Abstract
Neutrophil (PMN) transepithelial migration is dependent on the leukocyte β2 integrin CD11b/CD18, yet the identity of epithelial counterreceptors remain elusive. Recently, a JAM protein family member termed JAM-C was implicated in leukocyte adhesive interactions; however, its expression in epithelia and role in PMN-epithelial interactions are unknown. Here, we demonstrate that JAM-C is abundantly expressed basolaterally in intestinal epithelia and localizes to desmosomes but not tight junctions. Desmosomal localization of JAM-C was further confirmed by experiments aimed at selective disruption of tight junctions and desmosomes. In assays of PMN transepithelial migration, both JAM-C mAbs and JAM-C/Fc chimeras significantly inhibited the rate of PMN transmigration. Additional experiments revealed specific binding of JAM-C to CD11b/CD18 and provided evidence of other epithelial ligands for CD11b/CD18. These findings represent the first demonstration of direct adhesive interactions between PMN and epithelial intercellular junctions (desmosomes) that regulate PMN transepithelial migration and also suggest that JAM-C may play a role in desmosomal structure/function.
INTRODUCTION
Neutrophil (PMN) transepithelial migration, a key feature of many inflammatory diseases of epithelial lined organs, involves a multistep cascade of events with each stage governed by distinct mechanisms. The early stages of PMN transepithelial migration involve adhesion to the basolateral epithelial membrane and appear to be dependent on the leukocyte β2 integrin CD11b/CD18 (Mac-1, CR-3; Parkos et al., 1991). Although abundant data support a role of CD11b/CD18 in this process, the nature of epithelial counterreceptor(s) for this β2 integrin have remained elusive. Receptortargeted studies using CD11b/CD18 have demonstrated that this integrin has great promiscuity in ligand binding with more than 30 protein or nonprotein molecules reported to date. Examples of CD11b/CD18 ligands with characterized functions include intercellular adhesion molecules 1 (ICAM-1; Diamond et al., 1990, 1991; Larson and Springer, 1990), complement C3 fragment iC3b (Beller et al., 1982), fibrinogen (FBG; Altieri et al., 1988), heparin (Diamond et al., 1995), neutrophil elastase (Cai and Wright, 1996), neutrophil inhibitory factor (NIF; Moyle et al., 1994), and fucoidin (Zen et al., 2002). CD11b/CD18 also interacts with several plasma proteins and vascular endothelial cell receptors, including high-molecular-weight kininogen (Gustafson et al., 1989), factor X (Altieri et al., 1988), complement factor H (DiScipio et al., 1998), GPIb (Simon et al., 2000), uPAR (Simon et al., 2000), and E-selectin (Kotovuori et al., 1993). However, none of these molecules have been demonstrated to act as an epithelial adhesive counterreceptor for CD11b/CD18 in a physiolocally appropriate manner.
Recently, members of the junctional adhesion molecule family (JAMs) have been reported to have roles in leukocyte transmigration (Martin-Padura et al., 1998; Del Maschio et al., 1999). Although this is a growing family of proteins, to date there are four members that were originally designated as JAM or JAM-1 (Martin-Padura et al., 1998; Del Maschio et al., 1999; Ozaki et al., 1999; Liu et al., 2000; Sobocka et al., 2000), JAM-2 (Aurrand-Lions et al., 2000; Cunningham et al., 2000), JAM-3 (Arrate et al., 2001; Santoso et al., 2002), and JAM-4 (Hirabayashi et al., 2003). Very recently, new nomen-clature was proposed designating the above proteins as JAM-A, JAM-B, JAM-C, and JAM-D, respectively (Bazzoni, 2003). In general, JAM proteins are type I transmembrane receptors belonging to the immunoglobulin superfamily (IgSF; Aurrand-Lions et al., 2001; Chavakis et al., 2003). Current data supports a role of JAM molecules as cell-cell adhesive receptors through homophilic or heterophilic interactions between JAMs and other other integrins (Cunningham et al., 2000; Liang et al., 2002). Because of their unique localization at tight junctions (TJs) and lateral cell membranes, JAM proteins are attractive candidate receptors for leukocytes as they migrate across endothelial and epithelial monolayers. Indeed, studies on murine JAM-A by Dejana and coworkers (Martin-Padura et al., 1998; Del Maschio et al., 1999) demonstrated that anti–JAM-A antibody inhibited transendothelial migration of monocytes and PMN in vitro and in vivo. Subsequently, Ostermann et al. (2002) reported that JAM-A binds specifically to CD11a/CD18 and mediates T-cell interactions with endothelial cells. In a recent report by Santoso et al. (2002) platelets expressing JAM-C were shown to mediate neutrophil-platelet adhesion. Using endothelioma cells transfected with JAM-C, Johnson-Leger et al. (2002) reported that JAM-C was able to facilitate lymphocyte transendothelial migration. Although these latter observations suggest that JAM-C is an attractive candidate receptor for migrating PMN, the expression and biological function of JAM-C in epithelia is currently unknown.
In this study, we report that JAM-C is abundantly expressed in intestinal epithelial cells and, in contrast to other JAMs, is a novel component of epithelial desmosomes. Furthermore, we demonstrate that JAM-C is a ligand for CD11b/CD18 during PMN migration across epithelial monolayers. The significance of these findings in the context of mucosal inflammation and epithelial intercellular junctions is discussed.
MATERIALS AND METHODS
Chemicals and Antibodies
Human FBG was purchased from Sigma (St. Louis, MO). Goat anti-human JAM-3 antibody (hJ3G) was obtained from R&D Systems (Minneapolis, MN). mAb against human JAM-C (LUCA14) was a kind gift from Raven biotechnologies, inc. (South San Francisco, CA). Both of these antibodies bind to the extracellular domain of JAM-C, and LUCA14 blocks binding of JAM-B to JAM-C (unpublished data). Mouse antiserum against human JAM-C was raised by immuonizing mice with JAM-C/Fc chimera in our lab using methods as previously described (Liu et al., 2000). Functionally inhibitory anti-CD11b mAb (CBRM1/29, IgG1) was used as previously described (Balsam et al., 1998). A polyclonal antibody against human CD11b (R7928A) was raised by immunizing rabbit with a peptide DMMSEGGPPGAEPQ corresponding to the C terminus of CD11b subunit (Jesaitis et al., 1990). As controls, functionally inhibitory mAbs to CD11a (TS1/22, subclass IgG1; Sanchez-Madrid et al., 1982), CD11c (4G1; Stacker and Springer, 1991), and CD18 (TS1/18, subclass IgG1; Sanchez-Madrid et al., 1982) were also used. Anti-human JAM-A (clone J10.3, J10.4) and anti-ZO-1 antibodies were used as previously described (Liu et al., 2000; Nusrat et al., 2000; Ivanov et al., 2004). Anti-desmoplakin (DP) antibody was obtained from Serotex (Oxford, United Kingdom) and used as previously described (Arnemann et al., 1993). HRP-conjugated or Alexa Fluor 488 (495/519) and Alexa Fluor 568 (578/603)-conjugated secondary antibodies and TOPO were obtained from Molecular Probes (Eugene, OR). All other reagents were purchased from Sigma.
Cells
Human intestinal epithelial T84 cells (passages 62–70) were grown in a 1:1 mixture of DMEM and Ham's F-12 medium supplemented with 15 mM HEPES buffer (pH 7.5), 14 mM NaHCO3, 40 μg/ml penicillin, 8 μg/ml ampicillin, 90 μg/ml streptomycin, and 6% newborn calf serum. For transmigration experiments, cells were grown on collagen-coated, permeable polycarbonate filters (5-μm pore size; Costar, Cambridge, MA) as previously described (Parkos et al., 1996). PMN were isolated from the whole blood of normal human volunteers by ficoll/dextran sedimentation as previously described (Parkos et al., 1996). Isolated PMN were resuspended in modified HBSS devoid of calcium or magnesium (HBSS-; 4°C) at a concentration of 5 × 107 cells/ml and used within 4 h of isolation.
