Abstract
Pre-eclampsia (PE) is a complication of pregnancy that affects 5–8% of women after 20 weeks of gestation. It is usually diagnosed based on the de novo onset of hypertension and proteinuria. Preexisting hypertension in women developing PE, also known as superimposed PE on chronic hypertension (SPE), leads to elevated risk of maternal and fetal mortality. PE is associated with an altered microRNA (miRNA) expression pattern in the placenta, suggesting that miRNA deregulation is involved in the pathogenesis of PE. Whether and how the miRNA expression pattern is changed in the SPE placenta remains unclear. The present study analyzed the placental miRNA expression profile in pregnancies complicated by SPE. miRNA expression profiles in SPE and normal placentas were investigated using an Ion Torrent sequencing system. Sequencing data were processed using a comprehensive analysis pipeline for deep miRNA sequencing (CAP-miRSeq). A total of 22 miRNAs were identified to be deregulated in placentas from patients with SPE. They included 16 miRNAs previously known to be associated with PE and 6 novel miRNAs. Among the 6 novel miRNAs, 4 were upregulated (miR-518a, miR-527, miR-518e and miR-4532) and 2 downregulated (miR-98 and miR-135b) in SPE placentas compared with controls. The present results suggest that SPE is associated with specific alterations in the placental miRNA expression pattern, which differ from alterations detected in PE placentas, and therefore, provide novel targets for further investigation of the molecular mechanisms underlying SPE pathogenesis.
Keywords: placenta, microRNA, pre-eclampsia, superimposed pre-eclampsia on chronic hypertension, pregnancy complication, Ion Torrent sequencing
Introduction
Pre-eclampsia (PE) is a complication of pregnancy characterized by the onset of hypertension and proteinuria after 20 weeks of gestation. It affects 5–8% of pregnant women and is one of the predominant causes of maternal and neonatal mortality and morbidity worldwide (1,2). Women with chronic hypertension are 3–5 times more likely to develop PE than those with normal blood pressure (3). Superimposed PE on chronic hypertension (SPE) is associated with poor pregnancy out comes (3,4).
Despite the fact that the origin and pathogenesis of both PE and SPE have been extensively investigated (1,2,5–7), they remain unclear to date. Recent studies have reported the implication of microRNAs (miRNAs) in the development of PE (1,2). miRNAs are a class of 21–25-nucleotide noncoding single-stranded small RNAs. miRNAs negatively regulate gene expression by binding to the 3′-untranslated region of the target mRNAs and thus may possess important control functions in diverse biological processes (1,2,8). Aberrant expression of certain miRNAs has been identified in the placentas and blood from women with PE, suggesting that miRNA deregulation is involved in PE pathogenesis (1,2). Chang et al (9) reported that the top-ranked placental mRNA transcripts differed between patients with PE and those with SPE, suggesting that the pathogeneses of these two diseases are driven by different molecular mechanisms, which may include variations in the miRNA regulation network. Whether and how the miRNA expression pattern is changed in the placentas of patients with SPE is yet to be elucidated. In addition, the severity and frequent occurrence of SPE substantiates the requirement of the identification of candidate molecules which may provide novel insights into SPE pathogenesis.
In the present study, next generation sequencing (NGS) was performed and the placental miRNA expression profiles were compared between pregnancies of patients with SPE and normal pregnancies.
Materials and methods
Study groups and tissue samples
The study was performed on placenta samples collected from two groups of patients: Patients with SPE (n=5) and patients with normal pregnancies (control group; n=6). The mean age in the whole cohort was 31.9 ± 1.4 years. The mean age was 35.0±2.4 years in the SPE group and 29.3±0.6 years in the control group. The inclusion criterion for patients with SPE was the onset of proteinuria (≥300 mg of protein in a 24-h specimen) following the 20th week of gestation in women with documented chronic hypertension and no proteinuria prior to the 20th week of gestation, which was set by the National High Blood Pressure Education Program Working Group on High Blood Pressure in Pregnancy (10). The patients in the control group had normal pregnancies. Patients with chronic hypertension, cardiovascular disease, renal disease, hepatitis, diabetes, intrapartum infection or other pregnancy complications were excluded from the study.
All placentas were obtained by cesarean section in the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology (St. Petersburg, Russia). Placental samples were collected using the previously described systematic sampling technique to achieve uniformity and adequate sampling (11). Following dissection, tissue fragments were immediately placed in 0.9% NaCl precooled to 4°C. Subsequently, placenta villi samples (~30 mg) were selected and released from blood clots under the Leica M125 stereomicroscope (Leica Microsystems, Wetzlar, Germany) within 15 min of the cesarian section. All samples were stabilized in RNAlater (Qiagen, Inc., Valencia, CA, USA) and stored at −70°C until use.
The study was approved by the Institutional Review Board of the D.O. Ott Research Institute of Obstetrics Gynecology and Reproductology (St. Petersburg, Russia). Informed consent was signed by all patients prior to their inclusion in the study and to processing of their personal and medical data. The study was performed in accordance with the Declaration of Helsinki.
Small RNA isolation and library preparation for sequencing
Small RNA was extracted from placenta samples using PureLink miRNA Isolation kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol, and was then stored at −70°C until library preparation.
