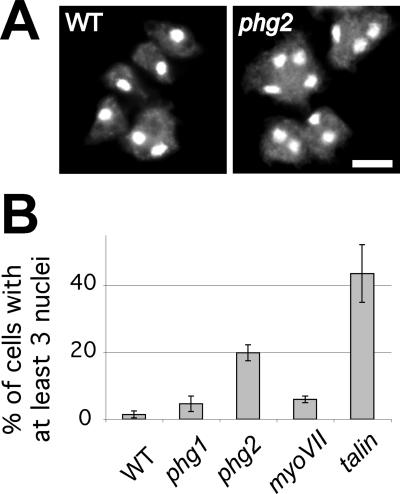
For staining nuclei, cells were grown for 3 d on a glass coverslip, fixed in HL5 containing 4% paraformaldehyde for 30 min, and postfixed overnight in 70% ethanol at 4°C. Coverslips were then incubated in PBS containing 100 μg/ml RNase A for 3 h at room temperature, and then in PBS containing 50 μg/ml propidium iodide for 1 h. They were then washed twice in PBS and mounted for observation. Cells were imaged using a Zeiss confocal microscope (LSM 510) with maximal pinhole aperture. The percentage of cells with at least three nuclei was counted (at least 200 cells per experiment, at least four independent experiments).
For scanning electron microscopy, cells were grown in HL5 and then incubated in HL5 or in PB for 1 h. The cells were then fixed using 2% glutaraldehyde in 200 mM phosphate buffer (pH 6.0) for 1 h. Cells were rinsed and postfixed in 1% osmium tetroxide in 100 mM phosphate buffer (pH 6.0) for 45min. The fixative was removed, and cells were progressively dehydrated through an ethanol series of 30–100% ethanol followed by incubation into hexamethyl disilane. After air drying, the cells were sputter-coated in gold and viewed on a Hitachi S800 scanning electron microscope.
Antibodies and Western Blot Analysis
Antibodies used here were the YC1 rabbit antipeptide antiserum to a Phg1 peptide (Cornillon et al., 2000) and the H161 monoclonal antibody to the p80 endosomal marker (Ravanel et al., 2001) described previously. Anti-talin ascitis 169.477.5 was a kind gift from Prof. G. Gerisch (Martinsried, Germany).
For Western blot analysis, 106 cells were washed once in PB, lysed in 50 μl in sample buffer (0.103 g/ml sucrose, 5 × 10-2 M Tris, pH 6.8, 5 × 10-3 M EDTA, 0.5 mg/ml bromphenol blue, 2% SDS), and 10 μl of each sample was run on either a 10% (for anti-Phg1 and anti-p80 blots) or a 7% (anti-talin blot) acrylamide gel, for 45 min and 1 h, respectively. Gels were then transferred onto a nitrocellulose BA 85 membrane (Schleicher & Schuell, Dassel, Germany) for 1 h (for anti-Phg1 and anti-p80 blots) or for 2 h (for anti-talin blot). Membranes were incubated with the YC1 antiserum (1/300, anti-Phg1), H161 monoclonal hybridoma supernatant (1/10, anti-p80), or 169.477.5 ascitis (1/1000, anti-talin), and then with a horseradish peroxidase-coupled donkey anti-rabbit Ig (for anti-Phg1 blot) or anti-mouse Ig (for anti-p80 and anti-talin blots) (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom), washed, and revealed by enhanced chemiluminescence (Amersham Biosciences UK).
Testing Protein–Protein Interactions by Two-Hybrid Assay
Two-hydrid assays were carried out using the Matchmaker LexA two-hybrid system (BD Biosciences Clontech, Palo Alto, CA). To test the ability of various small GTP-binding proteins to interact with the ras-binding domain (RBD) of Phg2, the DNA sequence encoding the putative RBD (residues 594–667) was fused to the DNA binding protein LexA in the expression vector pEG202. To test for interacting proteins, the DNA sequence encoding the constitutively active forms of Dictyostelium Rap1, RasG, or RasS proteins (Rap1(G12V), Ras G(G12T), and Ras S(G12T), respectively, kindly provided by Gerald Weeks (University of Columbia, Vancouver, Canada) were fused to the B42 activation domain in the vector pJG4–5 containing the inducible GAL1 promoter. Transformed EGY48 yeast expressing the plasmid p80-LacZ were tested for their ability to grow on selective plates (synthetic complete medium without leucine and containing galactose) and give a blue color on Xgal supplemented plates. For more accurate results, the β-galactosidase activity was determined in liquid conditions for a fixed number of yeast cells by using O-nitrophenyl β-d-galactopyranoside as a substrate. The background activity in cells expressing only the B42 activation domain fusion protein was subtracted. Expression of the B42 activation domain fusion protein in the presence of the DNA binding protein LexA fused to irrelevant sequences resulted in an identical background level of activation.
Phosphorylation Assay
GST and GST-Phg2 (kinase domain, residues 807-1101) proteins were produced in bacteria, partially purified on Sepharose-glutathione beads, and assayed for in vitro phosphorylation of various substrates in the presence of [γ-32P]ATP. Protein phosphorylation was assayed in a total volume of 20 μl containing 25 mM Tris-HCl, pH 7, 5 mM MgCl2, 5 mM MnCl2, 1 mM dithiothreitol, 1 mM EDTA, 10 μM and 250 μCi ml-1 [γ-32P]ATP (specific activity 4500 Ci mmol-1). After 10 min at 37°C, the reaction was stopped by adding 5 μl of a buffer containing 25% mercaptoethanol, 10% SDS, 400 mM Tris-HCl, pH 6.8, 50% glycerol, and 1% bromphenol blue according to Laemmli and heating the mixture at 100°C for 5 min. After electrophoresis, gels were soaked in 16% trichloroacetic acid for 10 min at 90°C. They were stained with Coomassie Blue, and radioactive proteins were visualized by autoradiography by using direct exposure films. No phosphorylation of exogenously added substrates (myelic basic protein, histones, and casein) was observed. However, an unknown 100-kDa bacterial protein was phosphorylated in the presence of glutathione S-transferase (GST)-Phg2, and not in the presence of GST (our unpublished observations).
To characterize the phosphorylated amino acid residues, protein samples were first subjected to SDS-PAGE and then electroblotted onto an Immobilon polyvinylidene difluoride membrane. Phosphorylated proteins bound to the membrane were detected by autoradiography. The 32P-labeled protein bands were excised individually from the Immobilon blot, wetted in methanol, rinsed in water, and finally hydrolyzed in 6 M HCl at 110°C for 1 h. The acid-stable amino acids thus liberated were separated by electrophoresis in the first dimension at pH 1.9 (800 V h) in 7.8% acetic acid and 2.4% formic acid, followed by ascending chromatography in the second dimension in 2-methyl-propanol:formic acid:water (8:3:4). After migration, radioactive molecules were detected by autoradiography using direct exposure MS films (Eastman Kodak, Rochester, NY). Authentic phosphoserine, phosphothreonine, and phosphotyrosine were run in parallel and visualized by staining with 0.05% ninhydrin in acetone.