Soluble Human JAM-C/Fc Chimera Preparation
cDNA encoding the human JAM-C extracellular domain (amino acid residues 1–242, including V- and C-Ig loop; Arrate et al., 2001; Liang et al., 2002; Santoso et al., 2002) was amplified by PCR using sense primer 5′-ATATAAGCTTTCAGCAACCCTCGACATG-3′ and antisense primer 5′-ATGCGGATCCGTCATAGACTTCCATCTC-3′ from a T84 cell cDNA library prepared as below. Briefly, RNA was isoloated from T84 cells using an RNeasy Mini-prep kit (Qiagen, Chatsworth, CA) and treated with RNase-free DNase (Promega, Madison, WI). After reverse transcription (Advantage RT-for-PCR kit; BD Biosciences Clontech, San Diego, CA) cDNA encoding the JAM-C extracellular domain was fused to a modified rabbit IgG1 Fc region using a BamHI digestion site. The cDNA was then cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA) and transfected into COS-7 using DEAE-Dextran (Liu et al., 2000). JAM-C/Fc was affinity purified from transfected cell culture supernatants by protein A-Sepharose (Sigma) and eluted with 100 mM glycine/HCl, pH 4.0, followed by neutralization, concentration, and dialysis. A second JAM-C/Fc chimera containing human IgG1 Fc fragment (Fc/JAM-C2) was purchased from R&D Systems (Minneapolis, MN). As a control, the Fc portion of fusion protein was also prepared. Briefly, cDNA encoding the human JAM-A signal peptide (amino acid residues 1–27; Liu et al., 2000) was amplified by PCR using sense primer 5′-ATATAAGCTTTCCTTCGGCGGCTGTTGT-3′ and antisense primer 5′-ATGCGGATCCGCCCAATGCCAGGGAGCA-3′ from T84 cell cDNA library. The cDNA encoding the JAM-A signal peptide was fused to a modified rabbit IgG1 Fc region and then cloned into pcDNA3.1 (Invitrogen). Fc peptide was affinity-purified from transfected COS-7 culture supernatants as described above.
Immunofluorescence
T84 monolayers cultured on permeable supports were fixed and permeabilized with cold ethanol (-20°C, 20 min) and labeled with primary antibodies followed by fluorescence-conjugated secondary antibodies using previously described methods (Nusrat et al., 2000; Edens et al., 2002). Monolayers were then mounted in ProLong antifading embedding solution (Molecular Probes) and analyzed using a Zeiss Laser Scanning microscope LSM510 (Zeiss Microimaging, Inc., Thornwood, NY). A 100× NA/1.3 Plan-Neofluar oil immersion lens (Zeiss) was used for all images. Images shown were representative of at least three experiments, with multiple images taken per slide. For Z-series, optical sections were recorded at 0.5-μm intervals. As a control for background labeling, monolayers were incubated with comparable concentrations of irrelevant IgG and secondary antibody. For tissue staining, 5 μm frozen tissue sections of human colon obtained from discarded surgical resection specimens within the Emory Pathology Department (Emory University, Atlanta, GA) were mounted on glass coverslips, air-dried, and fixed in 100% ethanol (-20°C, 20 min) before fluorescent labeling and analysis with confocal microscopy as above.
Desmosome and Tight Junction Disruption Experiments
Experiments aimed at selective disruption of desmosomes or tight junctions were performed to determine which structures JAM-C affiliates with. Because desmosomes are more resistant to short-term calcium-depletion than tight junctions (Watt et al., 1984; Mattey and Garrod, 1986; Sandig et al., 1990), calcium-depletion (Siliciano and Goodenough, 1988) was performed to disrupt epithelial monolayer tight junction structures. T84 monolayers were washed twice with cold HBSS- and incubated with calcium-free Eagle's minimum essential medium (S-MEM; Sigma) supplemented with 2 mM EGTA, 10 mM HEPES, 14 mM NaHCO3, 40 μg/ml penicillin, 90 μg/ml streptomycin, and 5% dialyzed newborn calf serum for 1 h at 37°C. Monolayers were then fixed and labeled for TJ markers and JAM-C and analyzed by confocal microscopy. In parallel, because desmosome structure is dependent on intermediate filaments (IFs) and IFs can be disrupted by acrylamide (Aggeler and Seely, 1990; Shabana et al., 1994; Salas, 1999), we studied the localization of JAM-C in T84 monolayers after acrylamide treatment. For acrylamide treatment, T84 monolayers were washed twice with HBSS and incubated with 5 mM acrylamide in DMEM for 2 h at 37°C. Calcium-depleted or acrylamide-treated T84 monolayers were then rinsed with cold HBSS- and fixed/permeabilized with cold ethanol (20 min, -20°C) followed by immunofluorescence labeling with antibodies against JAM-A, -C, ZO-1, and DP, respectively.
Immuno-gold Labeling and Electron Microscopy
To verify the desmosomal localization of JAM-C at an ultrastructural level, T84 monolayers were cooled on ice and permeabilzed with cold 0.05% saponin (ICN Biochemicals, Cleveland, OH) in HBSS for 10 min at 4°C. After blocking with 3% BSA in HBSS at 4°C monolayers were washed and incubated with LUCA-14 (1:50 dilution in HBSS with 3% BSA) for 1 h at 4°C. Unbound antibody was washed away and monolayers were incubated with gold-conjugated anti-mouse antibody in 3% BSA for 2 h at 4°C followed by fixation in 3.7% paraformaldehyde after removal of unbound antibody. Washed monolayers were then fixed in 2% glutaraldehyde for 1 h and washed, and silver enhancement was performed as previously described (Wageningen, The Netherlands; Sesack and Snyder, 1995). After osmification, monolayers were washed with PBS before dehydration and embedding in Eponate 12 resin (Ted Pella, Redding, CA). Ultrathin sections were cut at 70 nm and examined on a Hitachi H-7500 transmission electron microscope (Pleasanton, CA). Primary antibodies were omitted from negative control samples.
Transmigration Experiments
PMN transepithelial migration experiments were performed using confluent, high-resistance T84 cell monolayers as previously described (Parkos et al., 1996; Liu et al., 2001) with a minor modification. To assay PMN migration acoss T84 monolayers in the physiologically relevant basolateral to apical direction, monolayers were preincubated with antibodies (25 μg/ml in HBSS in the upper chamber) for 30 min at 37°C before addition of PMN suspension followed by time course migration assay as previously described (Liu et al., 2001). To assay PMN associated with epithelial monolayers, PMN transmigration was stopped at indicated time points, and monolayers were washed free of bound PMN, followed by solublization in 0.1 M citrate buffer, pH 4.0, containing 0.5% Triton X-100. Lysates were then assayed for myeloperoxidase (MPO) and compared with standards of known amounts of PMN (Parkos et al., 1996).
SDS-PAGE/Western Blot
T84 cell and PMN (1 × 106 cells) were solubilized in sample buffer and boiled under reducing conditions. Samples were separated by SDS-PAGE (12% acrylamide gel) and electrophoretically transferred onto nitrocellulose membranes. Membranes were blocked with 10% nonfat milk in TTBS (30 min) and incubated with anti–JAM-C antibody for 1 h. After washing with TTBS, nitrocellulose transfers were incubated with HRP-conjugated secondary antibody followed by ECL detection.
