Brazil has experienced an unprecedented epidemic of Zika virus (ZIKV), with ~30,000 cases reported to date. ZIKV was first detected in Brazil in May 2015 and cases of microcephaly potentially associated with ZIKV infection were identified in November 2015. Using next generation sequencing we generated seven Brazilian ZIKV genomes, sampled from four selflimited cases, one blood donor, one fatal adult case, and one newborn with microcephaly and congenital malformations. Phylogenetic and molecular clock analyses show a single introduction of ZIKV into the Americas, estimated to have occurred between May-Dec 2013, more than 12 months prior to the detection of ZIKV in Brazil. The estimated date of origin coincides with an increase in air passengers to Brazil from ZIKV endemic areas, and with reported outbreaks in Pacific Islands. ZIKV genomes from Brazil are phylogenetically interspersed with those from other South American and Caribbean countries. Mapping mutations onto existing structural models revealed the context of viral amino acid changes present in the outbreak lineage; however no shared amino acid changes were found among the three currently available virus genomes from microcephaly cases. Municipality-level incidence data indicate that reports of suspected microcephaly in Brazil best correlate with ZIKV incidence around week 17 of pregnancy, although this correlation does not demonstrate causation. Our genetic description and analysis of ZIKV isolates in Brazil provide a baseline for future studies of the evolution and molecular epidemiology in the Americas of this emerging virus.
Zika virus (ZIKV) is a single stranded, positive-sense RNA virus with a 10.7 kb genome encoding a single polyprotein that is cleaved into three structural proteins (C, prM/M, E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (1). ZIKV is a member of the family Flaviviridae, genus Flavivirus, and is transmitted among humans by Aedes mosquito species such as A. aegypti, A. albopictus, and A. africanus. The virus was first isolated in 1947 from a sentinel rhesus monkey in the Zika forest in Uganda (2) and is classified by sequence analysis into two genotypes, African and Asian (3). In humans, ZIKV infection typically causes a mild and self-limiting illness known as Zika fever (4) accompanied by maculopapular rash, headache, conjunctivitis and myalgia. In April 2007, a large epidemic of Asian genotype ZIKV was reported in Yap Island and Guam, Micronesia (5, 6). Between 2013–2014 the Asian genotype caused epidemics reported in several Pacific Islands, including French Polynesia (7), New Caledonia (8), Cook Islands (9), Tahiti (10), and Easter Island (11).
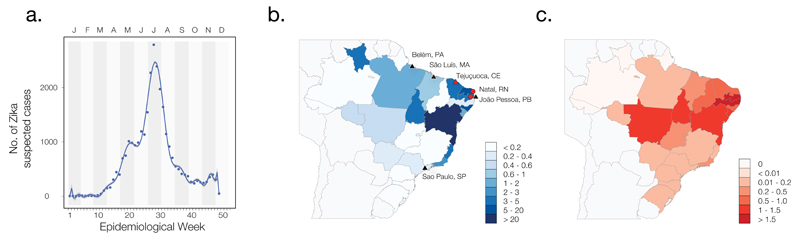
By May 2015, ZIKV was reported in Brazil (12) and subsequently in several countries of South and Central America, and the Caribbean. In Brazil nearly 30,000 cases of ZIKV infection had been notified by 30th Jan 2016 (supplementary materials section 1.4). Reported cases in Brazil indicate an epidemic peak in mid-July 2015 (Fig. 1A) and most Brazilian ZIKV cases (93%) were reported in Bahia state (Fig. 1B). ZIKV surveillance in Brazil began after the first reported Brazilian case and is conducted through the national Notifiable Diseases Information System (SINAN), which currently relies on passive case detection and reporting and therefore underestimates incidence (13). ZIKV is now widespread in Brazil, with autochthonous transmission and high incidence notified in 22 out of 27 administrative states (14). ZIKV infection during pregnancy has been hypothesized to cause microcephaly and congenital abnormalities (15–20). The detection of ZIKV in fetal brain tissue (17, 20) and amniotic fluid (21) supports the hypothesis that the virus is transmitted from mother-to-child (22) and the virus infects neural progenitor cells in vitro (23). In Brazil, between Nov 2015 and 30th Jan 2016, 4783 suspected cases of microcephaly were reported