Abstract
Optically controlled release of a DNA strand based on a nonradiative relaxation process of black hole quenchers (BHQs), which are a sort of dark quenchers, is presented. BHQs act as efficient energy sources because they relax completely via a nonradiative process, i.e., without fluorescent emission-based energy losses. A DNA strand is modified with BHQs and the release of its complementary strand is controlled by excitation of the BHQs. Experimental results showed that up to 50% of the target strands were released, and these strands were capable of inducing subsequent reactions. The controlled release was localized on a substrate within an area of no more than 5 micrometers in diameter.
OCIS codes: (350.5340) Photothermal effects, (350.4600) Optical engineering
1. Introduction
Biological systems are both complex and dynamic. While many sophisticated technologies have been developed to analyze and control the behavior of these systems, the determination and use of biological functions remains challenging [1, 2]. Optical methods are indispensable in the molecular/cell biology research at various scales, ranging from molecules and cells to organs, because they offer many valuable properties, including diverse light-matter interactions, remote and flexible accessibility to matter, spatial and temporal localization abilities, spatial parallelism, and ease of multiplexing. Among the available optical methods, one rapidly developing technology is optogenetics, in which light-sensitive ion channels such as channelrhodopsins are expressed genetically but artificially in living cells, and especially in neurons, and the activities of these cells are controlled by optical stimulation to enable the investigation of biological events [3]. Similar optical activation is achievable in other molecules, and it is an effective way to control associated biological events. Good examples of these methods include conditional gene editing and gene expression by optical regulation of clustered regularly interspaced short palindromic repeat (CRISPR) associated protein 9 (Cas9) activity [4], light-induced in vivo gene transfer using multifunctional nanocarriers [5], optical activation of transforming growth factor β (TGF-β) signal transduction assisted by carbon nanotubes [6], femtosecond-laser induced gene transcription by activation of the nuclear factor of activated T-cell (NFAT) proteins [7], and artificial targeted light-activated nanoscissors for use as a sequence-specific DNA cleavage nanomaterial [8]. Oligonucleotides are especially attractive for regulation of cell activities because they allow sequence-specific reactions to be designed. A photo-responsive antisense oligonucleotide capable of cross-linking to a target messenger ribonucleic acid (mRNA) is useful for the regulation of gene expression in living cells [9]. Light-triggered splice-switching oligonucleotides work well for regulation of alternative splicing pathways [10]. Notably, the above functions relating to oligonucleotides and other biomolecules can be localized via optical activation.
Controlled release of molecules is also an important process because it offers many potential applications, including drug delivery and therapy. To date, optically controlled release has been studied for a variety of molecules, including insulin [11], anticancer drugs [12], angiogenesis inhibitors [13], and DNA [14–16]. Controlled release of a single-stranded DNA (ss-DNA) can be realized by dehybridization of a double-stranded DNA (dsDNA). For example, the thermal energy produced by light irradiation can be used to induce dehybridization. Surface plasmon resonance on a metal nanoparticle or nanorod is a promising phenomenon that can provide increased local temperatures around the metal nanostructure. Light-induced dehybridization, or remotely controlled release of single-stranded DNA, has been demonstrated using gold nanoparticles and nanorods [16–18]. This method can be extended to the selective release of DNA strands using nanorods of various sizes, which are melted by ultrafast laser irradiation at different resonance wavelengths [19]. A metallic nanostructure, however, is usually larger in size than a DNA strand. Consequently, it is difficult to build a release system inside a single DNA structure to manipulate a specific strand selectively.
In this paper, we report on the optically controlled release of DNA based on a nonradiative relaxation process of dark quenchers. Dark quenchers are typically used in biomolecular sensing, and are also used in other applications such as nanoparticle aggregation [20]. This study proposes new usage of dark quenches. An excited dark quencher relaxes to its ground state or a lower energy state completely through a nonradiative process, and it thus produces thermal or photochemical energy that leads to dehybridization. We selected black hole quenchers (BHQs) from several types of available dark quenchers because they have high dissipation yield and their absorption spectrum is relatively broad. A BHQ, which is smaller than a DNA strand, can be attached at an arbitrary position in a DNA strand, meaning that the release system can be constructed inside a single DNA structure to manipulate a specific part of that structure. In addition, the method offers valuable extended functions. For example, using a BHQ as an acceptor in a fluorescence resonance energy transfer (FRET) system means that the release of DNA strands can be controlled using light at the absorption peak wavelength of the donor molecule. This provides wavelength selectivity for parallel or multiplexed control. We experimentally demonstrate the controlled release of DNA by excitation of BHQs, an ability to drive subsequent reactions with the released DNA, and localization of the controlled release process on a glass substrate.
2. Method
Atoms/molecules in an excited energy state return to their ground energy state or a lower-energy excited state through a radiative process such as fluorescence emission and/or a nonradiative process such as thermal relaxation or photochemical relaxation. Dehybridization of DNA, or the release of DNA, requires energy of some sort. This energy can be supplied via a nonradiative process. BHQs are expected to be efficient energy supply sources for DNA release because they relax completely through nonradiative processes. When a single molecule is modelled as a two-level system, the dissipated power, pd, is written as [21]
| (1) |
| (2) |
where pabs is the power absorbed by the molecule, ηd is the dissipation yield, Pex is the power of the excitation light, σ is the absorption cross-section, and A is the area of excitation. Pex at the saturated excitation state, , is written as
| (3) |
where h is Planck’s constant, ν is the frequency of the excitation light, and τ is the fluorescence lifetime. By assigning to Pex, Eq. (1) then becomes
| (4) |
A larger pd is obtained with a larger ηd and a smaller τ. Based on this relationship, we selected a BHQ as an energy source for the release of DNA among fluorescent or quencher molecules.