CD11b/CD18-JAM-C Binding Assay
The binding of JAM-C to CD11b/CD18 was studied using two reciprocal methods. First, we tested the binding of JAM-C/Fc chimera to immobilized CD11b/CD18. Functionally active CD11b/CD18 was purified to homogeneity from large quantities of human PMN (Diamond et al., 1995) and then added to 96-well flat-bottom microtiter plates at a concentration of 5 μg/ml in HBSS for protein immobilization as previously described (Zen et al., 2002). After blocking the wells with 1% BSA for 1 h, JAM-C/Fc chimera (10 μg/ml) was added and incubated for 1 h at 37°C in the presence or absence of inhibitors. Wells were washed three times with HBSS containing 1% BSA, and the bound JAM-C was detected by anti–JAM-C mAb followed by HRP-conjugated secondary antibody and colorimetric measurement. As a reciprocal method, CD11/CD18 was assayed for binding to immobilized JAM/Fc chimeras and other known CD11b binding proteins. After washing off unbound JAM/Fc chimeras and other CD11b ligands, protein-coated microtiter wells were blocked with 1% BSA and CD11b/CD18 (5 μg/ml) in HBSS containing 0.1% Triton X-100 was then added. After 1-h incubation at 37°C, wells were washed three times with HBSS containing 0.1% Triton X-100 followed by incubation with polyclonal antibody R7928A (1/200 dilution) and HRP-conjugated secondary antibody. Bound CD11b was detected colorimetrically by addition of substrate (ABTS). Wells coated with BSA only served as controls.
Cell Adhesion Assay
Adhesion assays of suspensions of T84 cells to purified CD11b/CD18 were performed using CD11b/CD18-coated microtiter plates as previously described (Balsam et al., 1998; Zen et al., 2002). For these assays, functionally active CD11b/CD18 was purified to homogeneity from large quantities of human PMN (Diamond et al., 1995) and then added to microtiter wells at a concentration of 5 μg/ml in HBSS for protein immobilization followed by blocking with 1% BSA. T84 confluent monolayers were dissociated from culture dishes by nonenzymatic cell dissociated solution (Sigma) followed by labeling with BCECF-AM (Molecular Probes; Balsam et al., 1998). Labeled T84 cells were added to CD11b/CD18-coated wells (∼2.5 × 105 cells/well in a total volume of 150 μl) in the absence or presence of inhibitors followed by stationary incubation (1 h, 37°C) to allow cell adhesion. The fluorescence intensity in each well was measured using a fluorescence microtiter plate reader at excitation/emission wavelengths of 485/535 nm (Millipore, Milford, MA) before and after washing. Cell adhesion was calculated as percentage of total applied cells (Zen et al., 2002). PMN adhesion to immobilized JAM-C/Fc or Fc only (as a control) was also measured in the presence of functionally inhibitory antibodies against CD11a (TS1/22), CD11b (CBRM1/29), CD11c (4G1), and CD18 (TS1/18). Briefly, microtiter plates were incubated with 10 μg/ml JAM-C/Fc or Fc in HBSS followed by block with 3% BSA. Isolated PMN were labeled with BCECF-AM (Molecular Probes) for 15 min at 37°C and ∼2.0 × 105 cells were added to JAM-C/Fc or Fc coated wells containing 150 μl HBSS and test antibodies (10 μg/ml). After incubation for 1 h at 37°C, the wells were washed, and adherent cells assayed in a fluorescence microtiter plate reader.
Statistics
Data are presented as the mean ± SE and were compared by Student's t test.
RESULTS
Identification of JAM-C as a Novel Component of Epithelial Cell Desmosomes
Human JAM-C cDNA predicts a 310-amino acid (aa) residue precursor protein with a putative 31-aa signal peptide, a 210-aa extracellular region containing two Ig domains, a 23-aa transmembrane domain, and a 46-aa cytoplasmic domain containing a PDZ-binding motif and a PKC phosphorylation site (Arrate et al., 2001; Liang et al., 2002). It has been reported that JAM-C is widely expressed and is found in the placenta, brain, kidney, and heart. At the cellular level, JAM-C has been shown to be expressed on endothelial cells, platelets, T-cells, and NK cells (Arrate et al., 2001; Johnson-Leger et al., 2002; Liang et al., 2002). In the present study, we found that JAM-C was expressed on human intestinal epithelial cells but not on human peripheral PMN. As shown in Figure 1A, cDNA encoding the human JAM-C extracellular domain (V- and C-type Ig domains and signal peptide) was amplified from a human intestinal epithelial cell (T84 cell) cDNA library but not from a human leukocyte cDNA library (BD Clontech, Palo Alto, CA). For comparison, the cDNA encoding human JAM-A extracellular domain was amplified from both T84 cell and leukocyte cDNA libraries. At the protein level, T84 cell monolayers were strongly labeled by anti–JAM-C antibody in a pattern characteristic of intercellular junction proteins (Figure 1B). However, freshly isolated human PMN that were brightly labeled by anti–JAM-A antibody were not labeled by anti–JAM-C antibody (Figure 1B). By Western blot, a ∼43-kDa JAM-C protein was in T84 cell lysates but not in lysates from PMN (Figure 1C). Similar cell labeling and blotting results were obtained using mouse antiserum generated against a recombinant form of human JAM-C extracellular domain (unpublished data). The results demonstrate expression of JAM-C in human intestinal epithelia but not in human peripheral PMN. The absence of JAM-C in PMN is also in agreement with a recent report by Santoso et al. (2002).
Figure 1.
JAM-C is expressed in human intestinal epithelial cells but not in neutrophils. (A) cDNAs encoding the extracellular domains of JAM-A and JAM-C were amplified from a T84 cell cDNA library and human leukocyte cDNA library, respectively. Note that both JAM-A and JAM-C were amplified from a T84 cell cDNA library, whereas only JAM-A was amplified from the leukocyte library. (B) Immunofluorescence labeling of T84 monolayers and PMN with anti–JAM-A or anti–JAM-C antibody. Bar, 10 μm. (C) Western blots of T84 cell lysates and PMN lysates with anti–JAM-A or anti–JAM-C antibody. Note the ∼43-kDa protein band was recognized in T84 cells but not PMN.
Expression of JAM-C in epithelial cells was further investigated in immunolabeling labeling experiments of natural human colonic epithelium from surgical resection specimens. The localization of JAM-C was compared with those of known TJ proteins, JAM-A (Martin-Padura et al., 1998) and ZO-1 (Stevenson et al., 1986; Anderson et al., 1988; Siliciano and Goodenough, 1988). As shown in Figure 2, JAM-C is abundantly expressed in human colonic epithelial cells (red). Double-labeling with anti–JAM-C/anti–JAM-A antibodies demonstrated that the majority of JAM-C (red) did not colocalize with JAM-A (green). Areas of apparent colocalization on the basolateral membrane could be attributed to a minor nontight junctional pool of JAM-A that has been previously reported on epithelial cells (Liu et al., 2000). Double-labeling with anti–JAM-C (red)/anti-ZO-1 (green) antibody demonstrated completely different labeling patterns. This pattern of labeling suggested that JAM-C is not localized to epithelial TJs. Because the subjunctional “beaded” staining pattern observed for JAM-C (Figure 1B) was similar to that observed for desmosomes, we compared the localization of JAM-C to that of desmoplakin (DP), a specific desmosomal marker. The localization pattern of JAM-C (red) was indistinguishable from that of DP (green), indicating that JAM-C colocalized with the epithelial desmosome marker and might be a desmosomal component.
Figure 2.
Localization of JAM-C in normal human colonic mucosa. Intestinal mucosa tissue sections were double labeled with anti–JAM-C antibody LUCA14 (in red) and anti–JAM-A antibody (in green), anti-ZO-1 antibody (in green) or antidesmoplakin (DP) antibody (in green), respectively. In some tissue stains, cell nuclei were labeled with TOPRO (in blue). Bars, 20 μm.
We also examined the expression and localization of JAM-C in T84 cell monolayers cultured on permeable supports by double immunolabeling and confocal fluorescence microsopy. As shown in the X-Z images in Figure 3, anti–JAM-C labeling is clearly observed below anti–JAM-A or anti-ZO-1 labeling, suggesting JAM-C is localized underneath epithelial TJs. En face images taken in the X-Y plane at the level of the apical junction complex demonstrate a general lack of colocalization of JAM-C with JAM-A or ZO-1. In contrast, there was striking colocalization between JAM-C (red) and desmoplakin (DP; green) in T84 cell monolayers.
