Abstract
Coherent fiber bundles can be used to relay the image plane from the distal tip of an endomicroscope to an external confocal microscopy system. The frame rate is therefore determined by the speed of the microscope’s laser scanning system which, at 10-20 Hz, may be undesirably low for in vivo clinical applications. Line-scanning allows an increase in the frame rate by an order of magnitude in exchange for some loss of optical sectioning, but the width of the detector slit cannot easily be adapted to suit different imaging conditions. The rolling shutter of a CMOS camera can be used as a virtual detector slit for a bench-top line-scanning confocal microscope, and here we extend this idea to endomicroscopy. By synchronizing the camera rolling shutter with a scanning laser line we achieve confocal imaging with an electronically variable detector slit. This architecture allows us to acquire every other frame with the detector slit offset by a known distance, and we show that subtracting this second image leads to improved optical sectioning.
OCIS codes: (170.2150) Endoscopic imaging, (110.0180) Microscopy
1. Introduction
In endoscopic microscopy, or ‘endomicroscopy’, coherent fiber bundle image guides provide a convenient means of relaying images from the tissue to external microscope optics [1–5]. This is particularly advantageous for confocal imaging because the laser scanning system can sit outside of the patient. This avoids the need for miniaturized fiber scanners or micro-mirrors [6–8], meaning that the probe itself can be entirely passive. These probes are small enough to deploy through the working channel of a standard endoscope, simplifying their use in the clinical workflow, and allowing interchange with biopsy forceps as required. As a result, fiber bundle endomicroscopy - and confocal fluorescence endomicroscopy in particular - is finding a wide range of clinical applications, including monitoring of Barrett’s esophagus [9], diagnosis of colorectal lesions [10], and investigation of pancreatic cysts [11].
Nevertheless, the costs and complexity of point-by-point confocal scanning, and the low frame rate of typically 10–20 Hz that it imposes, have motivated the development of alternative endomicroscope designs. Widefield epi-fluorescence illumination endomicroscopy (known as high resolution microendoscopy or HRME) [12] has been validated for a number of applications, including for diagnosis of esophageal [13], oral [14], cervical [15] and bowel [13] cancers. It has the advantage of significantly reduced complexity due to the use of incoherent, non-scanned illumination, but as a result does not offer optical sectioning. Topical staining is therefore used to minimize the contribution of out-of-focus fluorescence, allowing key features to be identified [16]. However, for many applications there is a clear decrease in image quality compared to confocal imaging [17], and HRME is not used with intravenous fluorescein, where the out-of-focus signal is significantly greater. Structured illumination techniques can be used to achieve optical sectioning under widefield illumination [17–20], but generally at the expense of introducing motion artifacts [19] and reducing the signal to noise ratio [17] and image bit depth.
If the point-by-point scanning and detector pinhole of a confocal endomicroscope are replaced by a scanning laser line and a detector slit, respectively, then the frame rate can be improved significantly [21–23]. This arrangement provides reduced optical sectioning, with a significant tail to the axial profile, but frame rates of up to 120 Hz have been reported [24]. The line-scanning mechanism maintains much of the complexity of point-scanning confocal endomicroscopy, and is again typically only used with topical stains. However, if the reduction in optical sectioning strength relative to full-confocal microscopy can be tolerated, then there are significant advantages to line-scanning, including a higher frame rate to assist mosaicking [24,25] and the possibility of multispectral imaging [26].
In previous work, the detector slit has been implemented both as a physical slit, with the line subsequently re-scanned onto a 2D camera [21–23], and by a 1D linear camera [24]. In this paper, we provide the first demonstration of line-scanning fiber bundle endomicroscopy using the rolling shutter of a CMOS scanner as a virtual detector slit. By electronically varying the width of this virtual slit it’s possible to adjust the optical sectioning strength to optimize sensitivity for specific imaging applications while maintaining a high frame rate limited only by the camera read-out time (120 Hz in this instance). This approach has recently been demonstrated (and now commercialized by Aeon Imaging) for bench-top confocal microscopy [27], with the commercial system using a spatial light modulator rather than a scanning mirror. Similar exploitation of the rolling shutter has also been explored in light sheet microscopy [28,29], but its applicability to fiber bundle microscopy has not yet been evaluated.
We also show that depth sectioning can be enhanced by subtracting a second image with the scanning line and the virtual slit offset. The second image contains a first order estimate of the out-of-focus light that has not been rejected by the slit, and hence, when subtracted from the standard image, reduces the residual out-of-focus background [30]. While this subtraction technique does not recover the full performance of a confocal endomicroscope, particularly with respect to noise, and is likely to be unsuitable for imaging using intravenous fluorescein, we demonstrate improved sectioning when imaging topically-stained ex vivo tissue.