electronically to the RESP database (www.resp.saude.gov.br; Ministry of Health, Brazil; see supplementary materials section 1.4) (Fig. 1C), although most suspected cases are still under investigation and a substantial proportion may represent misdiagnosis and over-reporting (24). Using the WHO guidelines for microcephaly diagnosis provided on the 4th March 2016 (25), we identified a total of 1118 suspected microcephaly cases suitable for analysis. The relationship between total per capita ZIKV incidence (Fig. 1B) and per capita suspected microcephaly cases (Fig. 1C) in each state is weak and only significant under non-parametric correlation (p < 0.01) (fig. S1A); noise and uncertainty probably affect both variables. However, the relation is strengthened if suspected microcephaly cases are measured per pregnancy (fig. S1B). For municipalities with reported ZIKV incidence and cases of suspected microcephaly, we used a simple linear model to link microcephaly cases as a function of past ZIKV incidence (supplementary materials section 1.5). Suspected microcephaly cases are best predicted by ZIKV incidence during week 17 of pregnancy on average (95% confidence interval of mean = +/−0.11 weeks), or week 14 for suspected severe microcephaly cases (+/−0.08 weeks), in general agreement with individual reports of the timing of ZIKV symptoms in mothers of infants with microcephaly (16, 19, 21). We stress that these results quantify only the correlation between ZIKV and suspected microcephaly and does not demonstrate a causal link. Work is ongoing to establish whether or not ZIKV is a causal factor in microcephaly and other conditions (15–17, 23, 26).
Fig. 1. Time series and cartography of reported Zika virus and microcephaly cases in Brazil.
(A) Number of suspected cases of ZIKV per week in 5596 municipalities in Brazil. The epidemic peaked from 12 to 18 July 2015 (n = 2791 cases). Letters indicate months. (B) Total incidence of ZIKV cases per 100,000 people in each federal state. Triangles indicate sampling locations of the sequences reported here; circles indicate locations of other genomes from Brazil [municipality of Natal in Rio Grande do Norte state (16) and an unknown municipality in Paraiba state (21)]. Red symbols indicate ZIKV genomes isolated from microcephaly cases. Federal states are indicated by 2-letter codes: PA: Para, MA: Maranhão, CE: Ceará, RN: Rio Grande do Norte, PB: Paraíba. Per capita incidences in each state were calculated using high-resolution gridded human population size datasets for Brazil (45). (C) Incidence of suspected microcephaly cases per 100,000 people in each federal state. Per capita incidences for each state were calculated as described for panel (B).
We used phylogenetic, epidemiological, and mobility data to quantify ZIKV evolution and explore the introduction of the virus to the Americas. As part of ongoing surveillance by the Brazilian Ministry of Health, national laboratories, and other institutions, we used next generation sequencing to generate seven complete ZIKV coding region sequences from samples collected during the outbreak, including one from a deceased newborn with microcephaly and congenital malformations collected in Ceará and one from a fatal adult case with lupus and rheumatoid disease from Maranhão State (Fig. 1B). None of the Brazilian patients reported overseas travel (information unavailable in one case) and one subject was a blood donor (supplementary materials section 2). A comparison of our genomes with other available Brazilian strains reveals that Brazilian ZIKV isolates differ at multiple nucleotide sites across the 10.3kb coding region. The ZIKV genome recovered from isolate ZIKSP, from São Paulo, had 32 nucleotide changes compared to the microcephaly case (BeH823339) and 34 to the fatal case from Maranhão (BeH818305). Isolates BeH819966 from Belém, BeH815744 from Paraíba, and BeH18995, from Belém had a maximum of 5 nucleotide changes.
