Abstract
In the present study, gene expression profiles of acute myeloid leukemia (AML) samples were analyzed to identify feature genes with the capacity to predict the mutation status of FLT3/ITD. Two machine learning models, namely the support vector machine (SVM) and random forest (RF) methods, were used for classification. Four datasets were downloaded from the European Bioinformatics Institute, two of which (containing 371 samples, including 281 FLT3/ITD mutation-negative and 90 mutation-positive samples) were randomly defined as the training group, while the other two datasets (containing 488 samples, including 350 FLT3/ITD mutation-negative and 138 mutation-positive samples) were defined as the test group. Differentially expressed genes (DEGs) were identified by significance analysis of the micro-array data by using the training samples. The classification efficiency of the SCM and RF methods was evaluated using the following parameters: Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and the area under the receiver operating characteristic curve. Functional enrichment analysis was performed for the feature genes with DAVID. A total of 585 DEGs were identified in the training group, of which 580 were upregulated and five were downregulated. The classification accuracy rates of the two methods for the training group, the test group and the combined group using the 585 feature genes were >90%. For the SVM and RF methods, the rates of correct determination, specificity and PPV were >90%, while the sensitivity and NPV were >80%. The SVM method produced a slightly better classification effect than the RF method. A total of 13 biological pathways were overrepresented by the feature genes, mainly involving energy metabolism, chromatin organization and translation. The feature genes identified in the present study may be used to predict the mutation status of FLT3/ITD in patients with AML.
Keywords: acute myeloid leukemia, feature gene, support vector machine, random forest, functional enrichment analysis
Introduction
Fms-like tyrosine kinase 3 (FLT3) is expressed in hematopoietic progenitor cells. In acute myeloid leukemia (AML), its most frequent mutation is an internal tandem duplication (FLT3/ITD), which has a prevalence of 30–35% (1). FLT3/ITD is a critical prognostic factor for patients with AML. Compared with carriers of wild-type FLT3, patients with the FLT3/ITD mutation have shorter overall survival time and disease-free survival time (2). Early diagnosis of FLT3/ITD allows for timely treatment of AML and thus benefits the clinical outcome.
Certain achievements have been made in revealing the role of the FLT3/ITD mutation in AML and several feature genes associated with the FLT3/ITD mutation have been identified. Chen et al (3) reported that signaling associated with the FLT3/ITD mutation includes the suppression of SHP-1. Furthermore, aberrant expression of CD7 in myeloblasts has been found to be highly associated with the FLT3/ITD mutation in AML (4). Okamoto et al (5) indicated that Lyn, an important component of the signal transduction pathway specific for FLT3/ITD, may be utilized as a therapeutic target for the treatment of AML in carriers of the FLT3/ITD mutation. Furthermore, PIM1, a serine/threonine kinase, has been found to be upregulated in FLT3-ITD mutation-positive AML and may be involved in FLT3-mediated leukemogenesis (6). Dalal et al (7) reported that CD56 can predict the presence of the FLT3-ITD mutation in AML.
In order to distinguish the FLT3/ITD mutation from the wild-type at the transcriptional level, the present study analyzed microarray gene expression data of AML samples. Feature genes were identified by a bioinformatics analysis and subsequent classification was performed by machine learning models, namely the support vector machine (SVM) and random forest (RF) methods. The classification efficiency of the two models was also evaluated. The feature genes identified in the present study may be used to predict the mutation status of FLT3/ITD in patients with AML.
Materials and methods
Microarray data and data pre-processing
Gene expression data of AML samples were downloaded from the European Bioinformatics Institute (EBI; http://www.ebi.ac.uk) (8). Four relevant data sets for patient cohorts with AML containing information on the FLT3/ITD mutation were obtained, which included a total of 859 AML samples (Table I). Two data sets (containing 371 samples, including 281 FLT3/ITD mutation-negative and 90 FLT3/ITD mutation-positive samples) were selected as the training group, while the other two data sets (containing 488 samples, including 350 FLT3/ITD mutation-negative samples and 138 FLT3/ITD mutation-positive samples) were used as the test group.
Table I.
