Abstract
Research in biosensing approaches as alternative techniques for food diagnostics for the detection of chemical contaminants and foodborne pathogens has increased over the last twenty years. The key component of such tests is the biorecognition element whereby polyclonal or monoclonal antibodies still dominate the market. Traditionally the screening of sera or cell culture media for the selection of polyclonal or monoclonal candidate antibodies respectively has been performed by enzyme immunoassays. For niche toxin compounds, enzyme immunoassays can be expensive and/or prohibitive methodologies for antibody production due to limitations in toxin supply for conjugate production. Automated, self-regenerating, chip-based biosensors proven in food diagnostics may be utilised as rapid screening tools for antibody candidate selection. This work describes the use of both single channel and multi-channel surface plasmon resonance (SPR) biosensors for the selection and characterisation of antibodies, and their evaluation in shellfish tissue as standard techniques for the detection of domoic acid, as a model toxin compound. The key advantages in the use of these biosensor techniques for screening hybridomas in monoclonal antibody production were the real time observation of molecular interaction and rapid turnaround time in analysis compared to enzyme immunoassays. The multichannel prototype instrument was superior with 96 analyses completed in 2 h compared to 12 h for the single channel and over 24 h for the ELISA immunoassay. Antibodies of high sensitivity, IC50's ranging from 4.8 to 6.9 ng/mL for monoclonal and 2.3–6.0 ng/mL for polyclonal, for the detection of domoic acid in a 1 min analysis time were selected. Although there is a progression for biosensor technology towards low cost, multiplexed portable diagnostics for the food industry, there remains a place for laboratory-based SPR instrumentation for antibody development for food diagnostics as shown herein.
Keywords: Surface plasmon resonance, Antibody, Biosensor, Domoic acid, Marine toxin, Shellfish
1. Introduction
Awareness of human exposure to a wide range of contaminants through the diet, originating from anthropogenic, natural sources and fraudulent practices along the food supply chain has been heightened with increasing media attention and consumer cognizance. The food supply chain can become contaminated at various points from environment to farm to fork by bacteria, viruses or chemicals present in the environment; the improper use of agrichemicals such as antibiotics and pesticides; the illegal use of growth promoting compounds in animal production; by-products of food processing techniques; and naturally occurring toxins such as phycotoxins and mycotoxins. Food contamination can cause serious acute and chronic health effects resulting in economic and political repercussions with subsequent “food scares” and recalls of products [1,9,16,18,19,25,30,31]. In order to monitor food for contamination and to ensure that unacceptable levels do not enter the human food chain, it is imperative that products are subjected to scrutiny, in real time at critical control points in the chain, and with respect to their safety in an efficient and rapid manner.
In recent years, advances in biosensor and nanotechnology for diagnostics have offered solutions, albeit mostly as research tools in portable, multiple analyte diagnostics for small numbers of samples. Relatively expensive biosensor technology (e.g., ≥$100K USD) for food analysis as a screening tool that is restricted to the laboratory setting is generally no longer deemed as cost-effective in comparison to the advances in state-of-the art confirmatory mass spectrometry methods for multiple analytes. These biosensors tend to be only suitably cost-effective when they have the ability to efficiently analyse a large number of samples for the presence of multiple chemical contaminants in food matrices with a relatively short turnaround time [28]. Additionally, biosensors may be used as an alternative to mass spectrometry as a way of diversifying resources and skills in a routine analytical laboratory, but employing both technologies still remains an innovative challenge.
Advances over the past two decades have led to the manufacture of a range of instrumentation employing a variety of detection principles. Modern biosensors offer the ability to be portable devices for regulatory bodies and the food industry alike to verify the safety and quality of products intended for human consumption along the food supply chain. It is expected that biosensors will have added advantages over traditional technologies, since they combine the high affinity of the biochemical interactions, resulting in high sensitivities and low limits of detection, with the possible miniaturisation, “point-of-use” portability and automation which make them interesting for in situ monitoring [3]. Importantly, as biosensors operate on the principle of measuring a biomolecular interaction of two components on a surface, the key component for food analysis still remains the biorecognition element for the interaction and detection with the target contaminant. Polyclonal antibodies, and more recently monoclonal antibodies that reduce the use of animals, still tend to be the most suitable biorecognition elements in commercial immunodiagnostics for food analysis. For the production and characterisation of biorecognition elements, the use of a laboratory based biosensing instrument that operates with a high degree of automation, user friendliness, accuracy and precision with real-time analysis is a vital tool for the robust screening and characterisation of antibody binding and matrix performance.
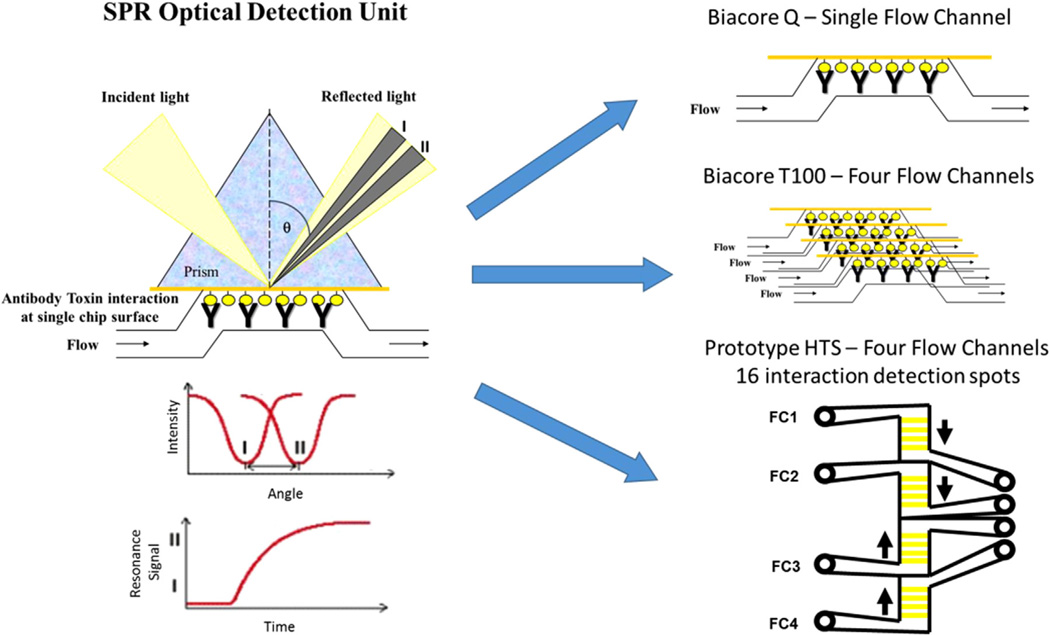
Surface plasmon resonance (SPR) biosensor technology has been the most successful immunosensor to date for food analysis and is primarily employed in a laboratory setting. SPR biosensors are affinity, optical sensors where the signal is based on measuring binding-induced changes in refractive index and this platform thus allows for label-free, real-time analysis (Fig. 1). Depending on SPR response to the binding of analytes to a surface, an analyte can be detected with a direct, sandwich, or inhibition assay. For small analytes (molecular weight <1000 Da), an inhibition assay is often used, where small analytes are premixed with antibodies and unbound antibody sites are captured by the small analytes immobilized on a sensor surface [43]. This approach has been used widely with SPR biosensors for the detection of aquatic toxins ([6,7,12,13,14,26,32,40,42]). Variations in SPR technology is available with different capabilities in analysis due to the utilisation of single or multiple flow channels (Fig. 1). Therefore, SPR biosensor technology enables simultaneous observations of molecular interactions on a surface containing multiple channels offering multiplexed evaluations in a single analysis [4,5,29].