RESULTS
PHG2, a Kinase Involved in Phagocytosis
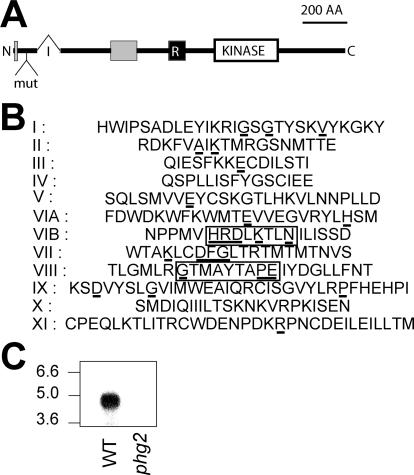
To identify new genes involved in phagocytosis, we performed a random insertional mutagenesis and selected mutants defective for phagocytosis of fluorescent latex beads. A mutant named phg2 was isolated from this screening, with a 90% reduction in phagocytosis of latex beads compared with wild-type cells. The coding sequence of PHG2 was determined and is almost identical to a predicted coding sequence in the Dictyostelium genome (DD186362C-CU). In the phg2 mutant, the mutagenic plasmid is inserted in the 5′ region of the PHG2 coding sequence, 177 nucleotides downstream from the start codon (Figure 1A). The Phg2 protein is relatively large (1389 amino acid residues) with several predicted functional domains. In particular, a kinase domain (position 802-1074) exhibits a characteristic set of 10 subdomains (Figure 1B), as well as a conserved kinase ATP-binding region (position 815–836, IGSGTYSKVYKGKYRDKFVAIK) (Falquet et al., 2002). The presence of a HRDLKTLN sequence (position 930–937) in subdomain VI and of a GT-MAYTAPE sequence (position 970–978) in subdomain VIII suggests strongly that Phg2 is a serine/threonine kinase rather than a tyrosine kinase. Phg2 also contains a putative RBD (position 594–667), as well as two proline-rich domains (positions 6–18 and 336–451), through which it may interact with other cellular proteins. To confirm that the phg2 mutant phenotype was due exclusively to an insertion in the PHG2 gene, we disrupted the PHG2 gene in the wild-type DH1 strain by homologous recombination. These cells showed a phagocytosis defect identical to the original phg2 mutant clone, and no longer express PHG2 mRNA (Figure 1C).
Figure 1.
Structure of the Phg2 protein. (A) Organization of the Phg2 protein. The position of an intron (I) in the genomic sequence is indicated (321 nucleotides downstream from the start codon). In the phg2 mutant, the mutagenic plasmid (mut) is inserted in the first exon, encoding the N-terminal portion of Phg2. The Phg2 protein contains a putative kinase domain (KINASE), a putative ras-binding domain (R), and two proline-rich domains (gray boxes). (B) Organization of the Phg2 kinase subdomains (residues 802-1074): the residues most conserved in kinase domains are underlined. Residues indicating a serine/threonine- rather than a tyrosine-kinase activity are boxed. (C) Cellular RNA was extracted from wild-type (WT) or phg2 mutant cells, migrated on an agarose gel, transferred to a nitrocellulose membrane, and revealed with a probe corresponding to the 3′ end of the PHG2 coding sequence. A 4.5-kb transcript was detected in wild-type but not in phg2 mutant cells. The sequence data are available from European Molecular Biology Laboratory database under accession no. AJ585374.
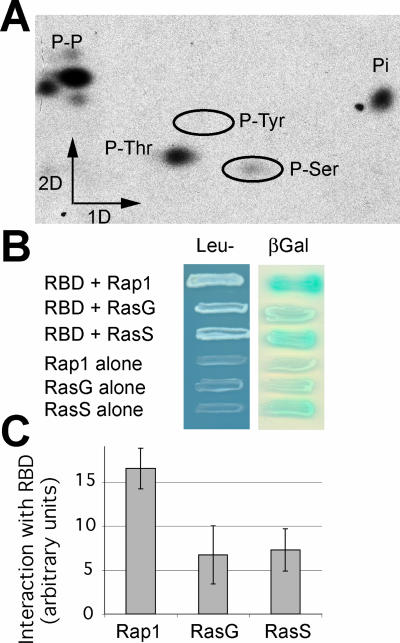
As a first step in verifying whether functional predictions based on the Phg2 sequence were founded, we tested the ability of Phg2 to function as a kinase in vitro. For this, the putative Phg2 kinase domain was fused to a GST protein and expressed in bacteria, purified, and its kinase activity tested by 32P incorporation. No kinase activity was detected when nonspecific substrates (myelic basic protein, histones, and casein) were used (our unpublished data). However, one of the bacterial proteins present in the preparation was specifically phosphorylated, demonstrating that the kinase domain of Phg2 is functional, and allowing us to test the nature of the phosphorylated amino acid residues. As seen in Figure 2A, only serine and threonine residues were phosphorylated. This result demonstrates that Phg2 is a serine/threonine kinase and suggests that it phosphorylates substrates with a high degree of specificity.
Figure 2.
Characterization of Phg2 kinase activity and ras-binding domain. (A) The Phg2 kinase domain fused to GST was produced in bacteria, partially purified, and used to phosphorylate in vitro a bacterial protein in the presence of [γ-32P]ATP. The phosphorylated protein was purified and hydrolyzed, and the phosphorylated amino acid residues were separated by electrophoresis in the first dimension and ascending chromatography in the second dimension. Phosphorylated serine and threonine residues were detected, as well as unhydrolyzed phosphopeptides (P-P) and Pi (Pi). (B) The ability of the putative Phg2 ras-binding domain to interact with the constitutively active forms of Rap1, RasG, or RasS was tested in a yeast two-hybrid system. Growth in Leu- medium as well as β-galactosidase activity indicated that an interaction took place between the Phg2 RBD and the ras proteins. (C) To evaluate the interaction between the Phg2 RBD and the ras proteins in a more accurate manner, the β-galactosidase activity was measured (e.g., Rap1 + Phg2 RBD), and the background (e.g., Rap1 alone) was subtracted. The average and SEM of four independent experiments are indicated. The Phg2 RBD interacts with all three ras proteins, and most strongly with Rap1.