Figure 3.
Localization of JAM-C in T84 cell monolayers cultured on permeable filters. T84 monolayers were double labeled with anti–JAM-C mAb (in red) and with anti–JAM-A (in green), anti-ZO-1 (in green) or anti-DP antibody (in green), respectively. To the right, from the apical (A) to basolateral (B) surface of T84 monolayers (as shown in X-Z images), JAM-C was generally found beneath JAM-A and ZO-1. However, JAM-C showed an identical labeling pattern as with DP. Bar, 20 μm.
To further investigate the possibility that JAM-C is a desmosomal protein and not affiliated with TJs, as is the case for JAM-A, experiments were performed to selectively disrupt desmosomes and TJs followed by immunofluorescence localization of JAM-C, TJ and desmosome proteins. Because epithelial desmosomes are more resistant to disruption by Ca2+ removal than TJs (Watt et al., 1984; Mattey and Garrod, 1986; Sandig et al., 1990), we depleted Ca2+ in culture medium and labeled T84 monolayers with antibodies against JAM-C and JAM-A, ZO-1 and DP, respectively. As shown in Figure 4 and consistent with our previous findings (Ivanov et al., 2004), depletion of Ca2+ for 1 h resulted in disruption and internalization of TJs as highlighted by JAM-A (green) and ZO-1 (green) staining. Note the lack of colocalization of JAM-C with either of the TJ markers. In contrast, the junctional staining pattern of JAM-C (red) was less affected by Ca2+ depletion and was indistinguishable from that observed for DP (green).
Figure 4.
Transient calcium-depletion disrupts epithelial TJs more rapidly than desmosomes and JAM-C. T84 cell monolayers were incubated with 2 mM EGTA in S-MEM for 1 h at 37°C and then fixed/permeabilized with cold ethanol followed by immunofluorescence labeling with antibodies against JAM-C (in red), JAM-A (in green), ZO-1 (in green), and DP (in green). Note that ZO-1 and JAM-A were significantly internalized, whereas JAM-C and DP remain colocalized and maintain an intercellular junctional staining pattern. Bar, 10 μm.
Because it is well known that perturbation of intermediate filaments (IFs) results in disruption of desmosome integrity (Fuchs and Cleveland, 1998; McMillan and Shimizu, 2001) but not TJs, in a parallel set of experiments, we used acrylamide to selectively disrupt IFs and desmosome structure (Aggeler and Seely, 1990; Shabana et al., 1994; Salas, 1999). As seen in Figure 5, disruption of epithelial IFs resulted in little change in TJ staining patterns of JAM-A and ZO-1. However, the crisp junctional staining patterns of JAM-C and DP became dispersed when compared with before treatment.
Figure 5.
Disruption of IFs distorts JAM-C localization in T84 monolayers. T84 monolayers were incubated with DMEM (control) or 5 mM acrylamide in DMEM for 2 h at 37°C and then fixed/permeabilized with cold ethanol. The monolayers were immunofluorescently labeled with antibodies against JAM-A, ZO-1, JAM-C, and DP, respectively. Note that the localization of JAM-A and ZO-1 was not significantly affected by acrylamide treatment, however, the labeling pattern of anti–JAM-C and anti-DP show distortion (blurring) of the junctional staining pattern. Bar, 10 μm.
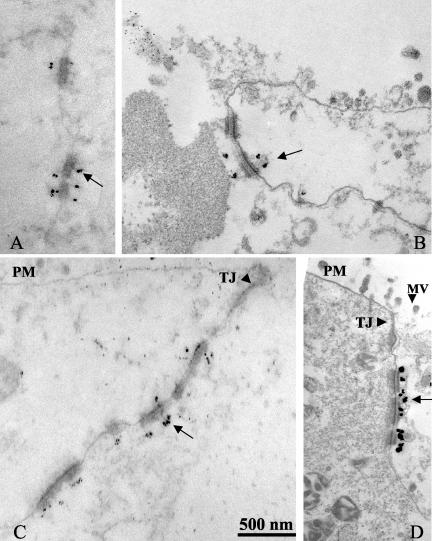
We also performed immunogold labeling of ultrathin sections of T84 monolayers to analyze JAM-C staining at the ultrastructual level. As shown in Figure 6, anti–JAM-C antibody labeling was concentrated at desmosomes (Figure 6, A–D), whereas no specific staining was found in TJs (Figure 6, C and D). Taken together, the above results indicate that JAM-C is not a TJ-associated protein but a novel desmosomal-associated protein in intestinal epithelia.
Figure 6.
Immunoelectron microscopy of JAM-C in T84 monolayers cultured on permeable supports. Monolayers were gently permeabilized using 0.1% saponin at 4°C and incubated with anti–JAM-C antibody followed by gold particle conjugated secondary antibody. Monolayers were then fixed and prepared for electron microscopy as detailed in MATERIALS AND METHODS. (A–D) Four electron micrographs of anti–JAM-C antibody/gold labeling in epithelial monolayers. Note that gold particles are concentrated at desmosomal structures (A–D, arrows), whereas microvilli (MV; D, arrowhead), plasma membrane (PM; C and D), and tight junction (TJ; C and D, arrowheads) are devoid of gold particle labeling. Bar, 0.5 μm.
JAM-C Regulates PMN Transepithelial Migration
Given the basolateral localization of JAM-C with desmosomes and reports of JAM-C involvement in leukocyte interactions with endothelia and platelets (Johnson-Leger et al., 2002; Santoso et al., 2002), we tested whether JAM-C plays a role in PMN transepithelial migration. Using T84 epithelial cell monolayers cultured on the underside of permeable supports, we tested whether anti–JAM-C antibodies had inhibitory effects on PMN transepithelial migration in the physiologically appropriate basolateral-to-apical (b-to-a) direction. For these experiments, inverted T84 monolayers were preincubated with JAM-C reagents for 30 min followed by assessment of the time-course transepithelial migration in the presence of antibodies/fusion proteins. As shown in Figure 7, PMN migration in the control group (no antibodies) or in the presence of isotype matched IgG, was rapid, with the majority of PMN migrating across T84 monolayers during the first 1 h (46.1 ± 4.7% for no antibody and 49.2 ± 6.1% for control IgG, respectively). Compared with the migration in control conditions, addition of anti–JAM-C antibodies hJ3G and LUCA14 resulted in significantly reduced PMN migration into the lower chamber during the early migration time periods. In particular, at 60-min incubation, both anti–JAM-C antibodies inhibited PMN transepithelial migration by ∼50% (21.2 ± 2.7% for LUCA14 and 24.8 ± 1.7 for hJ3G compared with 46.1 ± 4.7% for no antibody and 49.2 ± 6.1% for control IgG, respectively). Thus, the time course of total PMN migration demonstrated a decrease in the rate of b-to-a PMN transepithelial migration with anti–JAM-C treatment. Interestingly, anti–JAM-A antibody (J10.4) had no effect on the kinetics of PMN transepithelial migration, which is in agreement with our previous observations (Liu et al., 2000).
Figure 7.
Effects of JAM-C on PMN transepithelial migration in basolateral-to-apical (b-to-a) direction. The figure shows a time-course assay of PMN migration (b-to-a) across T84 monolayers (Liu et al., 2001) in the presence of a transepithelial gradient of fMLP (10-6 M). PMN migration into the lower chamber after each time point was quantified by MPO assay. For these experiments, antibodies (25 μg/ml each) were added to the upper assay chamber (basolateral epithelial surface) 30 min before addition of PMN. Data represent mean ± SE of three independent experiments performed in duplicate (p < 0.01).
In parallel experiments, we also measured the number of PMN associated with T84 monolayers at different time points during b-to-a transepithelial migration. As shown in Table 1, in the presence of no antibody (control) or anti–JAM-A mAb (J10.4), the number of monolayer-associated PMN decreased after 1 h. However, in the presence of anti–JAM-C antibody, the number of monolayer-associated PMN remained high after 1 h and is consistent with the reduced numbers of PMN observed to transmigrate into the lower chambers.