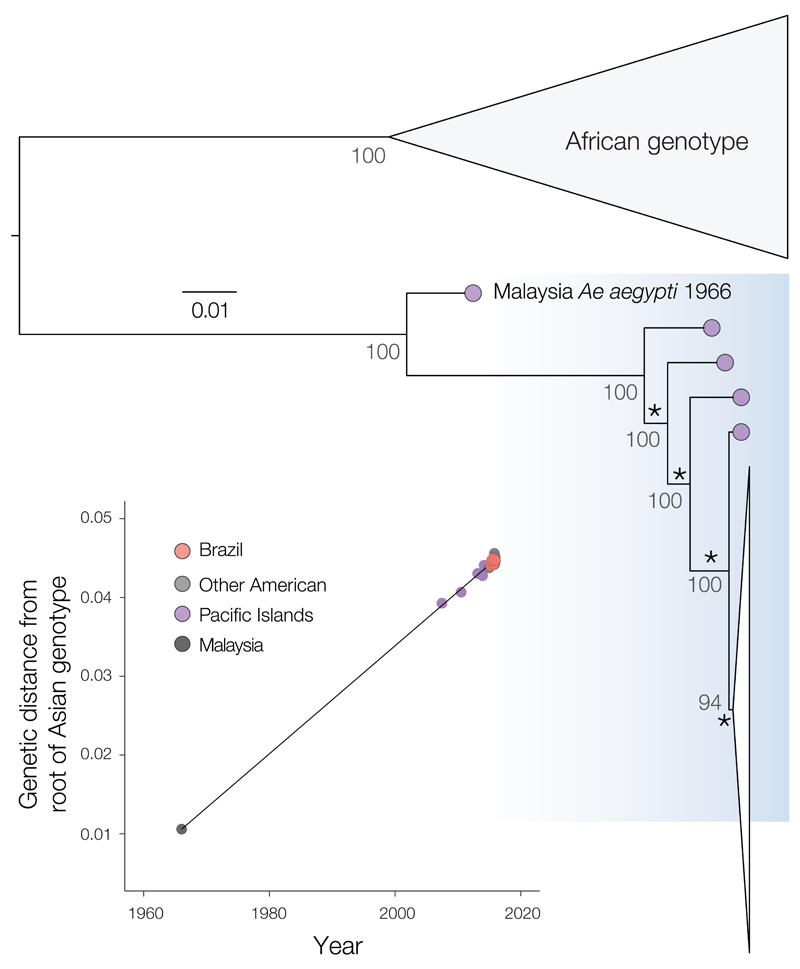
Maximum likelihood analysis of complete coding regions from our and other ZIKV genome sequences reveals that all viruses sampled in the Americas, including those from Brazil, form a robust monophyletic cluster (bootstrap score = 94%) within the Asian genotype (Fig. 2 and fig. S2) and share a common ancestor with the ZIKV strain that circulated in French Polynesia in November 2013 (Fig. 3). Previous analyses of outbreaks of related flaviviruses [e.g., (27, 28)] suggest that, to be informative, molecular epidemiological studies of the current ZIKV epidemic should use full or near-complete coding region sequences.
Fig. 2. Maximum likelihood phylogeny of ZIKV complete coding region sequences.
Bootstrap scores are shown next to well-supported nodes and the phylogeny was mid-point rooted. A fully annotated tree is provided in Fig. S2. The American ZIKV outbreak clade is drawn as a narrow white triangle and is shown in detail in Fig. 3. Asterisks highlight the four internal branches that are ancestral to the American ZIKV lineage (see main text and Fig. S3). Correlation between the sampling date of each sequence and the genetic distance of that sequence from the root of a maximum likelihood phylogeny of the Asian genotype (correlation coefficient R2 = 0.997). A molecular clock phylogeny of this data is shown in Fig. 3. The Malaysian strain (HQ234499) sampled in 1966 is the oldest representative of the Asian genotype and falls on the regression line, indicating that it does not appear to be unusually divergent for its age. A similar analysis with the HQ234499 strain excluded is shown in fig. S5C.
