Abstract
Beginning with the precedent of XIST RNA as a “chromosomal RNA” (cRNA), there is growing interest in the possibility that a diversity of non-coding RNAs may function in chromatin. We review findings which lead us to suggest that RNA is essentially a widespread component of interphase chromosomes. Further, RNA likely contributes to architecture and regulation, with repeat-rich “junk” RNA in euchromatin (ecRNA) promoting a more open chromatin state. Thousands of low-abundance nuclear RNAs have been reported, however it remains a challenge to determine which of these may function in chromatin. Recent findings indicate that repetitive sequences are enriched in chromosome-associated non-coding RNAs, and repeat-rich RNA shows unusual properties, including localization and stability, with similarities to XIST RNA. We suggest two frontiers in genome biology are emerging and may intersect: the broad contribution of RNA to interphase chromosomes and the distinctive properties of repeat-rich intronic or intergenic junk sequences that may play a role in chromosome structure and regulation.
Introduction
During development, genomic DNA becomes packaged differently in various cell-types, forming regions of active, open euchromatin and inactive, condensed heterochromatin. Traditionally studies of chromatin regulation have focused largely on the protein components, particularly histones, and their covalent modifications. However, the discovery of XIST RNA’s role in X-chromosome inactivation established that RNA can function in chromatin as a “master switch” for downstream modifications that collectively silence the chromosome. Here we will consider the potential for other types of “chromosomal RNAs” (cRNA), which we define as RNA which functions within an interphase chromosome, either at a specific locus or across a broader chromatin region. Among thousands of putative large non-coding RNAs (lncRNAs) with unknown function, several are suggested to reside in and function in chromatin, and many others are speculated to do so. However, the field now faces the challenge of determining whether these RNAs make a structural/functional contribution at specific chromatin sites, or simply localize there during their biogenesis. Based on recent findings, we postulate that the presence of RNA throughout the interphase chromosome is more the rule than the exception, not only on heterochromatin, but also through much or all of euchromatin. We suggest that RNA may prove to be essentially a fundamental component of interphase chromosomes. We further hypothesize that distinct properties of repetitive sequences (the vast junk of the genome), might play a major role in chromosome architecture and regulation, not only at the level of DNA, but in potentially ubiquitous, euchromatin-associated chromosomal RNAs (ecRNAs).
Insights from XIST RNA and a Paradox of the Barr body
XIST RNA is the paradigm for an RNA that binds and regulates chromatin (Figure 1A) [1–4] and is well reviewed elsewhere [5–7]. Here, we limit our discussion of XIST RNA and female X-inactivation to a few points related to the broader role of RNA in chromatin and the potential involvement of repeat-rich DNA and RNA. As previously reviewed [8], repeat elements in DNA have long been noted to correlate approximately with greater or lesser susceptibility of chromatin to be inactivated by XIST RNA, based largely on studies of X-autosome translocations, or more recently XIST-transgenes inserted into autosomes [9,10]. However, the more limited or variable inactivation seen in autosomal chromatin (in individuals or cultured cells) will in large part reflect selection against creating a functional monosomy for dosage-sensitive genes. A recent study thoroughly examined human autosomal inactivation in the context of trisomy, where there is no selection against silencing. Jiang et al. [11] showed that an XIST transgene inserted into one of three Chromosome 21s (in Down syndrome induced pluripotent stem cells) exhibited comprehensive spread of heterochromatic marks, and silencing of genes throughout essentially the whole targeted chromosome. Given that XIST RNA is not indifferent to the underlying DNA sequence (since some X-linked genes escape silencing) the ability of XIST RNA to inactivate this autosomal material as thoroughly (or better) than the X-chromosome [11] supports that the RNA spreads and acts via widely distributed sequence elements common to all chromosomes. Notably, repeat sequences are highly abundant elements common to all chromosomes, but their raison d’etre is largely unknown, and they are widely thought to be “junk”.
FIGURE 1.
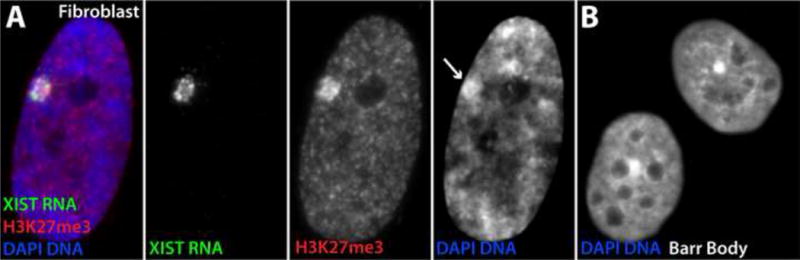
A) XIST RNA is expressed from and coats the silent interphase X-chromosome territory in cis, and can be seen as a large RNA signal in the three color image at left and the separated channel to the right. The RNA initiates a cascade of chromatin modifications at both protein and DNA levels (including histone modifications as seen in the three color image and separated channel at right) as well as architectural changes to form the heterochromatic Barr Body (see arrow in the separated DAPI channel at right). B) The Barr Body structure is striking in some cell preparations, including for this autosome (chromosome 4) carrying an XIST transgene.
A second point, regarding the structure of the Barr Body (BB), sets the stage for our discussion below of the potential role of abundant repeat-rich “junk” RNAs associated with euchromatin (ecRNAs). The BB has long been presumed (including by us) to reflect condensed, silenced genes of the inactive X-chromosome (Xi). However, we realized many years ago a paradox: since genic DNA is only a small subset of the genome and only a portion of genes are expressed in a given cell type, why would silencing <10% of the chromosome produce such a large, solid mass of dense DNA? (In the best preps this mass of condensed DNA is striking, as shown in Figure 1B). Observed differences in the organization of gene loci versus repeat-rich “junk” DNA suggested to us that the BB may actually not represent the entire Xi chromosome, but rather a condensed chromosome core enriched for silent, repeat-rich DNA ([12] reviewed in: [8]). Although a recent study by Teller et al. appears to show DNA sequence organization in the BB is random [13], this study used large pools of BACs (multiple Mb of DNA) rather than probes to specific genes, and thus in our view it remains plausible that the condensed BB forms from coalescence of silent “junk” DNA enriched for repetitive sequences.