Small RNA libraries were prepared using the Ion Total RNA-Seq kit v2 (Thermo Fisher Scientific, Inc.), following the manufacturer's protocol. Small RNA libraries were prepared using the Ion Total RNA-Seq kit version 2 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, for each sample, ~100 ng of small RNA was used as the starting template. RNA samples (3 µl) were mixed with the hybridization solution and adaptor mix, incubated at 65°C for 10 min and 16°C for 5 min. Then, ligation reagents were added and the samples were incubated at 16°C overnight. After ligation, reverse transcription master mix (RT) was added to synthesize single strand cDNA. The samples were incubated at 70°C for 5 min, cooled on ice, then RT enzyme was added and the samples were incubated at 42°C for 30 min. cDNA was size-selected using Purification Module containing magnetic beads, then eluted with 12 µl nuclease-free water. The purified cDNA samples were used as templates for subsequent PCR. Briefly, 6 µl cDNA samples were combined with PCR primers and Platinum PCR SuperMix High Fidelity reaction mix. Cycling conditions were as follows: 94°C for 2 min, 2 cycles of 94°C for 30 sec, 50°C for 30 sec, and 68°C for 30 sec, 16 cycles of 94°C for 30 sec, 62°C for 30 sec, and 68°C for 30 sec, then 68°C for 5 min. PCR products were purified using the Magnetic Bead Purification Module and eluted with 15 µl nuclease-free water. For assessment of the yield, size distribution and molar concentration of the amplified DNA libraries, the samples were run on 2200 TapeStation Instrument with High Sensitivity D1K ScreenTape and High Sensitivity D1K Reagents (all purchased from Agilent Technologies, Santa Clara, CA, USA). The quantity of library required for template preparation was determined according to the manufacturer's protocol (Ion Total RNA-Seq Kit v2, Thermo Fisher Scientific, Inc.).
Ion Torrent sequencing
Each library template was clonally amplified by emulsion polymerase chain reaction (PCR) on Ion Sphere Particles (ISPs) using the Ion One Touch 200 Template kit with the Ion One Touch 2 system (Thermo Fisher Scientific, Inc.). ISPs were prepared and cleaned according to the manufacturer's protocol. Quantification of recovered particles was performed using a Qubit 2.0 fluorometer and an Ion Sphere quality control kit (both from Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The optimal percentage of template-positive ISPs was considered to be 50–150. Relative fluorescent unit values obtained outside this range were not used in subsequent ISP enrichment. The ISPs were enriched using Dynabeads MyOne Strepavidin C1 beads (Thermo Fisher Scientific, Inc.). The ISPs were then loaded into Ion 318 Chip and sequenced on the Ion Torrent Personal Genome machine (Thermo Fisher Scientific, Inc.).
Statistical analysis
Statistical analysis was performed using Statistica 10.0 (StatSoft, Inc., Tulsa, OK, USA). The Mann-Whitney test was selected for the comparison of continuous variables and Fisher's exact test for the comparison of categorical variables. P<0.05 was considered to indicate a statistically significant difference.
Computational analysis
Sequencing data were processed using a comprehensive analysis pipeline for deep miRNA sequencing (CAP-miRSeq) (12). First, according to the CAP-miRSeq algorithm, pre-processing of read sequences was performed. The quality of the reads was checked by FastQC (12) and low quality bases were trimmed from the 3′ end using Cutadapt (13). Reads containing 17 bases or less were then discarded. All types of RNA were evaluated in the sequencing libraries. The remaining reads were aligned to the GENCODE (release 18) transcripts using Bowtie software (14). RNA quantification was performed using HTseq-count (12). The sequencing reads were then analyzed to identify miRNAs using Bowtie and the Human Genome RefSeq Hg19 or miRBase (15) as a reference. Subsequently, the expression levels of detected miRNAs were estimated using miRDeep2 (16), and differential analysis of miRNAs was conducted in SPE samples, which were compared with the control. For each dataset, counts were normalized to the total number of read sequences, and then the normalized number of counts for each RNA was compared between the patient groups. The normalization and differential expression was performed using Bioconductor edgeR (17). The false discovery rate (FDR) was calculated according to Benjamini's method (18). miRNAs were considered to be differentially expressed at a P<0.01 and FDR<0.05.
Results
Study groups
The clinical characteristics of the patients from the two groups were compared (Table I), and it was found that the patients' age, gestational age at delivery, fetal weight and length at birth did not differ between the SPE and control groups. Patient systolic and diastolic blood pressure prior to and during pregnancy, as well as levels of proteinuria, were shown to be significantly higher in the SPE group compared with the control group (P<0.05).
Table I.
Clinical characteristics of the patients with normal (control) and SPE pregnancies.
| Characteristic | Control (n=6) | SPE (n=5) |
|---|---|---|
| Maternal characteristic | ||
| Age, years | 29.3±0.6 | 35.0±2.4 |
| Ethnicity | Russian, 6/6 | Russian, 5/5 |
| Systolic blood pressure before pregnancy, mmHg | 109.2±4.5 | 131.0±4.0a |
| Diastolic blood pressure before pregnancy, mmHg | 65.8±3.7 | 86.0±2.4a |
| Systolic blood pressure at diagnosis, mmHg | 108.3±3.1 | 160.0±8.9a |
| Diastolic blood pressure at diagnosis, mmHg | 70.8±2.7 | 100.0±5.5a |
| Proteinuria at diagnosis, g/24 h | 0 | 0.6±0.2a |
| Pregnancy outcome | ||
| Mode of delivery | Cesarian section, 6/6 | Caesarian section, 5/5 |
| Gestational age at delivery, weeks | 38.8±0.2 | 36.3±1.4 |
| Fetal weight, g | 3,410.0±124.5 | 2724.0±490.4 |
| Fetal length, cm | 51.2±0.5 | 47.6±2.6 |
Continuous variables are presented as the mean ± standard error. P-values were calculated using the Mann-Whitney rank-sum test for continuous variables and Fisher's exact test for categorical variables.
P<0.05 vs. control. SPE, superimposed pre-eclampsia on chronic hypertension.
miRNA sequencing
The expression profiles of small RNAs in the placenta samples were compared between the SPE and control groups. For this purpose, 5 SPE and 6 control small RNA libraries were prepared using the Ion Torrent system. Sequencing reads obtained from 5 SPE and 6 control libraries were used to create two separate libraries: The SPE and control libraries. The small RNAs from the two libraries were a similar size of 19–24 nt. The total reads obtained from the SPE and control libraries were 1,502,484 and 2,241,561, respectively (Table II). After the read sequences had been pre-processed, 1,207,080 and 1,844,141 clean reads were selected from the SPE and control libraries, respectively, for further analysis (Table II).