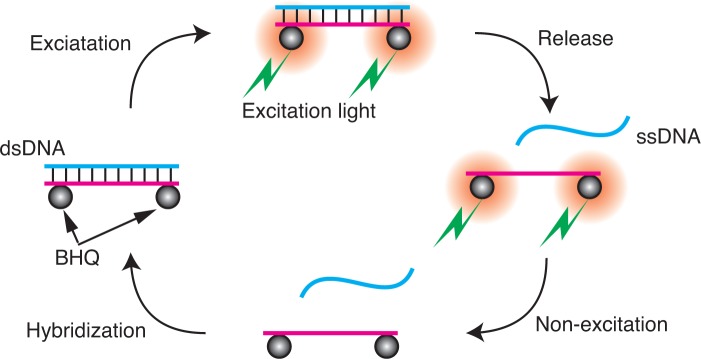
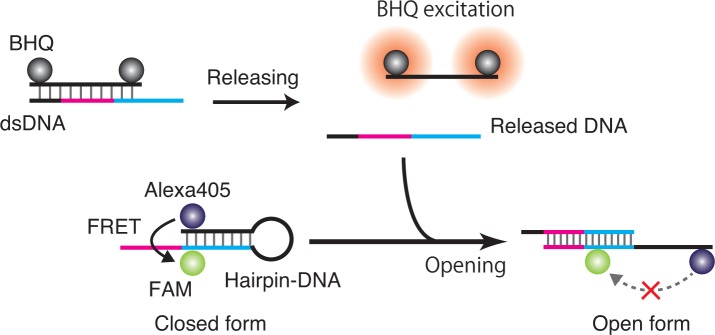
Figure 1 shows a schematic diagram of the process of optically controlled release of ssDNA using BHQs. A target ssDNA to be released is hybridized with another DNA strand that contains the complementary sequence to which the BHQs are attached to form dsDNA. When a BHQ is irradiated by the excitation light, it produces thermal and/or photochemical energy. Because of this thermal energy, the temperature around the BHQ increases, and the dsDNA subsequently dehybridizes to release the target strand. After the excitation stops, the strands can then hybridize again to return to their initial states. Photochemical relaxation of the excited BHQs may also contribute to the dehybridization of the dsDNA; however, clarification of this mechanism is not an objective of this work, and no further description of the mechanism is provided in this paper.
Fig. 1.
Schematic diagram of optically controlled release of ssDNA using BHQs.
Using this optical method, the release of ssDNA is induced in localized volumes at desired times using light. In addition, the most strongly affected part of a DNA nanostructure can be restricted because the size of a BHQ is sufficiently small to be comparable to a DNA strand. The released DNA is modification-free, and can ordinarily induce a subsequent reaction. This function is useful for the control of DNA-mediated biological events by indirect use of external optical signals. Because of the inherent design flexibility of the released DNA, various subsequent reactions can be considered.
The method also offers some extended applications. A BHQ is often used as an acceptor in a FRET system and thus a FRET-based DNA-release system can be constructed. A BHQ accepts excitation energy from various kinds of fluorescent molecules that act as donors. These excitation wavelengths are different for the individual fluorescent molecules, and therefore the selected release or multiplexed release of ssDNA will be achievable when the BHQ is used as an acceptor. In addition, FRET is an excellent probing system for detection of the states of DNA nanostructures [22, 23]. By combining a FRET-based DNA release system with a DNA nanostructure, ssDNA will be released, depending on its state. FRET is also useful in DNA-based computation applications [24, 25]. For example, we have demonstrated a molecular logic operation scheme called DNA scaffold logic, which is based on the self-assembly of fluorescent molecules on a DNA scaffold depending on a set of input molecules and is also based on cascaded FRET signaling [25, 26]. In the original DNA scaffold logic, the output signal is produced as a FRET signal by a reporter molecule on the scaffold. When using a BHQ as the reporter, the output signal provided is the state of the DNA release, i.e., it denotes whether ssDNA is released (when the output is ”1”) or not released (when the output is ”0”).