Hence, the “Barr Body Paradox” which puzzled us years ago could be explained by the congregation of silent, abundant repeat-rich DNA to form the condensed heterochromatic core of the inactive X chromosome. As will be further considered below, a key question is whether on an active chromosome these repeat-rich sequences produce RNA which helps to promote more open conformation of the chromosome.
A Few New Prospective “Chromosomal” RNAs: The Challenge is to Discriminate RNA Localization for Function versus Biogenesis
In recent years numerous studies have reported thousands of enormously diverse non-coding RNAs of various sizes in mammalian cells. Most have no known function(s), although some are hypothesized to play a role in chromatin-mediated gene regulation (reviewed in: [14–17]). A challenge for the field is to discriminate when RNA is with chromatin simply because it is synthesized there, or plays some role there. A common misconception is that an RNA that accumulates at a specific chromatin site indicates function there. However, pre/mRNAs are typically most concentrated at their site of synthesis, therefore all RNAs will generally exhibit some accumulation in cis at their gene locus.
Several criteria can be used to evaluate the likelihood an RNA remains with chromatin to contribute to structure/function, borrowing from the tools and lessons learned from XIST RNA. For example, the RNAs increased stability upon transcriptional inhibition may suggest an intimate relationship with chromatin (and will be further illustrated below). Similarly, if the RNA spreads across or “paints” a large chromatin region as XIST RNA does, this is a very important indication of chromatin-related function, but is not a requirement, as many RNAs that might function in chromatin could interact at a more local level. However, The size of the RNA accumulation at an active locus is also not necessarily indicative of function either, as this will vary not only with the gene size but also the relative kinetics of transcription, processing, and transport (e.g. [18,19]), and will be impacted by such things as detained or slow splicing introns [20]. In select cases a significant RNA accumulation may form over a chromatin region due to the sheer size of the gene being transcribed (e.g. the 2Mb dystrophin gene: Figure 2A)[19]. Other quite large RNA accumulations often may not actually be localized over chromatin at all, but rather transcripts can accumulate in non-chromatin nuclear domains for a variety of reasons. For example, some pre-mRNAs accumulate in Nuclear Speckles (aka SC35 Domains: accumulations of pre-mRNA splicing factors) adjacent to their gene where their post-transcriptional processing is completed (e.g. collagen: Figure 2B) [18]). Other nuclear lncRNAs (e.g. Neat-1 or Neat2/Malat1) also accumulate in non-chromatin domains in relation to their function (Figure 2C) ([21]; reviewed in: [22]). The task of discerning bona fide “chromosomal RNAs” (cRNAs) would be easier if an RNA remained on the chromosome through mitosis, but even XIST RNA detaches, typically in early prophase, although it can be glimpsed briefly on early mitotic chromosomes in some mouse cells (Figure 2D)[23]. We’ve shown that XIST RNA can be “forced” to stay on the mitotic chromosome by inhibition of Aurora B kinase [24], and this approach may be useful to show other chromosomal RNAs, provided their mitotic release is similarly controlled through chromatin phosphorylation by AURKB.
FIGURE 2.
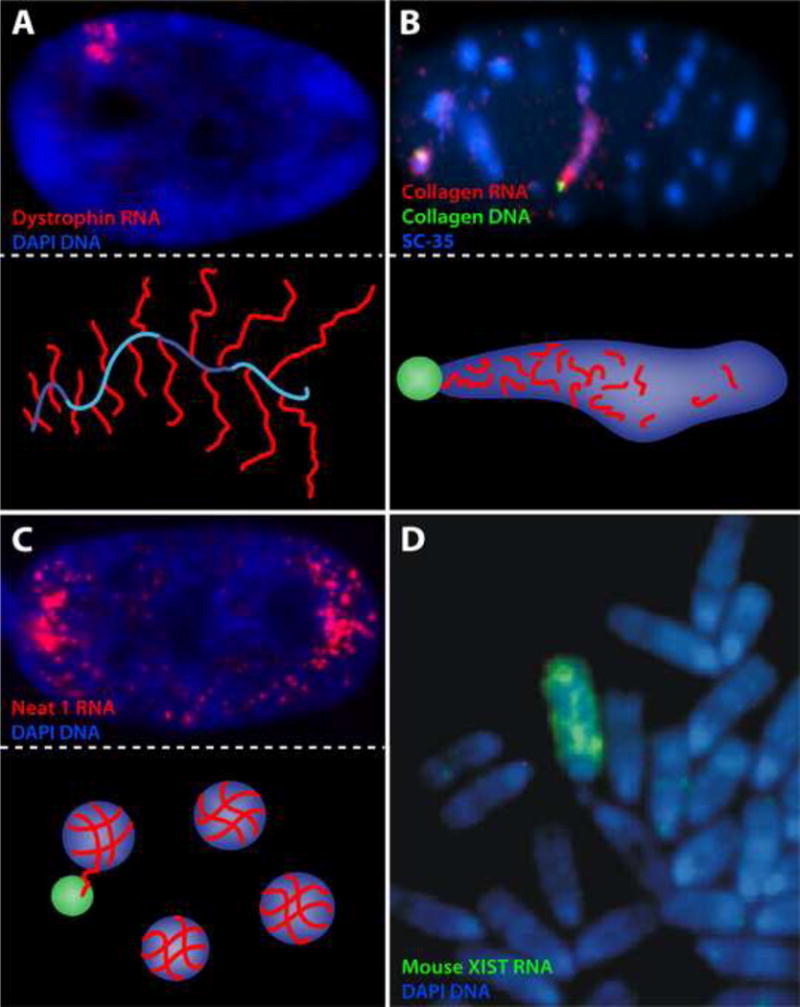
A) Photo: The large (2MB) dystrophin gene produces a substantial accumulation of nascent transcripts localized over chromatin. Illustration: Other analyses [19] showed this was a “tree” of multiple transcripts (red) in synthesis on the large gene (blue), producing what looks like a small fir tree (which can be often seen by electron microscope). B) Photo: Collagen pre/mRNAs (red), which require processing of 50 introns in a relatively small transcription unit, produce a large post-transcriptional accumulation in non-chromatin Nuclear Speckles (visualized using the spliceosome assembly factor SC35), which is adjacent to the Collagen genes (green) [18]. Illustration: Collagen pre-mRNA (red) emanates from the gene (green) and is processed in the Nuclear Speckle (blue). C) Photo: NEAT1 is an architectural-RNA that is required to form the underpinnings of non-chromatin nuclear structures termed paraspeckles, enriched for specific proteins. Illustration: The paraspeckles (blue) form on NEAT1 RNA (red) transcribed at the gene locus (green) but then move into the nucleoplasm [21]. D) Mouse Xist RNA in some mouse cell types is retained long enough to be glimpsed on mitotic chromosomes before it detaches, and can be seen to paint the whole Xi except the centromere.