Table II.
Characteristics of miRNA-sequencing libraries obtained from normal (control) and SPE placentas.
| Characteristic | Control (n=6) | SPE (n=5) |
|---|---|---|
| Total reads, n | 2,241,561 | 1,502,484 |
| Trimmed reads, n (%) | 100,821 (4.5) | 58,019 (3.9) |
| Too short after trimming (<17 bps), n (%) | 296,599 (13.2) | 236,655 (15.8) |
| Reads sent to aligner, n | 1,844,141 | 1,207,080 |
| Aligned reads, n (%) | 1,184,799 (64.2) | 827,455 (68.6) |
| Precursor miRNA reads, n | 9,060 | 7,576 |
| Mature miRNA reads, n | 805,102 | 606,821 |
| Known miRNAs with ≥5X coverage, n | 522 | 466 |
SPE, superimposed pre-eclampsia; miRNA, microRNA.
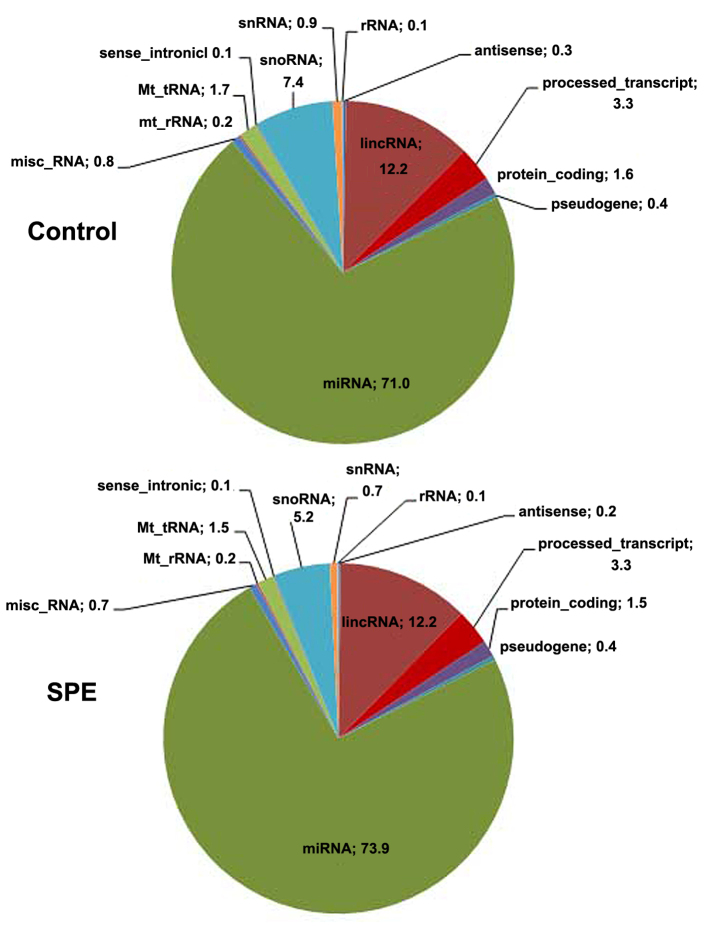
The relative proportions of different RNA categories were similar in both libraries, with miRNAs represented the largest fraction of all types of small RNAs (Fig. 1). The number of reads aligned to the reference genome, and to mature and precursor miRNAs, as well as the number of miRNAs reads with ≥5x coverage detected are provided in Table II. In total, 466 known miRNAs were detected in the SPE library and 522 in the control library (Table II).
Figure 1.
Placental RNA categories in normal pregnancies (control) and in pregnancies complicated by SPE. SPE, superimposed pre-eclampsia on chronic hypertension; miRNA, microRNA; rRNA, ribosomal RNA; Mt_rRNA, mitochondrial rRNA; Mt_tRNA, mitochondrial transfer RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; misc_RNA, miscellaneous RNA; lincRNA, long intergenic non-coding RNA.
miRNA expression
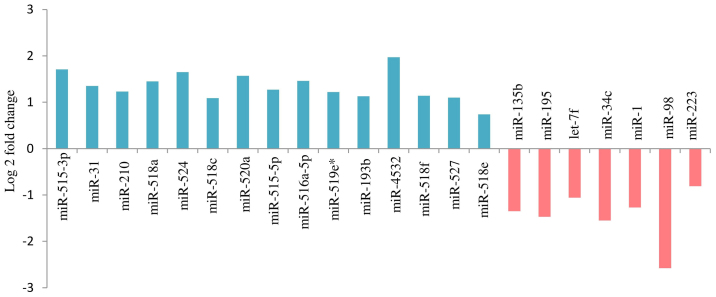
A total of 36 miRNAs (P<0.01) exhibited altered expression patterns in the SPE placentas, when compared with those in the control placentas. In 22 of the 36 miRNAs, the difference in expression was significant (FDR<0.05). Among them, 15 were upregulated and 7 downregulated in the SPE placentas, as compared with the control (Fig. 2).
Figure 2.
Deregulated placental miRNAs in pregnancies complicated by SPE. A total of 22 placental miRNAs were differentially expressed in pregnancies complicated by SPE compared with normal pregnancies (P<0.01, false discover rate<0.05). Upregulated miRNA expression is indicated by upward bars and downregulated by downward bars. Validation was performed using CAP-miRSeq. miRNA, microRNA; SPE, superimposed pre-eclampsia on chronic hypertension.
Discussion
The miRNA expression profiles were detected and compared between the SPE and normal placentas. The experimental approach followed in the present study was based on NGS using Ion Torrent Sequencer. In contrast to reverse transcription-quantitative PCR and microarray hybridization, NGS does not depend on the design of primers, can detect an unlimited number of miRNAs in one experiment, is suitable for quantitative evaluation of low-expressing transcripts and allows for a precise analysis of small numbers of valuable samples (7).