3. Experiments
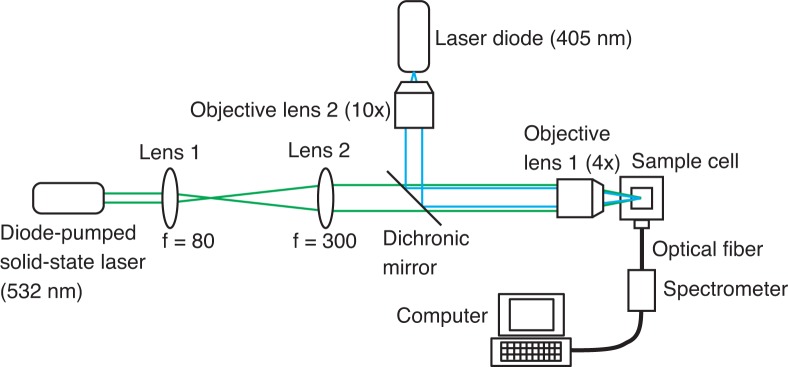
The method used for the controlled release of DNA strands using BHQs was demonstrated and characterized experimentally. Four types of BHQs, BHQ-0, BHQ-1, BHQ-2, and BHQ-3, are commercially available at the present. They have different absorption spectra, whose peak wavelengths are 495 nm, 534 nm, 579 nm, and 672 nm, respectively [27]. The full width at half maximum of absorption band is wider than 100 nm [28]. Given the choice of BHQ-0, BHQ-1, BHQ-2, and BHQ-3, we used BHQ-1 after consideration of the absorption spectrum of the BHQ, the wavelength of the BHQ-excitation laser, and the excitation and emission wavelengths of the fluorescence molecule used. In the following text, ”BHQ” means ”BHQ-1”. The sequences and modifications of the strands used in the experiments are summarized in Tab. 1. The DNA strands with Alexa 405 were purchased from Japan Bio Services Co., Ltd. and the other strands, including the strands with BHQs, were purchased from Tsukuba Oligo Service CO., Ltd. In the first experiment, a complementary pair of 10-base DNA strands, St and , were used. St is the target strand that is to be released. The BHQs were attached to at its ends. One of the BHQs was used as a quencher for Alexa 405 in addition to its function as an energy source. The state of the pair of strands, i.e., the hybridization state or the dehybridization state, was sensed as a FRET signal from Alexa 405. The experimental setup is shown in Fig. 2. A continuous-wave diode-pumped solid state laser (Spectra-Physics KK., Excelsior 532 Single Mode, wavelength: 532 nm) is used to excite the BHQs. The beam, which has a waist of 0.32 mm, is focused by objective lens 1 (OLYMPUS, UPlanFl 4×, NA of 0.13) on the sample plane. The power of the focused spot was 90 mW. A beam from a laser diode (RS Components Ltd., DL-405-0.14, wavelength: 405 nm) is focused by objective lens 1 and objective lens 2 (Melles Griot, 04 OAS 010 10×, NA of 0.25) to be used as the fluorescence excitation source, and the fluorescence signal from Alexa 405 is measured using a spectrometer (B& W TEK Inc., BTC112E).
Table 1.
Sequences and modifications of DNAs used in the experiments.
| Name | Sequence |
|---|---|
| St | 5′-ATACAAGATA-Alexa405-3′ |
| 5′-BHQ-1-TATCTTGTAT-BHQ-1-3′ | |
| 5′-ATACAAGATA-3′ | |
| 5′-TATCTTGTAT-3′ | |
| So | 5′-AGAGCGATACAAGATA-3′ |
| Sh | 5′-TCTTGTAT*CGCTCTGAAAGAGCGAT-Alexa405-3′ (*: FAM) |
| Si | 5′-Biotin-CAAAAATACAAGATA-Alexa405-3′ |
Fig. 2.
Experimental setup. f: focal length.
Three solutions were prepared; solution (i) contained strands (5 μM) and in an SSC buffer (NaCl 0.135 M, sodium citrate 0.0135 M), solution (ii) contained strand St (5 μM) alone in the SSC buffer, and solution (iii) contained the strands St (5 μM) and (5 μM) in the SSC buffer. Solutions (i) and (ii) were prepared for control experiments. Solution (i) was used to confirm that the BHQs emitted no fluorescence and solution (ii) was used to show that the fluorescence from Alexa405 provided no changes with and without the BHQ-excitation light. We tested only pairs of perfect complementary strands in the experiments for the following reasons. When using a pair of strands with a single or more mismatches, double-stranded DNAs become unstable or are not formed. In this case, it is difficult to know that the dehybridization is induced by the effect of BHQs or not. Furthermore, for achieving optically controlled release, it is important to maintain the non-release state during no BHQ excitation, in addition to release the target strand during BHQ excitation. From this point of view, perfect complementary strands are adequate to be used in practical use.
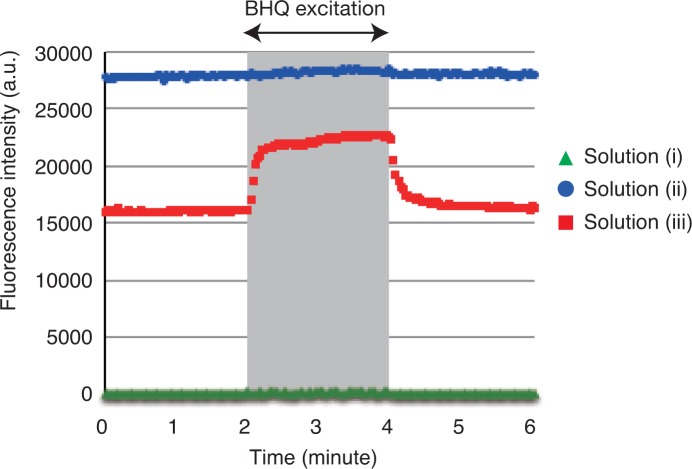
Figure 3 shows the time course of the fluorescence intensity at the emission peak wavelength (425 nm) of Alexa 405 that was measured around the two-minute excitation of the BHQs. The intensity capturing duration was two seconds for solution (i) and one second for solutions (ii) and (iii). Solution (i) contained BHQs, but no fluorescence molecules (Alexa405), and thus the measured intensity was around zero during the entire operation. Solution (ii) contained no BHQs, and thus the high fluorescence intensity was maintained during the entire operation. In contrast, in solution (iii), the fluorescence intensity was in the background level in the initial state because Alexa 405 was quenched by the BHQs. At the start of the excitation process, the fluorescence intensity increased, and it then decreased again immediately after the excitation stopped. The results indicate that the system is in the released state when and only when the BHQs are excited.