A few recent studies suggest particular lncRNAs bind and potentially regulate sites on chromatin, or even “paint” large regions, and we consider a few examples here. FIRRE RNA was reported to localize in cis and organize trans-chromosomal associations between its gene locus and several loci from other chromosomes [25], although a more recent report draws this into question [26]. In any finding based on precise “co-localization” of two loci, it is important to eliminate any potential for “bleed-through” of a very bright signal in the red channel which can produce a weak signal in the green channel, creating the appearance that two signals overlap. Bergmann et al. [26] find FIRRE RNA is actually widely distributed in the nucleus and its deletion affects regulation of many genes in stem cells, but not those reported by Hacisuleyman et al. [25]. They suggest the discrepancy may be partly due to low sensitivity of detection in the earlier study, emphasizing that hybridization efficiency is key to such studies. Despite remaining questions, the role of FIRRE RNA in nuclei remains of interest: in fact, a very recent study suggests it controls Xi localization to the nucleolus, a finding that bears further analysis [27].
Another interesting case of a prospective cRNA is XACT RNA, which was reported to coat the entire active X-chromosome(s) in pluripotent human cells, similar to XIST RNA’s relationship to the Xi [28]. Building on this observation, these authors further proposed that XACT expression results in reactivation of unstable X-inactivation [29]. While we also see that XACT RNA forms a fairly large accumulation from the active X-chromosome (Xa) in pluripotent cells (Figure 3A), the conclusion that this RNA coats essentially the whole Xa, similar to XIST RNA on Xi, requires better substantiation. The published images show XACT RNA overlapping only a modest portion of the DNA territory, which the authors argue was due to technical limitations of DNA/RNA detection. However we routinely see XIST RNA overlap almost all (~90%) [3] of a chromosome DNA paint (e.g. Figure 3B), but we see XACT RNA co-localize with a much smaller part of the substantially larger (decondensed) Xa DNA territory (Figure 3A & D and unpublished results). While the sizeable XACT RNA accumulation might reflect transcription from this large (~300kb) locus, further studies are needed to determine whether mature transcripts have a more extensive association with Xa chromatin, which is a critical point to support its proposed functional role as a cRNA associated with “activation”.
FIGURE 3.
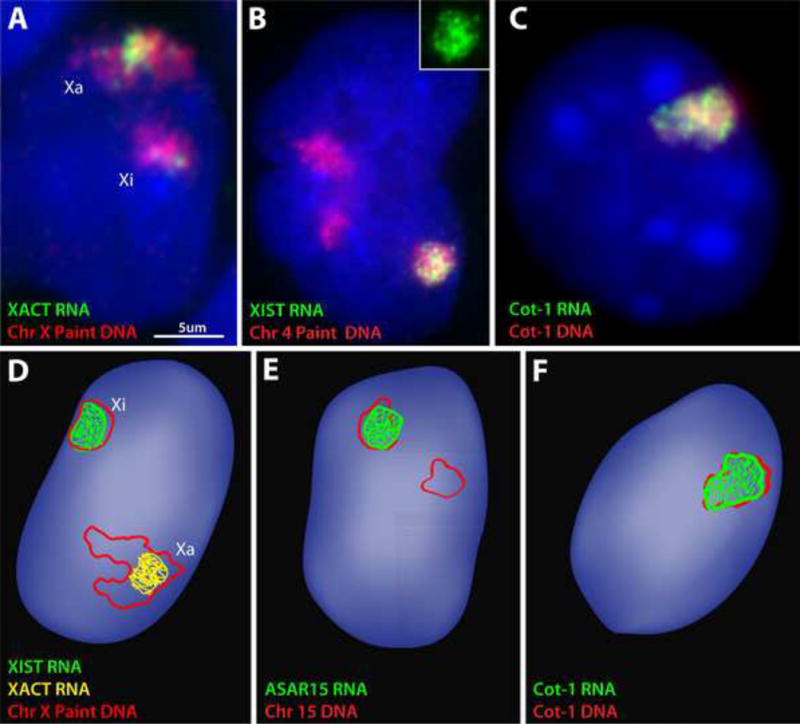
A) We find that XACT RNA produces a substantial accumulation (almost the same size as XIST RNA), but “paints” only a portion of the Xa territory. The Xa territory is very large and distended in pluripotent human stem cells (WIBR2 hESCs), and the XACT RNA signal would be much larger than the XIST RNA signal (which is coating a compacted Xi territory) if it painted the whole Xa territory. B) Even on a large autosome (Chromosome 4) carrying an XIST transgene, we consistently find that XIST RNA paints the vast majority of the chromosome territory. The green XIST RNA channel is shown alone in the insert. C) Cot-1 ecRNA signal consistently forms a tightly cis-limited RNA territory, with little to no “drift” of RNA away from the parent DNA territory (shown for a single human chromosome in a mouse/human hybrid cell). D) An illustration of the amount of interphase X-chromosome territory typically “painted” by XIST RNA (green) and XACT RNA (yellow) in a pluripotent cell (as seen by us and in published images) [28]. E) An illustration of the amount of interphase chromosome-15 territory typically “painted” by ASAR5 RNA (as seen in published images) [30]. F) An illustration of the amount of a human interphase chromosome territory typically “painted” by Cot-1 ecRNA in mouse/human hybrid cells (as seen in our published images) [39].
Another particularly promising prospect for an RNA that likely spreads across Chromosome 15 is ASAR15 (Figure 3E), discovered because disruption of this large locus impacts the replication timing and stability of the chromosome [30]. Several published images show an extensive overlap of ASAR15 RNA with chromosome 15 DNA (marked by many loci rather than a DNA library). In keeping with their hypothesis, the RNA is expressed from just one homologue and has a similarly long nuclear half-life to that of XIST RNA. It has been previously hypothesized that XIST-like autosomal RNAs existed to coordinate monoallelic gene expression [31,32], so ASAR15 is an exciting prospect that may provide a precedent for this.