A total of 22 miRNAs were found to be differentially expressed in the SPE placentas compared with the control placentas (Fig. 2). Of these 22 miRNAs, 11 were mapped to the C19MC cluster, the largest human miRNA gene cluster, whose expression is almost exclusively confined to the placenta (Table III). The cluster spans ~100 kb at chromosome 19q13.41 and contains 54 predicted miRNA genes, 43 of which have been cloned and sequenced (2). The precise biological function of the C19MC cluster is unknown. These results, in combination with findings from other miRNA studies of PE (2,19,20), suggested that the C19MC aberrant expression may comprise a contributing factor to the development of placental dysfunction. In the present study, all 11 differentially expressed miRNAs from the C19MC cluster were found to be overexpressed in SPE placentas. Eight of these miRNAs (miR-515-3p, miR-515-5p, miR-516a-5p, miR-518c, miR-518f, miR-519e*, miR-520a and miR-524) have been reported to be upregulated in placentas from patients with PE (19–22). The other 3 overexpressed miRNAs detected in the present study (miR-518a, miR-527 and miR-518e) had not been previously reported to be associated with PE, and could therefore be involved in pathogenic biochemical pathways specific to SPE (Table III).
Table III.
Deregulated microRNAs in SPE (present study) and in PE pregnancies (literature data).
| miRNA | Chromosome | SPE pregnancies (present study)
|
PE pregnancies (literature data)
|
||||
|---|---|---|---|---|---|---|---|
| Regulation of expression | Method | Tissue | Regulation of expression | Method | Refs. | ||
| miR-515-3p | 19, C19MC | Up | NGS | Placenta | – | Mcroarray | (52) |
| Plasma | Pregnancy-specific miRNA | RT-qPCR | (52) | ||||
| Placenta | Up | RT-qPCR-based array, not NGS | (20) | ||||
| Placenta | Up, severe PE | Mcroarray (not validated by RT-qPCR | (22) | ||||
| Plasma | Up, severe and mild PE | NGS (not checked by RT-qPCR) | (7) | ||||
| miR-515-5p | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Plasma | Pregnancy-specific miRNA | RT-qPCR | (52) | ||||
| Serum | Pregnancy-specific miRNA | RT-qPCR | (51) | ||||
| Placenta | Up | RT-qPCR-based array, not NGS | (20) | ||||
| miR-516a-5p | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Placenta | Up, severe PE | Microarray (not checked by RT-qPCR) | (21) | ||||
| miR-518a | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Serum | Pregnancy-specific miRNA | RT-qPCR | (51) | ||||
| miR-527 | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Serum | Pregnancy-specific miRNA | RT-qPCR | (51) | ||||
| miR-518c | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Plasma | Pregnancy-specific miRNA | RT-qPCR | (52) | ||||
| Placenta | Up | NGS and RT-qPCR-based array | (20) | ||||
| Plasma | Up, severe and mild PE | NGS (not checked by RT-qPCR) | (7) | ||||
| miR-518e | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Plasma | Pregnancy-specific miRNA | RT-qPCR | (52) | ||||
| Serum | Pregnancy-specific miRNA | RT-qPCR | (51) | ||||
| Placenta | Up | NGS, not RT-qPCR-based array | (20) | ||||
| Serum | Up | NGS | (53) | ||||
| miR-518f | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Placenta | – | NGS | (20) | ||||
| miR-519e* | 19, C19MC | Up | NGS | Serum | Pregnancy-specific miRNA | RT-qPCR | (51) |
| Placenta | Up, severe PE | Microarray (not checked by RT-qPCR) | (19) | ||||
| Placenta | Up, severe PE | Microarray (not validated by RT-qPCR) | (22) | ||||
| Placenta | Up | NGS and RT-qPCR-based array | (20) | ||||
| miR-520a | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Placenta | Up | NGS and RT-qPCR-based array | (20) | ||||
| miR-524 | 19, C19MC | Up | NGS | Placenta | – | Microarray | (52) |
| Serum | Pregnancy-specific miRNA | RT-qPCR | (51) | ||||
| Placenta | Up, severe PE | Microarray (not validated by RT-qPCR) | (21) | ||||
| Placenta | Up, severe PE | Microarray (validated by RT-qPCR) | (22) | ||||
| Placenta | Up | RT-qPCR-based array, not NGS | (20) | ||||
| miR-210 | 11 | Up | NGS | Placenta | – | RT-qPCR-based array | (48) |
| Placenta | Up | Microarray (validated by RT-qPCR) | (11) | ||||
| Placenta | Up, severe PE | Microarray (validated by RT-qPCR) | (19) | ||||
| Placenta | Up | RT-qPCR-based array | (28) | ||||
| Placenta | Up, severe PE | Microarray (validated by RT-qPCR) | (20) | ||||
| Placenta | Up | NGS and RT-qPCR-based array | (20) | ||||
| Placenta | Up, severe PE (basal plate, not in the chorionic plate) | RT-qPCR | (29) | ||||
| Placenta | Up, late onset PE | NGS (not checked by RT-qPCR) | (30) | ||||
| Plasma | Up | RT-qPCR | (8) | ||||
| Plasma | Up, severe PE | RT-qPCR | (22) | ||||
| Plasma | Up, severe and mild PE | RT-qPCR | (31) | ||||
| miR-195 | 17 | Down | NGS | Placenta | – | NGS | (20) |
| Placenta | – | RT-qPCR-based array | (48) | ||||
| Placenta | Down, severe PE | Microarray (validated by RT-qPCR) | (22) | ||||
| Placenta | Down, severe PE | RT-qPCR | (33) | ||||