Fig. 1.
Surface plasmon resonance (SPR) technology measures light reflection from the side of the chip opposite the flow channel. Upon antibody-toxin interaction at the chip surface, the change in reflected light angle (I to II) is detected. Each of the biosensors has different capacity for the simultaneous determination of toxin interaction on the surface due to the number of flow channels and detection points on each channel.
In this research, a Biacore SPR high throughput prototype instrument was employed for effective multiplexed analysis for evaluation of the marine toxin, antibody interaction. The system is designed primarily for rapid, simultaneous screening of multiple ligands (up to 16) by employing a 4 × 4 format with four 0.6 mm2 flow cell areas that each contain four SPR sensing spots. This flexible, multichannel format enables assays with either multi-ligand or multi-sample focuses. When a Series S CM5 chip with 16 immobilised ligands is docked, four independent parallel flow cells are formed by the sensor chip pressing against moulded channels on the integrated microfluidic cartridge (IFC), and the response is measured from four detection spots in each flow cell [4]. Variation in assay design offers alternative approaches for antibody selection, characterisation and target analysis which may be extremely beneficial when the target analyte is expensive or difficult to source ([5,41]).
Marine biotoxins are an exemplar case whereby the development of rapid methods including antibody-based assays have been restricted due to the limitations in and relatively expensive costs of many toxins. In order to investigate SPR biosensor technology for characterizing antibodies to small-molecule toxins, the cost-effective and easily obtainable domoic acid (DA) toxin is used as a model compound in this study. This neurotoxin is renowned to cause amnesic shellfish poisoning (ASP) illness in humans [20,24,33] and is predominately found in bivalve shellfish that filter feed on toxin producing diatom of the genus Pseudonitzschia. Thereby, shellfish are regulated with a limit of 20 µg DA per gram of tissue in the United States and European Union [11,38,39], and monitoring from both a public health perspective and the protection of the industry brand from toxic recalls is vital. Recent events, including the November 2015 closure of U.S. West Coast Dungeness crab harvesting due to high levels of DA in the waters and accumulation in the crabs [8], highlight the need for appropriate, rapid detection methods.
To this end, this study aims to demonstrate the (1) development of robust methods for antibody production for a small molecular weight toxin, (2) use of SPR biosensor technologies for antibody screening and selection and (3) investigate the use of a commercial, multi-channel SPR instrument (Biacore T100) for toxin immunoassay development and detection in complex matrices. While this work focuses on the model system of DA in shellfish matrices, the protocols discussed herein should have applicability to a large number of antibody/analyte systems.
2. Materials and methods
2.1. Instrumentation
An optical SPR Biosensor system (Biacore Q) and a high throughput multichannel prototype instrument [4] with Control and BIAevaluation software 4.1 were obtained from Biacore AB, GE Healthcare (Uppsala, Sweden). For additional assays, a four-channel SPR instrument (Biacore T100, GE Healthcare) was employed and controlled via Biacore T100 Control software with evaluation via Biacore T100 Evaluation software. Further data processing was performed with GraphPad Prism 5 (La Jolla, CA).
2.2. Materials
Biacore Q CM5 and Series S CM5 sensor chips, HBS-EP and HBS-EP+ buffers and amine coupling kits were obtained from Biacore AB (Uppsala, Sweden). Domoic acid (DA), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), bovine thyroglobulin (BTG), 2-(N-morpholino) ethanesulfonic acid (MES), 2,2′-(ethylenedioxy)bis(ethylamine), l-glutamic acid (GA), l-glutamine (GluNH2), kainic acid monohydrate (KA), aspartic acid (AA) and sodium acetate were purchased from Sigma-Aldrich Company Ltd. (Dorset, UK; St. Louis, MO, USA). The Quil A was from Invivogen (France) while the Pam3Cys-Ser(Lys)4-OH (PCSL) was acquired from EMC Micro-collections, GmbH, (Germany). Isotyping kits (Isostrip) were obtained from Roche (UK). Cell culture reagents were obtained from Invitrogen (UK). Borate buffer was procured from Pierce (Thermo Fisher Scientific, Rockford, IL, USA). Tetrodotoxin (TTX, Sankyo Co Ltd., Tokyo, Japan), saxitoxin (STX, National Institute of Standards and Technology (NIST), Gaithersburg, MD, USA), and okadaic acid (OA, National Research Council Canada, Ottawa, Ontario) were employed in cross-reactivity studies. All solvents were of LC grade and were obtained from Rathburn, Walkerburn, Strathclyde, UK.
2.3. Protein conjugate synthesis
Domoic acid was employed as the model small molecular weight compound to demonstrate the capability of the antibody production and instrumentation. Antibodies were raised to domoic acid, bovine thyroglobulin (BTG) protein conjugate. In brief, EDC (11.6 mg) and NHS (10.6 mg) in MES buffer were added to a solution of DA in 1.0 mL water and mixed for 15 min at room temperature. BTG (50 mg) was then added to the activated DA solution. The reaction mixture was incubated overnight at room temperature before purification by 24 h dialysis against saline (0.15 M sodium chloride). The purified immunogen was diluted with saline to a final concentration of 1 mg/mL protein and stored at −20 °C until used. Domoic acid, horse radish peroxidase (DA-HRP) was prepared as the reporter for ELISA in a similar manner as the BTG protein conjugate for immunisations.
2.4. Antibody production
Polyclonal antibodies were raised in rabbits by subcutaneous injection with 0.2 mg of the DA immunogen emulsified with Freund's complete adjuvant. Booster injections (0.2 mg of immunogen emulsified with Freund's incomplete adjuvant) were then administered on a fortnightly basis for 2 boosters and monthly for a further 3 booster immunisations. Test bleeds were collected ten days after each booster injection and monitored for the presence of antibodies by the SPR sensor. The antiserum was harvested 28 days after the final booster and stored frozen at −20 °C until required.
For the production of monoclonal antibodies a balb/c mouse was immunised with 0.02 mg of immunogen emulsified with Quil A for two immunisations on day 0 and 14. Three further booster immunisations (0.02 mg of immunogen emulsified with the adjuvant Pam3Cys-Ser-(Lys)4 (PCSL)) were then administered on a monthly basis. Test bleeds were collected ten days after each booster injection and monitored for the presence of antibodies by the SPR sensor. Three days after the fifth immunisation the mouse was sacrificed, and the spleen was removed for fusion with an SP2 myeloma cell line to produce monoclonal antibodies [15]. All protocols for antibody production performed using animals were conducted in accordance with a licence issued by the Department of Health, Social Services and Public Safety in Northern Ireland, UK under the animal scientific procedures Act 1986.