These are but a few recent examples of potential cRNAs which may interact with chromatin regions outside of their own immediate locus. However, there are thousands more “lncRNA” sequences in databases which are often very low abundance and may be primarily nuclear, but are yet to be examined.
Repeat-rich “Cot-1 RNA” as an abundant, stable component of euchromatic chromosome territories
Prompted by the paradox of the BB discussed above, we long ago performed an unusual experiment, using “Cot-1” DNA, the repetitive fraction of the genome that reanneals most rapidly in “Cot” (concentration X time) curves [33,34]. Rather than use unlabeled Cot-1 DNA to suppress cross-hybridization to unwanted repeats as is the norm, we labelled Cot-1 DNA as a probe for RNA FISH, to explore the extent of repeat-containing RNA in situ. Since repeat-rich intronic sequences are thought to degrade rapidly, we were struck by the striking abundance of repeat-rich RNA detected, restricted to the nucleus of all cell types examined. This repetitive “hnRNA” (heterogeneous nuclear RNA) was associated with euchromatin and absent over heterochromatin, making a useful assay to determine if a nuclear or chromosome region was transcriptionally active or not [35], and was widely adopted for this purpose. However, we long suspected the Cot-1 RNA signal was too abundant to comprise just intronic repeats in expressed genes, and may reflect something more fundamental to genome biology. Highly repetitive sequences comprise over half the human genome; apart from the tandem satellite repeats, there are simple sequence repeats (SSRs) and highly abundant SINEs and LINEs (short and long interspersed nuclear elements) and endogenous retroviruses (e.g. HERV-H) widely distributed through our genomes. Repeat sequences are rarely conserved, however we point out that lack of primary sequence conservation does not indicate lack of function (as is true for XIST RNA). Further, we emphasize that there is a highly non-random genomic organization of LINEs and SINEs in the genome [36–38] which is largely conserved, as is their unexplained abundance.
Given the prevalence of repeats in introns, the challenge was to discriminate whether Cot-1 RNA is a byproduct of genic transcription, or something more interesting. In our recent study [39], a series of findings support that “Cot-1 RNA” comprises a class of repeat-rich ecRNAs, and exhibits multiple characteristics similar to XIST, the precedent for a heterochromatin-associated chromosomal RNA (hcRNA). For example, Cot-1 ecRNA is strictly localized throughout the active interphase chromosome territory in cis (Figure 3C & F), and, importantly, does not “drift” into the rest of the nucleoplasm as do other RNAs, including pre/mRNAs and spliced introns. The Cot-1 ecRNA territory remains undiminished and tightly localized many (4–32) hours after transcriptional arrest, as well as after biochemical fractionation that removes >90% of DNA and nuclear proteins, or after mechanical disruption of nuclei. Yet, this tenacious binding can be released by expression of a mutant scaffold protein (SAF-A/hnRNP-U) that disrupts the nuclear scaffold. Like XIST hcRNA, Cot-1 ecRNAs are released from chromosomes in early prophase, as are a host of putative scaffold proteins. All of these finding suggest the repeat-rich Cot-1 ecRNA behaves as a stable structural component associated with the nuclear scaffold of active chromosomes.
The embedded nature of this repeat rich ecRNA in chromatin/nuclear structure required our testing and identification of the most effective extraction techniques prior to RNAseq analysis (in progress). However, in situ analysis using probes to specific repeats showed that the nucleoplasmic Cot-1 ecRNA signal is not due to satellite sequence RNAs, but contains Alu sequences (more prevalent in gene-rich regions) and showed unexpectedly abundant LINE-1 (L1) sequences as a substantial component. This L1 RNA has similar characteristics to Cot-1 ecRNA. Importantly, this abundant L1 RNA signal is from the numerous 5′ truncated L1s, distinct from the small number of full-length L1 elements capable of retrotransposition. Full-length L1s are inactivated early in development, although recent evidence indicates they can be activated in cancer [40,41], neurodegeneration (reviewed in: [42]) and during development in the mammalian brain ([43,44] reviewed in: [45,46]). However, this likely represents a separate phenomenon from the L1-rich Cot-1 ecRNA prevalent in all cells.
Repeat-rich RNA as a structural component which may promote more open euchromatin
The functional significance of Cot-1 ecRNAs is far from established, but evidence suggests the hypothesis that the presence of these ecRNAs on chromatin likely serves to promote a more open chromatin state (Figure 4A). This function does not arise through the simple act of transcribing Cot-1, since inhibition of ongoing RNAPII transcription (and all short-lived RNAs) for several hours had no visible impact on chromatin structure. A fundamental possibility suggested by this work is that the physical presence of RNA is required to promote open chromatin, since only the lack of stable RNAs (primarily Cot-1 RNA) in G1 daughter cells was associated with chromatin clumping and condensation. While transcriptional inhibition could impact other aspects of cell and nuclear function (e.g. suppressing production of key proteins, or induction of a stress response), this is not the case with RNAse experiments, which resulted in rapid chromatin collapse. This suggests that the physical presence of ecRNA with chromatin is needed to counter chromatin compaction [37].
FIGURE 4.