Fig. 3.
Fluorescence intensity of Alexa 405 measured around the two-minute excitation of the BHQs. The gray area indicates the BHQ-excitation period.
The ratio, α, of the released strands is estimated using the following equation:
| (5) |
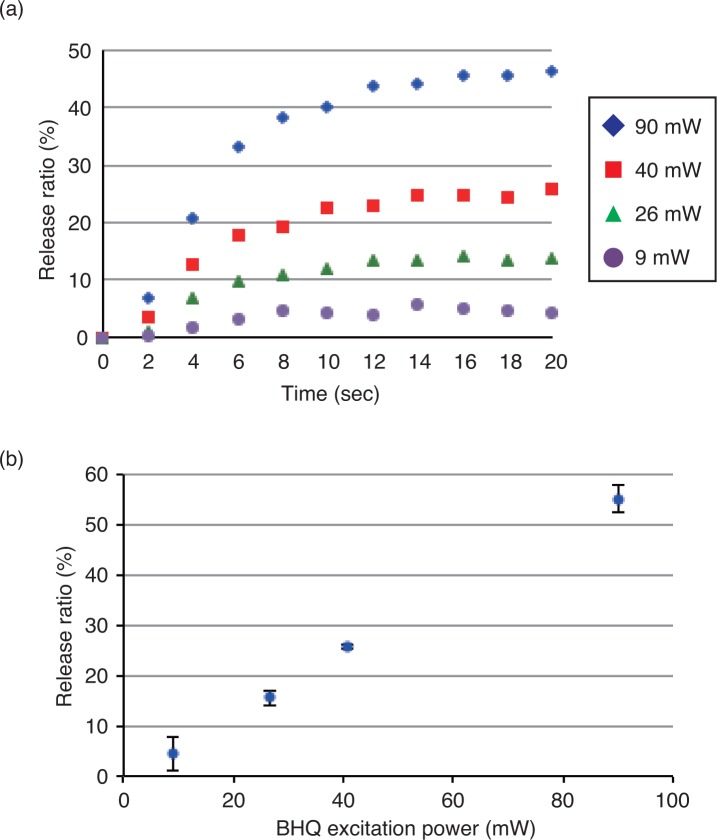
Here, Is is the fluorescence intensity that was measured for solution (ii), I is the corresponding intensity for solution (iii) during irradiation by the excitation light, and Id is the intensity for solution (iii) when there is no irradiation. To clarify the dependence of α on the power of the excitation light, we irradiated solution (iii) with various excitation powers. Figure 4(a) shows the time course of α after the excitation begins for various excitation powers. When calculating α, we obtained Is as the averaged intensity during the period from 0 to 6 min for solution (ii), Id as the averaged intensity during the period from 0 to 2 min for solution (iii), and I as the instantaneous intensity. The ratio α depends on the excitation power at any given time, which indicates that the number of released strands can be adjusted by tuning of the excitation power. Optically controlled release reached a near-equilibrium state 20 s or earlier after starting excitation. α at the near-equilibrium state is less than 100%. This is because the excitation beam irradiates only a part of a sample solution, and optically controlled release is not induced within the entire solution.
Fig. 4.
(a) Time course of the release ratio α for various excitation powers. (b) Variation of α due to the BHQ excitation power. Error bars indicate the standard deviation of three measurements.
The ratio α averaged over the period from 20 s to 2 min after starting the excitation (which is considered to be at the equilibrium state) for different excitation powers is shown in Fig. 4(b). The ratio α was measured three times at every excitation powers. In our setup, the sample solution is not entirely irradiated with the BHQ-excitation light, and the strands come into and go out of the irradiation area repeatedly by diffusion. The behavior of the strands changes owing to only a little difference of conditions including the temperature, the BHQ-excitation power, and the concentrations of the individual DNAs. This is a possible reason for measurement errors shown in Fig. 4(b). During the measurement, the fluctuation of the excitation power was no more than 1%. An approximately linear relationship is found between the excitation power and α, although modeling of this process is difficult at present. According to Ref. [21], the parameters τ and σ of the BHQ in Eq. (3) are estimated to be τ = 0.05 nsec and σ = 0.013 nm2, and then the saturated excitation power can be calculated to be 282 W. is much higher than the maximum power used in the experiment, and α will be increased by increasing the excitation power or using a higher-numerical aperture (NA) objective lens. Note that increasing the number of BHQs attached to each strand will be effective to enhance α with the same excitation power. For example, use of superquenchers, assemblies of multiple quenchers, is a possible approach to increase the number [29].