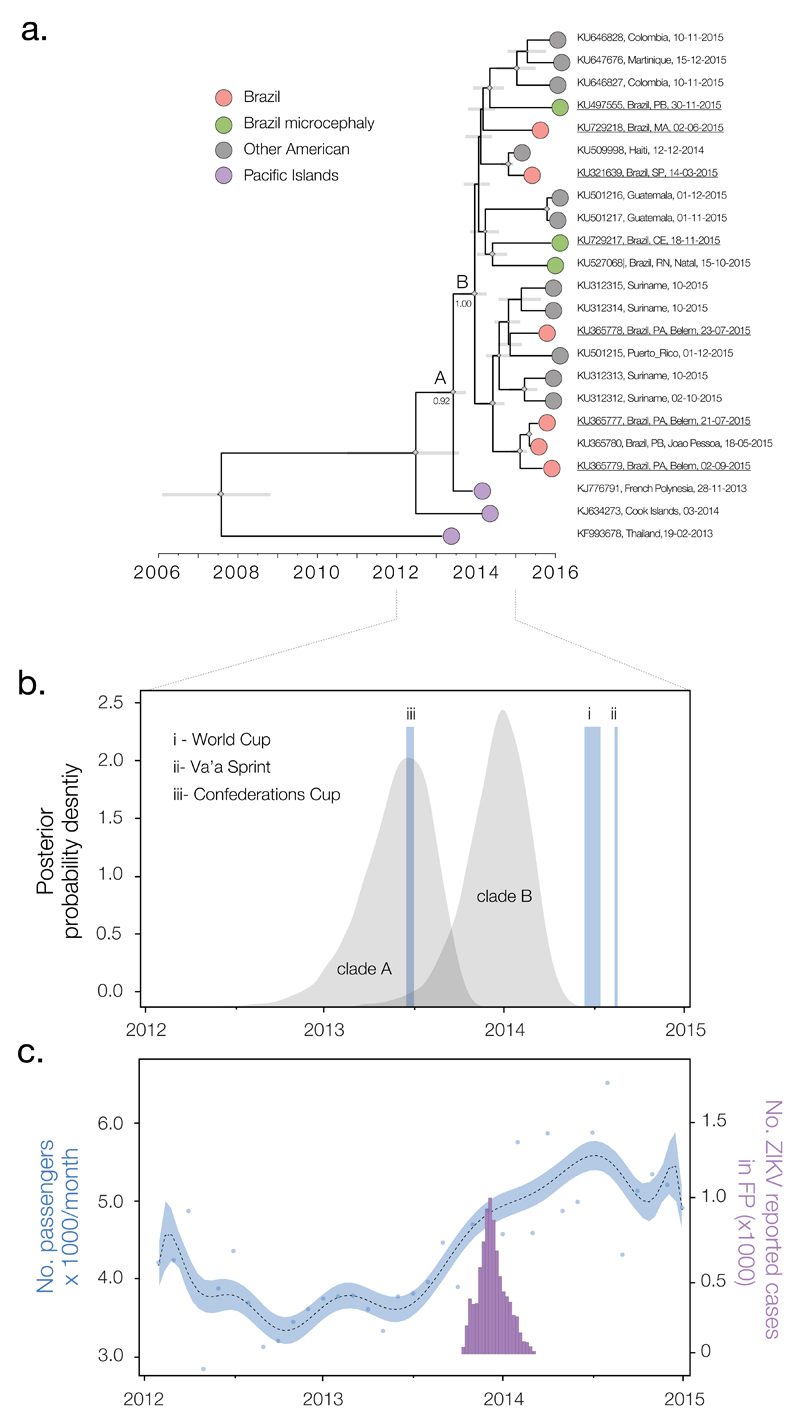
Fig. 3. Timescale of the introduction of ZIKV to the Americas.
(A) Molecular clock phylogeny of the ZIKV outbreak lineage estimated from complete coding region sequences, plus 6 sequences (KJ634273, KU312315, KU312314, KU212313, KU646828, and KU646827) longer than 1500nt (available data as of 7th March 2016). For visual clarity, three basal sequences, HQ23499 (Malaysia, 1966), EU545988 (Micronesia, 2007) and JN860885 (Cambodia, 2010) are not displayed here (see Fig. S3). Gray horizontal bars represent 95% Bayesian credible intervals for divergence dates. A and B denote clades discussed in main text and numbers next to them denote posterior probabilities. Diamond sizes represent, at each node, the posterior probability support of that node. Taxa are labeled with accession number, sampling location, and sampling date. Names of sequences generated in this study are underlined. (B) Posterior distributions of the estimated ages (TMRCAs) of clades A and B, estimated in BEAST software using the best-fitting evolutionary model (table S2). The time and duration of the three events (i-iii) discussed in the main text are shown. (C) Number of airline passengers from specific countries arriving in Brazil per month versus number of suspected cases of ZIKV in French Polynesia. The blue curve (left y axis) shows a polynomial fitting of the number of travelers (blue points) from countries with recorded ZIKV outbreaks between 2012 and 2015 (French Polynesia, Thailand, Indonesia, Malaysia, Cambodia, and New Caledonia) (supplementary materials section 6), aggregated across 20 Brazilian national airports. The purple bars represent weekly numbers of suspected ZIKV cases (right y axis) in French Polynesia (FP) from 30 October 2013 to 14 February 2014 (4).