The subtraction technique is a similar concept to that presented for a bench-top microscope by Poher et al. [30], in which the axial resolution was improved to that of a full-confocal system. The underlying theory of our approach is the same as that shown in Ref [30], and we refer the reader to this report for a mathematical treatment. Our more practical implementation, which is significantly different from the previous demonstration which used an LED array and a conventional camera (i.e. with no rolling shutter), allows for real-time use of the technique at 60 fps. This high speed is possible due to the versatility of the virtual slit approach, but would be difficult or impossible to achieve with the previously reported line-scanning endomicroscopy architectures.
2. Methods
To confirm the feasibility of the virtual slit scheme, and to characterize its performance, we developed the virtual slit line-scanning endomicroscopy system shown in Fig. 1. The output from a 50 mW, 488 nm laser (Vortran Stradus 488) passes through an excitation filter (Thorlabs FES0500) and then is expanded by a 2.5X telescope to a beam diameter of approximately 4 mm. This is then reflected off a galvanometer scanning mirror (Thorlabs GVS001) and enters a telescope consisting of a cylindrical lens (f = 50 mm) and an achromatic doublet (f = 50 mm). A dichroic mirror (Thorlabs MD498) reflects the beam onto a 10x plan infinity-corrected microscope objective (Thorlabs RMS10X) which focuses it to a line on the proximal face of the fiber bundle. The system is arranged so as to form an image of the scanning mirror on the back aperture of the objective, avoiding beam translation and clipping by the objective aperture.
Fig. 1.
Schematic of virtual slit line-scanning endomicroscope.
For the results presented below, we used a commercially available Cellvizio Gastroflex UHD probe (Mauna Kea Technologies) which consists of a 30,000-core Fujikura fiber bundle (FIGH-30-650S), with 2.9 µm core spacing, coupled to a proprietary micro-lens with a magnification of approximately 2.5, inside a plastic sheath with an outer diameter of approximately 1.4 mm. The distal lens is encased in a rigid tube of outer diameter 2.6 mm, and has a nominal working distance of approximately 50 µm. This probe is designed for use with the Cellvizio confocal laser endomicroscopy system, and so we built a custom adapter to mount the proximal end to the endomicroscopy system.
The probe transfers the laser line to the tissue, with some pixilation due to the fiber core pattern, and collects emitted fluorescence. During scanning, a time-averaged power of 1.6 mW is delivered to the tip of the probe. The maximum power, obtained when the line is at the centre of the fibre bundle, is 5.1 mW. The average line width at the tissue (measured by imaging the line onto a camera with a 20x objective) is 1.6 µm.
The proximal face of the bundle is imaged onto the CMOS camera (Point Grey Flea 3), via a fluorescence emission filter (Thorlabs FEL0500) and notch filter (Thorlabs NF488-15), using an achromatic doublet (f = 75 mm). The 600 µm-diameter active area of the bundle is imaged to a size of approximately 2.47 mm on the camera chip. As the camera has a pixel pitch of 3.63 µm, this provides 680 pixels across the bundle, and provides better than Nyquist sampling for the fiber cores. The camera can run at a frame rate of 120 Hz, which was used for all experiments reported below, and has a 12 bit analog to digital converter, although all images were scaled to 16 bits for transfer to the PC.
A virtual detector slit is provided by the rolling shutter of the camera, which is synchronized with the scanning laser line. Unlike when using a global shutter, wherein all lines of pixels on the camera are exposed simultaneously and read-out at the end of the exposure, with a rolling shutter the start of the exposure of each line is staggered. Each line therefore finishes its exposure at a different time, and is read-out immediately. Changing the exposure time changes how many lines are exposed simultaneously, and hence controls the width of the virtual slit. If reduced sufficiently, it can be used to ensure only a single line is being exposed at any one time.
For a camera line rate of (in our case equal to 130.7506 kHz), the number of concurrently exposed lines, , is given by
| (1) |
whereis the exposure time. For the Flea3 camera, the minimum allowable exposure is 7.63 µs, which results in a slit width of 1 pixel. The physical slit width, as projected onto the proximal face of the bundle, is therefore,
| (2) |
whereis the pixel size (3.63 µm in our case) andis the magnification factor between the camera and the bundle (4.11 in our case). The minimum slit width is therefore 1 pixel on the camera, or 0.88 µm on the proximal face of the bundle, adjustable in increments of 0.12 µm (equivalent to a change of 1 µs in exposure) up to a maximum which exceeds the diameter of the fiber bundle.