After ceasing excitation, the strand with the BHQs can hybridize again with the target strand because no physical or chemical change caused by the release process occurs in these strands, and the target DNA strand is then re-released by the excitation of the BHQs. Figure 5 shows the change that occurs in α during such repeated release and hybridization operations (five cycles). Each excitation term lasted for 1 min and the excitation power was 89.8 mW. During the calculation of α, we obtained Is as the averaged intensity during the period from 0 to 11 minutes for solution (ii), Id as the averaged intensity during the period from 0 to 1 min for solution (iii), and I is the instantaneous intensity. The fluorescence intensity varied periodically with following the BHQ excitation process. Dehybridization (release) and hybridization were both achieved reversibly and repeatedly based on the proposed method.
Fig. 5.
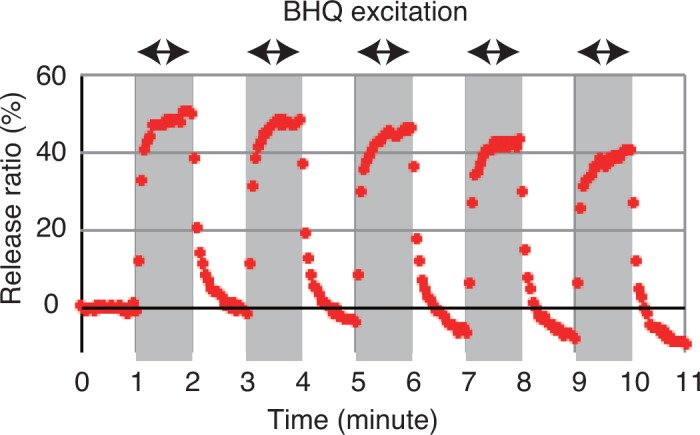
Variation of the release raito α during repeated operations. The grey areas indicate the periods of BHQ excitation.
A released DNA strand can ordinarily work in a subsequent reaction because the released strand itself is modification-free. To demonstrate this capability, we designed a scheme in which a hairpin DNA structure is opened by the released strand. The investigated scheme is shown in Fig. 6. The system contains a hairpin DNA with a sticky end, an opening strand that is released to open the hairpin structure, and a BHQ-attached strand that is partially complementary to the opening strand. By irradiating the BHQs with the excitation light, the opening strand is released; it then reacts with the hairpin DNA to open the hairpin structure. Strands , , So, and Sh, the sequences and modifications of which are shown in Table 1, were used: and have the same sequences with and without the BHQs, respectively, So is the opening strand, and Sh is the hairpin DNA. A FRET pair, consisting of Alexa 405 acting as a donor and the indicator dye FAM acting as an acceptor, were used to detect the form of the hairpin DNA. Three solutions containing an SSC buffer (NaCl 3 M, Sodium Citrate 0.3 M) were prepared: solution (iv) contained all the components (strands , So, and Sh), while solution (v) contained strands , So, and Sh, and solution (vi) only contained strand Sh. An operating cycle consists of a 5 min excitation and the measurement of the fluorescence intensity when the excitation stops. The cycle was repeated six times. For solution (iv), the fluorescence intensity was also measured without excitation of the BHQs.
Fig. 6.
Scheme of reaction for opening of hairpin DNA using optically-released ssDNA.
Figure 7 shows the measured fluorescence intensities for the individual cycles. The intensity is normalized with respect to the initial intensity. The highest intensity was obtained for solution (iv) with excitation, while little increase in intensity was measured for solution (iv) without application of the excitation. The results show that the hairpin DNA opens following the release of the opening strand by BHQ excitation. The intensities for solutions (v) and (vi) increase with time, although they are not expected to increase. One possible reason for this is photobleaching of FAM caused by irradiation by the BHQ-excitation light. FAM is the acceptor of the FRET pair that is used to detect the form of the hairpin DNA, and the FRET signal increases because of the photobleaching of FAM. However, the increase in the intensity for solution (iv) with excitation is greater than those for solutions (v) and (vi). These results demonstrate that the reaction required to open the hairpin DNA is driven based on the optically controlled release of DNA. Note that this method is not limited to the reaction of opening hairpin DNA, and it can be applied to other reactions through specific design.
Fig. 7.
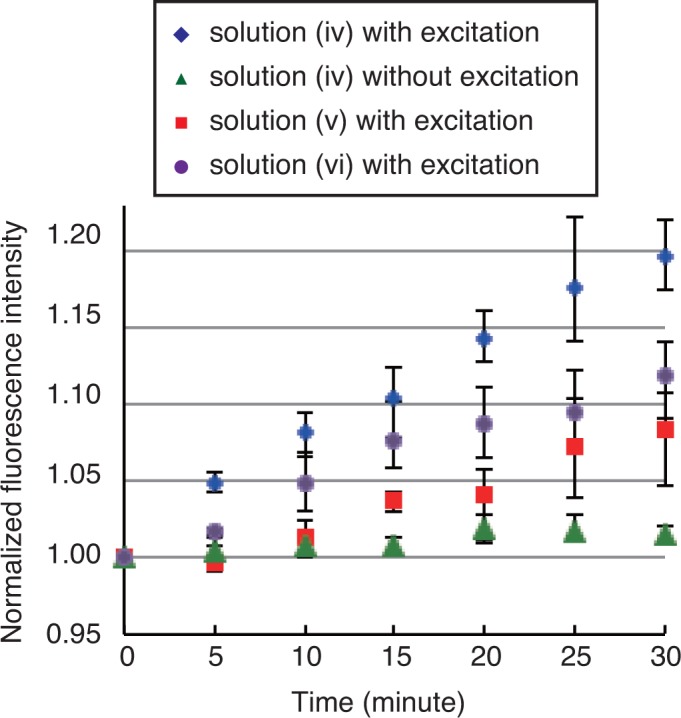
Fluorescence intensity measured during reaction to open the hairpin DNA based on optically controlled release of the opening DNA. Error bars indicate the maximum and minimum values among the three measurements.