Microarray data sets used in the present study.
| Data set ID | Total samples (n) | FLT3/ITD mutation negative (n) | FLT3/ITD mutation positive (n) | Undetermined samples (n) |
|---|---|---|---|---|
| Training sets | ||||
| E-GEOD-61804 | 325 | 243 | 50 | 32 |
| E-GEOD-34860 | 78 | 38 | 40 | 0 |
| Total | 403 | 281 | 90 | 32 |
| Testing sets | ||||
| E-GEOD-17855 | 237 | 189 | 48 | 0 |
| E-GEOD-15434 | 251 | 161 | 90 | 0 |
| Total | 488 | 350 | 138 | 0 |
The raw data were pre-processed using the affy package (9) in R (www.r-project.org), including data format conversion, filling in missing values (using median gene expression), background correction using the MAS method and normalization with the quantiles method (10).
Screening of differentially expressed genes (DEGs)
Microarray data from wild-type and FLT3/ITD mutation-positive AML samples were screened for DEGs using the significance analysis of microarray method in R (11). The false discovery rate (FDR) was estimated using the permutation method with P<0.05 (12,13) and |log2(fold change)|>1 set as the thresholds.
Prediction of mutation status of AML samples
The ability of DEGs to predict the FLT3/ITD mutation status in AML samples was examined using two methods: SVM and random forest.
SVM is a classification technique based on the structural risk minimization principle (14). The SVM classifier was constructed via the SVM function in the e1071 package of R with the non-linear radial basis function as the kernel and penalty functions set at 1,000.
RF utilizes multiple classification and regression trees to classify samples (15). The function randomForest from the randomForest package in R was adopted to classify AML samples from the training group.
A leave-one-out cross validation method was performed to evaluate the classification efficiency of the two methods. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) (16) and area under the receiver operating characteristic (ROC) curve (17) were calculated. The classification efficiency for the training group, test group and the combined group were evaluated individually. Whenever the construed SVM or RF classifier produced a high reliability, the DEGs collected from the training sets were considered as feature genes for distinguishing wild-type from FLT3/ITD-mutation positive samples.
Functional enrichment analysis
Functional enrichment analysis of the feature genes was performed using the Database for Annotation, Visualization and Integration Discovery (http://david.abcc.ncifcrf.gov/) (18,19). P<0.5 and FDR<0.1 were set as the cut-off values to screen out significantly over-represented biological pathways.
Results
Screening for DEGs
A total of 585 DEGs were identified in FLT3/ITD mutation-positive samples from the training group, comprising 580 upregulated and 5 downregulated genes compared with those in the FLT3/ITD mutation-negative AML samples.
Sample classification using SVM or RF classifier
Classification of AML samples with regard to their FLT3/ITD mutation status depending on their gene expression profiles was performed using the SVM and RF methods (Fig. 1).
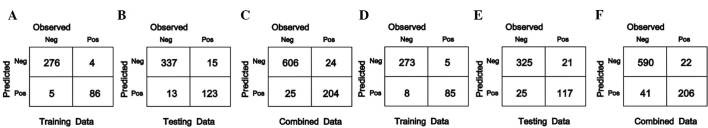
Figure 1.
Classification results of acute myeloid leukemia samples using the SVM and RF methods. Classification according to the SVM method for (A) the training group, (B) the test group and (C) the combined group. Classification according to the RF method for (D) the training group, (E) the test group and (F) the combined group. SVM, support vector machine; RF, random forest; Neg, negative; Pos, positive.
For the 371 AML samples from the training group, 276 and 273 mutation-negative samples, as well as 86 and 85 mutation-positive samples were correctly classified using the SVM and RF method, respectively. The accuracy rates were 97.57 and 96.5%, respectively.
Among the 488 AML samples from the test group, 337 and 325 mutation-negative samples, as well as 123 and 117 mutation-positive samples were correctly classified by using the SVM and RF method, respectively, and the accuracy rates were 94.26 and 90.57%.
For the 859 AML samples from the combined group, 606 and 590 mutation-negative samples, as well as 204 and 206 mutation-positive samples were correctly classified by using the SVM and RF method, respectively, with accuracy rates of 94.3% and 92.67%.
According to above classification results (Fig. 2), the classification using the SVM method had a better accuracy rate than that of the RF method. However, the accuracy rates were >90%, suggesting a good classification ability of these two method based on the DEGs identified.