Next, we used the yeast two-hybrid system to test for interactions between the putative Phg2 RBD and three members of the Dictyostelium ras family (Figure 2B). Growth in Leu- medium as well as production of β-galactosidase indicated that the Phg2 RBD can interact with the constitutively active forms of Rap1, RasS, and RasG. For a more quantitative evaluation, β-galactosidase activity was measured in liquid conditions (Figure 2C), and the background subtracted (e.g., Rap1 + Phg2 RBD minus Rap1 alone). These tests also detected an interaction between the Phg2 RBD and the three ras proteins tested, with a particularly prominent interaction with Rap1, the level of specific signal being 4 times higher than the background in this case. These results confirm that the putative RBD of Phg2 has the ability to interact with members of the ras family, although detailed biochemical experiments will be necessary to determine which member(s) of the ras family interact with Phg2 in vivo.
Mutant phg1, phg2, myoVII, and Talin Cells Exhibit Similar Adhesion Defects
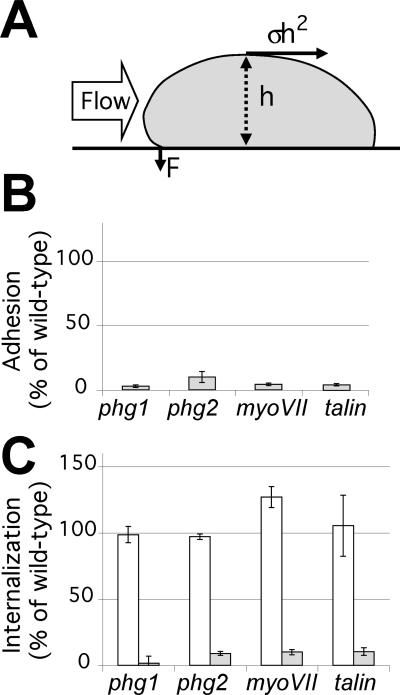
Previous analysis showed that phg1 mutant cells have a strong defect in phagocytosis because they adhere less effectively to particles, and this adhesion defect is also observed when cells adhere to a glass surface (Cornillon et al., 2000). As detailed above, several other gene products also play a critical role in cellular adhesion in Dictyostelium, in particular myosin VII and talin (Titus, 1999; Niewohner et al., 1997; Tuxworth et al., 2001). To compare their role in adhesion with the function of Phg2, we generated the corresponding disruption mutants in our Dictyostelium laboratory strain (DH1–10), and measured the adhesion of phg1, phg2, myoVII, and talin cells to a glass substrate in HL5 medium. As discussed previously (Cornillon et al., 2000), in these conditions glass is coated with components of the HL5 medium and constitutes a very hydrophilic surface. For this experiment, cells attached to a glass substratum were subjected to a flow of medium, and the speed of the flow necessary to detach 50% of the cells was determined (Figure 3A). It was previously shown that the cells are peeled away from the substrate by the flow when it applies a critical stress. The strength necessary to detach the cells can be extrapolated and represents the strength of the cellular adhesion to the substrate (Decave et al., 2002a,b). As seen in Figure 3B, all four mutants tested showed a strong adhesion defect. All four mutants were also severely impaired in phagocytosis of latex beads in HL5 medium (Figure 3C), presumably because they adhere less efficiently to these particles. As would be expected, uptake of fluid phase, which is not dependent on cellular adhesion, was normal in all mutants (Figure 3C).
Figure 3.
Adhesion and phagocytosis defects in phg1, phg2, myoVII, and talin mutant cells. (A) Schematic showing a side view of a cell attached to a substrate and exposed to a flux of medium. Cell-substrate adhesion was calculated by determining the speed of a flow of medium necessary to detach 50% of the cells (Decave et al., 2002b). In these conditions, the strength exerted by the flux of medium on the cell is σh2, and its mechanical moment (σh3) is balanced by the adhesive force (F). (B) Adhesion of mutant cells to a glass substrate in HL5 medium, expressed as a percentage of wild-type adhesion in the same experiment. In these conditions, the cell morphology is identical in wild-type and mutant cells (Figure 4), allowing direct comparison of the results obtained. (C) Cells were incubated for 20 min either with latex beads (gray bars), or with fluorescein isothiocyanate-dextran in HL5 medium (white bars). Results are expressed as the percentage of internalization by wild-type cells in the same experiment. Indicated values are the mean and SEM of at least four independent experiments.
Interestingly, as previously reported for phg1 mutant cells (Cornillon et al., 2000), the ability of all four mutants to adhere to a more hydrophobic surface (glass in phosphate buffer) was only slightly affected (107, 97, 89, and 74% of wild-type adhesion for phg1, phg2, myoVII, and talin null cells, respectively), indicating that a different adhesion machinery acts in phosphate buffer, largely independent of the four gene products studied here. As discussed previously (Cornillon et al., 2000), this might reflect the fact that the cellular physiology is different in phosphate buffer or that different receptors are involved in binding to more hydrophobic surfaces. Phg1, Phg2, myosin VII, and talin are essential for cellular adhesion in HL5 but not in phosphate buffer, suggesting that these four proteins are involved in the same cellular adhesion processes.
Phg2, Myosin VII, and Talin Are Involved in the Organization of the Actin Cytoskeleton
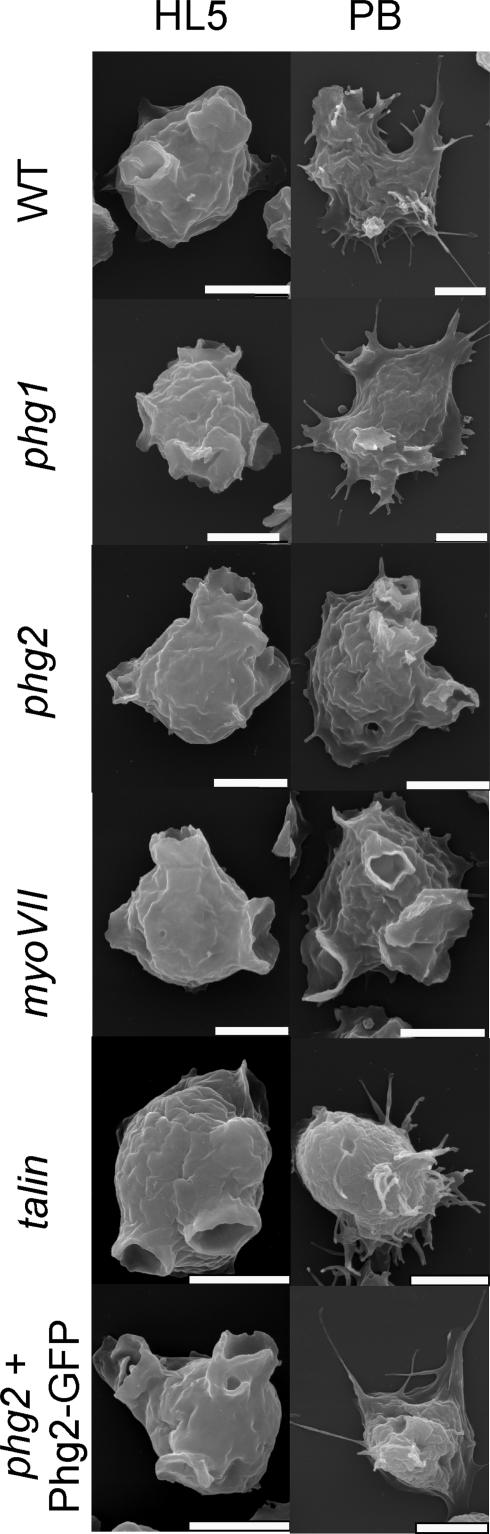
Previous reports have shown that myosin VII and talin, both actin-binding proteins, are involved in the organization of the actin cytoskeleton (Niewohner et al., 1997; Tuxworth etal., 2001). Accordingly, alterations in actin-dependent processes such as the control of cell shape or cytokinesis were reported in the myoVII and talin mutant cells. To evaluate the role of Phg1, myosin VII, talin, and Phg2 in the function of the actin cytoskeleton in our laboratory strain, we first examined the morphology of the cells by scanning electron microscopy. Dictyostelium cells are round in HL5 medium and have mostly macropinocytic cups at their surface (Figure 4). Cells of all four mutants showed a similar morphology in HL5 medium (Figure 4). In phosphate buffer, wild-type cells flatten onto the substrate and extend thin filopodia. Phg1 null cells also flattened and produced filopodial extensions in these conditions. Phg2 and myoVII mutant cells, however, did not noticeably change shape in phosphate buffer and did not form filopodia (Figure 4). Membrane extensions were seen in talin mutant cells but these were unusually short for filopodia, and the cells did not flatten. Expression of a Phg2-GFP fusion protein (see below) complemented the morphological defect observed in phg2 mutant cells (Figure 4).