| Placenta | Down, severe PE | Microarray (not checked by RT-qPCR) | (19) | ||||
| miR-223 | X | Down | NGS | Placenta | – | RT-qPCR-based array | (48) |
| Placenta | Down, severe PE | Microarray (not checked by RT-qPCR) | (19) | ||||
| Placenta | Down, severe PE | Microarray (validated by RT-qPCR) | (22) | ||||
| Placenta (FFPE) | Down | PNA-based microarray (not checked by RT-qPCR) | (34) | ||||
| Placenta | Down, early-onset PE | NGS (validated by RT-qPCR) | (30) | ||||
| Placenta | Down | NGS, not RT-qPCR-based array | (20) | ||||
| Plasma | Down, severe and mild PE | NGS (not checked by RT-qPCR) | (7) | ||||
| Serum | Down | NGS | (53) | ||||
| miR-34c | 11 | Down | NGS | Placenta | – | RT-qPCR-based array | (48) |
| Plasma | Pregnancy-specific miRNA | RT-qPCR-based array | (48) | ||||
| Placenta | Down | Microarray (not checked by RT-qPCR) | (11) | ||||
| Placenta | Down, severe PE | Microarray (not validated by RT-qPCR) | (21) | ||||
| miR-1 | 18 | Down | NGS | Placenta | – | Microarray | (52) |
| Placenta | Down | Microarray (validated by RT-qPCR) | (11) | ||||
| Placenta | Down, severe PE | Microarray (not checked by RT-qPCR) | (19) | ||||
| miR-193b | 16 | Up | NGS | Placenta | Up | NGS and RT-qPCR-based array | (20) |
| Placenta | Up, severe PE | Microarray (validated by RT-qPCR) | (22) | ||||
| let-7f | 9 | Down | NGS | Placenta | – | NGS | (20) |
| Placenta | Up, severe PE | Microarray (not checked by RT-qPCR) | (54) | ||||
| Serum | Down | NGS | (53) | ||||
| Plasma | Down, severe and mild PE | NGS (not checked by RT-qPCR) | (7) | ||||
| miR-31 | 9 | Up | NGS | Placenta | – | RT-qPCR-based array | (48) |
| Placenta | Up | NGS, not RT-qPCR- array | (20) | ||||
| Placenta | Up, severe PE | Microarray (not validated by RT-qPCR) | (22) | ||||
| miR-98 | X | Down | NGS | Placenta | – | RT-qPCR-based array | (48) |
| Placenta | Up | NGS, not RT-qPCR-array | (20) | ||||
| miR-135b | 1 | Down | NGS | Placenta | – | Microarray | (52) |
| Placenta | – | NGS | (20) | ||||
| Placenta | – | RT-qPCR-based array | (48) | ||||
| Plasma | Pregnancy-specific miRNA | RT-qPCR-based array | (48) | ||||
| miR-4532 | 20 | Up | NGS | – | – | – | – |
PE, pre-eclampsia; SPE, superimposed PE on chronic hypertension; NGS, next-generation sequencing; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; FFPE, formalin-fixed, paraffin-embedded tissue.
Notably, the C19MC cluster is regulated by genomic imprinting with only the paternally inherited allele being expressed in the placenta, while the maternal one displays a methylation imprint (23,24). The C19MC expression pattern in human cells, including placental cells, has been shown to be associated with the methylation status of the distal CpG-rich region (23). Demethylating agents induce the upregulation of miRNAs from the C19MC cluster in cancer cells (23). These data suggest that the PE/SPE-associated deregulation of the C19MC cluster could be initiated at the beginning of the pregnancy (during preimplantation development), or even earlier (during gametogenesis), when the genome-wide DNA methylation reprogramming through demethylation and remethylation occurs (25–27).
Of the 22 deregulated miRNAs detected in the present study, 11 are located outside the C19MC cluster and are mapped to chromosomes 1, 9, 11, 16, 17, 20 and X. Among these miRNAs, 8 have been previously reported to be deregulated in the PE placenta (miR-210, miR-195, miR-223, miR-1, miR-34c, miR-193b, miR-let-7f and miR-31), while for the remaining 3 (miR-98, miR-135b and miR-4532), PE-associated aberrant expression has not been previously reported in the literature (Table III).
The present results on miRNA deregulation (up or down) in SPE placentas are largely consistent with the data reported by previous studies focusing on expression in placentas from patients with PE (11,19–22,28–32), suggesting that miRNA-mediated molecular mechanisms of SPE and PE pathogeneses are similar (Table III). A number of biochemical pathways of miRNA-mediated PE pathogenesis have been suggested. The overexpression of miR-210, repeatedly shown in PE placentas (11,19,20,22,28–30), may be provoked by hypoxia, which in turn can result from impaired placentation in PE and SPE pregnancies (19). Ectopic expression of miR-210 inhibits the migration and invasion capability of trophoblast cells by targeting Ephrin-A3 and Homeobox-A9, which are responsible for cell migration and vascular remodeling (33). Aberrant miR-210 expression may also contribute to PE and, thus, to SPE by interfering with potassium channel modulatory factor 1-mediated signaling in the placenta (29). The expression of another miR-210 relevant target, hydroxysteroid (17-b) dehydrogenase 1 (HSD17B1) has been shown to be decreased in the placentas from patients with PE (20). Notably, another upregulated miRNA in the placentas of the SPE group detected in the present study, miR-518c, has been experimentally demonstrated to target HSD17B1 (20), thus suggesting its possible involvement in PE.