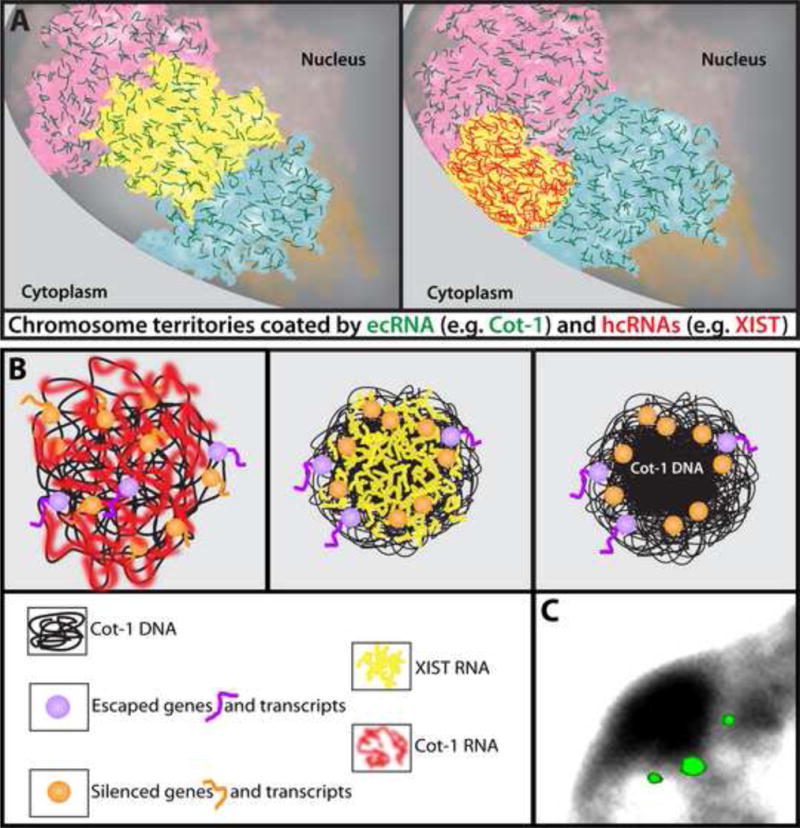
A) Model for the widespread presence of RNA bound throughout much of interphase chromatin, with two broad types of “chromosomal RNAs” (cRNAs): euchromatin-associated chromosomal RNAs (ecRNAs, green) present on active chromatin, such as Cot-1 ecRNAs, and heterochromatin-associated chromosomal RNAs (hcRNAs), such as XIST RNA (red). Left: All three chromosomes (pink, yellow and blue) are active and coated with ecRNA. Right: When XIST hcRNA coats the previously active yellow chromosome, ecRNAs are lost, and this potentially contributes to chromosome condensation. Whether there are other hcRNAs associated with other facultative heterochromatin at the nuclear periphery remains to be determined (note: Cot-1 ecRNAs are not found in silenced chromosomal regions up against the nuclear periphery). B) Left: Cot-1 ecRNA (red) is expressed from active chromosomes (e.g. Xa) which may help maintain an open chromatin state. Middle: When XIST RNA (yellow) first coats the X-chromosome, Cot-1 ecRNA is silenced, which may facilitate the compaction of Cot-1 DNA into the core of the chromosome territory forming the BB. Right: Genes position at the periphery of this condensed repeat-rich core to facilitate efficient silencing, while genes that escape silencing may be more peripheral. C) Simultaneous hybridization to three silenced genes on the inactive X-chromosome position just outside the condensed BB similar to the illustration at right above [12].
Relevant to this, it is important to note that when XIST RNA first coats and silences the X-chromosome, Cot-1 RNA is also lost across the bulk of the chromosome and Cot-1 DNA then condenses to form the Barr Body (Figure 4B) ([12], reviewed in [8]). Findings further suggest that XIST-mediated silencing of Cot-1 and genes may be spatially [12] and temporally [47] separable. While we believe such observations have profound implications for chromosome structure and the “repeat genome”, this needs to be looked at more closely for more genes, potentially specific “Cot-1” sequences, and preferably during human initiation of Xi (where condensation of DNA in the BB is more readily apparent). The model we envision is one in which many silenced genes position at the immediate outer periphery of a condensed core of repeat-rich DNA(Figure 4B–C), which we note could prove mechanistically similar to positioning of autosomal genes adjacent to heterochromatic mouse chromocenters for silencing [48].
Earlier studies had forwarded what was a controversial concept that a non-chromatin nuclear matrix or scaffold of insoluble proteins bound with RNA provides a structural underpinning of the nucleus necessary for transcription, splicing, and DNA replication [49–54], which is disrupted by RNAse treatment [55]. Many chromatin and scaffold proteins (e.g. SAF-A/hnRNPU, HP1 & CBX-7) are capable of binding both RNA and DNA, which make them a perfect “bridge”, or appear to require RNA in order to bind chromatin. We speculate that once this structural RNA is embedded in chromatin/scaffold proteins it becomes very stable, like Cot-1 and XIST RNA. The physical presence of this RNA may somehow provide the “water” to keep the chromatin/scaffold “sponge” from shrinking (condensing). In its absence, the chromatin/scaffold proteins can no longer interact with each other or with DNA, and the structure collapses. Hence ecRNAs may bridge canonical chromatin with non-chromatin elements of interphase chromosome structure and provide support to open up chromatin.
Chromosomal RNAs, Cot-1 RNA, and a Flood of Low-abundance non-coding nuclear RNAs
Transcriptome studies document a vast “dark matter” of nuclear RNA, including thousands of often very low abundance lncRNAs, enhancer RNAs (eRNAs), and intergenic transcripts, with recent efforts aiming to discriminate the chromatin bound fraction [56,57]. It was reported that chromatin bound RNAs are “tethered” via RNAPII [57], which would not be unexpected for RNAs at their transcription site. Hence the challenge still remains to discriminate whether such RNAs are with chromatin simply due to synthesis or they contribute to chromatin function.
Since repeats will be prevalent in this morass of RNA species ([58,59] reviewed in [60]), a collection of very low abundance nuclear ncRNAs, as well as pre-mRNAs, may contribute at some level to the robust Cot-1 RNA discussed above. However, pre-mRNAs and many lncRNAs and eRNAs are reported to be short lived following transcriptional inhibition [61–63], whereas Cot-1 ecRNA signal persists for long periods after transcription arrest with several different inhibitors [39]. Interestingly, intronic RNAs (which would be repeat rich) were found to accumulate in xenopus oocyte nuclei, and these were also shown to be exceptionally stable [64], whereas most introns of pre-mRNAs in mammalian nuclei are not.
Many of the repetitive sequences detected may also be part of longer intergenic transcripts. It’s been shown that truncated L1 elements contain non-canonical promoters which can drive expression of neighboring sequences [65] as can LTRs of endogenous retroviruses (ERV) [66] [67] [68], hence these could drive intergenic (repeat) transcription as well. A recent study found that osmotic stress in mammalian cells results in active genes continuing transcription into the downstream intergenic region (which we note would also be repeat rich) [69], and the authors suggest that this RNA may function to prevent chromatin collapse similar to the structural role suggested for Cot-1 ecRNAs [39]. However, what “promoter” (lnRNAs, eRNAs, genic or intergenic element, etc) produces Cot-1 ecRNA in non-stressed cells is still unknown.