Figure 4.
The control of cell shape is altered in phg2, myoVII, and talin mutant cells. Wild-type and mutant cells were incubated for 1 h in either HL5 medium or PB, and then fixed and processed for scanning electron microscopy. The cell shape of phg2, myoVII, and talin mutant cells in PB was altered compared with wild-type or phg1 cells. Bar, 5 μm.
Next, to examine the role of Phg1, Phg2, myosin VII, and talin in cytokinesis, we stained cell nuclei and counted the percentage of multinucleated cells (at least three nuclei per cell). Phg2 and talin mutant populations show a high proportion of multinucleated cells (Figure 5), suggesting defects at some stage in the process of cell division. Only minor increases in the number of multinucleated cells were observed in phg1 and myoVII mutant cells. Together, these results indicate that in addition to its role in adhesion, the Phg2 kinase, like myosin VII and talin, is involved in controlling the actin cytoskeleton during cell shape changes and cytokinesis.
Figure 5.
Phg2 and talin mutant cells are defective in cytokinesis. (A) Wild-type and mutant cells were grown on coverslips, fixed, treated with RNAse A, and their nuclei stained with propidium iodide. Wild-type and phg2 cells were photographed under a fluorescence microscope. Bar, 20 μm. (B) The percentage of cells containing at least three nuclei was counted (mean and SEM of at least four independent experiments). Multinucleated cells are unusually frequent in phg2 and talin null cells.
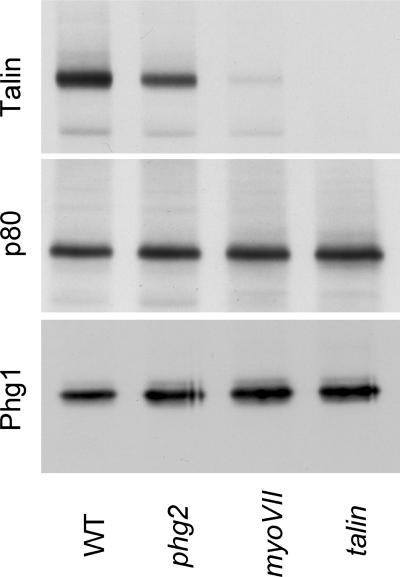
To investigate the possibility of a functional link between the Phg1, Phg2, myosin VII, and talin proteins, we examined the expression of several proteins in the mutant lines. The levels of Phg1 and of the endosomal p80 protein were not strongly affected in all three mutant cells (Figure 6). This indicates that the adhesion defects observed in phg2, myoVII, and talin mutant cells cannot be accounted for by a loss of Phg1 expression. A strong decrease in the level of talin protein was seen in myoVII mutant cells (Figure 6), and this might explain at least partly the phenotype of myoVII mutants. On the contrary, the slight but reproducible decrease in the amount of talin in phg2 mutant cells is unlikely to account significantly for the phenotype of phg2 mutant cells (see DISCUSSION). In addition, these data suggest a putative functional link between talin, myosin VII, and Phg2, the absence of Phg2 or myosin VII having a direct or indirect effect on the amount of talin in the cell. Detailed biochemical analysis will be necessary to establish whether Phg2, myosin VII, and talin interact functionally to control cell-substrate adhesion and the actin cytoskeleton.
Figure 6.
Levels of talin protein are lower in phg2 and myoVII null cells. Cell lysates from wild-type or mutant cells were separated on an SDS-polyacrylamide gel (105 cells/lane), transferred to a nitrocellulose membrane, and the amount of talin (top gel), p80 (medium), and Phg1 (bottom gel) was assessed by Western blot. Whereas the amounts of Phg1 or p80 protein (an endosomal marker) were not affected, the amount of talin was reduced in phg2 and myoVII mutant cells. This experiment was repeated three times with identical results.
Intracellular Distribution of Phg2
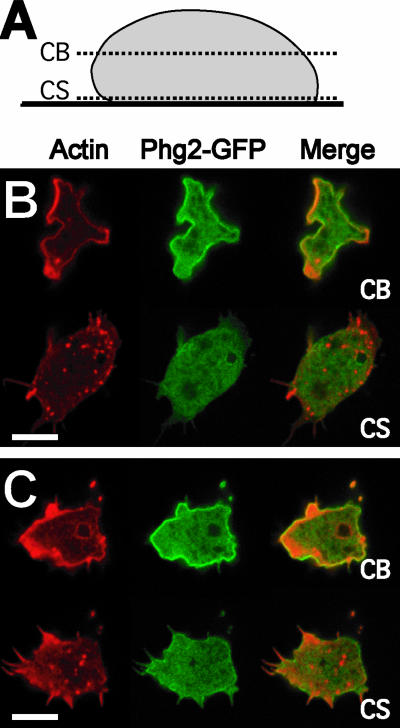
To gain a better view of the role of Phg2, we expressed a Phg2-GFP fusion protein in phg2 null cells. In all respects, these cells behaved like wild-type cells: phagocytosis of latex beads was normal (95% of wild-type phagocytosis), as were cell shape (Figure 4) and actin organization (our unpublished data, but compare, for example, Figure 7 with Figure 8). Cells expressing the Phg2-GFP protein (in green) attached to a glass coverslip for 10 min and were fixed, their actin cytoskeleton was labeled (in red), and imaged by confocal microscopy at the level where cells establish a contact with the substratum (CS), or within the cell body (CB) (Figure 7). The actin labeling within the cell body was essentially cortical, with sites of intense staining corresponding to forming macropinocytic cups or pseudopods. Within the same sections, Phg2-GFP was seen in the cytosol, but largely accumulated at the level of the plasma membrane. The fusion protein did not strongly accumulate at sites of abundant actin or on the membrane of any intracellular compartment. At the site of contact with the substratum, the actin cytoskeleton was polymerized in small punctate structures reminiscent of focal adhesion sites seen in mammalian cells. Near this membrane, Phg2-GFP, however, was evenly distributed with no accumulation in foci or in extending filopods (Figure 7). For simplicity, actin foci at the site of contact with the substratum are referred to here as actin focal sites, and their nature and function are discussed in more detail in the Discussion.