2.5. Biosensor chip production
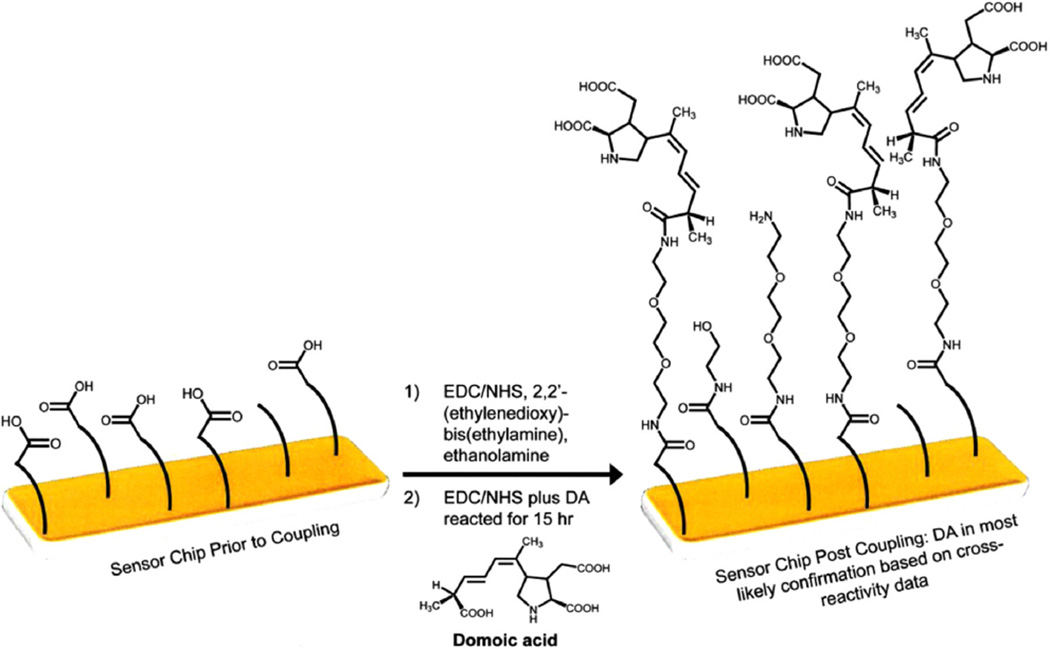
Domoic acid was immobilised onto the CM5 surface of the sensor chip (Fig. 2). Briefly, the chip surface was activated by contact with 50 µL of a 1:1 mixture of 0.4 M EDC: 0.2 M NHS for 20 min The reactants were removed and 50 µL of 2,2′-(ethylenedioxy)bis(ethylamine) was added and allowed to remain in contact with the sensor chip surface for 1 h; the chip surface was then washed with deionised water prior to the addition of 50 µL of 1M ethanolamine solution for 20 min. EDC (1.5 mg) and NHS (0.9 mg) were dissolved in 50 µL pH 4.7 MES buffer. This mixture (50 µL) was added to 250 µL of 2 mg/mL DA in water. An aliquot (50 µL) of this reactive mixture was added to the chip surface and allowed to react overnight. The chip surface was washed repeatedly with deionised water, dried using a stream of nitrogen gas and stored desiccated at +4 °C when not in use.
Fig. 2.
Chip surface chemistry for the immobilisation of domoic acid onto the surface of CM5 SPR biosensor chips.
For the Biacore T100 instrument, a partial in-instrument and partial on-bench coupling method was used. Specifically, the Biacore amine kit, standard amine wizard protocol, and pH 8.5, 50 mM borate buffer were used to link 2,2′-(ethylenedioxy)bis (ethylamine) to flow cells 2–4 with flow cell 1 left as a reference surface (no coupling). Following ethanolamine inactivation, the sensor chip was removed and DA was coupled to the surface in the same manner as described above. After reacting for 15 h, the sensor chip was rinsed/dried/stored following the same procedure as above.
2.6. Comparison of hybridoma screening by SPR biosensors and ELISA
In SPR biosensor analysis, equal volumes of hybridoma supernatant and HBS-EP pH 7.4 were mixed and tested for binding to DA immobilised on the chip surface. For a positive control, the final mouse bleed, at 1:300 in HAT media, was diluted 1:1 with HBS-EP pH 7.4. A negative control was prepared by 1:1 mixing of HAT media with HBS-EP buffer. Samples were injected over the chip surface at a flow rate of 20 µL/min for 1 min on both the Biacore Q and prototype high throughput Biacore SPR for comparison. The chip surface was regenerated after each sample injection with 50 mM sodium hydroxide at a flow rate of 20 µL/min for 1 min
An antigen capture ELISA assay was also utilised for the initial hybridoma screening. In brief, 25 µL of hybridoma supernatant were mixed with 75 µL of phosphate buffer pH 7.2, and the antibody allowed to immobilise on 96 well Falcon plates overnight at room temperature. The media was tapped out and DA-HRP prepared in 0.1% BSA solution was added to the plates and incubated at 4 °C overnight. The plates were washed ten times with ELISA wash solution (0.15 M saline with 0.125% Tween 20). Subsequently, TMB-E substrate (100 µL) was added, the colour development was stopped after 15 min by adding 50 µL of 2.5 M H2SO4, and the optical densities were read immediately at 450 nm using a Tecan Safire plate reader.
Controls were included within each assay type alternating between negative and positive. A positive was deemed as such for any sample that gave a signal equal or greater than the final heart bleed positive control, as lower would be interpreted as non-specific binding. Positive binders were further checked for specificity of binding by competition in the assay with and without DA (100 ng/mL), where DA specific binders were characterised by a reduction of signal when DA was present. Both SPR biosensor and ELISA assays yielded first screen hybridomas displaying suitable binding and inhibition. These were then cloned twice for single colony selection whereby clones were then selected for antibody production.
2.7. Monoclonal antibody
On selection of the best monoclonal antibody, the cell line was grown, and antibody was concentrated in cell culture media using a CELLine 1000 bioreactor (IBS Integra Biosciences, UK), followed by purification via affinity chromatography using a protein G-sepharose gel column (MAbTrap Kit). Dialysis of the antibody over 24 h in 0.15 M saline (3 × 4 L) was performed. The protein concentration and isotyping of each monoclonal antibody was determined at A280 nm and using a mouse monoclonal antibody isotyping kit (Roche Diagnostics), respectively.
2.8. Biosensor characterisation of monoclonal and polyclonal antibodies with Biacore Q
Initially, various parameters were investigated in the development of the Biacore Q biosensor assay including: binder dilution, ratio of binder to standard, injection volume, and contact time. The final assay consisted of: 1:200 antibody in HBS-EP mixed with the sample, contact time of 60 s with 15 s dissociation, regeneration with 50 mM HCl with 0.5% SDS for 60 s, flow rate of 25 µL/min, and analysis temperature at 25 °C. Prior to use, the sample chip was conditioned via the regeneration solution for three cycles. The sensitivity of the monoclonal antibody to DA was assessed using the Biacore Q biosensor. A stock solution of DA (1 mg/mL) was prepared and calibrants at 0, 1, 2.5, 5.0, 10, 25 and 100 ng/mL were prepared in pH 7.4 HBS-EP buffer. These working standards were used to produce calibration curves based on dose-response of the biosensor to compare the antibody performance.
2.9. Transferability to a Biacore T100 assay
The monoclonal antibody showing the best sensitivity on the Biacore Q instrument was evaluated in higher-throughput assays for the detection of domoic acid in shellfish on the commercial, Biacore T100 instrument. Previous studies have shown that the Q and T100 instrumentation, while having variations in surface formation and sensor response, allow for ease of method transferability [17]. For the Biacore T100 inhibition assay, assay parameters were adjusted to be consistent with the Biacore Q assay as outlined previously. In the Biacore T100 assay, varying dilutions of binder were evaluated to explore the maximum response and sensitivity of the assay.