Table 1.
PMN association with T84 monolayers during transepithelial migration
| PMN associated with monolayers
|
|||
|---|---|---|---|
| Migration time (min) | No Ab | +J10.4 | +LUCA14 |
| 15 | 31.5 ± 3.7 | 32.7 ± 4.5 | 34.1 ± 3.7 |
| 30 | 37.4 ± 2.4 | 37.7 ± 3.9 | 51.7 ± 6.3a |
| 60 | 12.7 ± 4.1 | 17.3 ± 4.9 | 30.3 ± 9.1a |
| 90 | 7.1 ± 3.9 | 6.9 ± 1.3 | 20.5 ± 6.7a |
PMN migration across T84 monolayers (from basolateral to apical direction, b-to-a) was terminated at various time points and PMN within monolayers were quantified by MPO assay (n = 3).
p < 0.005 compared with no Ab group
JAM-C Serves as an Adhesive Ligand on Epithelial Cells for CD11b/CD18
Given our observations above and a recent report of interactions between JAM-C on platelets and neutrophils via CD11b/CD18 (Santoso et al., 2002), we performed experiments to determine whether JAM-C represents an epithelial ligand for CD11b/CD18 during PMN transepithelial migration. To define the interaction between human epithelial JAM-C and CD11b/CD18, we performed protein-protein binding assays using purified CD11b/CD18 and JAM-C/Fc chimeras. As shown in Figure 8A, CD11b/CD18 specifically bound to immobilized JAM-C/Fc chimeras, and the binding was comparable to the binding of CD11b/CD18 to one of its known ligands, FBG (Altieri et al., 1988). No binding interaction between CD11b/CD18 and soluble JAM-A/Fc chimera was detected, indicating that our above results were not due to binding interactions with the Fc portion of chimeras. We also immobilized CD11b/CD18 onto microtiter plates and probed with JAM-C/Fc chimera in the presence of antibodies and inhibitors. In these experiments, binding was detected using a goat anti-rabbit Fc antibody. As shown Figure 8B, the binding of JAM-C/Fc chimera to CD11b/CD18 was blocked by anti–JAM-C antibodies (hJ3G and LUCA14) but was not affected by anti–JAM-A antibody. The binding was also blocked by CBRM1/29 or FBG, suggesting that binding between JAM-C and CD11b is mediated by the I domain of CD11b. We also tested the effect of EDTA on binding because adhesive properties of the I domain are Mg2+ dependent (Michishita et al., 1993). As can be seen, chelation of the divalent cations with EDTA also blocked the binding of JAM-C/Fc chimera to CD11b/CD18.
Figure 8.
Direct binding of human JAM-C to CD11b/CD18. (A) JAM-A/Fc, JAM-C/Fc, fibrinogen, and BSA were immobilized in microtiter wells to which was added purified CD11b/CD18 diluted in HBSS. After a 1-h incubation at 37°C, bound CD11b/CD18 was detected by anti-CD11b antibody (LM2/1) followed by HRP-conjugated goat anti-mouse (Fab′) antibody (detailed in MATERIALS AND METHODS). (B) Purified CD11b/CD18 was immobilized in microtiter wells followed by addition of JAM-C/Fc chimera in the presence of different antibodies/inhibitors. After a 1-h incubation at 37°C, JAM-C/Fc bound to CD11b was detected by HRP-conjugated antirabbit Fc antibody. JAM-A/Fc served as a control in the assay. Data represent mean ± SE of three independent experiments performed in duplicate (p < 0.01).
To further define the specificity of JAM-C binding to CD11b/CD18, we assessed adhesion of fluorescently labeled PMN to immobilized JAM-C/Fc in the presence of a variety of inhibitory anti-β2 integrin antibodies with defined epitopes on CD11a, CD11b, CDllc, and CD18, respectively. For these assays, 50–60% of added PMN adhered to JAM-C/Fc coated microtiter plates with <10% of PMN adherent to wells coated with Fc only. Adhesion of PMN to JAM-C/Fc–coated plates was reduced to background levels in the presence of anti-CD11b (CBRM1/29), anti-CD18 (TS1/18), and anti–JAM-C, whereas anti-CD11a (TS1/22) and anti-CD11c (4G1) antibodies had no effect (unpublished data). Taken together, these data demonstrate that JAM-C binds specifically and selectively to CD11b to mediate PMN adhesion.
Additional experiments were performed to evaluate the role of JAM-C in T84 cell adhesion to immobilized CD11b/CD18. Previously, we have reported that T84 cells strongly adhere to immobilized CD11b/CD18 in a specific manner (Balsam et al., 1998; Zen et al., 2002). Thus, we asked whether the adhesion of T84 cells to CD11b/CD18 is mediated through interactions with JAM-C. For these experiments, T84 cell monolayers were elicited using a nonenzymatic cell dissociation solution (Sigma) and fluorescently labeled with BCECF as previously described (Zen et al., 2002). Suspensions of labeled cells were then applied to microtiter wells coated with CD11b/CD18 in the presence or absence of antibodies or Fc chimeras and assayed for adhesion.
As shown in Figure 9, ∼50% of applied T84 cells were adherent to CD11b/CD18 after a 1-h adhesion assay (51.2 ± 6.5%). Specificity of T84 cell adherence to CD11b/CD18 was confirmed by near complete inhibition with anti-CD11b mAb CBRM1/29 (5.6 ± 1.3%, p < 0.005). As can be seen, both anti–JAM-C antibodies hJ3G and LUCA14 significantly inhibited T84 cell adhesion (21.4 ± 3.1% for hJ3G and 23.1 ± 2.9% for LUCA14, respectively) but to a lesser extent than that observed with anti-CD11b. Adhesion was not affected by anti–JAM-A antibody (J10.4). Soluble JAM-C/Fc chimeras (JAM-C /Fc and JAM–C2/Fc) also reduced the adhesion of T84 cells to CD11b/CD18, presumably by competition for CD11b binding (19.3 ± 2.1% for JAM-C/Fc and 21.4 ± 2.9% for JAM-C2/Fc, respectively; p < 0.01). No inhibition of adhesion was observed with JAM-A/Fc. Taken together, these results indicate that JAM-C mediates T84 cell adhesion to CD11b/CD18. Residual adhesion of T84 cells in the presence of anti–JAM-C or JAM-C/Fc is consistent with the existence of additional epithelial ligands for CD11b/CD18.
Figure 9.
Adhesion of T84 cells to immobilized CD11b/CD18. Suspensions of T84 cells were fluorescently labeled with BCECF-AM and added to CD11b/CD18-coated microtiter wells in the presence or absence of different inhibitors for a 1-h incubation at 37°C as described in MATERIALS AND METHODS. Bound cells were then determined by fluorescence intensity before and after three washes and cell adhesion calculated by the fluorescence ratio as detailed in MATERIALS AND METHODS. Data represent mean ± SE of three independent experiments performed in duplicate (p < 0.01).
DISCUSSION
JAM-C Is a Novel Desmosomal-associated Membrane Protein of Human Epithelial Cells
The JAM family of proteins have generally been reported to localize to intercellular junctions of polarized endothelial and epithelial cells with some members also expressed on circulating blood cells (Martin-Padura et al., 1998; Liu et al., 2000; Arrate et al., 2001; Aurrand-Lions et al., 2001; Santoso et al., 2002). Most JAM proteins contain a PDZ domain binding motif at the C-terminus, which is thought to mediate junctional localization through interactions with PDZ domain containing TJ scaffolding proteins (Liu et al., 2000; Amieva et al., 2003). At the cellular level, JAM-C has been reported to be expressed in endothelial cells of high endothelial venules of human tonsil, platelets, T cells, and NK cells (Arrate et al., 2001; Johnson-Leger et al., 2002; Liang et al., 2002).