Figure 7.
Intracellular localization of Phg2-GFP. A construct expressing Phg2 fused to the green fluorescent protein (Phg2-GFP) was introduced into phg2 mutant cells, where it restored a wild-type phenotype. (A) The cells were fixed and stained with TRITC-labeled phalloidin (red). Confocal pictures were taken within the cell body (CB) or just at the level of the contact with the substrate (CS) as shown in the diagram. (B and C) Two distinct cells were imaged. Phg2-GFP (green) accumulated at the plasma membrane but also was detected in the cytosol. No enrichment of Phg2-GFP was detected in actin-rich foci near the site of contact with the slide or in actin-rich locations at the periphery of the cell body. Bar, 10 μm.
Figure 8.
Polymerization of actin in focal sites is affected in phg2 mutant cells. Wild-type (WT) and mutant cells were allowed to adhere to a glass coverslip for either 2 or 10 min in phosphate buffer, and then fixed and stained with phalloidin. The actin cytoskeleton just above the plane of contact with the coverslip was imaged by confocal microscopy as described in Figure 7 (CS). After 10 min, unusually large accumulations of actin were seen in phg2 mutant cells. Confocal images taken within the cell body (CB) showed that the actin staining was essentially identical in all four mutants compared with wild-type (our unpublished data). Bar, 10 μm.
Phg2 Is Involved in the Modeling of Actin Focal Sites
To further investigate the link between cellular adhesion and signaling to the actin cytoskeleton, we examined the organization of the actin cytoskeleton at the cell-substrate contact site in mutant cells. For this, cells were allowed to attach to a glass coverslip in phosphate buffer, and their actin cytoskeleton was labeled and imaged by confocal microscopy. As reported above, in these conditions all mutants adhere to the substrate with an efficiency close to that of wild-type cells. This ensures that any differences observed are not a secondary effect of an adhesion defect. In optical sections taken at the plane of the cell-substrate contact, changes in the pattern of actin labeling were observed at various times after adhesion. After 2 min, wild-type cells had readily adhered to the glass substrate and the actin cytoskeleton was polymerized in focal sites (Figure 8). After 10 min of adhesion, the cells were more spread out, and actin labeling at focal sites was weaker. Occasionally, more intensely labeled actin structures could be seen at the cell periphery. After 2 min of adhesion, the localization of actin in mutant cells was not markedly different from wild-type cells. However after 10 min, actin organization was drastically altered in phg2 null cells, with very intensely labeled actin-containing structures present in the central part of the cell-substrate contact zone (Figure 8). In phg1, myoVII, and talin mutant cells, the actin cytoskeleton was not different from wild-type cells. Within the cell body, the actin cytoskeleton looked identical in all four mutant strains and in wild-type cells at all times, with cortical actin labeling and a few areas of intense labeling (Figure 7; our unpublished data). Phg2, therefore, seems to play a specific role in signaling actin polymerization/depolymerization at places where the amoeba comes into direct contact with a substrate.
Phg2 Is Required for Cell Motility and Mechanotransduction
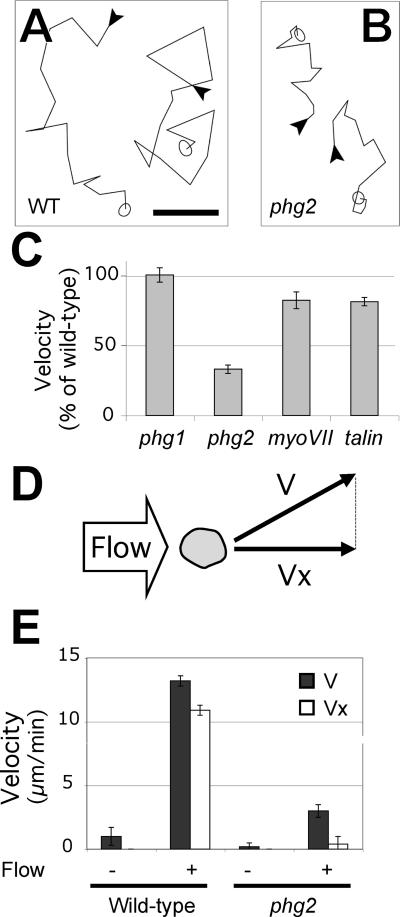
Cultured Dictyostelium cells are constantly moving in a non-oriented manner. Signaling pathways that control the actin cytoskeleton are essential for this type of cellular motility, which involves the formation and retraction of pseudopods and rapid adhesion and detachment cycles (Friedl et al., 2001). We first measured the ability of wild-type and mutant cells to move on a substrate by recording their random motility. In HL5 moderate motility defects were observed for phg1, phg2, and myoVII mutants (respectively, 77, 40, and 56% of wild-type motility), whereas talin mutant cells behaved identically to wild-type cells (97% of wild-type motility). However, whether these results indicate a direct role for the corresponding gene products in cellular motility is unclear because all four mutants show strong adhesion defects in these conditions. We therefore repeated these observations in phosphate buffer where mutant and wild-type cells show virtually identical adhesion. In these conditions, no defect in motility was seen for phg1 cells, and only a slight defect was observed for myoVII and talin cells (Figure 9). Phg2 null cells, however, show a threefold reduction in random motility compared with wild-type cells (Figure 9). Phg2 therefore plays a specific role in cellular motility in addition to its more general function in the organization of the actin cytoskeleton.
Figure 9.