The camera is operated in free-run mode, which allows the full frame-rate to be achieved, and generates a pulse on its strobe output pin at the start of each frame acquisition. The pulse triggers the analog output of a DAQ board (National Instruments USB-6211) which is programmed to send a ramp-shaped voltage signal to the galvo mirror on each trigger, with some user-specified delay. By adjusting this delay, as well as the slope and offset of the voltage ramp, it is then possible to ensure that the scanning line of the camera rolling shutter line readout is aligned with the position of the scanning laser line. Fine adjustment of the ramp slope ensures that the virtual slit is aligned with the laser line throughout the acquisition of each frame. The relative rotation of the camera or the cylindrical lens must also be adjusted to ensure that the laser line is rotationally aligned with the pixel line, as this will otherwise lead to non-uniformity across the image.
An initial calibration of the voltage signal sent to the galvo scanner was made by driving the galvo to two arbitrary positions, and recording the positions of the lines in pixels. This allows for conversion between galvo voltage and the position of the scanning line in camera pixels and hence, through knowing the line rate of the camera, provides an estimate of the required laser line scanning speed (in V/s). The exposure time was then set to 26 µs, and an offset to the linear voltage ramp was applied and adjusted by hand until the image brightness was maximized. Finally, the voltage ramp slope and offset were adjusted iteratively until a uniform, bright image of the bundle was obtained.
Prior to imaging, a background calibration is made by recording 100 frames with the tip of the probe covered. The mean of these images is then subtracted from subsequent image frames. A circular area of interest of 660 pixels diameter (582 µm on the proximal face of the bundle, 232 µm on the tissue) is taken to remove artifacts from the edges of the fiber bundle. Finally, a Gaussian filter (σ = 1.6 pixels, 1.4 µm on the bundle) is applied to remove the fiber core pattern.
To implement the subtraction-imaging system, the system is set to capture pairs of images in which the ramp pattern is offset by some fixed amount. The slit is therefore misaligned with the laser line by a fixed amount in every other frame. This is achieved by adjusting the voltage pattern sent to the galvo scanner so that the ramp is offset by the required amount. In practice, we generate a single waveform that provides the ramps for a pair of aligned and offset images, and trigger this on every other camera strobe pulse. The image frames acquired with the offset virtual slit are then subtracted from the normal image frames prior to other processing.
3. Results
We measured the optical sectioning performance of the system in reflection mode, as this allowed a mirror to be used as a thin target, avoiding the need for a sub-axial-resolution fluorescent layer. The system was converted to reflection mode by removing the emission filters, while the dichroic was left in place to act as an attenuator to prevent saturation of the camera. The probe was fixed to a translation stage, initially placed in contact with the mirror, and then moved away from it at 30 µm s−1, giving one image frame per 0.25 µm of axial shift. The mean pixel value from a 50x50 pixel region of interest at the centre of each image was taken as the intensity at that depth position. The background signal level, mainly due to reflections from the fiber bundle, was determined by acquiring an image frame with the mirror removed, and this value was then subtracted from each point on the profile.
For virtual slit widths ranging from 1 to 400 µm, Fig. 2(a) shows the axial distance from focus at which the collected intensity drops to 50% of the at-focus value. The best value obtained was approximately 4.5 µm, suggesting an axial resolution of 9 µm for a two-sided-profile. The continuous transition between confocal and non-confocal regimes is indicated by the linear relationship for widths between 3 and 100 µm (coefficient of regression = 0.49 µm/µm, intercept = 3.15 µm, R2 = 1). The relationship becomes non-linear at large and small slit widths. At large widths this is because the slit is now larger than the bundle diameter, while at small widths (below approximately 3 µm) the slit is smaller than the fiber core spacing. Examples of full sectioning profiles are provided in Fig. 2(b), where long tails can be observed, even for the smallest slit widths. Note that the focus position is some finite distance from the tip of the probe, and that the profiles are only plotted from the peak intensity onwards. Hence ‘0 depth’ is the focus position rather than the surface of the probe.
Fig. 2.
Optical sectioning performance of the virtual slit line-scanning endomicroscope. (a) Distance from focus at which collected intensity drops by 50% (i.e. half-width half-maximum) for a range of slit widths (note that the x-axis is log scale); (b) axial sectioning profiles for selected slit widths. The dashed line in (a) shows a linear fit to the data-points between 3 µm and 100 µm.
It should be noted that the slit width does not affect the lateral resolution, which is determined by the fibre core spacing and magnification of the distal lens. If the resolution is taken to be twice the core spacing, then the lateral resolution here is 2.3 µm. An example image of a USAF lateral resolution target acquired using the same probe can be seen in Ref [24].
Figure 3 shows the effect of changing the virtual slit width on images of a lens tissue paper phantom and ex vivo porcine esophageal mucosa tissue, both stained with acriflavine 0.02%. All images were acquired with the same hardware gain, but for display each image has been scaled so that the maximum pixel value is 255. The mean background pixel values, which are subtracted from each image prior to display, ranged from 1250 (2 µm slit) to 3680 (100 µm slit), compared to mean pixel values of 2194 (2 µm slit) and 21400 (100 µm slit) for a 100 x 100 pixel region in the centre of the esophageal images. This shows that the background signal can be considered largely ‘in-focus’ and that it will contribute significantly more to noise at smaller slit widths.