As discussed above, recent evidence suggests that the presence of RNA on euchromatin following transcription arrest counters a tendency for chromatin to condense, yet the RNA sequence itself may not matter, or it might only need to match the DNA locus in cis. However, since comparison to a chromosome library of unique sequences suggested the majority of RNA sequences accumulated on an interphase chromosome are repetitive [39], and there is an unexplained non-random pattern of repeat sequences in chromosomal domains, the repeat sequences could prove key to regional chromosome regulation. While still speculative, we consider that the unusual physical properties of repeat sequences provide unique “architectural” potential; i.e. they can form structures of a non-canonical nature that we’ve barely begun to imagine, such as inter-strand connections to make an RNA or RNA-protein lattice [21,70]. For example, some simple SSRs form triplex or Z-DNA [71], and interspersed repeats can form hairpins, triplex DNA and single-strand or multi-strand G-quadruplexes (reviewed in: [72,73]). G-quadruplexes at the DNA level are reported to play roles in regulating chromatin structure and expression (reviewed in: [74]), and such sequences are predicted to be enriched in the 3′ UTR of L1 ([75], reviewed in:[76]), and thus in Cot-1 ecRNA, which might also form such structures. In addition, many repeat sequences retain protein binding and regulatory sequences from the parent element, hence repeat containing ecRNAs can still bind specific RNA binding proteins [77,78], in a manner which may also impact chromatin packaging.
Finally, we consider that the presence of ecRNA on chromatin may serve to promote a more open state by interacting with transcription factors, or potentially even chromatin remodelers. It is noteworthy that some transcription factors are reported to bind RNA as well as DNA [79] and two very recent studies [80,81] provide evidence that ncRNAs associated with gene transcription were found to retain a transcription factor (e.g. YY1) at the transcription site. This supports a model in which some transcription supports continued expression from that locus, in a manner which we suggest may not be through simple binding of a factor on that one gene, but potentially by supporting a more complex RNA-protein structure (aka: scaffold, matrix, lattice)(Kolpa et al, submitted), that influences the architecture of the immediate chromosomal region. An extension of this is that we propose ecRNA promotes an open chromatin state not only near canonical promoters, but in intergenic regions around active genes. The absence of most or all ecRNA from chromatin may be necessary to form condensed chromatin for mitosis or in heterochromatin domains. Hence, repeat-rich ecRNAs may be present throughout the “junk” DNA of the active X chromosome prior to XIST inducing heterochromatin formation and gene silencing (Figure 4B), which would explain the paradox of the Barr body, as discussed above.
Conclusion
RNA’s role in chromatin is a growing area of interest but still is largely restricted to particular RNAs at particular sites, thought of as specialized instances. We suggest, based on recent evidence as well as earlier studies, that RNA is prevalent throughout interphase chromatin, not just because it is made there, but because RNAs, even following transcription arrest, make a major structural contribution to the chromatin state. Hence cRNAs are not just involved in inducing heterochromatin (e.g. XIST hcRNA), but the presence of cRNA in euchromatin may be important to counter a natural tendency for chromatin and the nuclear scaffold to condense (e.g. Cot-1 ecRNA). Moreover, the vast diversity of transcripts with unexplained function are rich in various classes of repeat sequences. The possibility should be given more consideration that the repeat elements that comprise half our genome are not trivial bystanders, but may be a significant common theme with a biological role, with the potential to contribute to chromosome architecture in ways we have only begun to imagine. Finally, we want to acknowledge work dating back 50 years which reported that repeated sequences are prevalent and expressed in a development and cell type specific manner [82] [34] [33], and further suggested a role for repeats in genome regulation [83] [33].
Acknowledgments
Our research is supported by the NIH in grants to JBL; GM053234 and GM107604. We thank Meg Byron for excellent assistance in composing the figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 2.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 3.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132(3):259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386(6622):275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 5.Gendrel AV, Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol. 2014;30:561–580. doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- 6.Cerase A, Pintacuda G, Tattermusch A, Avner P. Xist localization and function: new insights from multiple levels. Genome Biol. 2015;16:166. doi: 10.1186/s13059-015-0733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sado T, Brockdorff N. Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110325. doi: 10.1098/rstb.2011.0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall LL, Lawrence JB. XIST RNA and architecture of the inactive X chromosome: implications for the repeat genome. Cold Spring Harb Symp Quant Biol. 2010;75:345–356. doi: 10.1101/sqb.2010.75.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bala Tannan N, Brahmachary M, Garg P, Borel C, Alnefaie R, Watson CT, Thomas NS, Sharp AJ. DNA methylation profiling in X;autosome translocations supports a role for L1 repeats in the spread of X chromosome inactivation. Hum Mol Genet. 2014;23(5):1224–1236. doi: 10.1093/hmg/ddt553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotton AM, Chen CY, Lam LL, Wasserman WW, Kobor MS, Brown CJ. Spread of X-chromosome inactivation into autosomal sequences: role for DNA elements, chromatin features and chromosomal domains. Hum Mol Genet. 2014;23(5):1211–1223. doi: 10.1093/hmg/ddt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, Pearl JR, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500(7462):296–300. doi: 10.1038/nature12394. This study has implications for chromosomal trisomy in Down Syndrome that are unrelated to the focus of this review, however it also makes a fundamental point for chromosome biology. It provides the most comprehensive analysis to date of the extent to which XIST RNA can silence an autosome. Importantly, this is examined in under conditions that do not select against autosomal inactivation. By targeting a large XIST gene into one Chromosome 21 in trisomy 21 stem cells, results using several methods show remarkably comprehensive silencing of the chromosome 21 by XIST RNA. This supports that most sequences in the genome are capable of being induced to form facultative heterochromatin by XIST RNA, supporting other evidence that XIST RNA may act via repeat elements common throughout all chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci U S A. 2006;103(20):7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teller K, Illner D, Thamm S, Casas-Delucchi CS, Versteeg R, Indemans M, Cremer T, Cremer M. A top-down analysis of Xa- and Xi-territories reveals differences of higher order structure at >/= 20 Mb genomic length scales. Nucleus. 2011;2(5):465–477. doi: 10.4161/nucl.2.5.17862. [DOI] [PubMed] [Google Scholar]