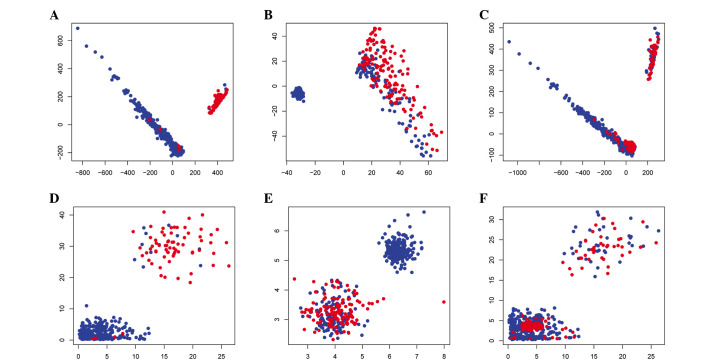
Figure 2.
Scatter diagrams showing the classification results. Blue dots indicate FLT3/ITD mutation-negative samples and red dots indicate FLT3/ITD mutation-positive samples. Classification according to the SVM method for (A) the training group, (B) the test group and (C) the combined group. Classification according to the RF method for (D) the training group, (E) the test group and (F) the combined group. SVM, support vector machine; RF, random forest.
Classification efficiency
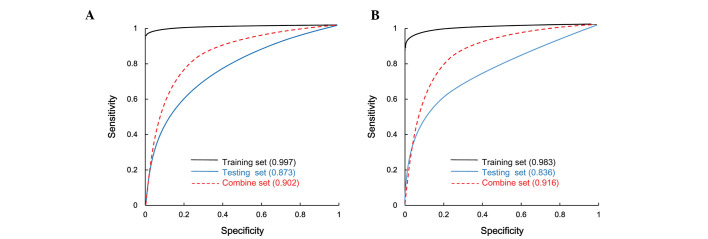
Five parameters were calculated to evaluate the classification efficiency: Rate of correct prediction, sensitivity, specificity, PPV, NPV (Table II) and the area under the ROC curve (Fig. 3). For the SVM and RF methods, the rate of correct prediction, specificity and PPV were >90%, while the sensitivity and NPV were >80%, with the SVM method producing a slightly better classification efficiency than the RF method.
Table II.
Classification effects of SVM method and RF method.
| Method | No. of samples | Correct rate | Sensitivity | Specificity | PPV | NPV | AUROC |
|---|---|---|---|---|---|---|---|
| SVM | |||||||
| Training group | 371 | 0.9757 | 0.9556 | 0.9822 | 0.9451 | 0.9857 | 0.997 |
| Test group | 488 | 0.9426 | 0.8913 | 0.9629 | 0.9044 | 0.9574 | 0.876 |
| Combined | 859 | 0.9430 | 0.8947 | 0.9604 | 0.8908 | 0.9619 | 0.902 |
| RF | |||||||
| Training group | 371 | 0.9650 | 0.9444 | 0.9715 | 0.9140 | 0.9820 | 0.983 |
| Test group | 488 | 0.9057 | 0.8478 | 0.9286 | 0.8239 | 0.9393 | 0.818 |
| Combined | 859 | 0.9267 | 0.9035 | 0.9350 | 0.8340 | 0.9656 | 0.916 |
PPV, positive predictive value; NPV, negative predictive value; AUROC, area under the receiver operating characteristic curve; SVM, support vector machine; RF, random forest.
Figure 3.
Receiver operator characteristic curves generated using (A) the support vector machine method and (B) the random forest method.
The feature genes identified were not only suitable for correct predictions of the FLT3/ITD mutation status of AML samples in the training group, but also in the test group and the combined group, suggesting that these DEGs may be utilized for distinguishing FLT3/ITD mutation-negative AML samples from mutation-positive samples. It was indicated that the DEGs identified in the present study are feature genes of the FLT3/ITD mutation, including IDH1, SUZ12, BCORL1, RUVBL2, JMJD1C, TOP2A, DAPK3, RPS15, RPS16, RPS9, EIF2α, EIF4E, EIF3B, EIF3 K, EIF3 L and EIF1B.