Fig. 3.
Endomicroscopy images of lens tissue paper stained with acriflavine (left images) and porcine esophageal mucosa tissue stained with acriflavine 0.02% (right images) for virtual detector slit widths of (a) 2 µm, (b) 3 µm, (c) 6 µm, (d) 12 µm, (e) 24 µm, and (f) 100 µm (as measured on the proximal face of the bundle). The insets in (a) and (b) show small regions with contrast adjusted to demonstrate slightly increased noise when using the 2 µm slit.
The transition between confocal and non-confocal imaging can be seen as the slit width is increased, and is particularly apparent between 12 and 24 µm. It can be seen that the images corresponding to a 2 µm slit exhibit no greater optical sectioning than those with a width of 3 µm, which is expected given the results above. However, reducing the width below 3 µm results in lowering of the signal collected, demonstrated by a slight but noticeable increase in visibility of camera read-out noise. Since these images were acquired in fluorescence mode, unlike the reflectance mode profiles used to generate Fig. 2, we expect the sectioning performance to be somewhat different, and indeed the qualitative change up to a slit width of 12 µm is rather small. The choice of optimal slit width should therefore be made based on assessment of image quality rather than nominal values predicted by Fig. 2.
As the slit width is increased, the system gradually transitions to a non-sectioning microscope, in which out-of-focus blur becomes more significant and eventually begins to conceal detail in the images. However, a larger slit could be useful when imaging weakly fluorescent samples, where the signal to noise ratio of the full sectioning images becomes very low, similar to how larger pinholes are used in confocal microscopy. For example, for the esophageal images, the average intensities across the whole field of view (normalized so that the 2 µm image has an intensity of 1) are (a) 1, (b) 1.4, (c), 2.7 (d), 5.0, (e) 8.2, and (f) 17.0. The virtual slit allows a quick transition between these different modes, or even the collection of a stack of images acquired with different slit widths.
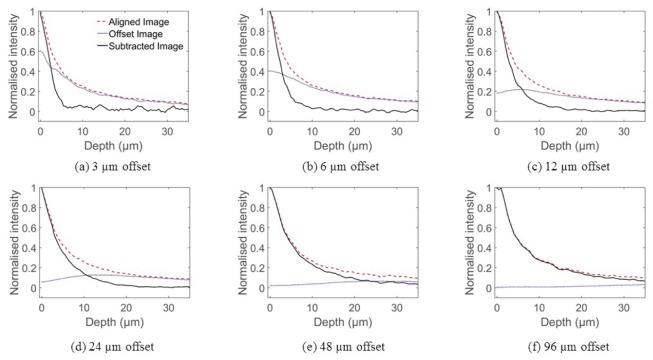
Figure 4 demonstrates the effect of subtracting an image with an offset slit. The axial profiles were generated in the same way as those shown in Fig. 2, except that every other frame was acquired with the virtual slit offset from the laser line by a known distance. A 3 µm slit width was used, and profiles are shown for misalignments of 3, 6, 12, 24, 48 and 96 µm, as measured on the proximal face of the bundle. Both the conventional profile, the profile generated from the offset images, and the profile resulting from subtraction are shown. It can be seen that, for all but the largest slit offsets, the aligned and offset profiles are similar far from focus, while at focus the aligned profile has significantly greater intensity. This illustrates why subtracting the offset images from the aligned images leads to enhanced optical sectioning.
Fig. 4.
Axial sensitivity profiles for the subtraction technique, with slit offsets set to (a) 3 µm, (b) 6 µm, (c) 12 µm, (d) 24 µm, (e) 48 µm and (f) 96 µm. Each plot shows profiles acquired with the aligned slit (dashed red line), with the offset slit (dotted blue line) and the result of subtracting the offset profile from the aligned profile (solid black line). The aligned and subtracted image profiles were individually normalized, while the offset image profile is shown on the same scale as the aligned image profile.
For a small slit offset, the subtraction results in a reduction in the 50% fall-off distance of approximately 30%. However, the subtraction also introduces significant noise, since the two profiles are similar in amplitude, leading to a low signal level in the subtracted profile. As the offset is increased to 12 and 24 µm, the noise is reduced, and while there is a less significant improvement in the 3 dB fall-off, there is still a significant reduction in the tail of the subtracted profiles. For larger offsets, the level of improvement in sectioning and the noise are both reduced further.
The effect of the subtraction technique on images is shown in Fig. 5, using similar samples to those used for Fig. 3. The results of using slit offsets of 3, 6, 12, 24, and 48 µm are shown. For small offsets, the aligned and misaligned images are very similar, and so subtraction leads to a very noisy image. For 12 and 24 µm offsets, there is a significant reduction of the residual out-of-focus signal compared to the conventional image. As the offset is increased further, the effect becomes less apparent. Some minor artifacts of the subtraction process are visible, including a slight shadow underneath the strands of tissue.