To verify the controlled release process in a local space, a DNA release system was constructed on a substrate and subsequently demonstrated. The system was immobilized on a glass substrate via a biotin-streptavidin bond [30]. The immobilization method is described as follows. Biotinylated Bovine Serum Albumin (BioVision, 7099-5, 12 biotin/BSA) was adsorbed on the substrate by a hydrophobic interaction, and then streptavidin (Thermo Fisher Scientific) was bound to the biotin. Subsequently, the biotinylated DNA strand Si was bound to the streptavidin, and the strand that was used as the target strand to be released was added to allow it to hybridize with Si. The sequences and the modifications of the strands used are shown in Table 1. Figure 8 shows the experimental setup used. Similar to Fig. 2, the diode-pumped solid-state laser was used for BHQ excitation and the laser diode was used for fluorescence excitation. The beam waist of the BHQ-excitation light behind the objective lens was 0.32 mm wide. The beam diameter (1/e2 of the peak intensity) of the spot that was focused by the objective lens (Olympus, water immersion, LUMPlanFl, NA of 1.00) is calculated to be 3.8 μm. The fluorescence image was captured using a charge-coupled device (CCD) camera (Roper Scientific, CoolSNAP fx Monochrome) through a bandpass filter (transmission range: 415 nm – 460 nm).
Fig. 8.
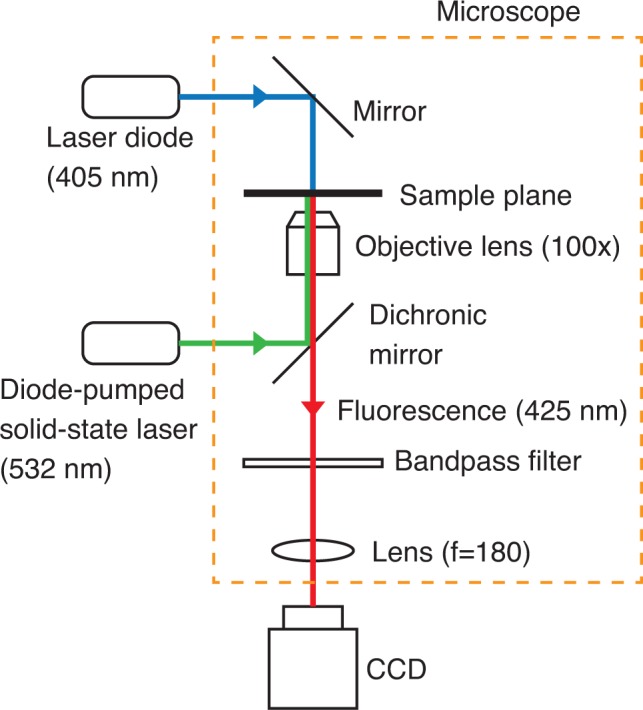
Experimental setup used for controlled release on a glass substrate.
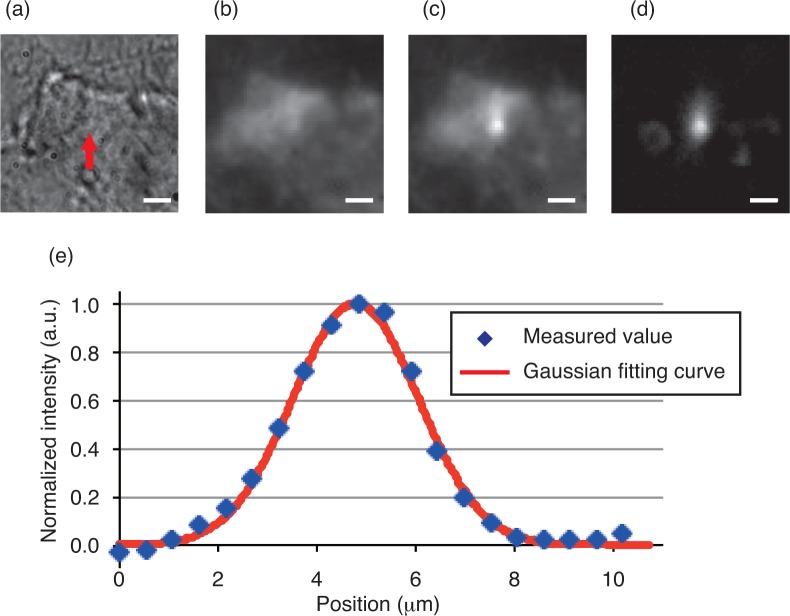
The fluorescence images were captured before and after 10-sec periods of irradiation with the BHQ excitation light. To increase the signal-to-noise ratio, an 8 × 8 binning operation was introduced for the fluorescence observation. Figures 9(a)–(d) show the captured and processed images. Figure 9(d) shows that the fluorescence intensity increased around the irradiation spot. Figure 9(e) shows the intensity profile of a cross-section that includes the maximum intensity point in Fig. 9(d) and the Gaussian fitting curve. The fluorescence spot width was measured to be 5.1 μm. DNA release was achieved within a local area with a diameter of 5 micrometers. The results here suggest that controlled DNA release can be realized at multiple positions or in separate areas with resolution of 5 micrometers or better by simply shaping the excitation beam.