In the present study, we demonstrate that human JAM-C is strongly expressed in human intestinal epithelia. In cultured epithelial cells or natural human colonic epithelium, JAM-C is expressed at desmosomes but not at TJs, as it is for JAM-A. The desmosomal localization of JAM-C was confirmed by electron microscopy after immunogold labeling and in experiments targeting selective disruption of TJs and desmosomes. It is not clear how JAM-C is targeted to desmosomes or whether JAM-C is directly linked to IFs, even though disruption of IFs with acrylamide was observed to distort the junctional staining pattern of JAM-C. JAM-C has been reported to associate with scaffolding proteins such as ZO-1 and PAR-3 in a PDZ domain–dependent manner (Ebnet et al., 2003). However, the role of scaffold protein association in regulating JAM-C desmosomal localization remains to be determined. Because other JAM family protein members also possess PDZ domain–binding motifs and bind to ZO-1 and PAR-3 but have different cellular localization, it is likely that other factors contribute to JAM-C localization to desmosomes, such as binding to undefined desmosomal scaffolding protein(s) or association with known desmosomal components such as DP and desmosomal cadherins including desmoglein (Troyanovsky et al., 1994) and desmocollins (Nuber et al., 1996). By analyzing cluster formation of desmoglein 2 and desmosome structure in cultured epithelial cells, Koeser et al. (2003) reported that desmosomal proteins desmoplakin, plakoglobin, and plakophilin 2 were all necessary and each plaque protein played a unique role in de novo formation of desmosomes. As a novel component of epithelial cell desmosomal structure, JAM-C may also participate in the assembly and organization of epithelial desmosomes and directly interact with other desmosomal components. However, further studies are clearly necessary to answer these questions.
JAM-C–binding Interactions with CD11b/CD18 as One of Multiple Steps Involved in PMN Transepithelial Migration
Using T84 cell monolayers cultured on permeable supports as model epithelia, we have shown that JAM-C plays a role in regulating PMN transepithelial migration through direct binding interactions with CD11b/CD18. Because JAM-C has not been shown to bind to itself in a homophilic manner (Arrate et al., 2001) as has been shown for JAM-A (Bazzoni et al., 2000; Liang et al., 2000) and JAM-B (Cunningham et al., 2000), it is unlikely that the inhibitory effects of JAM-C blockade on PMN transmigration that we observe are mediated by disruption of epithelial JAM-C/JAM-C interactions. It has been reported that JAM-B (VE-JAM/JAM-2) binds to JAM-C in heterophilic manner to mediate cell-cell adhesion (Arrate et al., 2001; Liang et al., 2002). However, this binding interaction does not play a role in PMN transepithelial migration because PMN do not express JAM-B (VE-JAM/JAM-2; Arrate et al., 2001; Liang et al., 2002). Furthermore, our protein binding data suggests that JAM-C binds to the I domain of CD11b in a divalent cation–dependent manner.
Analysis of the kinetics of PMN migration (Figure 7) across monolayers indicate that anti–JAM-C antibodies decreased the rate of PMN migration but did not block transmigration in a sustained manner. Given that treatment with anti–JAM-C antibody did not reduce the numbers of PMN associated with monolayers at early stages of transmigration (Table 1), it is likely that JAM-C is not involved in initial adhesion of PMN to epithelial monolayers but plays a role in subsequent adhesion-dependent transmigration. This is consistent with the requirement of preincubation of T84 monolayers with anti–JAM-C (25 μg/ml) before transmigration assays, presumably to allow for antibody access to subjunctional desmosomes.
Our findings suggest that JAM-C mediates CD11b/CD18-dependent PMN transepithelial migration at points subsequent to initial adhesion. Because PMN migrate under physiologically relevant conditions in the basolateral to apical direction, initial adhesive interactions between PMN and epithelia must be mediated by epithelial molecules located at the basal aspect of the epithelium, which would be distant from more laterally positioned desmosomes where JAM-C is localized. Indeed, our adhesion data in Figure 9 and our published report demonstrating fucosylated proteoglycan ligands for CD11b/CD18 on T84 cells (Zen et al., 2002) strongly support the existence of other epithelial ligands that could mediate initial adhesion of PMN to epithelial cells.
The current concept of PMN transepithelial migration is one that comprises multiple sequential steps or stages beginning with initial adherence of PMN to the basal aspect of the epithelium followed by migration along the epithelial paracellular space and across the apical junction complex (Zen and Parkos, 2003). This latter process would require, at least, transient opening of TJs to allow leukocytes passage to the luminal or apical epithelial surface (Edens et al., 2002; Zen and Parkos, 2003). Our finding of participation of a desmosomal component (JAM-C) in regulating PMN transepithelial migration suggests that transepithelial migration may also require transient opening of desmosomes. Perhaps the strategic localization of desmosomes along the lateral cellular membrane serves as “rungs on a ladder” for PMN as they climb up the interepithelial space. Thus, our data adds to the concept that PMN transepithelial migration is a highly regulated multistep process that includes not only initial adhesive interactions but subsequent adhesive/deadhesive processes along the migration route in which JAM-C plays an important role. Interestingly, our data suggests that CD11b/CD18 not only plays a key role in initial adhesion of PMN to epithelia but also serves as an important adhesion molecule during subsequent transmigration. In addition, because PMN transepithelial migration most often occurs in an environment enriched in inflammatory cytokines that have been shown to alter expression of epithelial surface proteins (Colgan et al., 1993), it is likely that JAM-C–mediated regulation of PMN transmigration is altered under inflammatory conditions. Clearly more studies are necessary to shed light on this important question.
From these observations, we can add important new information to a model of physiologically directed PMN transepithlelial migration that involves multiple epithelial ligands for CD11b/CD18, one of which is JAM-C (Figure 10). In this model, PMN initially adhere to the basal aspect of the epithelium via, as of yet uncharacterized, CD11b/CD18 counterreceptor(s). After initial adhesion, PMN migrate along the basolateral epithelial membrane in the paracellular space where cell-cell signaling events occur that regulate the rate of migration through CD47-SIRPα interactions (Parkos et al., 1996; Liu et al., 2001, 2002) and result in enhanced permeability through as of yet undefined mechanisms (Edens et al., 2002). Along the lateral epithelial membrane, migrating PMN then encounter desmosomes where JAM-C binding to CD11b/CD18 serves as a foothold to facilitate subsequent migration across the apical junction complex and into the lumen. It is worth noting that the above model requires additional adhesive steps for a migrating PMN to successfully reach the apical epithelial surface. Although current data suggest the presence of these ligands, further studies, including in vivo models, are necessary to identify them and determine their role(s) in regulating PMN transepithelial migration.
Figure 10.
Model of JAM-C regulation of CD11b/CD18-mediated PMN transepithelial migration. TJ, tight junction. N, nuclei.
Acknowledgments
We thank Ken Mandell for preparing JAM-A/Fc chimera and Drs. Andrei Ivanov and Ann M. Hopkins for their helpful discussion in experiment design. We also thank Susan Voss for her assistance in cell culture and Hong Yi (Emory School of Medicine Microscopy Core, Atlanta, GA) for her expertise in EM experiments. This work was supported by an National Institutes of Health (NIH) DDRDC Grant DK-77640 (tissue culture and morphology) and by NIH Grants HL 54229 (C.A.P.), HL 72124 (C.A.P.), DK 61379 (C.A.P.) and DK 59888 (A.N.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-04-0317. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-04-0317.
Abbreviations used: PMN, polymorphonuclear leukocyte; JAM, junctional adhesion molecule; TJ, tight junction; DP, desmoplakin; IFs, intermediate filaments; MPO, myeloperoxidase; HBSS-, Hanks' balanced salt buffer devoid of Ca2+ and Mg2+; FBG, fibrinogen; fMLP, formylmethionylleucylphenylalanine; RT-PCR, reverse transcript-polymerase chain reaction.