2.10. Cross-reactivity
The cross-reactivity of seafood toxins that coexist with DA (e.g., TTX, STX, and OA) and compounds that have similar chemical structure to DA (e. g., GA, GluNH2, KA, and AA) were evaluated. Ten-fold dilutions in buffer of 10,000 ng/mL of each compound down to 0.1 ng/mL were analysed with the anti-DA and SPR assay on the Biacore T100. The curves were fit, and cross-reactivity was determined from a comparison of the IC50 values for each toxin curve to the DA standard curve IC50. Additionally, each cross-reactor at 1000 ng/mL was tested with the addition of 9.9 ng/mL DA to determine if additive or subtractive effects were present in these mixtures.
2.11. Matrix evaluation
DA is a known contaminant of shellfish [35]. To further develop the assay, shellfish matrices were prepared and spiked with DA to evaluate assay performance. Specifically, a modified NSSP method was used for the preparation of shellfish samples [2]. First, 4 mL of methanol in water (50% (v/v)) was added to 1 g of homogenized shellfish tissue (aliquoted from a pool of 12 homogenized organisms). For the recovery evaluation, 20 µg DA (20 µL of 1 mg/mL standard) was added to the homogenized tissue, the sample briefly vortex, and then allowed to sit for 2 min prior to addition of the solvent. Following a 1.5 min vortex, each sample was roller-mixed for 30 min and then centrifuged at 3000g for 10 min at room temp. The supernatant was decanted and used for further studies with 5 µL supernatant added to 1995 µL HBS-EP+ for the Biacore T100 assay.
3. Results and discussion
3.1. Immunisation results
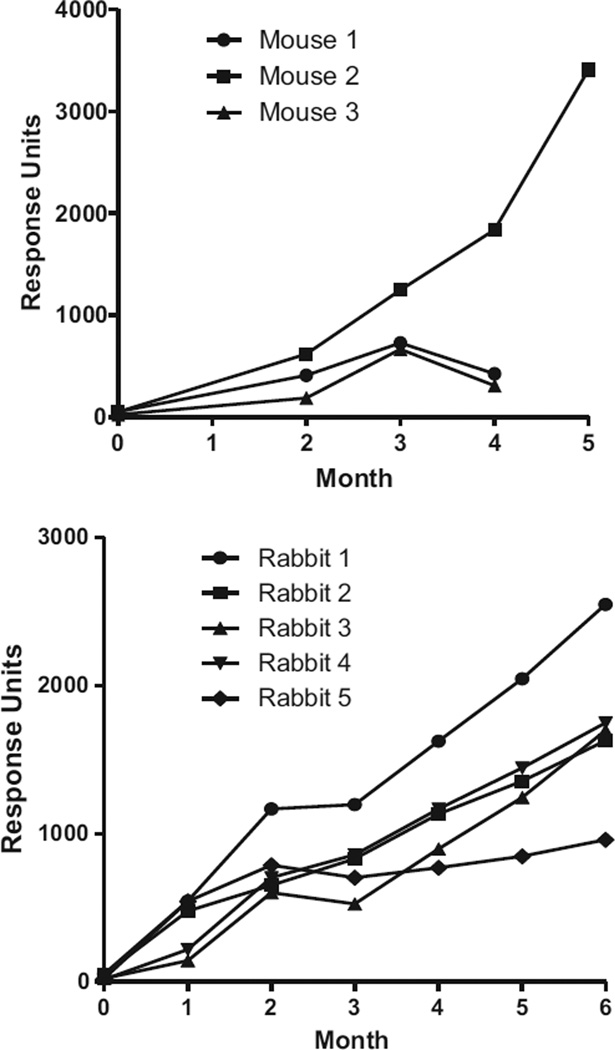
Small molecular weight molecules or haptens such as domoic acid have limited immunogenicity and must be conjugated to a protein carrier to add immunogenicity. Previous studies indicated that the affinity of the produced antibody for the hapten changed according to the protein carrier used in the conjugation [10,22]. In previous studies monoclonal antibodies for domoic acid have been raised to BSA, OVA, HGG and KLH for the immunisation process ([21,22,23,37]) with success primarily for those conjugated to BSA and KLH. However, bovine thyroglobulin, an established carrier protein for producing specific antibodies, had not previously been used for DA. In this work, the immunogen DA-BTG successfully produced an immune response in both mice and rabbits with increasing titres observed compared to before immunisation (Fig. 3). For the five rabbits over the 6 month period, the trend in increasing response is typical for the immunisation schedule with a slight dip in response following the switch in adjuvant from Freunds complete to incomplete (between months 2 and 3). For the three immunised mice at month four, one mouse (Mouse 2) showed an increase in response and had higher response than the rabbit counterparts and was thus selected for a further immunisation prior to harvesting the spleen for the fusion process. Compared to the first monoclonal antibody produced for DA [22] this work showed that the conjugation ratio of toxin to protein can be reduced to 50% and the dose of immunogen to 20%. This may be crucial for other marine toxins when limited supplies of toxin are available for antibody production.
Fig. 3.
Evaluation of antibody titres for mice and rabbits each month over the immunisation period illustrated by the SPR response value obtained by the analysis of each sera at 1 in 50 dilution mixed in HBS-EP buffer each month.
3.2. Comparison of techniques for hybridoma screening
Mouse 2 was selected for fusion, as the titre for this mouse had the highest binding in the titre, SPR assays. The fusion process was successful, and the first line screening of hybridomas for antibodies against DA was compared by single channel Biacore Q biosensor, multiplex prototype SPR sensor, and ELISA. With these methods, 44 hybridomas screened positive by ELISA, 42 by the single channel biosensor and 43 by the multichannel biosensor, although each method had different positive hybridomas. On comparison, 22 out of 919 hybridomas (2.3%) screened positive for all methods. Due to the multi spot (4 spots per channel) and four results per sample nature of the Biacore prototype [4], this method offered increased sensitivity and repeatability of selection compared to the single channel Biacore Q. This prototype instrument can screen 96 samples (4 × 24) in 2 h from loading of the instrument, whereas the Biacore Q takes over 12 h for the same number of samples. Additionally, one ELISA plate, due in part to the overnight coating, takes a minimum of two days. Further, the re-usability of the Biacore chips offered 4-fold reduction in the use of toxin compared to the ELISA screening procedure.
This efficiency in analysis time for routine monitoring for shellfish toxins is an advantage that was highlighted in addition to the use of this prototype instrument for the simultaneous, multiplex detection of marine toxins [5]. This system can therefore be extremely beneficial if there are large sample numbers to be analysed for the molecular interaction with the same target, but it would not be viably cost effective for small sample numbers. For hybridoma screening for antibody selection, this type of multiplexed, prototype instrument is very valuable, as the interactions can still be monitored in real-time and decisions can be made on the same day to proceed with the workload in hybridoma selection processes.