Fig. 5.
Subtraction technique applied to lens tissue paper phantom and porcine esophageal tissue. (a) Conventional line-scanning imaging without subtraction; (b-f) after subtraction of images with slit misalignments of (b) 3 µm, (c) 6 µm, (d) 12 µm, (e) 24 µm, and (f) 48 µm.
Since the technique involves subtraction, there is a loss of effective bit depth in the images. For the esophageal images, the mean pixel value (over a 100x100 pixel central region) of the aligned image shown in Fig. 3(a) is 3000, while the same means for the offset images are (b) 2710, (c) 2270, (d) 1810, (e) 1520, and (f) 1280. The equivalent means for the final images are therefore (b) 290, (c) 730, (d) 1190, (e) 1480, and (f) 1720. This means that approximately (b) 90%, (c) 76%, (d) 60%, (e) 51%, and (f) 43% of the bit depth is lost due to use of the subtraction technique.
While we envisage the virtual slit line scanning endomicroscope being used primarily with topical stains, as with other line-scanning endomicroscopes, we performed additional experiments to evaluate its potential to reject very large background signals of the kind that could be expected when imaging using intravenous (IV) fluorescein. While IV fluorescein images cannot be simulated accurately ex vivo, we performed a simple demonstration by soaking bovine adipose tissue in a solution of sodium fluorescein for 10 minutes, and then injecting the solution beneath the surface of the tissue using a needle and syringe. Lens tissue paper was then stained for 2 minutes with acriflavine 0.02% and placed on top of the tissue. Figure 6 shows images acquired with (a) the virtual slit at 400 µm, (b) the virtual slit at 3 µm, and (c) the virtual slit at 3 µm and the subtraction technique used with an offset of 12 µm. For the 400 µm slit width, which gives essentially a non-sectioned image, it was necessary to reduce the laser power from 50 mW to 4 mW to prevent saturation of the camera.
Fig. 6.
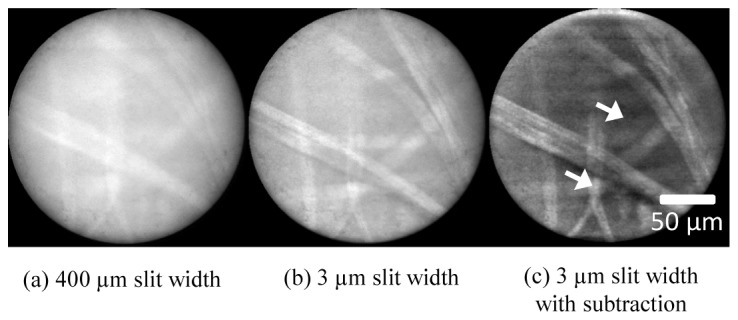
Demonstration of performance of sectioning and subtraction technique under conditions of a large background signal. Lens tissue paper, stained with acriflavine, was placed over bovine adipose tissue stained with fluorescein. (a) Conventional (non-subtracted) image with a slit width of 400 μm; (b) conventional (non-subtracted) image with a slit width of 3 μm; (c) subtraction imaging with slit width of 3 μm and offset of 12 μm.
The improvement when the slit width is reduced to 3 µm is clear, although the background signal remains high. The subtraction technique reduces the background further and some features of the tissue paper become clearer. However, the image also appears noisy, even in this relatively undemanding task of imaging a bright, well-defined, single-layer object. A ‘signal-to-background’ measurement was made by taking the ratio of the averages over 10x10 pixels from a region of tissue paper and a region between tissue strands (marked by arrows in Fig. 6). The signal to background ratio increases from 0.69 in the 400 µm slit image (a), to 0.72 in the 3 µm slit image (b), and to 1.12 in the subtraction technique image (c).
As for all multi-frame techniques, this subtraction approach is subject to motion artifacts. However, the misaligned image should contain predominantly out-of-focus contributions, and hence there should be some tolerance to small motions. To test this, we used a translation stage to move the probe across the surface of the tissue paper at velocities of 0.1, 1 and 2 mm/s. Extracted frames covering approximately the same area are shown in Fig. 7. There are no apparent artifacts at lower velocities, and artifacts remain relatively minor even at 1 mm/s. We also tested the technique while using the probe for freehand imaging of the esophageal tissue; a video is available in the supplementary materials (Visualization 1 (3.4MB, MP4) ), and an example pair of frames are shown in Fig. 7(e) and Fig. 7(f). Again, motion artifacts do not appear to be a significant limiting factor.
Fig. 7.