We used a phylogenetic molecular clock approach to further explore the molecular epidemiology of ZIKV in the Americas. A strong correlation between genetic divergence and sampling time within the outbreak lineage (Fig. 2, inset) shows this approach is appropriate provided that whole genomes are used. The estimated time-scaled phylogeny (Fig. 3A) again contains a well-supported clade of American ZIKV strains (denoted B; posterior probability, PP = 1.00) that share a common ancestor (denoted A) with the French Polynesia lineage (PP = 0.92). Within the American ZIKV lineage (clade B), Brazilian isolates are interspersed among isolates from elsewhere in the Americas. The mingling of ZIKV genomes from different countries reveals ZIKV movement within the Americas since its introduction to the continent. Two observations suggest that the common ancestor of the American ZIKV lineage existed in Brazil. First, Brazil was the first country in the Americas to detect ZIKV (29) and second, Brazilian strains are phylogenetically more diverse within clade B than those from elsewhere. However, these observations may reflect differences in surveillance intensity among countries and more data are required before we can exclude the scenario that ZIKV was introduced to Brazil multiple times from other locations. Although two of three ZIKV-associated microcephaly isolates group together in the phylogeny, there is no reason to posit that this lineage is associated with increased disease severity.
Estimated rates of ZIKV molecular evolution are consistent among different evolutionary models and vary from 0.98 × 10−3 to 1.06 × 10−3 nucleotide substitutions per site per year (table S3). Although this rate is high compared to whole genome rates for other flaviviruses [e.g., (28)], it is consistent with retrospective analyses of previous epidemics, which show that evolutionary rate estimates decline as the epidemic progresses (30, 31). Hence, this result should not be interpreted as implying that ZIKV in the Americas is unusually mutable. We estimate that the date of the most recent common ancestor (TMRCA) of all Brazilian genomes (clade B) is Aug 2013 to Apr 2014 (95% Bayesian credible intervals, BCIs; point estimate = mid Dec 2013; Fig. 3B). The common ancestor of the French Polynesian and America lineages (clade A) was dated to Dec 2012 to Sep 2013 (BCIs; point estimate = late May 2013; Fig. 3B). The posterior distribution for the age of clade B encompasses the recorded duration of the ZIKV outbreak in 3 of 5 island groups of French Polynesia (4) (Fig. 3C). Divergence date estimates are robust among different combinations of prior distributions, molecular clock models, and coalescent models (supplementary materials sections 4 and 5), and are more likely to shift into the past than toward the present as virus genomes accumulate through time (30).
To explore possible routes of entry of ZIKV in Brazil, we collated airline flight data from all countries with reported ZIKV outbreaks between 2012 and end of 2014. From late 2012 we find an increase in the number of travellers arriving in Brazil from these countries, rising from 3775 passengers per month in early 2013 to 5754 passengers per month a year later (Fig. 3C). This increase in visitors to Brazil from ZIKV-affected countries coincides with the period during which ZIKV is estimated to have entered the Americas (i.e., between the TMRCAs of clades A and B) (Fig. 3B and supplementary materials section 5). If the ZIKV epidemic in Brazil did indeed arise from a single introduction then the virus must have circulated in the country for at least 12 months prior to the first case being reported in May 2015. ZIKV clinical symptoms may be confused with those caused by dengue and chikungunya viruses, two endemic and epidemic viruses that co-circulate and share mosquito vectors with ZIKV in Brazil (27, 32, 33). Reliable differential diagnosis is possible only by using improved surveillance and laboratory diagnostics, which are now being implemented throughout the country.
There are two published hypotheses for how ZIKV came to be introduced into Brazil, during (i) the 2014 World Cup soccer tournament (Jun 12th - Jul 13th) (29) or (ii) the Va’a canoe event held in Rio de Janeiro between 12-17 Aug 2014 (34). Alternatively, introduction could have occurred during (iii) the 2013 Confederations Cup soccer tournament (15th–30th Jun 2013). Events (ii) and (iii) notably included competitors from French Polynesia. Our results suggest that the introduction of ZIKV to the Americas predated events (i) and (ii). Although the molecular clock dates are more consistent with the Confederations cup, that event ended before ZIKV cases were first reported in French Polynesia (4). Consequently, we believe that large-scale patterns in human mobility will provide more useful and testable hypotheses about viral introduction and emergence (33, 35, 36) than ad hoc hypotheses focused on specific events.