3.3. Monoclonal isotyping via the SPR biosensor
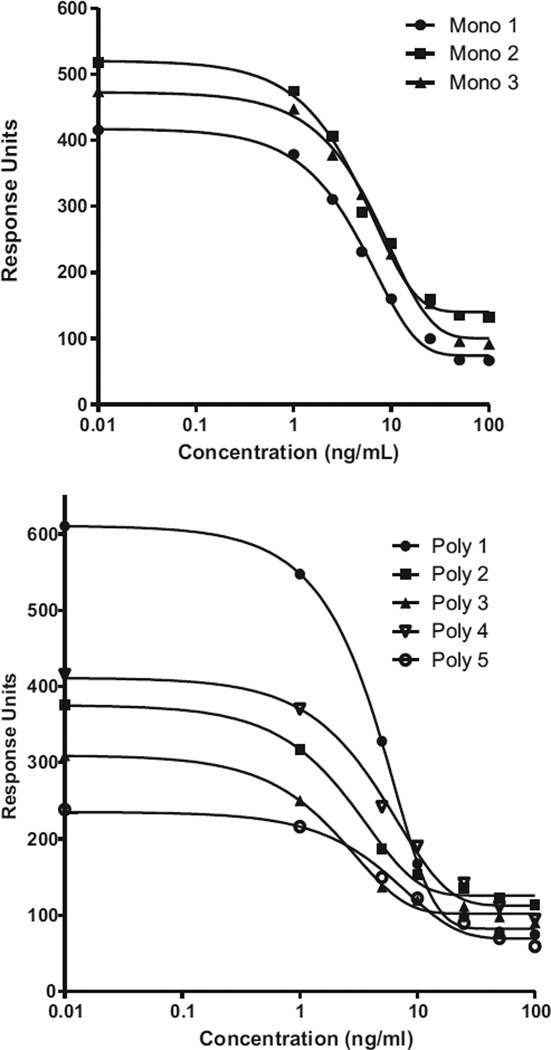
Following cloning, three hybridomas from the original Mouse 2 antibody were selected and monoclonal antibodies purified with protein concentrations Mono 1 = 1.16 mg/mL, Mono 2 = 0.86 mg/mL, and Mono 3 = 1.35 mg/mL. On characterisation by isotyping, the heavy and light chains of the antibodies were IgG2a and k, respectively. The calibration curves (Fig. 4) and associated figures of merit (Table 1) for the three monoclonal antibodies and five polyclonal (rabbit) antibodies as run on the Biacore Q showed good sensitivity for the detection of domoic acid with an average IC50 at 5.6 ± 1.2 ng/mL for the three monoclonal antibodies and 4.2 ± 1.6 ng/mL for the five polyclonal antibodies. The sensitivity of the antibodies as assessed in the performance of the SPR biosensor assay was comparable to those used in different systems [21,37] and improved by at least a factor of >100 for the monoclonal antibody compared to a previous SPR biosensor assays by other researchers [23]. It is important to note that for each of the antibody calibration curves there was a degree of non-specific binding on the chip surface of approximately 75–100 RU as seen in the high concentration binding levels and that the dynamic range for each of the curves varied. For example, Poly 5 has the lowest binding response but a large linear range (13.4 ng/mL based on IC80 minus the IC20). Poly 3 has a mid-binding response and the smallest linear range (4.9 ng/mL), whereas Poly 1 has the highest binding response with a mid-range linearity of 7.7 ng/mL. When choosing the best antibodies, these elements are taken into account and were utilised to identify Mono 2, as the antibody further studied due to good binding to the DA-chip of approx. 525 response units, 9.7 ng/mL dynamic range, and longevity of supply and stability due to the monoclonal nature.
Fig. 4.
Concentration analysis curves of response versus concentration for mouse derived monoclonal antibodies and each rabbit polyclonal antibody (1–5) (n = 2 for each data point with standard deviation less than the data point).
Table 1.
Figures of merit for the monoclonal (Mono) and polyclonal (Poly) antibodies developed to the DA-BTG conjugate as analysed by the Biacore Q.
| Domoic acid concentration(ng/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Mono 1 | Mono 2 | Mono 3 | Poly 1 | Poly 2 | Poly 3 | Poly 4 | Poly 5 | |
| IC20 | 1.6 | 1.6 | 2.2 | 1.7 | 0.9 | 0.7 | 1.5 | 1.8 |
| IC50 | 5.0 | 4.8 | 6.9 | 4.7 | 2.8 | 2.3 | 5.1 | 6.0 |
| IC80 | 11.8 | 11.3 | 16.6 | 9.4 | 6.9 | 5.6 | 12.8 | 15.2 |
3.4. Transferability to T100 Biacore
Domoic acid, surface plasmon resonance assays have been performed by a number of groups, thus setting the stage for rapid, robust immunosensors for DA in shellfish. In Stevens et al. [34], a portable SPR biosensor was developed for detection of DA down to 3 ppb in diluted clam extract with a surface capable of at least 50 regenerations; however, this prototype SPR instrumentation had high standard deviations which could limit the utility for quantification. An immunosensor developed with a polyclonal antibody researched by Traynor et al. [36] on a single-channel instrument allowed for up to 200 sample injections and shellfish DA concentrations compared well with LC-MS results. Additionally, assays in buffer have been developed by a number of groups including those focusing on surface chemistry and assay development [43], DA antibody screening [23], and novel molecularly imprinted polymer surfaces [27]. By employing these newly produced antibodies and designing a robust surface and assay on a commercial instrument, we are advancing biosensor technology to a potentially more broadly applicable platform for detecting DA in shellfish matrices.
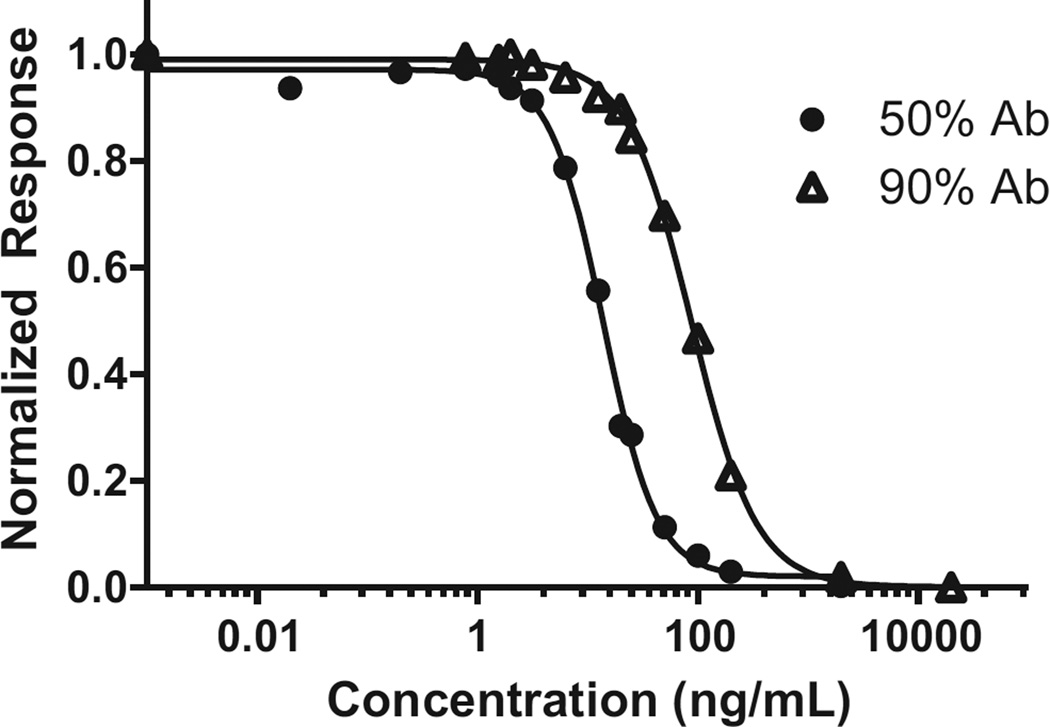
As shown, Mono 2 showed marginally better sensitivity on the Biacore Q instrument and was used for the comparison and evaluation on the Biacore T100. When using 50% 1:200 Ab mixed with the DA solutions, a raw response of 330 RU was observed for 0 ng DA/mL. For a 90% 1:200 Ab assay, the raw response was 650 RU. Upon normalization to these values, a curve fit via a log (inhibitor) versus response, four parameter fit was obtained (GraphPad Prism v.5.02, La Jolla, CA, USA). As expected, and shown in Fig. 5, the curve shapes are similar for each antibody percent but are shifted along the x-axis with the dynamic range for 50% Ab from 6.0 to 31.2 ng/mL and that for the 90% Ab 33.1–226.6 ng/mL. Therefore, this approach of increasing the percentage of antibody can be used to desensitise the assay where necessary. Based on the extraction procedure used herein, 12.5 ng DA/mL is equivalent to the United States and European Union guidance levels of 20 µg DA/gram tissue. In order to obtain the best quantitative capability around this level, the 50% Ab mixture with an IC50 of 13.7 ng/mL was used for further assay evaluation. Additionally, this sensor surface for the T100 assay can be used for at least 311 cycles (per flow cell) with a maximum loss of 25% of the surface activity (~330 to ~250 RU). This signal decrease, however, did not result in a change to the normalized calibration curve, thus highlighting the robust and reusable nature of the SPR assay.