The activin A receptor, type IIA (ACVR2A), a candidate gene for PE predicted by a genome-wide linkage study (34), is regulated by miRNA. miR-195 has been shown to repress ACVR2A and promote the invasion of trophoblast cells (31). The decrease in miR-195 expression that was detected in the placenta from patients with SPE in the present study and in those from patients with PE from other studies (19,22,31), results in an increase in ACVR2A. The latter can provoke inhibition of trophoblast invasion and insufficient remodeling of the spiral arteries, thus influencing the central pathophysiological features of PE (31) and SPE. mRNA of ACVR2A may also be a target for the miR-223 (miRDB database; http://mirdb.org/), which was reported to be downregulated in placentas from patients with PE (19,20,22,30,32) and SPE, as shown in the present study. An important biological role of ACVR2A, miR-195 and miR-223 in the establishment of pregnancy renders these molecules a focal point of interest for further studies.
miR-34c, downregulated in placentas from patients with PE (11,21) and SPE, normally mediates the p53-dependent suppression of endometrial cell proliferation. Thus, it may potentially contribute to PE and SPE development through the deregulation of the cell cycle (11). The downregulation of miR-1 may affect the risk of PE development through its effect on calcium signaling (11) and/or through its influence on the expression of metallopeptidase inhibitor 3, which is involved in the regulation of trophoblast invasion (35,36). The upregulation of miR-193b may decrease the expression of plasminogen activator, urokinase gene, encoding urokinase-type plasminogen activator (37), which results in the reduction of fibrinolytic activity in placenta vessels, promoting the development of PE (38,39). The participation of poorly expressed miR-let-7f in the development of PE has not yet been demonstrated; however, the role of let-7 family members in proliferation, apoptosis and inflammation (40), suggests their contribution to the onset of PE through the deregulation of these processes. The PE-associated miR-31 overexpression (20), which was also observed in SPE placentas of the present study, can contribute to placenta dysfunction in several ways. For eample, through targeting factor-inhibiting hypoxia-inducible factor gene, miR-31 can activate hypoxia pathways (41), suggesting an involvement of this particular miRNA in the hypoxic response. Integrin-β3, which has been found to be significantly decreased in placentas from patients with PE (42), is known to be another target for miR-31 in cancer cells (43).
In the present study, 3 deregulated miRNAs (miR-98, miR-135b and miR-4532), which were located outside the C19MC cluster and had not been previously reported to be deregulated in placentas from patients with PE, were identified in placentas from patients with SPE. These findings suggest that the aforementioned miRNAs may be of significance in the development of SPE. miR-135b and miR-98 were downregulated in the placentas from the SPE group, while miR-4532 was upregulated in the same samples. Several predicted targets of these miRNAs could be associated with SPE pathogenesis. Interleukin-6, the validated target of miR-98 in melanoma cells (44), has been reported to be significantly increased in maternal and umbilical serum of PE patients (45). The target for miR-135b predicted by the miRDB database corresponds to rho-associated coiled-coil protein kinase 2 (ROCK2) known to be elevated in PE placentas (46). ROCK2 has been mapped within the PE linkage peak on chromosome 2p25 in genome-wide linkage screening (47). miR-135b is expressed in placenta tissue and found in the maternal circulation (48), confirming its feasible functional significance in pregnancy. miR-4532 may contribute to SPE by downregulating collagen, type I, α 1 (COL1A1), a component of the extracellular matrix (49). The expression of COL1A1 mRNA has been shown to be lower in early-onset PE placentas than in the gestational age-matched control (49). In their candidate gene study, Goddard et al (50) reported that single nucleotide polymorphism variants in the COL1A gene are associated with a risk of PE. These findings are consistent with our hypothesis that miR-98, miR-135b, and miR-4532 are associated with SPE pathogenesis. Direct functional studies of these miRNA interactions with relevant targets could lead to a greater understanding of the molecular mechanisms underlying this association.
In conclusion, the present results provided novel insights in to the pathogenesis of SPE. It was found that the expression pattern of 22 miRNAs was significantly altered in SPE placentas. Of these 22 miRNAs, 16 had already been described to be associated with PE, and 6 had not previously been reported. These findings indicated that miRNA-mediated biochemical pathways of SPE pathogenesis largely overlap with those of PE; however, the detection of these 6 SPE-specific deregulated miRNAs suggested the existence of specific differences in the pathological mechanisms between PE and SPE. The differentially expressed miRNAs may serve as targets for the prevention and treatment of SPE. Notably, 12 of these miRNAs (miR-515-3p, miR-515-5p, miR-518a, miR-518e, miR-527, miR-518c, miR-519e*, miR-524, miR-210, miR-223, let-7f and miR-135b) have been previously reported to circulate in the blood of pregnant women (7,8,22,33,48,51–54), which makes them potential biomarkers of SPE; however, this requires further investigation.
Acknowledgments
Equipment from the Biobank and resource centre of the Development of Molecular and Cellular Technology, St. Petersburg State University was used in the present study. The collecting of tissue samples and small RNA isolation were supported by the Russian Federation President (grant no. 16.120.11.5773 MC). Library preparation and Ion Torrent sequencing were supported by the Russian Scientific Foundation (grant no. 14 50 00069). Computational analysis was supported by the Russian Federation Government (grant no. 074 U01). Dr Olga A. Efimova and Mr. Andrei V. Tikhonov are grantees of RF President scholarship (grant nos. SP-1405.2015.4 and SP-127.2015.4).