The ZIKV genome we obtained from a microcephaly case in Ceará Brazil contains eight amino acid changes not observed in any other complete genome in our dataset. However, none of these mutations are shared with either of two recently published genomes from microcephaly cases (16, 21). Thus, if a causal link between Asian lineage ZIKV and microcephaly is confirmed, it is possible that putative viral genetic determinants of disease will be found among the amino acid changes that occur on the ZIKV phylogeny branches ancestral to the French Polynesian and American ZIKV lineages (i.e., the two lineages associated with reports of microcephaly, Guillain-Barré syndrome and congenital abnormalities) (37). Phylogenetic character mapping using parsimony reveals 11 amino acid changes on the four internal branches (labeled with asterisks in Fig. 2; fig. S3) leading to these two lineages. We identified the structures of homologous proteins most closely related to ZIKV proteins (supplementary materials section 7) and used them to map 7 of the 11 amino acid changes in a structural context, to five proteins: the pr-peptide region of prM [changes Val123→Ala123 (V123A) and S139N (S, Ser; N, Asn)], NS1 (A982V), the RNA helicase [NS3; N1902H and Y2086H (H, His; Y, Tyr)], the FtsJ-like methyl transferase domain [NS5; M2634V (M, Met)], and the thumb domain of RNA-directed RNA polymerase (NS5; M3392V) (fig. S7). None of these mutations are predicted to substantially affect the physicochemical properties of the protein environment, except possibly Y2086H (in the helicase; Fig. S8), which may increase the hydrophilicity of the region. The remaining four amino acid changes could not be accurately mapped due to the absence of suitable related X-ray structures (supplementary materials section 7). Notably, none of the observed changes map to the E glycoprotein ectodomain, the primary target of humoral immune responses against flaviviruses (38, 39). Factors other than viral genetic differences may be important for the proposed pathogenesis of ZIKV; hypothesized factors include co-infection with chikungunya virus (40), previous infection with dengue virus (41), or differences in human genetic predisposition to disease.
Besides vector-borne and mother-to-child transmission, Zika virus may also spread via sexual contact (42, 43) and blood transfusion (44). The evidence of ZIKV in blood donors raises the possibility of ZIKV transmission through transfusion and indicates that it may be prudent to consider the screening of blood donors.
Supplementary Material
Acknowledgments
We thank Xavier de Lamballerie and John Lednicky for permission to include their unpublished ZIKV genomes in our analysis. We thank the Death Verification Service (SVO), Central Laboratories of Public Health (LACEN) and health departments of the Ceará State and Maranhão State, Brazil for collaboration. OGP is supported by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 614725-PATHPHYLODYN. JL is supported by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 268904-DIVERSITY. OGP received consulting fees from Metabiota Inc. between 2015-2016. This study is made possible in part by the generous support of the American people through the United States Agency for International Development (USAID) Emerging Pandemic Threats Program. The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the United States Government. SIH is funded by a Senior Research Fellowship from the Wellcome Trust (#095066), and grants from the Bill and Melinda Gates Foundation (OPP1119467, OPP1093011, OPP1106023, and OPP1132415). MRTN is funded as an associated Researcher in Public Health by the Evandro Chagas Institute, Brazilian Ministry of Health and as Researcher in Scientific productivity by CNPq (Brazilian National Council for Scientific and Technological Development) grant numbers 302032/2011-8, 200024/2015-9, and supported in part by the National Institute of Science and Technology for Viral Hemorrhagic Fevers. R.T. is funded by grant R24 AT 120942 from the U.S. National Institutes of Health. S.C.H. is supported by a Wellcome Trust grant (102427). T.A.B. and I.R. are supported by grants from the UK Medical Research Council (MR/L009528/1) and Wellcome Trust (090532/Z/09/Z). PFCV is supported by CNPq-National Agency for Scientific and Technologic Development (grants 573739/2008–0, 301641/2010-2, and 457664/2013-4). All samples were obtained from persons visiting local clinics or hospitalized by the Brazilian Ministry of Health personnel as part of dengue, chikungunya, and Zika fever surveillance activities. In these cases, patient consent is oral and not recorded. The study was authorized by the Coordination of the National Program for Dengue, Chikungunya, and Zika Control coordinated by Brazil’s Ministry of Health. The data are available at DRYAD: DOI: doi:10.5061/dryad.6kn23. The new ZIKV genomes reported in this study are deposited in GenBank under the accession numbers KU321639, KU365777 to KU365780, KU729217, and KU729218.