Fig. 5.
Biacore T100 calibration curves of DA inhibition assay with new DA binding protein (Mono 2). (n = 3 for each data point with standard deviation less than the data point).
3.5. Cross-reactivity
The potential seafood toxin cross-reactors STX, TTX, and OA as well as the structurally similar GA, GluNH2, and AA did not show cross-reactivity to the anti-DA (0% cross-reactivity), while kainic acid showed 24% cross-reactivity in comparison to domoic acid. Additionally, when mixtures of DA plus the cross-reactors were run, there was no significant difference in signal to that of 10 ng/mL DA alone, except in the case of KA plus DA. This reactivity is solely due to the aforementioned binding of KA to the surface creating additional signal. These effects are not unexpected, as DA and KA molecules are very similar in structure. Additionally, these results potentially indicate that the antibody is most reactive towards this area of the DA molecule. As KA is associated with a natural marine acid present in some seaweed and is not expected to occur with DA in shellfish samples, there should not be concern about false-positives in naturally contaminated seafood samples.
3.6. Matrix evaluation and extraction recovery
Six matrices were chosen for evaluation, as these could potentially be contaminated with domoic acid: mussels, surfclams, oysters, scallops (gonad and viscera portions), and quahogs. The oysters (16 oz. wild caught with water added) and clams were purchased from a local vendor while the mussels, scallops and quahogs were harvested in 2007 from waters off the New England Coast of the United States and were determined in-house to be DA-free due to being at or below the blank measurement when using an ELISA.
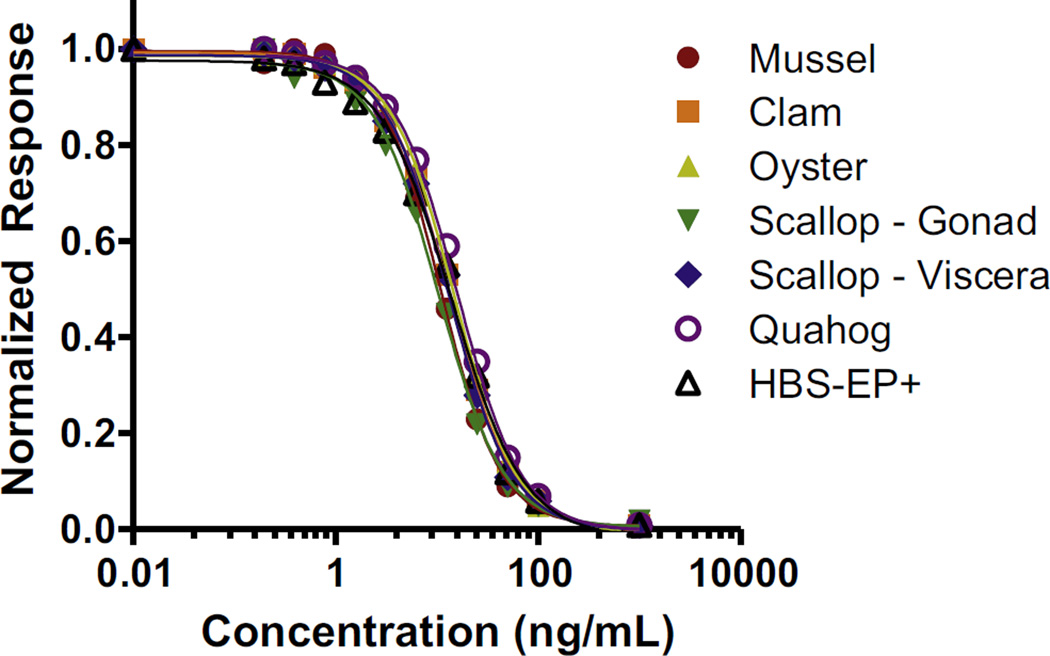
The raw response for the six matrices (0 ng DA/mL) and HB-SEP+ was between 308.2 and 310.3 RU on Day 1 and 283.3–296.2 RU on Day 2. The values are slightly different between days, likely due to minor differences in antibody dilution, but are less than 2% different within a day. When normalized to their respective 0 ng DA/mL response, nearly identical overlap of the curves for each matrix are observed (Fig. 6). This excellent overlap with the HBS-EP+ curve indicates that there are no obvious matrix effects. As there are no interferences from mussel, clam, oyster, scallop (gonad or viscera), and quahog matrices when using the 1 in 400 dilution necessary to have the action level near the centre of the linear range, there is no need to matrix match for the DA assay. As such, analysis can be significantly streamlined by having a singular buffer curve for quantitation of a variety of shellfish samples. To further evaluate this and determine DA recovery from the shellfish samples, an HBS-EP+ calibration curve was used to back calculate the concentration in the 20 µg DA per 1 g tissue spikes. Each sample was analysed (n = 3 per analysis) in duplicate. These initial studies show promise for this simple extraction method and the use of biosensor assays with this new antibody, as the extraction is efficient for each matrix (76–109%) as shown in Table 2. Additionally, there was little variation in the day one versus day two measurements as shown in Table 2 where the %CVs ranged from 2.01% to 6.45% for the six matrices. This further elucidates the robust nature of the SPR biosensor to yield repeatable results in sample analysis.
Fig. 6.
Biacore T100 calibration curves for DA-spiked shellfish matrices in comparison to DA-spiked buffer (n = 3 for each data point with standard deviation less than the data point).
Table 2.
Recovery analysis from shellfish matrices and the SPR assay. Concentration day one (Conc 1) is back calculated from GraphPad Prism and an HBS-EP+ curve (N = 3 for all measurements) and similarly repeated on day 2 for Conc 2.