Fig. 9.
(a) Bright field image of substrate. The position of the focused spot is indicated by an arrow. Fluorescence images captured (b) before and (c) after 10 s period of excitation, and (d) the difference image between them. Scale bar: 5 μm. (e) Intensity profile of the fluorescence distribution shown in (d).
4. Conclusions
We studied the optically controlled release of DNA strands via the nonradiative relaxation of excited BHQs. The experimental results showed that the target DNA strands were released with ratios of up to 50%, and this ratio was controllable by tuning of the power of the BHQ-excitation light. The released strands were free from modifications, meaning that they were hybridized again with their complementary strands reversibly and repeatedly. The strands were able to induce an ordinary reaction to open a hairpin DNA as an example for subsequent reactions. We also showed that the controlled release of the target DNA could be achieved within a local area with a diameter of 5 micrometers. BHQs are often used as acceptors in FRET systems, and various fluorescent molecules can be used as donors because of the broad absorption spectra of the BHQs. This indicates that the method is extendable to the selective and multiplexed control of DNA strand release.
References and links
- 1.Larance M., Lamond A. I., “Multidimensional proteomics for cell biology,” Nat. Rev. Mol. Cell Biol. 16, 269–280 (2015). 10.1038/nrm3970 [DOI] [PubMed] [Google Scholar]
- 2.Nandagopal N., Elowitz M. B., “Synthetic Biology: Integrated Gene Circuits,” Science 333(6047), 1244–1248 (2011). 10.1126/science.1207084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deisseroth K., “Optogenetics,” Nat. Methods 8(1), 26–29 (2011). 10.1038/nmeth.f.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemphill J., Borchardt E. K., Brown K., Asokan A., Deiters A., “Optical Control of CRISPR/Cas9 Gene Editing,” J. Am. Chem. Soc. 137(17), 5642–5645 (2015). 10.1021/ja512664v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nomoto T., Fukushima S., Kumagai M., Machitani K., Arnida, Matsumoto Y., Oba M., Miyata K., Osada K., Nishiyama N., Kataoka K., “Three-layered polyplex micelle as a multifunctional nanocarrier platform for light-induced systemic gene transfer,” Nat. Commun. 5, 3545 (2014). 10.1038/ncomms4545 [DOI] [PubMed] [Google Scholar]
- 6.Lin L., Liu L., Zhao B., Xie R., Lin W., Li H., Li Y., Shi M., Chen Y.-G., Springer T. A., Chen X., “Carbon nanotube-assisted optical activation of TGF-β signaling by near-infrared light,” Nat. Nanotechnol. 10(5), 465–471 (2015). 10.1038/nnano.2015.28 [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., He H., Li S., Liu D., Lan B., Hu M., Cao Y., Wang C., “All-optical regulation of gene expression in targeted cells,” Sci. Rep. 4, 5346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai T.-L., Shieh D.-B., Yeh C.-S., Tzeng Y., Htet K., Chuang K.-S., Hwu J. R., Sui W.-C., “The down regulation of target genes by photo activated DNA nanoscissors,” Biomaterials 31(25), 6545–6554 (2010). 10.1016/j.biomaterials.2010.04.058 [DOI] [PubMed] [Google Scholar]
- 9.Sakamoto T., Shigeno A., Ohtaki Y., Fujimoto K., “Photo-regulation of constitutive gene expression in living cells by using ultrafast photo-cross-linking oligonucleotides,” Biomater. Sci. 2(9), 1154–1157 (2014). 10.1039/C4BM00117F [DOI] [PubMed] [Google Scholar]
- 10.Hemphill J., Liu Q., Uprety R., Samanta S., Tsang M., Juliano R. L., Deiters A., “Conditional Control of Alternative Splicing through Light-Triggered Splice-Switching Oligonucleotides,” J. Am. Chem. Soc. 137(10), 3656–3663 (2015). 10.1021/jacs.5b00580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broichhagen J., Schönberger M., Cork S. C., Frank J. A., Marchetti P., Bugliani M., Shapiro A. M. J., Trapp S., Rutter G. A., Hodson D. J., Trauner D., “Optical control of insulin release using a photoswitchable sulfonylurea,” Nat. Commun. 5, 5116 (2014). 10.1038/ncomms6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hossion A. M. L., Bio M., Nkepang G., Awuah S. G., You Y., “Visible Light Controlled Release of Anticancer Drug through Double Activation of Prodrug,” ACS Med. Chem. Lett. 4(1), 124–127 (2013). 10.1021/ml3003617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huu V. A. N., Luo J., Zhu J., Zhu J., Patel S., Boone A., Mahmoud E., McFearin C., Olejniczak J., Lux C. G., Lux J., Fomina N., Huynh M., Zhang K., Almutairi A., “Light-responsive nanoparticle depot to control release of a small molecule angiogenesis inhibitor in the posterior segment of the eye,” J. Control. Release 200, 71–77 (2015). 10.1016/j.jconrel.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han G., You C.-C., Kim B., Turingan R. S., Forbes N. S., Martin C. T., Rotello V. M., “Light-regulated release of DNA and its delivery to nuclei by means of photolabile gold nanoparticles,” Angew. Chem. Int. Ed. Engl. 45(19), 3165–3169 (2006). 10.1002/anie.200600214 [DOI] [PubMed] [Google Scholar]