Effect of motion on subtraction technique. (a) Conventional (non-subtracted) image of lens tissue paper while probe was moving at 0.l mm/s; (b) subtraction imaging while moving at 0.1 mm/s; (c) subtraction imaging while moving at 1 mm/s; (d) subtraction imaging while moving at 2 mm/s; (e) conventional imaging of porcine esophagus with freehand moving probe, extracted from video in Visualization 1 (3.4MB, MP4) ; (f) subtracted image at same point, extracted from video in Visualization 1 (3.4MB, MP4) .
4. Discussion
The axial and lateral resolution performance of this system is essentially the same as the line-scanning (or ‘slit-scanning’) fiber bundle endomicroscopes reported previously (assuming a suitable choice of slit width). It could be used with both fluorescence and reflectance mode systems, and in principle with different fiber bundles or indeed GRIN lens systems. While the vast majority of clinical endomicroscopy studies have been performed using a point-scanning confocal laser endomicroscope, the success in clinical trials of the non-sectioning endomicroscopes [13] suggests that partially-sectioning line scanning endomicroscopy will have a range of potential applications. Several studies have suggested that some optical sectioning is beneficial even when using topical staining [16], but further work will be required to assess the effect on sensitivity and specificity of line-scanning verses non-sectioning endomicroscopy.
However, the system retains some limitations relative to a conventional point-scanning confocal endomicroscope. Without the subtraction imaging, axial resolution is poorer, and the significant tail to the sectioning profile, even at the smallest slit widths, will mean that some out-of-focus background will remain. This is likely to be particularly troublesome when imaging tissue stained with fluorescein. The subtraction mode imaging helps to remove some of this background, but as it relies on numerical subtraction rather than physical rejection of the unwanted signal, it leads to higher noise, as in structured illumination schemes. Therefore, it is unclear whether the system would allow imaging of in vivo fluorescein-stained tissue, and evaluating this will be the subject of future work.
Nevertheless, for imaging topically stained tissue, the virtual slit line-scanning endomicroscopy system allows for much higher frame rate imaging than is typical for confocal endomicroscopy. We demonstrated 120 Hz here, and this could potentially be higher if suitable rolling shutter CMOS cameras are available. The benefits of a high frame rate for mosaicking have already been shown for a limited set of ex vivo conditions, but further work will be required to assess the practical clinical advantage. It should be noted that the high frame rate is not specifically a feature of the virtual slit architecture, and we achieved the same speed using a line-scanning system using a linear camera [24]. However, in this other system, the slit width was fixed by the vertical size of the pixels on the linear camera. The maximum slit width in a system using a linear camera is limited by the aspect ratio of the pixels and the need to have sufficient pixel density for Nyquist sampling in the lateral direction. For example, if square pixels are used, then in order to have two pixels per core spacing, the maximum slit width is half a core-spacing. The re-scanning architecture of Sabharwal et al. [21] could also be run at a higher frame rate using a high speed camera, but the slit width is again fixed by a physical slit.
The advantage of the approach reported here over alternative line-scanning systems is two-fold. Firstly, the slit width could, if desired, be dynamically adjusted to suit different imaging conditions, or for use with different probes such as bundles with larger core spacing (either to increase the field of view or make use of more flexible leached bundles) or GRIN relays. The virtual slit can also be aligned electronically by adjusting the delay between the camera exposure and the laser scanning, removing the need for periodic manual or motorized alignment of a physical slit.
The second advantage concerns the possibility of acquiring a second image frame with the slit misaligned. As has been shown, this allows for rejection of the long tail of collected out-of-focus light that is a negative consequence of using line (rather than point) illumination and detection. The technique is inherently noisier then true point-scanning confocal endomicroscopy, and so may still not be suitable for use with intravenous fluorescein. The technique also reduces the frame rate to 60 Hz and is somewhat susceptible to motion artifacts at higher probe velocities, although we consider it to be usable in typical imaging situations. In principle, it would be possible to acquire both images simultaneously by splitting the image across two cameras. This would eliminate motion artifacts and allow running at the full frame-rate of the cameras, but would also lead to a small reduction in the signal-to-noise ratio.
5. Conclusion
Endoscopic microscopy is proving itself as a valuable technique across a range of medical specialties, and the technology remains under active development in both academia and industry. By offering high frame rate imaging with optical sectioning, the work presented here potentially opens up new avenues of research into techniques for rapidly surveying larger areas of topically-stained tissue. Using the rolling shutter of a CMOS camera as a virtual detector slit for a line-scanning endomicroscope, we obtain a system that is both simple and versatile. Further work will now be needed to compare the image quality provided by virtual slit line-scanning endomicroscopy to that of point-scanning and non-sectioning endomicroscopy in vivo, and to explore the potential benefits of high frame rate imaging.
Acknowledgments
We would like to thank Petros Giataganas, Khushi Vyas and many other members of the Hamlyn Centre for useful discussions. This work was supported by EPSRC Grant EP/I027769/1: SMART Endomicroscopy.