| Sample | Calculated (ng/mL) | 1 g | 4 mL | µg/g tissue | Spiked | % Recovery | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Conc 1 | Conc 2 | AVG | SD | %CV | Tissue | Extract volume | ||||
| Clam | 8.54 | 9.14 | 8.84 | 0.42 | 4.77 | 1.02 | 4.4 | 15.3 | 20 | 76 |
| Mussel | 11.7 | 12.82 | 12.26 | 0.79 | 6.45 | 1.02 | 4.5 | 21.6 | 20 | 108 |
| Oyster | 8.54 | 8.81 | 8.68 | 0.19 | 2.17 | 0.99 | 4.4 | 15.4 | 20 | 77 |
| Viscera | 9.87 | 10.52 | 10.2 | 0.46 | 4.50 | 1.00 | 4.4 | 17.9 | 20 | 90 |
| Gonad | 10.58 | 10.89 | 10.73 | 0.22 | 2.01 | 0.99 | 4.4 | 19.1 | 20 | 95 |
| Quahog | 12.49 | 13.23 | 12.86 | 0.52 | 4.08 | 1.01 | 4.3 | 21.9 | 20 | 109 |
4. Conclusions
This work illustrated the application of three different SPR optical biosensors (high throughput Biacore prototype, Biacore Q and Biacore T100) in antibody production for their development and characterisation using domoic acid as a model target. The conjugation and immunisation protocol developed herein for domoic acid applied lower toxin to protein ratios (50% less than previous methods) and thereby reduced the dose of immunogen by 20%. For antibody development and characterisation, the optical SPR biosensor offered the set-up of a simple assay design for real time evaluation of sensitivity, cross reactivity with structurally related compounds, as well as the effects of different shellfish matrices on the antibody-analyte interaction. The multichannel biosensors offer enhanced speed for screening for suitable binders during monoclonal antibody selection. The assays developed herein for DA are, to our knowledge, the fastest and most sensitive SPR assays and can be used for over 300 cycles with triplicate measurement per cycle. This study highlights that automated SPR biosensors can be effective tools for antibody characterisation, and these antibodies can then be employed for sensitive, robust DA detection in the diagnostics environment. Methods developed herein can be employed for further development of well-performing antibodies for toxin detection.
Acknowledgments
This research was funded in part by the European Commission through the FP7 European projects; MIDTAL, Grant no. 201724, MicroAqua, Grant no. 265409 and the Advanced ASSET Project, Grant no. 1210/101154811/A1, partly funded through InvestNI and from the European Sustainable Programme 2007–2013 under the European Regional Development Fund (“ERDF”). Thank you to Dr. Stacey DeGrasse for fruitful conversations and Stephen Conrad for maintaining the U.S. FDA/CFSAN seafood sample inventory and to Dr Shauna Devlin for assistance in the monoclonal antibody fusion procedure.
References
- 1.An Y, Jiang LP, Cao J, Geng CY, Zhong LF. Sudan I induces genotoxic effects and oxidative DNA damage in HepG2 cells. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2007;627:164–170. doi: 10.1016/j.mrgentox.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 2.AOAC International, AOAC Official Method 2006.02, Domoic Acid Toxins in Shellfish. Biosense ASP ELISA – a Competitive Direct Enzyme-Linked Immunosorbent Asssay. 2007 [PubMed] [Google Scholar]
- 3.Campàs M, Prieto-Simón B, Marty JL. Biosensors to detect marine toxins: assessing food safety. Talanta. 2007;72:884–895. doi: 10.1016/j.talanta.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 4.Campbell K, McGrath T, Sjölander S, Hanson T, Tidare M, Jansson O, Moberg A, Mooney M, Elliott C, Buijs J. Use of a novel micro-fluidic device to create arrays for multiplex analysis of large and small molecular weight compounds by surface plasmon resonance. Biosens. Bioelectron. 2011;26:3029–3030. doi: 10.1016/j.bios.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Campbell K, McNamee SE, Huet AC, Delahaut P, Vilarino N, Botana LM, Poli M, Elliott CT. Evolving to the optoelectronic mouse for phycotoxin analysis in shellfish. Anal. Bioanal. Chem. 2014;406(27):6867–6881. doi: 10.1007/s00216-014-8156-2. [DOI] [PubMed] [Google Scholar]
- 6.Campbell K, Stewart LD, Doucette GJ, Fodey TL, Haughey SA, Vilariño N, Kawatsu K, Elliott CT. Assessment of specific binding proteins suitable for the detection of paralytic shellfish poisons using optical biosensor technology. Anal. Chem. 2007;79(15):5906–5914. doi: 10.1021/ac070342o. [DOI] [PubMed] [Google Scholar]
- 7.Campbell K, Barnes P, Haughey SA, Higgins C, Kawatsu K, Vasconcelos V, Elliott CT. Development and single laboratory validation of an optical biosensor assay for tetrodotoxin detection as a tool to combat emerging risks in European seafood. Anal. Bioanal. Chem. 2013;405(24):7753–7763. doi: 10.1007/s00216-013-7106-8. [DOI] [PubMed] [Google Scholar]
- 8.Chemical & Engineering News. Neurotoxin Scuttles Crab Season. 2015 Nov 12;93(45):8. [Google Scholar]
- 9.Chen J. A worldwide food safety concern in 2008—melamine contaminated infant formula in China caused urinary tract stones in 290,000 children in China. Chin. Med. J. 2009;122:243–244. doi: 10.3901/jme.2009.07.243. [DOI] [PubMed] [Google Scholar]
- 10.Chu FS, Huang X. Production and characterization of antibodies against neosaxitoxin. J. AOAC Int. 1992;75:341–345. [Google Scholar]
- 11.Commission Decision of 15 March 2002 Establishing Special Health Checks for the Harvesting and Processing of Certain Bivalve Molluscs with a Level of Amnesic Shellfish Poison (ASP) Exceeding the Limit Laid Down by Council Directive 91/492/EEC Official Journal. 2002;L075:65–66. [Google Scholar]
- 12.Devlin S, Meneely JP, Greer B, Campbell K, Vasconcelos V, Elliott CT. Production of a broad specificity antibody for the development and validation of an optical SPR screening method for free and intracellular microcystins and nodularin in cyanobacteria cultures. Talanta. 2014;122:8–15. doi: 10.1016/j.talanta.2013.12.065. [DOI] [PubMed] [Google Scholar]
- 13.Fonfría ES, Vilariño N, Campbell K, Elliott C, Haughey SA, Ben-Gigirey B, Vieites JM, Kawatsu K, Botana LM. Paralytic shellfish poisoning detection by surface plasmon resonance-based biosensors in shellfish matrixes. Anal. Chem. 2007;79(16):6303–6311. doi: 10.1021/ac070362q. [DOI] [PubMed] [Google Scholar]
- 14.Garibo D, Campbell K, Casanova A, De La Iglesia P, Fernández-Tejedor M, Diogène J, Elliott CT, Campàs M. SPR immunosensor for the detection of okadaic acid in mussels using magnetic particles as antibody carriers. Sens. Actuators B: Chem. 2014;190:822–828. [Google Scholar]
- 15.Galfre G, Milstein C. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzym. 1981;72:3–46. doi: 10.1016/0076-6879(81)73054-4. [DOI] [PubMed] [Google Scholar]