- 14.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 15.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–18. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohmdorfer G, Wierzbicki AT. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015;25(10):623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson C, Primorac D, McKinstry M, McNeil J, Rowe D, Lawrence JB. Tracking COL1A1 RNA in osteogenesis imperfecta. splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J Cell Biol. 2000;150(3):417–432. doi: 10.1083/jcb.150.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144(4):617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29(1):63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chujo T, Yamazaki T, Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbagrm.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Smith KP, Byron M, Clemson CM, Lawrence JB. Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: enrichment in gene rich bands. Chromosoma. 2004;113(6):324–335. doi: 10.1007/s00412-004-0325-1. [DOI] [PubMed] [Google Scholar]
- 24.Hall LL, Byron M, Pageau G, Lawrence JB. AURKB-mediated effects on chromatin regulate binding versus release of XIST RNA to the inactive chromosome. J Cell Biol. 2009;186(4):491–507. doi: 10.1083/jcb.200811143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21(2):198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Bergmann JH, Li J, Eckersley-Maslin MA, Rigo F, Freier SM, Spector DL. Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 2015;25(9):1336–1346. doi: 10.1101/gr.189027.114. Here the authors analyzed the lncRNA transcriptome of mouse ESCs and neural progenitors cells (NPCs) and identify a large number of uncharacterized lncRNAs. They further characterize 11 annotated lncRNAs by RNA FISH and by depleting the RNA. A significant focus is on Firre RNA, which had been suggested in other study to orchestrate interchromosomal contacts between several genes. The findings of this study contrast substantially with those previously reported. However, the authors suggest that FIRRE RNA may still have an interesting role in nuclear structure related to regulation of genes involved in RNA metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, Moore JM, Filippova GN, Xu J, Liu Y, Noble WS, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallot C, Huret C, Lesecque Y, Resch A, Oudrhiri N, Bennaceur-Griscelli A, Duret L, Rougeulle C. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat Genet. 2013;45(3):239–241. doi: 10.1038/ng.2530. [DOI] [PubMed] [Google Scholar]
- 29.Vallot C, Ouimette JF, Makhlouf M, Feraud O, Pontis J, Come J, Martinat C, Bennaceur-Griscelli A, Lalande M, Rougeulle C. Erosion of X Chromosome Inactivation in Human Pluripotent Cells Initiates with XACT Coating and Depends on a Specific Heterochromatin Landscape. Cell Stem Cell. 2015;16(5):533–546. doi: 10.1016/j.stem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 30*.Donley N, Smith L, Thayer MJ. ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet. 2015;11(1):e1004923. doi: 10.1371/journal.pgen.1004923. Although the function is currently unknown, this study provides the first example of a ncRNA that appears to paint an autosomal chromosome territory, reminiscent to XIST RNA, and with a similar long nuclear half-life. The authors imply that this ASAR15 RNA may be an autosomal ncRNAs that functions something like XIST RNA that may help coordinate non-imprinted monoallelic gene expression, as had been previously proposed by Andy Chess. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318(5853):1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 32.Zwemer LM, Zak A, Thompson BR, Kirby A, Daly MJ, Chess A, Gimelbrant AA. Autosomal monoallelic expression in the mouse. Genome Biol. 2012;13(2):R10. doi: 10.1186/gb-2012-13-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Davidson EH, Britten RJ. Regulation of gene expression: possible role of repetitive sequences. Science. 1979;204(4397):1052–1059. doi: 10.1126/science.451548. Using DNA from the most highly repetitive fraction of the genome (Cot-1) as a probe for in situ analysis, the authors found that highly stable repeat-rich RNA was the predominant RNA associated with an interphase chromosome territory, as contrasted with the non-repeat sequences accumulated on chromosomes. Several findings support that repeat-rich RNAs are not just a byproduct of gene transcription, but appear abundant and stably associated with active chromosomes at interphase. Preliminary evidence indicates that the presence of this and potentially other RNAs on euchromatin may be needed to maintain an open chromatin structure. [DOI] [PubMed] [Google Scholar]
- 34.Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- 35.Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci U S A. 2002;99(13):8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen TL, Manuelidis L. SINEs and LINEs cluster in distinct DNA fragments of Giemsa band size. Chromosoma. 1989;98(5):309–316. doi: 10.1007/BF00292382. [DOI] [PubMed] [Google Scholar]
- 37.Holmquist GP. Chromosome bands, their chromatin flavors, and their functional features. Am J Hum Genet. 1992;51(1):17–37. [PMC free article] [PubMed] [Google Scholar]
- 38.Korenberg JR, Rykowski MC. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988;53(3):391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- 39.Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, Fackelmayer FO, Lawrence JB. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156(5):907–919. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aschacher T, Wolf B, Enzmann F, Kienzl P, Messner B, Sampl S, Svoboda M, Mechtcheriakova D, Holzmann K, Bergmann M. LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene. 2015 doi: 10.1038/onc.2015.65. [DOI] [PubMed] [Google Scholar]
- 41.Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, Sinha S, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153(1):101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szafranski K, Abraham KJ, Mekhail K. Non-coding RNA in neural function, disease, and aging. Front Genet. 2015;6:87. doi: 10.3389/fgene.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 44.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468(7322):443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson SR, Morell S, Faulkner GJ. L1 retrotransposons and somatic mosaicism in the brain. Annu Rev Genet. 2014;48:1–27. doi: 10.1146/annurev-genet-120213-092412. [DOI] [PubMed] [Google Scholar]
- 46.Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15(8):497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20(16):2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91(6):845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 49.Berezney R. Fractionation of the nuclear matrix. I Partial separation into matrix protein fibrils and a residual ribonucleoprotein fraction. J Cell Biol. 1980;85(3):641–650. doi: 10.1083/jcb.85.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bynum JW, Volkin E. Chromatin-associated RNA: differential extraction and characterization. Biochim Biophys Acta. 1980;607(2):304–318. doi: 10.1016/0005-2787(80)90083-0. [DOI] [PubMed] [Google Scholar]
- 51.Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102(5):1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fey EG, Ornelles DA, Penman S. Association of RNA with the cytoskeleton and the nuclear matrix. J Cell Sci Suppl. 1986;5:99–119. doi: 10.1242/jcs.1986.supplement_5.6. [DOI] [PubMed] [Google Scholar]