Biological pathways of feature genes
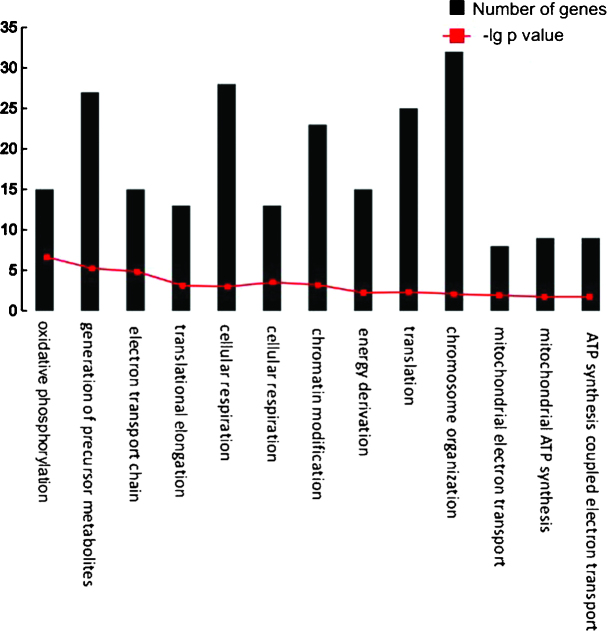
A total of 13 biological pathways were over-represented by the feature genes (Table III). The number of genes in each biological pathway is shown in Fig. 4. Several pathways were associated with energy metabolism, including oxidative phosphorylation, mitochondrial electron transport and mitochondrial adenosine triphosphate (ATP) synthesis. Furthermore, chromatin organization, chromosome organization and translation were significantly overrepresented.
Table III.
Significantly over-represented biological pathways in feature genes.
| Term | Count | P-value | FDR |
|---|---|---|---|
| GO:0006119 - Oxidative phosphorylation | 15 | 3.80×10−6 | 0.008857 |
| GO:0006091 - Generation of precursor metabolites | 27 | 1.50×10−5 | 0.017414 |
| GO:0022900 - Electron transport chain | 15 | 2.25×10−5 | 0.017423 |
| GO:0045333 - Cellular respiration | 13 | 8.00×10−5 | 0.045815 |
| GO:0016568 - Chromatin modification | 23 | 1.11×10−4 | 0.050724 |
| GO:0006414 - Translational elongation | 13 | 1.19×10−4 | 0.045367 |
| GO:0006325 - Chromatin organization | 28 | 1.40×10−4 | 0.045654 |
| GO:0006412 - Translation | 25 | 2.62×10−4 | 0.073784 |
| GO:0015980 - Energy derivation | 15 | 2.89×10−4 | 0.072430 |
| GO:0051276 - Chromosome organization | 32 | 3.37×10−4 | 0.075984 |
| GO:0006120 - Mitochondrial electron transport | 8 | 4.01×10−4 | 0.081888 |
| GO:0042775 - Mitochondrial ATP synthesis | 9 | 4.64×10−4 | 0.086660 |
| GO:0042773 - ATP synthesis-coupled electron transport | 9 | 4.64×10−4 | 0.086660 |
FDR, false discovery rate; GO, gene ontology; ATP, adenosine triphosphate.
Figure 4.
Biological pathways and corresponding numbers of feature genes. ATP, adenosine triphosphate.
Discussion
In the present study, a total of 585 feature genes were identified to be differentially expressed between FLT3/ITD mutation-positive and wild-type AML samples from the training group (two data sets). Two methods, SVM and RF, were adopted to classify AML samples from the training group and the test group (two further data sets). The accuracy rates were >90% using either method on either group of data sets. SVM produced a slightly more accurate classification than RF. It was indicated that the feature genes identified in the present study may be used to predict the FLT3/ITD mutation status in patients with AML. Functional enrichment analysis was also performed for the feature genes. Energy metabolism, chromatin organization and translation were significantly overrepresented.
Mitochondria are important organelles regulating the energy levels, metabolism and apoptosis in cells, which can in turn affect cell differentiation and proliferation. Therefore, they mitochondria have important roles in the pathogenesis of AML (20). Inhibition of mitochondrial translation has been suggested as a potential therapeutic strategy for AML (21). Yamaguchi et al (22) reported that a mutation in IDH1, which has an important role in the citrate circle, has an adverse effect in patients with AML.
Several genes associated with chromatin organization also participate in the development of AML. SUZ12 encodes a subunit of polycomb repressive complex 2, which was shown to drive aberrant self-renewal in a mouse model of AML (23). Tiacci et al (24) found that BCORL1 has a role AML. Zagaria et al (25) reported that the BCOR gene was dysregulated in AML is due to chromosomal translocation. RUVBL2 is a critical mediator of oncogenesis caused by the MLL-AF9 fusion gene and is a potential therapeutic target for MLL-AF9-associated leukemia (26). In addition, Sroczynska et al (27) found that JMJD1C is required for leukemia maintenance, and that depletion of JMJD1C impaired the expansion and colony formation of human leukemic cell lines. Amplification of TOP2A was found identified in myelodysplastic syndrome transforming to AML (28). DAPK3 was indicated to have a role in the induction of apoptosis, and that CpG island methylation of this gene, leading to its dysregulation, is implicated in AML (29).