Mutant phg2 cells show defects in motility. (A and B) Cells were allowed to settle onto a plastic substrate in phosphate buffer and their movement recorded at 4-min intervals for 60 min. Two typical traces showing movement of wild-type (A) or phg2 cells (B) are shown. The beginning of each trace is indicated with an arrowhead, and the amoeba is represented at the end of the trace. Bar, 50 μm. (C) The instantaneous speed was measured for at least 30 individual cells from each mutant population and expressed as a percentage of motility of wild-type cells (6.5 μm/min in these conditions). The mean and SEM are indicated. (D and E) Mutant phg2 cells are defective for shear-flow induced motility. (D) Schematic representation of a top view of the experimental setup. A slow lateral flow of medium exerts mechanical forces on Dictyostelium cells and induces their movement in the direction of the flow. The average cell speed (V) was determined with or without a flow of medium, as well as its projection along the flow axis (Vx) (E) V (full bars) and Vx (open bars) are indicated for wild-type and phg2 mutant cells. In the lateral flow chamber, the cells adhere on glass and this accounts for the lower motility of unstimulated cells compared with cells adhering on plastic.
We also observed the behavior of phg2 cells in conditions where motility is stimulated by a controlled flow of medium (Figure 9D) (Decave et al., 2003). This flow is not sufficient to detach the cells, but induces a marked increase in cell motility (Figure 9E). This mechanotransduction process is similar to the behavior of vascular endothelial cells where motility induced by shear stress allows cells to migrate in an oriented manner to repair a wounded endothelium (Davies, 2002). By calculating the speed of the cells along the axis of the flow (Vx), stimulated cells were observed to move mainly in the direction of the flow (Vx ≈ V). When phg2 cells were analyzed in this setup, cellular motility without stimulation (V0) was weaker, as reported above. Shear stress did induce an increase in cellular motility, but the movement elicited was weaker than for wild-type cells (Figure 9E), and not oriented (Vx << V). However the amplitude of the response (V/V0) was similar in wild-type and phg2 cells (13 and 15 for wild-type and phg2, respectively). Thus, whereas phg2 cells do react to shear stress, their motility in these conditions remains weaker than that observed in wild-type cells. This result indicates that the motility defect observed in phg2 null cells can be seen in stimulated as well as non-stimulated cells and most likely reflects a general defect in the ability of the mutant cells to move on a substrate.
DISCUSSION
Phg2, a Novel Kinase Involved in Adhesion and Phagocytosis
This study was aimed at identifying new proteins controlling cellular adhesion and phagocytosis and at determining their role in various aspects of cellular physiology. For this, we performed a random insertional mutagenesis and selected mutants defective for phagocytosis of fluorescent latex beads in HL5 medium. An initial screen led to the isolation of the phg1 mutant (Cornillon et al., 2000). The Phg1 protein was shown to play a role in adhesion to the phagocytosed particles, the first step in the phagocytosis process (Cornillon et al., 2000). A second screen, reported here, led to the isolation of the phg2 mutant, with a 90% reduction in phagocytosis of latex beads compared with wild-type cells and no defect in macropinocytosis. The phagocytosis defect observed in phg2 mutants is also due to a defect in cell adherence to hydrophilic substrates (latex beads or glass coated with HL5 medium). Interestingly, adhesion to hydrophobic substrates is not affected in the phg2 mutant. In natural conditions amoebae presumably encounter hydrophilic (wet soil) and hydrophobic (the waxy surface of a leaf) environments, and it seems that Phg2 (as well as Phg1, myosin VII, and talin) is only required for adhesion under certain conditions.
Based on analysis of the primary sequence and on biochemical evidence, Phg2 is a serine/threonine kinase and can bind to members of the ras family. It also contains two proline-rich putative interaction domains. Ras proteins have been implicated in the regulation of the actin cytoskeleton both in Dictyostelium and in mammalian cells (Lim et al., 2002). In particular, rasG mutant cells have aberrant shapes, are defective in cytokinesis, and show unusually high amounts of actin polymerized at the site of contact with the substrate (Tuxworth et al., 1997), a phenotype similar to that of phg2 mutant cells. Similarly, overexpression of Rap1, with which Phg2 interacts most strongly, leads to spreading and flattening of cells and redistribution of actin (Rebstein et al., 1993). It will be interesting to determine which ras protein(s) interacts with Phg2 in living cells.
Based on the presence of a kinase domain, of a RBD and of several protein–protein interaction domains, the closest homologues of Phg2 in mammalian cells are the Rho kinases (ROCKs), although there are some differences in the organization of the functional domains (for review, see Riento and Ridley, 2003). ROCKs are serine/threonine kinases activated by binding to a small GTP-binding protein (Rho) and also contain several other protein-interacting domains. Several other kinases involved in the control of cellular adhesion in mammalian cells, such as FAK (Schwartz, 2001), also contain one kinase domain and several putative protein–protein interaction domains like Phg2. Parallels between mammalian kinases involved in cellular adhesion and Phg2 also extend to function, because several of them (ROCKs and FAK) are involved in controlling the actin cytoskeleton, focal adhesion sites, and cell motility (Schwartz, 2001; Riento and Ridley, 2003).
Phg2 Plays a Specific Role in the Organization of the Actin Cytoskeleton
Further analysis of the phenotype of phg2 mutant cells revealed defects in the control of cell shape. Because, in Dictyostelium, cell shape largely reflects the organization of the underlying actin cytoskeleton, this observation establishes a functional link between Phg2 and the actin cytoskeleton. Similarly, cytokinesis, another actin-dependent process, is altered in phg2 mutants. As described previously (Niewohner et al., 1997; Titus, 1999), inactivation of the genes encoding talin and myosin VII, two proteins linked to the actin cytoskeleton, created similar defects, suggesting a functional link between Phg2, talin and myosin VII. A link between the three proteins is also suggested by the fact that a decreased amount of talin is detected in phg2 and myoVII mutant cells. However, the phenotype of phg2 mutants could not be accounted for entirely by a decrease in the level of talin, because the amount of talin is only slightly decreased in phg2 mutant cells, and because phg2 mutant cells present phenotypes not observed in talin mutant cells. In addition it is unlikely that Phg2 forms a stable complex with either myosinVII or talin, because Phg2 is evenly distributed at the cell membrane, whereas myosin VII and talin concentrate at sites of active actin remodeling, i.e., at the tip of filopods and at the leading edge of migrating cells (Kreitmeier et al., 1995; Tuxworth et al., 2001). Thus, an interaction between Phg2 and these cytoskeletal proteins could only be transient or indirect.
One novel feature of the phg2 mutant is the presence of huge actin-containing structures at the site of contact with the substrate. Because this phenotype was not observed in myoVII or talin mutants, this defect can be distinguished from the other actin defects observed (aberrant cell shape and cytokinesis). This suggests an additional and specific role for Phg2 in limiting the polymerization of actin at focal sites, or inducing its depolymerization. Motile cells continuously establish new points of contact with the substrate, and lose their previous site of attachment. Phg2 might play a role in motility by allowing the local reorganization of the actin cytoskeleton, and local detachment of the cells.