- 16.Gossner CME, Schlundt J, Ben Embarek P, Hird S, Lo-Fo-Wong D, Beltran JJO, Teoh KN, Tritscher A. The melamine incident: implications for international food and feed safety. Environ. Health Perspect. 2009;117:1803–1808. doi: 10.1289/ehp.0900949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haughey SA, Campbell K, Yakes BJ, Prezioso SM, DeGrasse SL, Kawatsu K, Elliott CT. Comparison of biosensor platforms for surface plasmon resonance based detection of paralytic shellfish toxins. Talanta. 2011;85(1):519–526. doi: 10.1016/j.talanta.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 18.Holt D, Harvey D, Hurley R. Chloramphenicol toxicity, adverse drug reaction. Toxicol. Rev. 1993;12:83–95. [PubMed] [Google Scholar]
- 19.Hoogenboom R, Traag W, Fernandes A, Rose M. European developments following incidents with dioxins and PCBs in the food and feed chain. Food Control. 2015;50:670–683. [Google Scholar]
- 20.Iverson F, Truelove J. Toxicology and seafood toxins: domoic acid. Nat. Toxins. 1994;2:334–339. doi: 10.1002/nt.2620020514. [DOI] [PubMed] [Google Scholar]
- 21.Kania M, Kreuzer M, Moore E, Pravada M, Hock B, Guilbault G. Development of polyclonal antibodies against domoic acid for their use in electrochemical biosensors. Anal. Lett. 2003;36(9):1851–1863. [Google Scholar]
- 22.Kawatsu K, Hamano Y, Noguchi T. Production and characterization of a monoclonal antibody against domoic acid and its application to enzyme immunoassay. Toxicon. 1999;37:1579–1589. doi: 10.1016/s0041-0101(99)00106-3. [DOI] [PubMed] [Google Scholar]
- 23.Le Berre M, Kane M. Biosensor-based assay for domoic acid: comparison of performance using polyclonal, monoclonal, and recombinant antibodies. Anal. Lett. 2006;39(8):1587–1598. [Google Scholar]
- 24.Lelong A, Hegaret H, Soudant P, Bates SS. Pseudo-nitzschia (Bacillariophyceae) species, domoic acid and amnesic shellfish poisoning: revisiting previous paradigms. Phycologia. 2012;51(2):168–216. [Google Scholar]
- 25.Lewis L, Onsongo M, Njapau H, Schurz-Rogers H, Luber G, Kieszak S, Nyamongo J, Backer L, Dahiye AM, Misore A, Decock K, Rubin C, Kenya I. Aflatoxicosis, Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicosis in eastern and central Kenya. Environ. Health Perspect. 2005;113:1763–1767. doi: 10.1289/ehp.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llamas N, Stewart L, Fodey T, Higgins HC, Velasco MLR, Botana, Elliott CT. Development of a novel immunobiosensor method for the rapid detection of okadaic acid contamination in shellfish extracts. Anal. Bioanal. Chem. 2007;389:581–585. doi: 10.1007/s00216-007-1444-3. [DOI] [PubMed] [Google Scholar]
- 27.Lotierzo M, Henry OYF, Piletsky S, Tothill I, Cullen D, Kania M, Hock B, Turner APF. Surface plasmon resonance sensor for domoic acid based on grafted imprinted polymer. Biosens. Bioelectron. 2004;20(2):145–152. doi: 10.1016/j.bios.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 28.McGrath TF, Elliott C, Fodey TL. Biosensors for the analysis of microbiological and chemical contaminants in food. Anal. Bioanal. Chem. 2012;403(1):75–92. doi: 10.1007/s00216-011-5685-9. [DOI] [PubMed] [Google Scholar]
- 29.McNamee SE, Elliott CT, Delahaut P, Campbell K. Multiplex biotoxin surface plasmon resonance method for marine biotoxins in algal and seawater samples. Environ. Sci. Pollut. Res. Int. 2013;20(10):6794–6807. doi: 10.1007/s11356-012-1329-7. [DOI] [PubMed] [Google Scholar]
- 30.McKay C, Scharman EJ. Intentional and Inadvertent Chemical Contamination of food, water, and medication. Emerg. Med. Clin. North Am. 2015;33(1):153–177. doi: 10.1016/j.emc.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Salleras L, Dominguez A, Mata E, Taberner JL, Moro I, salva P. Epidemiologic study of an outbreak of clenbuterol poisoning in Catalonia, Spain. Public Health Rep. 1995;110:338–342. [PMC free article] [PubMed] [Google Scholar]
- 32.Reverté L, De La Iglesia P, Del Río V, Campbell K, Elliott CT, Kawatsu K, Katikou P, Diogène J, Campàs M. Detection of tetrodotoxins in puffer fish by a self-assembled monolayer-based immunoassay and comparison with surface plasmon resonance, LC-MS/MS, and mouse bioassay. Anal. Chem. 2015;87(21):10839–10847. doi: 10.1021/acs.analchem.5b02158. [DOI] [PubMed] [Google Scholar]
- 33.Scholin CA, Gulland F, Doucette GJ, Benson S, Busman M, Chavez FP, Cordaro J, DeLong R, De Vogelaere A, Harvey J, Haulena M, Lefebvre K, Lipscomb T, Loscutoff S, Lowenstine LJ, Marin R, Miller PE, McLellan WA, Moeller PDR, Powell CL, Rowles T, Silvagni P, Silver M, Spraker T, Trainer V, Van Dolah FM. Mortality of sea lions along the central California coast linked to a toxic diatom bloom. Nature. 2000;403(6765):80–84. doi: 10.1038/47481. [DOI] [PubMed] [Google Scholar]
- 34.Stevens RC, Soelberg SD, Eberhart BTL, Spencer S, Wekell JC, Chinowsky TM, Trainer VL, Furlong CE. Detection of the toxin domoic acid from clam extracts using a portable surface plasmon resonance biosensor. Harmful Algae. 2007;6(2):166–174. [Google Scholar]
- 35.Tasker RA. Domoic acid and other amnesic toxins: toxicological profile. In: Gopalakrishnakone P, Haddad V Jr, Kem WR, Tubaro A, Kim E, editors. Marine and Freshwater Toxins. Dordrecht Netherlands: Springer Science + Business Media Dordrecht; 2015. pp. 1–16. [Google Scholar]
- 36.Traynor IM, Plumpton L, Fodey TL, Higgins C, Elliott CT. Immunobiosensor detection of domoic acid as a screening test in Bivalve Molluscs: comparison with liquid chromatography-based analysis. J. AOAC Int. 2006;89(3):868–872. [PubMed] [Google Scholar]
- 37.Tsao ZJ, Liao YC, Liu BH, Su CC, Yu FY. Development of a monoclonal antibody against domoic acid and its application in enzyme-linked immunosorbent assay and colloidal gold immunostrip. J. Agric. Food Chem. 2007;55(13):4921–4927. doi: 10.1021/jf0708140. [DOI] [PubMed] [Google Scholar]
- 38.U.S. Food and Drug Administration. Fish and fisheries products hazards and controls guidance. Chapter 6: Natural Toxins (Online) 〈 http://www.fda.gov/Food/GuidanceRegulation/GuidanceDocumentsRegulatoryInformation/Seafood/ucm2018426.htm〉.
- 39.U.S. Food and Drug Administration. Bad bug book: handbook of foodborne pathogenic microorganisms and natural toxins. Chapter Title: Shellfish Toxins (PSP, DSP, NSP, ASP, AZP) (Online) 2012:200. 〈 http://www.fda.gov/downloads/Food/FoodborneIllnessContaminants/UCM297627.pdf〉.
- 40.Yakes BJ, Prezioso S, Haughey SA, Campbell K, Elliott CT, DeGrasse SL. An improved immunoassay for detection of saxitoxin by surface plasmon resonance biosensors. Sens. Actuators B Chem. 2011;156(2):805–811. [Google Scholar]
- 41.Yakes BJ, Kanyuck KM, DeGrasse SL. First report of a direct surface plasmon resonance immunosensor for a small molecule seafood toxin. Anal. Chem. 2014;86(18):9251–9255. doi: 10.1021/ac502271y. [DOI] [PubMed] [Google Scholar]
- 42.Yakes BJ, Handy SM, Kanyuck KM, DeGrasse SL. Improved screening of microcystin genes and toxins in blue-green algal dietary supplements with PCR and a surface plasmon resonance biosensor. Harmful Algae. 2015;47:9–16. [Google Scholar]
- 43.Yu QM, Chen SF, Taylor AD, Homola J, Hock B, Jiang SY. Detection of low-molecular-weight domoic acid using surface plasmon resonance sensor. Sens. Actuator B-Chem. 2005;107(1):193–201. [Google Scholar]