References
- Aggeler, J., and Seely, K. (1990). Cytoskeletal dynamics in rabbit synovial fibroblasts: I. Effects of acrylamide on intermediate filaments and microfilaments. Cell Motil. Cytoskel. 16, 110-120. [DOI] [PubMed] [Google Scholar]
- Altieri, D.C., Bader, R., Mannucci, P.M., and Edgington, T.S. (1988). Oligo-specificity of the cellular adhesion receptor Mac-1 encompasses an inducible recognition specificity for fibrinogen. J. Cell Biol. 107, 1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieva, M.R., Vogelmann, R., Covacci, A., Tompkins, L.S., Nelson, W.J., and Falkow, S. (2003). Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300, 1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.M., Stevenson, B.R., Jesaitis, L.A., Goodenough, D.A., and Mooseker, M.S. (1988). Characterization of ZO-1, a protein component of the tight junction from mouse liver and Madin-Darby canine kidney cells. J. Cell Biol. 106, 1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnemann, J., Sullivan, K.H., Magee, A.I., King, I.A., and Buxton, R.S. (1993). Stratification-related expression of isoforms of the desmosomal cadherins in human epidermis. J. Cell Sci. 104(Pt 3), 741-750. [DOI] [PubMed] [Google Scholar]
- Arrate, M.P., Rodriguez, J.M., Tran, T.M., Brock, T.A., and Cunningham, S.A. (2001). Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J. Biol. Chem. 276, 45826-45832. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions, M., Johnson-Leger, C., Wong, C., Du Pasquier, L., and Imhof, B.A. (2001). Heterogeneity of endothelial junctions is reflected by differential expression and specific subcellular localization of the three JAM family members. Blood 98, 3699-3707. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions, M.A., Duncan, L., Du Pasquier, L., and Imhof, B.A. (2000). Cloning of JAM-2 and JAM- 3, an emerging junctional adhesion molecular family? Curr. Top. Microbiol. Immunol. 251, 91-98. [DOI] [PubMed] [Google Scholar]
- Balsam, L.B., Liang, T.W., and Parkos, C.A. (1998). Functional mapping of CD11b/CD18 epitopes important in neutrophil-epithelial interactions: a central role of the I domain. J. Immunol. 160, 5058-5065. [PubMed] [Google Scholar]
- Bazzoni, G. (2003). The JAM family of junctional adhesion molecules. Curr. Opin. Cell Biol. 15, 525-530. [DOI] [PubMed] [Google Scholar]
- Bazzoni, G., Martinez-Estrada, O.M., Mueller, F., Nelboeck, P., Schmid, G., Bartfai, T., Dejana, E., and Brockhaus, M. (2000). Homophilic interaction of junctional adhesion molecule. J. Biol. Chem. 275, 30970-30976. [DOI] [PubMed] [Google Scholar]
- Beller, D.I., Springer, T.A., and Schreiber, R.D. (1982). Anti-Mac-1 selectively inhibits the mouse and human type three complement receptor. J. Exp. Med. 156, 1000-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, T.Q., and Wright, S.D. (1996). Human leukocyte elastase is an endogenous ligand for the integrin CR3 (CD11b/CD18, Mac-1, alpha M beta 2) and modulates polymorphonuclear leukocyte adhesion. J. Exp. Med. 184, 1213-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavakis, T., Preissner, K.T., and Santoso, S. (2003). Leukocyte trans-endothelial migration: JAMs add new pieces to the puzzle. Thromb. Haemost. 89, 13-17. [PubMed] [Google Scholar]
- Colgan, S.P., Parkos, C.A., Delp, C., Arnaout, M.A., and Madara, J.L. (1993). Neutrophil migration across cultured intestinal epithelial monolayers is modulated by epithelial exposure to IFN-gamma in a highly polarized fashion. J. Cell Biol. 120, 785-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, S.A., Arrate, M.P., Rodriguez, J.M., Bjercke, R.J., Vanderslice, P., Morris, A.P., and Brock, T.A. (2000). A novel protein with homology to the junctional adhesion molecule. Characterization of leukocyte interactions. J. Biol. Chem. 275, 34750-34756. [DOI] [PubMed] [Google Scholar]
- Del Maschio, A. et al. (1999). Leukocyte recruitment in the cerebrospinal fluid of mice with experimental meningitis is inhibited by an antibody to junctional adhesion molecule (JAM). J. Exp. Med. 190, 1351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond, M.S., Alon, R., Parkos, C.A., Quinn, M.T., and Springer, T.A. (1995). Heparin is an adhesive ligand for the leukocyte integrin Mac-1 (CD11b/CD1). J. Cell Biol. 130, 1473-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond, M.S., Staunton, D.E., de Fougerolles, A.R., Stacker, S.A., Garcia-Aguilar, J., Hibbs, M.L., and Springer, T.A. (1990). ICAM-1 (CD54): a counter-receptor for Mac-1 (CD11b/CD18). J. Cell Biol. 111, 3129-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond, M.S., Staunton, D.E., Marlin, S.D., and Springer, T.A. (1991). Binding of the integrin Mac-1 (CD11b/CD18) to the third immunoglobulin-like domain of ICAM-1 (CD54) and its regulation by glycosylation. Cell 65, 961-971. [DOI] [PubMed] [Google Scholar]
- DiScipio, R.G., Daffern, P.J., Schraufstatter, I.U., and Sriramarao, P. (1998). Human polymorphonuclear leukocytes adhere to complement factor H through an interaction that involves alphaMbeta2 (CD11b/CD18). J. Immunol. 160, 4057-4066. [PubMed] [Google Scholar]
- Ebnet, K. et al. (2003). The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PARi-3, a possible role for JAMs in endothelial cell polarity. J. Cell Sci. 116, 3879-3891. [DOI] [PubMed] [Google Scholar]
- Edens, H.A., Levi, B.P., Jaye, D.L., Walsh, S., Reaves, T.A., Turner, J.R., Nusrat, A., and Parkos, C.A. (2002). Neutrophil transepithelial migration: evidence for sequential, contact-dependent signaling events and enhanced paracellular permeability independent of transjunctional migration. J. Immunol. 169, 476-486. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and Cleveland, D.W. (1998). A structural scaffolding of intermediate filaments in health and disease. Science 279, 514-519. [DOI] [PubMed] [Google Scholar]
- Gustafson, E.J., Lukasiewicz, H., Wachtfogel, Y.T., Norton, K.J., Schmaier, A.H., Niewiarowski, S., and Colman, R.W. (1989). High molecular weight kininogen inhibits fibrinogen binding to cytoadhesins of neutrophils and platelets. J. Cell Biol. 109, 377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi, S., Tajima, M., Yao, I., Nishimura, W., Mori, H., and Hata, Y. (2003). JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol. Cell. Biol. 23, 4267-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, A.I., Nusrat, A., and Parkos, C.A. (2004). Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol. Biol. Cell 15, 176-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesaitis, A.J., Buescher, E.S., Harrison, D., Quinn, M.T., Parkos, C.A., Livesey, S., and Linner, J. (1990). Ultrastructural localization of cytochrome b in the membranes of resting and phagocytosing human granulocytes. J. Clin. Invest. 85, 821-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Leger, C.A., Aurrand-Lions, M., Beltraminelli, N., Fasel, N., and Imhof, B.A. (2002). Junctional adhesion molecule-2 (JAM-2) promotes lymphocyte transendothelial migration. Blood 100, 2479-2486. [DOI] [PubMed] [Google Scholar]
- Koeser, J., Troyanovsky, S.M., Grund, C., and Franke, W.W. (2003). De novo formation of desmosomes in cultured cells upon transfection of genes encoding specific desmosomal components. Exp. Cell Res. 285, 114-130. [DOI] [PubMed] [Google Scholar]
- Kotovuori, P. et al. (1993). The vascular E-selectin binds to the leukocyte integrins CD11/CD18. Glycobiology 3, 131-136. [DOI] [PubMed] [Google Scholar]