Actin-rich foci were previously observed in the ventral region of migrating Dictyostelium cells (Yumura and Kitanishi-Yumura, 1990; Bretschneider et al., 2004; Uchida and Yumura, 2004). They correspond to regions of close contact between the cell membrane and the substrate where actin and other cellular components are enriched, in particular the Arp2/3 complex. Analysis of their dynamics revealed that actin foci are immobile structures, but present a high turnover because they form and disappear within 10–20 s (Bretschneider et al., 2004; Uchida and Yumura, 2004). These structures were often studied in cells starved for rather prolonged periods of time (e.g., 6 h), to observe their dynamics in cells expressing cAMP receptors and engaged in chemotaxis (Bretschneider et al., 2002; Soll et al., 2002), but their dynamics seem very similar in unstarved cells (Bretschneider et al., 2004), as used in this study. Actin-rich foci in Dictyostelium resemble focal adhesion sites seen in mammalian cells, which also are characterized by the accumulation of actin and many proteins involved in cellular adhesion (membrane receptors, signal transduction molecules, and actin-binding proteins). However a detailed comparison is made difficult by the molecular and structural diversity of structures observed in mammalian cells (Zamir and Geiger, 2001). Interestingly, mammalian cells where both alleles of FAK have been inactivated show abnormal intracellular signaling, defects in cell migration (in particular in response to shear stress; Li et al., 2002), and an increased number of focal adhesion sites (Ilic et al., 1995), suggesting a role for FAK in the control of the stability of focal adhesion sites. Although Phg2 is not concentrated in actin foci, it might play a role akin to that of FAK in controlling the dynamics of actin focal sites.
A Hierarchy in the Functions of Phg1, Phg2, Myosin VII, and Talin
Many proteins implicated in cellular adhesion also are linked to other functions such as the organization of the actin cytoskeleton and cell motility. In the course of this study, we compared the phenotypes of phg1, phg2, myoVII, and talin mutant cells and in doing so we determined the relative importance of each gene product in various cellular functions. Targeted inactivation of genes encoding Phg1, Phg2, talin, or myosin VII results in similar defects in cellular adhesion. In a subset of these mutant cells (phg2, talin, and myoVII), additional defects were observed revealing a role in the control of cell shape and cytokinesis, presumably due to anomalies in the organization of the actin cytoskeleton. In addition, the Phg2 protein plays a unique role in cellular motility and in the modeling of actin-rich focal sites. These data establish a hierarchy among these four gene products, summarized in Figure 10, and which can only emerge from a detailed comparative analysis. Note that to allow meaningful comparisons, all mutants analyzed here were derived from the same wild-type strain in our laboratory, because it has been reported previously that the effect of a mutation can differ widely in different Dictyostelium strains (see, for example, Chen and Katz, 2000). These results provide a framework for comparative analysis of the role of other genes in adhesion, cytoskeleton organization, and motility, provided that the corresponding mutants are obtained in the same strain and analyzed in a similar manner.
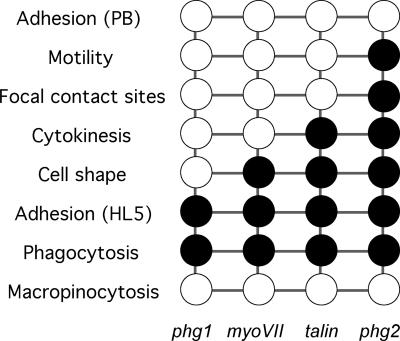
Figure 10.
Hierarchy in the roles of Phg1, myosin VII, talin, and Phg2 in various cellular processes. For each mutant cell line, a black circle indicates a defect for a particular function. Minor defects or a wild-type phenotype are represented by a white circle.
This comparative analysis reveals a central role for Phg2 in cellular physiology because it is involved in cell-substrate adhesion, the control of cell shape, cytokinesis, organization of focal actin sites, and cell motility. This functional complexity is reflected by the presence of several functional domains in the Phg2 protein. The ability to complement the inactivation of the endogenous PHG2 with a transfected plasmid expressing Phg2 opens the way for a detailed molecular analysis of Phg2 function. In this system, the role of each structural domain of Phg2 in each of the functions listed above can be determined. Molecular dissection of Phg2 will allow a better understanding of the manner in which kinase proteins are involved in the physiology of motile and adhering cells.
Acknowledgments
We thank Steve J. Charette for critical reading of the manuscript. This work was supported by a START fellowship of the Fonds National Suisse dela Recherche Scientifique and a grant from the Fondation Gabriella Giorgi-Cavaglieri (both to P.C.); by a grant from the Ministèredela Recherche and the Centre National dela Recherche Scientifique and a grant from the Institut dela Matièrecondensée (Grenoble) (both to F.B.); and by a grant from the Association pour la Recherche contre le Cancer and a grant from the Fondation pour la Recherche Médicale (both to F.L.). S.F. and J.D. are recipients of a MENRT fellowship.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-12-0908. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-12-0908.
Abbreviations used: FAK, focal adhesion kinase; PB, phosphate buffer; RBD, ras-binding domain; ROCK, rho-activated kinase.