- 15.Lia L., Tong R., Chu H., Wang W., Langer R., Kohane D. S., “Aptamer photoregulation in vivo,” PNAS 111(48), 17099–17103 (2014). 10.1073/pnas.1420105111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barhoumi A., Huschka R., Bardhan R., Knight M. W., Halas N. J., “Light-induced release of DNA from plasmon-resonant nanoparticles: Towards light-controlled gene therapy,” Chem. Phys. Lett. 482(4–6), 171–179 (2009). 10.1016/j.cplett.2009.09.076 [DOI] [Google Scholar]
- 17.Yamashita S., Fukushima H., Akiyama Y., Niidome Y., Mori T., Katayama Y., Niidome T., “Controlled-release system of single-stranded DNA triggered by the photothermal effect of gold nanorods and its in vivo application,” Bioorg. Med. Chem. 19(7), 2130–2135 (2011). 10.1016/j.bmc.2011.02.042 [DOI] [PubMed] [Google Scholar]
- 18.Lee S. E., Liu G. L., Kim F., Lee L. P., “Remote Optical Switch for Localized and Selective Control of Gene Interference,” Nano Lett. 9(2), 562–570 (2009). 10.1021/nl802689k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wijaya A., Schaffer S. B., Pallares I. G., Hamad-Schifferli K., “Selective Release of Multiple DNA Oligonucleotides from Gold Nanorods,” ACS Nano 3(1), 80–86 (2009). 10.1021/nn800702n [DOI] [PubMed] [Google Scholar]
- 20.Lovell J. F., Jin H., Ng K. K., Zheng G., “Programmed Nanoparticle Aggregation Using Molecular Beacons,” Angew. Chem. Int. Ed. 49, 7917–7919 (2010). 10.1002/anie.201003846 [DOI] [PubMed] [Google Scholar]
- 21.Gaiduk A., Yorulmaz M., Ruijgrok P. V., Orrit M., “Room-Temperature Detection of a Single Molecule’s Absorption by Photothermal Contrast,” Science 330(6002), 353–356 (2010). 10.1126/science.1195475 [DOI] [PubMed] [Google Scholar]
- 22.Bae W., Kim K., Min D., Ryu J.-K., Hyeon C., Yoon T.-Y., “Programmed folding of DNA origami structures through single-molecule force control,” Nat. Commun. 5, 5654 (2014). 10.1038/ncomms6654 [DOI] [PubMed] [Google Scholar]
- 23.Andersen E. S., Dong M., Nielsen M. M., Jahn K., Subramani R., Mamdouh W., Golas M. M., Sander B., Stark H., Oliveira C. L. P., Pedersen J. S., Birkedal V., Besenbacher F., Gothelf K. V., Kjems J., “Self-assembly of a nanoscale DNA box with a controllable lid,” Nature 459(7243), 73–76 (2009) . 10.1038/nature07971 [DOI] [PubMed] [Google Scholar]
- 24.Pistol C., Dwyer C., Lebeck A. R., “Nanoscale Optical Computing Using Resonance Energy Transfer Logic,” IEEE Micro 28(6), 7–18 (2008). 10.1109/MM.2008.91 [DOI] [Google Scholar]
- 25.Nishimura T., Ogura Y., Tanida J., “Fluorescence resonance energy transfer-based molecular logic circuit using a DNA scaffold,” Appl. Phys. Lett. 101(23), 233703 (2012). 10.1063/1.4769812 [DOI] [Google Scholar]
- 26.Nishimura T., Fujii R., Ogura Y., Tanida J., “Optically controllable molecular logic circuits,” Appl. Phys. Lett. 107(1), 013701 (2015). 10.1063/1.4926361 [DOI] [Google Scholar]
- 27.Johansson M. K., “Choosing reporter-quencher pairs for efficient quenching through formation of intramolecular dimers,” in Fluorescent Energy Transfer Nucleic Acid Probes: Designs and Protocols, Didenko V. V., ed. (Humana Press, 2006). 10.1385/1-59745-069-3:17 [DOI] [PubMed] [Google Scholar]
- 28.Marras S. A. E., Kramer F. R., Tyagi S., “Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes,” Nucleic Acids Res., 30(21), e122 (2002). 10.1093/nar/gnf121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovell J. F., Chen J., Huynh E., Jarvi M. T., Wilson B. C., Zheng G., “Facile Synthesis of Advanced Photodynamic Molecular Beacon Architectures,” Bioconjugate Chem. 21, 1023–1025 (2010). 10.1021/bc100178z [DOI] [PubMed] [Google Scholar]
- 30.Li J., Tan W., Wang K., Xiao D., Yang X., He X., Tang Z., “Ultrasensitive Optical DNA Biosensor Based on Surface Immobilization of Molecular Beacon by a Bridge Structure,” Anal. Sci. 17(10), 1149–1153 (2001). 10.2116/analsci.17.1149 [DOI] [PubMed] [Google Scholar]