- Larson, R.S., and Springer, T.A. (1990). Structure and function of leukocyte integrins. Immunol. Rev. 114, 181-217. [DOI] [PubMed] [Google Scholar]
- Liang, T.W. et al. (2002). Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM 2 interacts with T, NK, and dendritic cells through JAM 3. J. Immunol. 168, 1618-1626. [DOI] [PubMed] [Google Scholar]
- Liang, T.W. et al. (2000). Characterization of huJAM: evidence for involvement in cell-cell contact and tight junction regulation. Am. J. Physiol. Cell. Physiol. 279, C1733-C1743. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Buhring, H.J., Zen, K., Burst, S.L., Schnell, F.J., Williams, I.R., and Parkos, C.A. (2002). Signal regulatory protein (SIRPalpha), a cellular ligand for CD47, regulates neutrophil transmigration. J. Biol. Chem. 277, 10028-10036. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Merlin, D., Burst, S.L., Pochet, M., Madara, J.L., and Parkos, C.A. (2001). The role of CD47 in neutrophil transmigration. Increased rate of migration correlates with increased cell surface expression of CD47. J. Biol. Chem. 276, 40156-40166. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Nusrat, A., Schnell, F.J., Reaves, T.A., Walsh, S., Pochet, M., and Parkos, C.A. (2000). Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci. 113(Pt 13), 2363-2374. [DOI] [PubMed] [Google Scholar]
- Martin-Padura, I. et al. (1998). Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142, 117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattey, D.L., and Garrod, D.R. (1986). Splitting and internalization of the desmosomes of cultured kidney epithelial cells by reduction in calcium concentration. J. Cell Sci. 85, 113-124. [DOI] [PubMed] [Google Scholar]
- McMillan, J.R., and Shimizu, H. (2001). Desmosomes: structure and function in normal and diseased epidermis. J. Dermatol. 28, 291-298. [DOI] [PubMed] [Google Scholar]
- Michishita, M., Videm, V., and Arnaout, M.A. (1993). A novel divalent cation-binding site in the A domain of the beta 2 integrin CR3 (CD11b/CD18) is essential for ligand binding. Cell 72, 857-867. [DOI] [PubMed] [Google Scholar]
- Moyle, M. et al. (1994). A hookworm glycoprotein that inhibits neutrophil function is a ligand of the integrin CD11b/CD18. J. Biol. Chem. 269, 10008-10015. [PubMed] [Google Scholar]
- Nuber, U.A., Schafer, S., Stehr, S., Rackwitz, H.R., and Franke, W.W. (1996). Patterns of desmocollin synthesis in human epithelia: immunolocalization of desmocollins 1 and 3 in special epithelia and in cultured cells. Eur. J. Cell Biol. 71, 1-13. [PubMed] [Google Scholar]
- Nusrat, A., Parkos, C.A., Verkade, P., Foley, C.S., Liang, T.W., Innis-Whitehouse, W., Eastburn, K.K., and Madara, J.L. (2000). Tight junctions are membrane microdomains. J. Cell Sci. 113(Pt 10), 1771-1781. [DOI] [PubMed] [Google Scholar]
- Ostermann, G., Weber, K.S., Zernecke, A., Schroder, A., and Weber, C. (2002). JAM-1 is a ligand of the beta(2) integrin LFA-1 involved in transendothelial migration of leukocytes. Nat. Immunol. 3, 151-158. [DOI] [PubMed] [Google Scholar]
- Ozaki, H., Ishii, K., Horiuchi, H., Arai, H., Kawamoto, T., Okawa, K., Iwamatsu, A., and Kita, T. (1999). Cutting edge: combined treatment of TNF-alpha and IFN-gamma causes redistribution of junctional adhesion molecule in human endothelial cells. J. Immunol. 163, 553-557. [PubMed] [Google Scholar]
- Parkos, C.A., Colgan, S.P., Liang, T.W., Nusrat, A., Bacarra, A.E., Carnes, D.K., and Madara, J.L. (1996). CD47 mediates post-adhesive events required for neutrophil migration across polarized intestinal epithelia. J. Cell Biol. 132, 437-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkos, C.A., Delp, C., Arnaout, M.A., and Madara, J.L. (1991). Neutrophil migration across a cultured intestinal epithelium. Dependence on a CD11b/CD18-mediated event and enhanced efficiency in physiological direction. J. Clin. Invest. 88, 1605-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas, P.J. (1999). Insoluble gamma-tubulin-containing structures are anchored to the apical network of intermediate filaments in polarized CACO-2 epithelial cells. J. Cell Biol. 146, 645-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Madrid, F., Krensky, A.M., Ware, C.F., Robbins, E., Strominger, J.L., Burakoff, S.J., and Springer, T.A. (1982). Three distinct antigens associated with human T-lymphocyte-mediated cytolysis: LFA-1, LFA-2, and LFA-3. Proc. Natl. Acad. Sci. USA 79, 7489-7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandig, M., Hergott, G.J., and Kalnins, V.I. (1990). Effects of trypsin and low Ca2+ on zonulae adhaerentes between chick retinal pigment epithelial cells in organ culture. Cell Motil. Cytoskel. 17, 46-58. [DOI] [PubMed] [Google Scholar]
- Santoso, S., Sachs, U.J., Kroll, H., Linder, M., Ruf, A., Preissner, K.T., and Chavakis, T. (2002). The junctional adhesion molecule 3 (JAM-3) on human platelets is a counterreceptor for the leukocyte integrin Mac-1. J. Exp. Med. 196, 679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesack, S.R., and Snyder, C.L. (1995). Cellular and subcellular localization of syntaxin-like immunoreactivity in the rat striatum and cortex. Neuroscience 67, 993-1007. [DOI] [PubMed] [Google Scholar]
- Shabana, A.H., Oboeuf, M., and Forest, N. (1994). Cytoplasmic desmosomes and intermediate filament disturbance following acrylamide treatment in cultured rat keratinocytes. Tissue Cell 26, 43-55. [DOI] [PubMed] [Google Scholar]
- Siliciano, J.D., and Goodenough, D.A. (1988). Localization of the tight junction protein, ZO-1, is modulated by extracellular calcium and cell-cell contact in Madin-Darby canine kidney epithelial cells. J. Cell Biol. 107, 2389-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, D.I. et al. (2000). Platelet glycoprotein ibalpha is a counterreceptor for the leukocyte integrin Mac-1 (CD11b/CD18). J. Exp. Med. 192, 193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobocka, M.B. et al. (2000). Cloning of the human platelet F11 receptor: a cell adhesion molecule member of the immunoglobulin superfamily involved in platelet aggregation. Blood 95, 2600-2609. [PubMed] [Google Scholar]
- Stacker, S.A., and Springer, T.A. (1991). Leukocyte integrin P150,95 (CD11c/CD18) functions as an adhesion molecule binding to a counter-receptor on stimulated endothelium. J. Immunol. 146, 648-655. [PubMed] [Google Scholar]
- Stevenson, B.R., Siliciano, J.D., Mooseker, M.S., and Goodenough, D.A. (1986). Identification of ZO- 1, a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 103, 755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky, S.M., Troyanovsky, R.B., Eshkind, L.G., Krutovskikh, V.A., Leube, R.E., and Franke, W.W. (1994). Identification of the plakoglobin-binding domain in desmoglein and its role in plaque assembly and intermediate filament anchorage. J. Cell Biol. 127, 151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt, F.M., Mattey, D.L., and Garrod, D.R. (1984). Calcium-induced reorganization of desmosomal components in cultured human keratinocytes. J. Cell Biol. 99, 2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zen, K., Liu, Y., Cairo, D., and Parkos, C.A. (2002). CD11b/CD18-dependent interactions of neutrophils with intestinal epithelium are mediated by fucosylated proteoglycans. J. Immunol. 169, 5270-5278. [DOI] [PubMed] [Google Scholar]
- Zen, K., and Parkos, C.A. (2003). Leukocyte-epithelial interactions. Curr. Opin. Cell Biol. 15, 557-564. [DOI] [PubMed] [Google Scholar]