References
- Bracco, E., Pergolizzi, B., Peracino, B., Ponte, E., Balbo, A., Mai, A., Ceccarelli, A., and Bozzaro, S. (2000). Cell-cell signaling and adhesion in phagocytosis and early development of Dictyostelium. Int. J. Dev. Biol. 44, 733-742. [PubMed] [Google Scholar]
- Bretschneider, T., Diez, S., Anderson, K., Heuser, J., Clarke, M., Muller-Taubenberger, A., Kohler, J., and Gerisch, G. (2004). Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr. Biol. 14, 1-10. [DOI] [PubMed] [Google Scholar]
- Bretschneider, T., Jonkman, J., Kohler, J., Medalia, O., Barisic, K., Weber, I., Stelzer, E.H., Baumeister, W., and Gerisch, G. (2002). Dynamic organization of the actin system in the motile cells of Dictyostelium. J. Muscle Res. Cell Motil. 23, 639-649. [DOI] [PubMed] [Google Scholar]
- Caterina, M.J., Milne, J.L., and Devreotes, P.N. (1994). Mutation of the third intracellular loop of the cAMP receptor, cAR1, of Dictyostelium yields mutants impaired in multiple signaling pathways. J. Biol. Chem. 269, 1523-1532. [PubMed] [Google Scholar]
- Chen, C.F., and Katz, E.R. (2000). Mediation of cell-substratum adhesion by RasG in Dictyostelium. J. Cell. Biochem. 79, 139-149. [PubMed] [Google Scholar]
- Cornillon, S., Dubois, A., Bruckert, F., Lefkir, Y., Marchetti, A., Benghezal, M., De Lozanne, A., Letourneur, F., and Cosson, P. (2002). Two members of the beige/CHS (BEACH) family are involved at different stages in the organization of the endocytic pathway in Dictyostelium. J. Cell Sci. 115, 737-744. [DOI] [PubMed] [Google Scholar]
- Cornillon, S., Olie, R.A., and Golstein, P. (1998). An insertional mutagenesis approach to Dictyostelium cell death. Cell Death Differ. 5, 416-425. [DOI] [PubMed] [Google Scholar]
- Cornillon, S., Pech, E., Benghezal, M., Ravanel, K., Gaynor, E., Letourneur, F., Bruckert, F., and Cosson, P. (2000). Phg1p is a nine-transmembrane protein superfamily member involved in Dictyostelium adhesion and phagocytosis. J. Biol. Chem. 275, 34287-34292. [DOI] [PubMed] [Google Scholar]
- Davies, P.F. (2002). Multiple signaling pathways in flow-mediated endothelial mechanotransduction: PYK-ing the right location. Arterioscler Thromb. Vasc. Biol. 22, 1755-1757. [DOI] [PubMed] [Google Scholar]
- Decave, E., Garrivier, D., Brechet, Y., Bruckert, F., and Fourcade, B. (2002a). Peeling process in living cell movement under shear flow. Phys. Rev. Lett. 89, 108101. [DOI] [PubMed] [Google Scholar]
- Decave, E., Garrivier, D., Brechet, Y., Fourcade, B., and Bruckert, F. (2002b). Shear flow-induced detachment kinetics of Dictyostelium discoideum cells from solid substrate. Biophys. J. 82, 2383-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decave, E., Rieu, D., Dalous, J., Fache, S., Brechet, Y., Fourcade, B., Satre, M., and Bruckert, F. (2003). Shear flow-induced motility of Dictyostelium discoideum cells on solid substrate. J. Cell Sci. 116, 4331-4343. [DOI] [PubMed] [Google Scholar]
- Eichinger, L., and Noegel, A.A. (2003). Crawling into a new era-the Dictyostelium genome project. EMBO J. 22, 1941-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet, L., Pagni, M., Bucher, P., Hulo, N., Sigrist, C.J., Hofmann, K., and Bairoch, A. (2002). The PROSITE database, its status in 2002. Nucleic Acids Res. 30, 235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey, P., Stephens, S., Titus, M.A., and Chisholm, R.L. (2002). SadA, a novel adhesion receptor in Dictyostelium. J. Cell Biol. 159, 1109-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl, P., Borgmann, S., and Brocker, E.B. (2001). Amoeboid leukocyte crawling through extracellular matrix: lessons from the Dictyostelium paradigm of cell movement. J. Leukoc. Biol. 70, 491-509. [PubMed] [Google Scholar]
- Han, Y.H., Chung, C.Y., Wessels, D., Stephens, S., Titus, M.A., Soll, D.R., and Firtel, R.A. (2002). Requirement of a vasodilator-stimulated phosphoprotein family member for cell adhesion, the formation of filopodia, and chemotaxis in Dictyostelium. J. Biol. Chem. 277, 49877-49887. [DOI] [PubMed] [Google Scholar]
- Ilic, D., Furuta, Y., Kanazawa, S., Takeda, N., Sobue, K., Nakatsuji, N., Nomura, S., Fujimoto, J., Okada, M., and Yamamoto, T. (1995). Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 377, 539-544. [DOI] [PubMed] [Google Scholar]
- Kreitmeier, M., Gerisch, G., Heizer, C., and Muller-Taubenberger, A. (1995). A talin homologue of Dictyostelium rapidly assembles at the leading edge of cells in response to chemoattractant. J. Cell Biol. 129, 179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Butler, P., Wang, Y., Hu, Y., Han, D.C., Usami, S., Guan, J.L., and Chien, S. (2002). The role of the dynamics of focal adhesion kinase in the mechanotaxis of endothelial cells. Proc. Natl. Acad. Sci. USA 99, 3546-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, C.J., Spiegelman, G.B., and Weeks, G. (2002). Cytoskeletal regulation by Dictyostelium Ras subfamily proteins. J. Muscle Res. Cell Motil. 23, 729-736. [DOI] [PubMed] [Google Scholar]
- Manstein, D.J., Schuster, H.P., Morandini, P., and Hunt, D.M. (1995). Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene 162, 129-134. [DOI] [PubMed] [Google Scholar]
- Niewohner, J., Weber, I., Maniak, M., Muller-Taubenberger, A., and Gerisch, G. (1997). Talin-null cells of Dictyostelium are strongly defective in adhesion to particle and substrate surfaces and slightly impaired in cytokinesis. J. Cell Biol. 138, 349-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel, K., de Chassey, B., Cornillon, S., Benghezal, M., Zulianello, L., Gebbie, L., Letourneur, F., and Cosson, P. (2001). Membrane sorting in the endocytic and phagocytic pathway of Dictyostelium discoideum. Eur. J. Cell Biol. 80, 754-764. [DOI] [PubMed] [Google Scholar]
- Rebstein, P.J., Weeks, G., and Spiegelman, G.B. (1993). Altered morphology of vegetative amoebae induced by increased expression of the Dictyostelium discoideum ras-related gene rap1. Dev. Genet. 14, 347-355. [DOI] [PubMed] [Google Scholar]
- Riento, K., and Ridley, A.J. (2003). Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol. Cell. Biol. 4, 446-456. [DOI] [PubMed] [Google Scholar]
- Schwartz, M.A. (2001). Integrin signaling revisited. Trends Cell Biol. 11, 466-470. [DOI] [PubMed] [Google Scholar]
- Soll, D.R., Wessels, D., Heid, P.J., and Zhang, H. (2002). A contextual framework for characterizing motility and chemotaxis mutants in Dictyostelium discoideum. J. Muscle Res. Cell Motil. 23, 659-672. [DOI] [PubMed] [Google Scholar]
- Titus, M.A. (1999). A class VII unconventional myosin is required for phagocytosis. Curr. Biol. 9, 1297-1303. [DOI] [PubMed] [Google Scholar]
- Tuxworth, R.I., Cheetham, J.L., Machesky, L.M., Spiegelmann, G.B., Weeks, G., and Insall, R.H. (1997). Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 138, 605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth, R.I., Weber, I., Wessels, D., Addicks, G.C., Soll, D.R., Gerisch, G., and Titus, M.A. (2001). A role for myosin VII in dynamic cell adhesion. Curr. Biol. 11, 318-329. [DOI] [PubMed] [Google Scholar]
- Uchida, K.S., and Yumura, S. (2004). Dynamics of novel feet of Dictyostelium cells during migration. J. Cell Sci. 117, 1443-1455. [DOI] [PubMed] [Google Scholar]
- Yumura, S., and Kitanishi-Yumura, T. (1990). Fluorescence-mediated visualization of actin and myosin filaments in the contractile membrane-cytoskeleton complex of Dictyostelium discoideum. Cell Struct. Funct. 15, 355-364. [DOI] [PubMed] [Google Scholar]
- Zamir, E., and Geiger, B. (2001). Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 114, 3583-3590. [DOI] [PubMed] [Google Scholar]