Abbreviations
- SPE
superimposed PE on chronic hypertension
- PE
pre-eclampsia
References
- 1.Zhao Z, Moley KH, Gronowski AM. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin Biochem. 2013;46:953–960. doi: 10.1016/j.clinbiochem.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 2.Chen DB, Wang W. Human placental microRNAs and preeclampsia. Biol Reprod. 2013;88:130. doi: 10.1095/biolreprod.113.107805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perni U, Sison C, Sharma V, Helseth G, Hawfield A, Suthanthiran M, August P. Angiogenic factors in superimposed preeclampsia: A longitudinal study of women with chronic hypertension during pregnancy. Hypertension. 2012;59:740–746. doi: 10.1161/HYPERTENSIONAHA.111.181735. [DOI] [PubMed] [Google Scholar]
- 4.Genest DS, Falcao S, Michel C, Kajla S, Germano MF, Lacasse AA, Vaillancourt C, Gutkowska J, Lavoie JL. Novel role of the renin-angiotensin system in preeclampsia superimposed on chronic hypertension and the effects of exercise in a mouse model. Hypertension. 2013;62:1055–1061. doi: 10.1161/HYPERTENSIONAHA.113.01983. [DOI] [PubMed] [Google Scholar]
- 5.Glotov AS, Vashukova YS, Glotov OS, Nasykhova YuA, Mazur AM, Kurilov RV, Pekhov VM, Khrameyeva YeE, Ivashchenko TE, Baranov VS. Study of the population frequencies of gene polymorphisms, associated with preeclampsia. Russian Journal of Genetics: Applied Research. 2014;4:388–396. [Google Scholar]
- 6.Glotov AS, Tiys ES, Vashukova ES, Pakin VS, Demenkov PS, Saik OV, Ivanisenko TV, Arzhanova ON, Mozgovaya EV, Zainulina MS, et al. Molecular association of pathogenetic contributors to pre-eclampsia (pre-eclampsia associome) BMC Syst Biol. 2015;9(Suppl 2):S4. doi: 10.1186/1752-0509-9-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Ge Q, Guo L, Lu Z. Maternal plasma miRNAs expression in preeclamptic pregnancies. Biomed Res Int. 2013;2013:970265. doi: 10.1155/2013/970265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunel T, Zeybek YG, Akçakaya P, Kalelioğlu I, Benian A, Ermis H, Aydınlı K. Serum microRNA expression in pregnancies with preeclampsia. Genet Mol Res. 2011;10:4034–4040. doi: 10.4238/2011.November.8.5. [DOI] [PubMed] [Google Scholar]
- 9.Chang SD, Chao AS, Peng HH, Chang YL, Wang CN, Cheng PJ, Lee YS, Chao A, Wang TH. Analyses of placental gene expression in pregnancy-related hypertensive disorders. Taiwan J Obstet Gynecol. 2011;50:283–291. doi: 10.1016/j.tjog.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Report of the National high blood pressure education program working group on high blood pressure in pregnancy. Am J Obstet Gynecol. 2000;183:S1–S22. doi: 10.1067/mob.2000.107928. No authors listed. [DOI] [PubMed] [Google Scholar]
- 11.Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204:178.e12–178.e21. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Z, Evans J, Bhagwate A, Middha S, Bockol M, Yan H, Kocher JP. CAP-miRSeq: A comprehensive analysis pipeline for microRNA sequencing data. BMC Genomics. 2014;15:423. doi: 10.1186/1471-2164-15-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 14.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozomara A, Griffiths-Jones S. MiRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database Issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 19.Zhu X, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200:661.e1–661.e7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 20.Ishibashi O, Ohkuchi A, Ali MM, Kurashina R, Luo SS, Ishikawa T, Takizawa T, Hirashima C, Takahashi K, Migita M, et al. Hydroxysteroid (17-β) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: A novel marker for predicting preeclampsia. Hypertension. 2012;59:265–273. doi: 10.1161/HYPERTENSIONAHA.111.180232. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Feng L, Zhang H, Hachy S, Satohisa S, Laurent LC, Parast M, Zheng J, Chen DB. Preeclampsia up-regulates angiogenesis-associated microRNA (i.e., miR-17, -20a and -20b) that target ephrin-B2 and EPHB4 in human placenta. J Clin Endocrinol Metab. 2012;97:E1051–E1059. doi: 10.1210/jc.2011-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu P, Zhao Y, Liu M, Wang Y, Wang H, Li YX, Zhu X, Yao Y, Wang H, Qiao J, et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension. 2014;63:1276–1284. doi: 10.1161/HYPERTENSIONAHA.113.02647. [DOI] [PubMed] [Google Scholar]
- 23.Tsai KW, Kao HW, Chen HC, Chen SJ, Lin WC. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics. 2009;4:587–592. doi: 10.4161/epi.4.8.10230. [DOI] [PubMed] [Google Scholar]
- 24.Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefèvre A, Coullin P, Moore GE, Cavaillé J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19:3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 25.Pendina AA, Efimova OA, Fedorova ID, Leont'eva OA, Shilnikova EM, Lezhnina JG, Kuznetzova TV, Baranov VS. DNA methylation patterns of metaphase chromosomes in human preimplantation embryos. Cytogenet Genome Res. 2011;132:1–7. doi: 10.1159/000318673. [DOI] [PubMed] [Google Scholar]
- 26.Efimova OA, Pendina AA, Tikhonov AV, Fedorova ID, Krapivin MI, Chiryaeva OG, Shilnikova EM, Bogdanova MA, Kogan IY, Kuznetzova TV, et al. Chromosome hydroxymethylation patterns in human zygotes and cleavage-stage embryos. Reproduction. 2015;149:223–233. doi: 10.1530/REP-14-0343. [DOI] [PubMed] [Google Scholar]
- 27.Tang W W, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic J, Mittal P. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:261.e1–261.e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Luo R, Shao X, Xu P, Liu Y, Wang Y, Zhao Y, Liu M, Ji L, Li YX, Chang C, et al. MicroRNA-210 contributes to preeclampsia by downregulating potassium channel modulatory factor 1. Hypertension. 2014;64:839–845. doi: 10.1161/HYPERTENSIONAHA.114.03530. [DOI] [PubMed] [Google Scholar]