Translation was also significantly overrepresented in the feature genes identified by the present study. Wang et al (30) indicated that silencing of RPS14 inhibits the proliferation of AML cells via activating p53. It is likely that RPS15, RPS16, RPS9 and other members of the RPS family may exert similar roles. In addition, EIF2α and EIF4E have been implicated in AML (31,32). The roles of EIF3B, EIF3 K, EIF3 L and EIF1B in AML may be worth investigating.
In conclusion, the present study identified a number of feature genes that may be used to distinguish FLT3/ITD mutation-positive AML samples from FLT3 wild-type samples. Several of the feature genes identified have been previously implicated in AML. The computational tools developed in the present study may aid in the clinical detection of FLT3/ITD mutation-positive AML for possible early and targeted treatment of these patients.
References
- 1.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Li H, Wang MN, Lu D, Bassi R, Wu Y, Zhang H, Balderes P, Ludwig DL, Pytowski B, et al. Suppression of leukemia expressing wild-type or ITD-mutant FLT3 receptor by a fully human anti-FLT3 neutralizing antibody. Blood. 2004;104:1137–1144. doi: 10.1182/blood-2003-07-2585. [DOI] [PubMed] [Google Scholar]
- 3.Chen P, Levis M, Brown P, Kim KT, Allebach J, Small D. FLT3/ITD mutation signaling includes suppression of SHP-1. J Biol Chem. 2005;280:5361–5369. doi: 10.1074/jbc.M411974200. [DOI] [PubMed] [Google Scholar]
- 4.Rausei-Mills V, Chang KL, Gaal KK, Weiss LM, Huang Q. Aberrant expression of CD7 in myeloblasts is highly associated with de novo acute myeloid leukemias with FLT3/ITD mutation. Am J Clin Pathol. 2008;129:624–629. doi: 10.1309/NRTX9AKXHR5JBT93. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto M, Hayakawa F, Miyata Y, Watamoto K, Emi N, Abe A, Kiyoi H, Towatari M, Naoe T. Lyn is an important component of the signal transduction pathway specific to FLT3/ITD and can be a therapeutic target in the treatment of AML with FLT3/ITD. Leukemia. 2007;21:403–410. doi: 10.1038/sj.leu.2404547. [DOI] [PubMed] [Google Scholar]
- 6.Fathi AT, Arowojolu O, Swinnen I, Sato T, Rajkhowa T, Small D, Marmsater F, Robinson JE, Gross SD, Martinson M, et al. A potential therapeutic target for FLT3-ITD AML: PIM1 kinase. Leuk Res. 2012;36:224–231. doi: 10.1016/j.leukres.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalal BI, Mansoor S, Manna M, Pi S, Sauro GD, Hogge DE. Detection of CD34, TdT, CD56, CD2, CD4 and CD14 by flow cytometry is associated with NPM1 and FLT3 mutation status in cytogenetically normal acute myeloid leukemia. Clin Lymphoma Myeloma Leuk. 2012;12:274–279. doi: 10.1016/j.clml.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Stoesser G, Baker W, van den Broek A, Camon E, Garcia-Pastor M, Kanz C, Kulikova T, Leinonen R, Lin Q, Lombard V, et al. The EMBL nucleotide sequence database. Nucleic Acids Res. 2002;30:21–26. doi: 10.1093/nar/30.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, Smeekens SP. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- 10.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S. A comprehensive evaluation of SAM, the SAM R-package and a simple modification to improve its performance. Bmc Bioinformatics. 2007;8:230. doi: 10.1186/1471-2105-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the false discovery Rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 13.Benjamini, Yoav Discovering the false discovery rate. Journal of the Royal Statistical Society. 2010;72:405–416. doi: 10.1111/j.1467-9868.2010.00746.x. [DOI] [Google Scholar]
- 14.Duval B, Hao JK. Advances in metaheuristics for gene selection and classification of microarray data. Brief Bioinform. 2010;1:127–141. doi: 10.1093/bib/bbp035. [DOI] [PubMed] [Google Scholar]