References and links
- 1.Gmitro A. F., Aziz D., “Confocal microscopy through a fiber-optic imaging bundle,” Opt. Lett. 18(8), 565–567 (1993). 10.1364/OL.18.000565 [DOI] [PubMed] [Google Scholar]
- 2.Goualher G. L., Perchant A., Genet M., Cav C., Viellerobe B., Abrat B., Ayache N., “Towards Optical Biopsies with an Integrated Fibered Confocal Fluorescence Microscope,” MICCAI 2004, 761–768 (2004). [Google Scholar]
- 3.Sung K.-B., Richards-Kortum R., Follen M., Malpica A., Liang C., Descour M., “Fiber optic confocal reflectance microscopy: a new real-time technique to view nuclear morphology in cervical squamous epithelium in vivo,” Opt. Express 11(24), 3171–3181 (2003). 10.1364/OE.11.003171 [DOI] [PubMed] [Google Scholar]
- 4.Muldoon T. J., Pierce M. C., Nida D. L., Williams M. D., Gillenwater A., Richards-Kortum R., “Subcellular-resolution molecular imaging within living tissue by fiber microendoscopy,” Opt. Express 15(25), 16413–16423 (2007). 10.1364/OE.15.016413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jabbour J. M., Saldua M. A., Bixler J. N., Maitland K. C., “Confocal Endomicroscopy: Instrumentation and Medical Applications,” Ann. Biomed. Eng. 40(2), 378–397 (2012). 10.1007/s10439-011-0426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polglase A. L., McLaren W. J., Skinner S. A., Kiesslich R., Neurath M. F., Delaney P. M., “A fluorescence confocal endomicroscope for in vivo microscopy of the upper- and the lower-GI tract,” Gastrointest. Endosc. 62(5), 686–695 (2005). 10.1016/j.gie.2005.05.021 [DOI] [PubMed] [Google Scholar]
- 7.Dickensheets D. L., Kino G. S., “Micromachined scanning confocal optical microscope,” Opt. Lett. 21(10), 764–766 (1996). 10.1364/OL.21.000764 [DOI] [PubMed] [Google Scholar]
- 8.Piyawattanametha W., Ra H., Qiu Z., Friedland S., Liu J. T. C., Loewke K., Kino G. S., Solgaard O., Wang T. D., Mandella M. J., Contag C. H., “In vivo near-infrared dual-axis confocal microendoscopy in the human lower gastrointestinal tract,” J. Biomed. Opt. 17(2), 021102 (2012). 10.1117/1.JBO.17.2.021102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canto M. I., Anandasabapathy S., Brugge W., Falk G. W., Dunbar K. B., Zhang Z., Woods K., Almario J. A., Schell U., Goldblum J., “In vivo endomicroscopy improves detection of Barrett’s esophagus related neoplasia: a multicenter international randomized controlled trial (with video),” Gastrointest. Endosc. 79(2), 211–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiesslich R., Burg J., Vieth M., Gnaendiger J., Enders M., Delaney P., Polglase A., McLaren W., Janell D., Thomas S., Nafe B., Galle P. R., Neurath M. F., “Confocal laser endoscopy for diagnosing intraepithelial neoplasias and colorectal cancer in vivo,” Gastroenterology 127(3), 706–713 (2004). 10.1053/j.gastro.2004.06.050 [DOI] [PubMed] [Google Scholar]
- 11.Konda V. J., Meining A., Jamil L. H., Giovannini M., Hwang J. H., Wallace M. B., Chang K. J., Siddiqui U. D., Hart J., Lo S. K., Saunders M. D., Aslanian H. R., Wroblewski K., Waxman I., “A pilot study of in vivo identification of pancreatic cystic neoplasms with needle-based confocal laser endomicroscopy under endosonographic guidance,” Endoscopy 45(12), 1006–1013 (2013). 10.1055/s-0033-1344714 [DOI] [PubMed] [Google Scholar]
- 12.Pierce M., Yu D., Richards-Kortum R., “High-resolution fiber-optic microendoscopy for in situ cellular imaging,” J. Vis. Exp. 47, 8–11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierce M. C., Vila P. M., Polydorides A. D., Richards-Kortum R., Anandasabapathy S., “Low-cost endomicroscopy in the esophagus and colon,” Am. J. Gastroenterol. 106(9), 1722–1724 (2011). 10.1038/ajg.2011.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vila P. M., Park C. W., Pierce M. C., Goldstein G. H., Levy L., Gurudutt V. V., Polydorides A. D., Godbold J. H., Teng M. S., Genden E. M., Miles B. A., Anandasabapathy S., Gillenwater A. M., Richards-Kortum R., Sikora A. G., “Discrimination of benign and neoplastic mucosa with a high-resolution microendoscope (HRME) in head and neck cancer,” Ann. Surg. Oncol. 