- 53.Herman R, Weymouth L, Penman S. Heterogeneous nuclear RNA-protein fibers in chromatin-depleted nuclei. J Cell Biol. 1978;78(3):663–674. doi: 10.1083/jcb.78.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang RC, Bonner J. Histone-bound RNA, a component of native nucleohistone. Proc Natl Acad Sci U S A. 1965;54(3):960–967. doi: 10.1073/pnas.54.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nickerson JA, Krochmalnic G, Wan KM, Penman S. Chromatin architecture and nuclear RNA. Proc Natl Acad Sci U S A. 1989;86(1):177–181. doi: 10.1073/pnas.86.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caudron-Herger M, Cook PR, Rippe K, Papantonis A. Dissecting the nascent human transcriptome by analysing the RNA content of transcription factories. Nucleic Acids Res. 2015;43(14):e95. doi: 10.1093/nar/gkv390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Werner MS, Ruthenburg AJ. Nuclear Fractionation Reveals Thousands of Chromatin-Tethered Noncoding RNAs Adjacent to Active Genes. Cell Rep. 2015;12(7):1089–1098. doi: 10.1016/j.celrep.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13(11):R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su M, Han D, Boyd-Kirkup J, Yu X, Han JD. Evolution of Alu elements toward enhancers. Cell Rep. 2014;7(2):376–385. doi: 10.1016/j.celrep.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 60.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22(5):885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoon JH, Kim J, Gorospe M. Long noncoding RNA turnover. Biochimie. 2015;117:15–21. doi: 10.1016/j.biochi.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gardner EJ, Nizami ZF, Talbot CC, Jr, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26(22):2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41(5):563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 66.Sokol M, Jessen KM, Pedersen FS. Human endogenous retroviruses sustain complex and cooperative regulation of gene-containing loci and unannotated megabase-sized regions. Retrovirology. 2015;12:32. doi: 10.1186/s12977-015-0161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67*.Fort A, Yamada D, Hashimoto K, Koseki H, Carninci P. Nuclear transcriptome profiling of induced pluripotent stem cells and embryonic stem cells identify non-coding loci resistant to reprogramming. Cell Cycle. 2015;14(8):1148–1155. doi: 10.4161/15384101.2014.988031. Here the authors examined RNA sequences from nuclear and cytoplasmic RNA fractions from 11 different human and mouse stem cell lines before and after differentiation. They found most of the transcriptome is comprised of nuclear restricted, repeat-rich, non-annotated transcripts (NASTs). They are expressed at low levels from remnants of TEs and are fairly short (but are not made into small RNAs), and exhibit active promoter and enhancer histone marks. This may help be related to other findings regarding Cot-1 RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, Liu ET. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18(11):1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69*.Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA. Widespread Inducible Transcription Downstream of Human Genes. Mol Cell. 2015;59(3):449–461. doi: 10.1016/j.molcel.2015.06.016. The authors examined cytoplasmic, soluble nuclear, and chromatin RNA fractions, and report that many active genes produce 3′ intergenic transcripts that are strongly enriched in the chromatin fraction, and are continuous with the upstream gene for at least 5 kb. These transcripts show increased length and relative copy number under osmotic stress, and suggest these transcripts are a significant source of intergenic transcription in general and of osmotic stress-inducible transcription in particular. Transcription downstream of active genes may be related to Cot-1 RNA and the potential role of maintaining open chromatin state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21(22):4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McNeil JA, Smith KP, Hall LL, Lawrence JB. Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res. 2006;16(4):477–484. doi: 10.1101/gr.4627606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kejnovsky E, Tokan V, Lexa M. Transposable elements and G-quadruplexes. Chromosome Res. 2015 doi: 10.1007/s10577-015-9491-7. [DOI] [PubMed] [Google Scholar]
- 73*.Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43(18):8627–8637. doi: 10.1093/nar/gkv862. Here the authors searched for potential G-quadruplex forming sequences (PQSs) in human LINE-1, HERV, SVA and Alu elements, as these sequences have unique regulatory properties that may impact chromatin packaging. They found the most PQSs are in the 3′ UTR of L1 elements and SVA elements, and the LTR of ERVs, but none within Alu. The four main classes of elements studied carried almost half (49%) of total predicted PQSs in the human genome. While this study focuses on DNA, we suggest that these G-quadruplex forming sequences in the 850,000 L1 copies in the human genome may have functional implications in RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simone R, Fratta P, Neidle S, Parkinson GN, Isaacs AM. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015;589(14):1653–1668. doi: 10.1016/j.febslet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 75.Lexa M, Steflova P, Martinek T, Vorlickova M, Vyskot B, Kejnovsky E. Guanine quadruplexes are formed by specific regions of human transposable elements. BMC Genomics. 2014;15:1032. doi: 10.1186/1471-2164-15-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kejnovsky E, Lexa M. Quadruplex-forming DNA sequences spread by retrotransposons may serve as genome regulators. Mob Genet Elements. 2014;4(1):e28084. doi: 10.4161/mge.28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelley DR, Hendrickson DG, Tenen D, Rinn JL. Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol. 2014;15(12):537. doi: 10.1186/s13059-014-0537-5. A collection of 75 CLIP-Seq experiments on 51 RNA binding proteins in human cells have previously been used to examine RNA/protein interactions of unique sequences, but here the authors used the same data to examine the binding properties of repeat derived sequences that are usually tossed. They found widespread RBP interactions with TE-derived sequences and discovered TE-specific motifs that clustered on TE consensus sequences, and suggest a global role for TEs in shaping post-transcriptional RNA-protein regulatory networks in the human genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24(12):1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30(19):4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160(4):607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81*.Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015 doi: 10.1126/science.aad3346. The authors found that some transcription factors (YY1) bind ncRNAs from promoter and enhancer regions of genes that contain binding sequences for these TFs, but some (Oct4) did not. YY1 binding to DNA is stabilized by binding to ncRNA, supporting a model in which ncRNA expression supports continued gene expression from the immediate vicinity through the retention of TFs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165(3891):349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 83.Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci U S A. 2010;107(46):19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]