- 15.Díaz-Uriarte R, Alvarez de Andrés S. Gene selection and classification of microarray data using random forest. Bmc Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croshaw R, Shapiro-Wright H, Svensson E, Erb K, Julian T. Accuracy of clinical examination, digital mammogram, ultrasound and MRI in determining postneoadjuvant pathologic tumor response in operable breast cancer patients. Ann Surg Oncol. 2011;18:3160–3163. doi: 10.1245/s10434-011-1919-5. [DOI] [PubMed] [Google Scholar]
- 17.Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 18.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basak NP, Banerjee S. Mitochondrial dependency in progression of acute myeloid leukemia. Mitochondrion. 2015;21:41–48. doi: 10.1016/j.mito.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Schimmer AD, Skrtić M. Therapeutic potential of mitochondrial translation inhibition for treatment of acute myeloid leukemia. Expert Rev Hematol. 2012;5:117–119. doi: 10.1586/ehm.12.8. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi S, Iwanaga E, Tokunaga K, Nanri T, Shimomura T, Suzushima H, Mitsuya H, Asou N. IDH1 and IDH2 mutations confer an adverse effect in patients with acute myeloid leukemia lacking the NPM1 mutation. Eur J Haematol. 2014;92:471–477. doi: 10.1111/ejh.12271. [DOI] [PubMed] [Google Scholar]
- 23.Shi J, Wang E, Zuber J, Rappaport A, Taylor M, Johns C, Lowe SW, Vakoc CR. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras (G12D) acute myeloid leukemia. Oncogene. 2013;32:930–938. doi: 10.1038/onc.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiacci E, Grossmann V, Martelli MP, Kohlmann A, Haferlach T, Falini B. The corepressors BCOR and BCORL1: Two novel players in acute myeloid leukemia. Haematologica. 2012;97:3–5. doi: 10.3324/haematol.2011.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zagaria A, Anelli L, Casieri P, Coccaro N, Tota G, Minervini CF, Minervini A, Impera L, Brunetti C, Orsini P, et al. BCOR gene dysregulation due to chromosomal translocation in acute myeloid leukemia: A new mechanism based on long non-coding RNA dislocation? Leuk Lymphoma. 2014;55:2199–2201. doi: 10.3109/10428194.2013.874009. [DOI] [PubMed] [Google Scholar]
- 26.Osaki H, Walf-Vorderwülbecke V, Mangolini M, et al. The AAA+ ATPase RUVBL2 is a critical mediator of MLL-AF9 oncogenesis. Leukemia. 2013;27:1461–1468. doi: 10.1038/leu.2013.42. [DOI] [PubMed] [Google Scholar]
- 27.Sroczynska P, Cruickshank VA, Bukowski JP, Miyagi S, Bagger FO, Walfridsson J, Schuster MB, Porse B, Helin K. shRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood. 2014;123:1870–1882. doi: 10.1182/blood-2013-08-522094. [DOI] [PubMed] [Google Scholar]
- 28.Martinsubero JI, Harder L, Gesk S, Schoch R, Novo FJ, Grote W, Calasanz MJ, Schlegelberger B, Siebert R. Amplification of ERBB2, RARA, and TOP2A genes in a myelodysplastic syndrome transforming to acute myeloid leukemia. Cancer Genet Cytogenet. 2001;127:174–176. doi: 10.1016/S0165-4608(00)00431-3. [DOI] [PubMed] [Google Scholar]
- 29.Aggerholm A, Hokland P. DAP-kinase CpG island methylation in acute myeloid leukemia: Methodology versus biology? Blood. 2000;95:2997–2998. [PubMed] [Google Scholar]
- 30.Wang L, Luo J, Nian Q, Xiao Q, Yang Z, Liu L. Ribosomal protein S14 silencing inhibits growth of acute myeloid leukemia transformed from myelodysplastic syndromes via activating p53. Hematology. 2014;19:225–231. doi: 10.1179/1607845413Y.0000000127. [DOI] [PubMed] [Google Scholar]
- 31.Hariri F, Arguello M, Volpon L, Culjkovic-Kraljacic B, Nielsen TH, Hiscott J, Mann KK, Borden KL. The eukaryotic translation initiation factor eIF4E is a direct transcriptional target of NF-κB and is aberrantly regulated in acute myeloid leukemia. Leukemia. 2013;10:2047–2055. doi: 10.1038/leu.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH, Jr, Borden KL. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): A proof-of-principle clinical trial with ribavirin. Blood. 2009;2:257–260. doi: 10.1182/blood-2009-02-205153. [DOI] [PubMed] [Google Scholar]