19(11), 3534–3539 (2012). 10.1245/s10434-012-2351-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierce M. C., Guan Y., Quinn M. K., Zhang X., Zhang W.-H., Qiao Y.-L., Castle P., Richards-Kortum R., “A pilot study of low-cost, high-resolution microendoscopy as a tool for identifying women with cervical precancer,” Cancer Prev. Res. (Phila.) 5(11), 1273–1279 (2012). 10.1158/1940-6207.CAPR-12-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koucky M. H., Pierce M. C., “Axial response of high-resolution microendoscopy in scattering media,” Biomed. Opt. Express 4(10), 2247–2256 (2013). 10.1364/BOE.4.002247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keahey P. A., Tkaczyk T. S., Schmeler K. M., Richards-Kortum R. R., “Optimizing modulation frequency for structured illumination in a fiber-optic microendoscope to image nuclear morphometry in columnar epithelium,” Biomed. Opt. Express 6(3), 870–880 (2015). 10.1364/BOE.6.000870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bozinovic N., Ventalon C., Ford T., Mertz J., “Fluorescence endomicroscopy with structured illumination,” Opt. Express 16(11), 8016–8025 (2008). 10.1364/OE.16.008016 [DOI] [PubMed] [Google Scholar]
- 19.Ford T. N., Lim D., Mertz J., “Fast optically sectioned fluorescence HiLo endomicroscopy,” J. Biomed. Opt. 17(2), 021105 (2012). 10.1117/1.JBO.17.2.021105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyrish M., Dobbs J., Jain S., Wang X., Yu D., Richards-Kortum R., Tkaczyk T. S., “Needle-based fluorescence endomicroscopy via structured illumination with a plastic, achromatic objective,” J. Biomed. Opt. 18(9), 096003 (2013). 10.1117/1.JBO.18.9.096003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabharwal Y. S., Rouse A. R., Donaldson L., Hopkins M. F., Gmitro A. F., “Slit-scanning confocal microendoscope for high-resolution in vivo imaging,” Appl. Opt. 38(34), 7133–7144 (1999). 10.1364/AO.38.007133 [DOI] [PubMed] [Google Scholar]
- 22.Makhlouf H., Gmitro A. F., Tanbakuchi A. A., Udovich J. A., Rouse A. R., “Multispectral confocal microendoscope for in vivo and in situ imaging,” J. Biomed. Opt. 13(4), 044016 (2008). 10.1117/1.2950313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanbakuchi A. A., Rouse A. R., Udovich J. A., Hatch K. D., Gmitro A. F., “Clinical confocal microlaparoscope for real-time in vivo optical biopsies,” J. Biomed. Opt. 14(4), 044030 (2009). 10.1117/1.3207139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes M., Yang G.-Z., “High speed, line-scanning, fiber bundle fluorescence confocal endomicroscopy for improved mosaicking,” Biomed. Opt. Express 6(4), 1241–1252 (2015). 10.1364/BOE.6.001241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giataganas P., Hughes M., Yang G.-Z., “Force adaptive robotically assisted endomicroscopy for intraoperative tumour identification,” Int. J. CARS 10(6), 825–832 (2015). 10.1007/s11548-015-1179-0 [DOI] [PubMed] [Google Scholar]
- 26.Rouse A. R., Gmitro A. F., “Multispectral imaging with a confocal microendoscope,” Opt. Lett. 25(23), 1708–1710 (2000). 10.1364/OL.25.001708 [DOI] [PubMed] [Google Scholar]
- 27.Mei E., Fomitchov P. A., Graves R., Campion M., “A line scanning confocal fluorescent microscope using a CMOS rolling shutter as an adjustable aperture,” J. Microsc. 247(3), 269–276 (2012). 10.1111/j.1365-2818.2012.03642.x [DOI] [PubMed] [Google Scholar]
- 28.de Medeiros G., Norlin N., Gunther S., Albert M., Panavaite L., Fiuza U.-M., Peri F., Hiiragi T., Krzic U., Hufnagel L., “Confocal multiview light-sheet microscopy,” Nat. Commun. 6, 8881 (2015). 10.1038/ncomms9881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumgart E., Kubitscheck U., “Scanned light sheet microscopy with confocal slit detection,” Opt. Express 20(19), 21805–21814 (2012). 10.1364/OE.20.021805 [DOI] [PubMed] [Google Scholar]
- 30.Poher V., Kennedy G. T., Manning H. B., Owen D. M., Zhang H. X., Gu E., Dawson M. D., French P. M., Neil M. A., “Improved sectioning in a slit scanning confocal microscope,” Opt. Lett. 33(16), 1813–1815 (2008). 10.1364/OL.33.001813 [DOI] [PubMed] [Google Scholar]