- 30.Weedon-Fekjær MS, Sheng Y, Sugulle M, Johnsen GM, Herse F, Redman CW, Lyle R, Dechend R, Staff AC. Placental miR-1301 is dysregulated in early-onset preeclampsia and inversely correlated with maternal circulating leptin. Placenta. 2014;35:709–717. doi: 10.1016/j.placenta.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Bai Y, Yang W, Yang HX, Liao Q, Ye G, Fu G, Ji L, Xu P, Wang H, Li YX, Peng C. Downregulated miR-195 detected in preeclamptic placenta affects trophoblast cell invasion via modulating ActRIIA expression. PLoS One. 2012;7:e38875. doi: 10.1371/journal.pone.0038875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi SY, Yun J, Lee OJ, Han HS, Yeo MK, Lee MA, Suh KS. MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta. 2013;34:799–804. doi: 10.1016/j.placenta.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Fei M, Xue G, Zhou Q, Jia Y, Li L, Xin H, Sun S. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: New insights into molecular mechanisms for the disease. J Cell Mol Med. 2012;16:249–259. doi: 10.1111/j.1582-4934.2011.01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moses EK, Fitzpatrick E, Freed KA, Dyer TD, Forrest S, Elliott K, Johnson MP, Blangero J, Brennecke SP. Objective prioritization of positional candidate genes at a quantitative trait locus for pre-eclampsia on 2q22. Mol Hum Reprod. 2006;12:505–512. doi: 10.1093/molehr/gal056. [DOI] [PubMed] [Google Scholar]
- 35.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 36.Xiang Y, Zhang X, Li Q, Xu J, Zhou X, Wang T, Xing Q, Liu Y, Wang L, He L, Zhao X. Promoter hypomethylation of TIMP3 is associated with pre-eclampsia in a Chinese population. Mol Hum Reprod. 2013;19:153–159. doi: 10.1093/molehr/gas054. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda Y, Tanji E, Makino N, Kawata S, Furukawa T. MicroRNAs associated with mitogen-activated protein kinase in human pancreatic cancer. Mol Cancer Res. 2012;10:259–269. doi: 10.1158/1541-7786.MCR-11-0035. [DOI] [PubMed] [Google Scholar]
- 38.Buchholz T, Lohse P, Rogenhofer N, Kosian E, Pihusch R, Thaler CJ. Polymorphisms in the ACE and PAI-1 genes are associated with recurrent spontaneous miscarriages. Hum Reprod. 2003;18:2473–2477. doi: 10.1093/humrep/deg474. [DOI] [PubMed] [Google Scholar]
- 39.Lam KK, Chiu PC, Chung MK, Lee CL, Lee KF, Koistinen R, Koistinen H, Seppala M, Ho PC, Yeung WS. Glycodelin-A as a modulator of trophoblast invasion. Hum Reprod. 2009;24:2093–2103. doi: 10.1093/humrep/dep205. [DOI] [PubMed] [Google Scholar]
- 40.Zhang XD, Zhang YH, Ling YH, Liu Y, Cao HG, Yin ZJ, Ding JP, Zhang XR. Characterization and differential expression of microRNAs in the ovaries of pregnant and non-pregnant goats (Capra hircus) BMC Genomics. 2013;14:157. doi: 10.1186/1471-2164-14-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY, Wu KJ, Chiou SH, Lin SC, Chang KW. MiR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–1644. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y, Damsky CH, Fisher SJ. Preeclampsia is associated with failure of human cytotrophoblasts to mimic a vascular adhesion phenotype: One cause of defective endovascular invasion in this syndrome? J Clin Invest. 1997;99:2152–2164. doi: 10.1172/JCI119388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. MiR-31 is a broad regulator of β1-integrin expression and function in cancer cells. Mol Cancer Res. 2011;9:1500–1508. doi: 10.1158/1541-7786.MCR-11-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F, Li XJ, Qiao L, Shi F, Liu W, Li Y, Dang YP, Gu WJ, Wang XG, Liu W. MiR-98 suppresses melanoma metastasis through a negative feedback loop with its target gene IL-6. Exp Mol Med. 2014;46:e116. doi: 10.1038/emm.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tosun M, Celik H, Avci B, Yavuz E, Alper T, Malatyalioğlu E. Maternal and umbilical serum levels of interleukin-6, interleukin-8 and tumor necrosis factor-alpha in normal pregnancies and in pregnancies complicated by preeclampsia. J Matern Fetal Neonatal Med. 2010;23:880–886. doi: 10.3109/14767051003774942. [DOI] [PubMed] [Google Scholar]
- 46.Ark M, Yilmaz N, Yazici G, Kubat H, Aktaş S. Rho-associated protein kinase II (rock II) expression in normal and preeclamptic human placentas. Placenta. 2005;26:81–84. doi: 10.1016/j.placenta.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Peterson H, Laivuori H, Kerkelä E, Jiao H, Hiltunen L, Heino S, Tiala I, Knuutila S, Rasi V, Kere J, Kivinen K. ROCK2 allelic variants are not associated with pre-eclampsia susceptibility in the Finnish population. Mol Hum Reprod. 2009;15:443–449. doi: 10.1093/molehr/gap032. [DOI] [PubMed] [Google Scholar]
- 48.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 49.He P, Shao D, Ye M, Zhang G. Analysis of gene expression identifies candidate markers and pathways in pre-eclampsia. J Obstet Gynaecol. 2015;35:578–584. doi: 10.3109/01443615.2014.990430. [DOI] [PubMed] [Google Scholar]
- 50.Goddard KA, Tromp G, Romero R, Olson JM, Lu Q, Xu Z, Parimi N, Nien JK, Gomez R, Behnke E, et al. Candidate-gene association study of mothers with pre-eclampsia and their infants, analyzing 775 SNPs in 190 genes. Hum Hered. 2007;63:1–16. doi: 10.1159/000097926. [DOI] [PubMed] [Google Scholar]
- 51.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miura K, Miura S, Yamasaki K, Higashijima A, Kinoshita A, Yoshiura K, Masuzaki H. Identification of pregnancy-associated microRNAs in maternal plasma. Clin Chem. 2010;56:1767–1771. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 53.Yang Q, Lu J, Wang S, Li H, Ge Q, Lu Z. Application of next-generation sequencing technology to profile the circulating microRNAs in the serum of preeclampsia versus normal pregnant women. Clin Chim Acta. 2011;412:2167–2173. doi: 10.1016/j.cca.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 54.Hu Y, Li P, Hao S, Liu L, Zhao J, Hou Y. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin Chem Lab Med. 2009;47:923–929. doi: 10.1515/CCLM.2009.228. [DOI] [PubMed] [Google Scholar]