References and Notes
- 1.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/S0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 2.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 3.Faye O, Faye O, Diallo D, Diallo M, Weidmann M, Sall AA. Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol J. 2013;10:311. doi: 10.1186/1743-422X-10-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ioos S, Mallet HP, Leparc Goffart I, Gauthier V, Cardoso T, Herida M. Current Zika virus epidemiology and recent epidemics. Med Mal Infect. 2014;44:302–307. doi: 10.1016/j.medmal.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 6.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLOS Negl Trop Dis. 2012;6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao-Lormeau VM, Roche C, Teissier A, Robin E, Berry AL, Mallet HP, Sall AA, Musso D. Zika virus, French polynesia, South Pacific, 2013. Emerg Infect Dis. 2014;20:1085–1086. doi: 10.3201/eid2006.140138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupont-Rouzeyrol M, O’Connor O, Calvez E, Daurès M, John M, Grangeon JP, Gourinat AC. Co-infection with Zika and dengue viruses in 2 patients, New Caledonia, 2014. Emerg Infect Dis. 2015;21:381–382. doi: 10.3201/eid2102.141553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyke AT, Daly MT, Cameron JN, Moore PR, Taylor CT, Hewitson GR, Humphreys JL, Gair R. Imported Zika virus infection from the Cook Islands into Australia, 2014. PLOS Curr. 2014 doi: 10.1371/currents.outbreaks.4635a54dbffba2156fb2fd76dc49f65e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wæhre T, Maagard A, Tappe D, Cadar D, Schmidt-Chanasit J. Zika virus infection after travel to Tahiti, December 2013. Emerg Infect Dis. 2014;20:1412–1414. doi: 10.3201/eid2008.140302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tognarelli J, Ulloa S, Villagra E, Lagos J, Aguayo C, Fasce R, Parra B, Mora J, Becerra N, Lagos N, Vera L, et al. A report on the outbreak of Zika virus on Easter Island, South Pacific, 2014. Arch Virol. 2016;161:665–668. doi: 10.1007/s00705-015-2695-5. [DOI] [PubMed] [Google Scholar]
- 12.Hennessey M, Fischer M, Staples JE. Zika virus spreads to new areas – region of the Americas, May 2015–January 2016. MMWR Morb Mortal Wkly Rep. 2016;65:55–58. doi: 10.15585/mmwr.mm6503e1. [DOI] [PubMed] [Google Scholar]
- 13.Silva MMO, Rodrigues MS, Paploski IAD, Kikuti M, Kasper AM, Cruz JS, Queiroz TL, Tavares AS, Santana PM, Araújo JMG, Ko AI, et al. Accuracy of dengue reporting by national surveillance system, Brazil. Emerg Infect Dis. 2016;22:336–339. doi: 10.3201/eid2202.150495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ministério da Saúde do Brasil. Boletim Epidemiológico 47:7: Semana epidemiológica (SE) 04 (30/01/2016) Secretaria de Vigilância em Saúde. 2016 (in Portuguese) available at http://portalsaude.saude.gov.br/index.php/situacao-epidemiologica-dados-dengue. [Google Scholar]
- 15.ECDC. Microcephaly in Brazil potentially linked to the Zika virus epidemic. ECDC. 2015 [Google Scholar]
- 16.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 17.Schuler-Faccini L, Ribeiro EM, Feitosa IM, Horovitz DD, Cavalcanti DP, Pessoa A, Doriqui MJ, Neri JI, Neto JM, Wanderley HY, Cernach M, et al. Brazilian Medical Genetics Society–Zika Embryopathy Task Force, Possible association between Zika virus infection and microcephaly - Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:59–62. doi: 10.15585/mmwr.mm6503e2. [DOI] [PubMed] [Google Scholar]
- 18.Ventura CV, Maia M, Ventura BV, Linden VV, Araújo EB, Ramos RC, Rocha MA, Carvalho MD, Belfort R, Jr, Ventura LO. Ophthalmological findings in infants with microcephaly and presumable intra-uterus Zika virus infection. Arq Bras Oftalmol. 2016;79:1–3. doi: 10.5935/0004-2749.20160002. [DOI] [PubMed] [Google Scholar]
- 19.Martines RB, Bhatnagar J, Keating MK, Silva-Flannery L, Muehlenbachs A, Gary J, Goldsmith C, Hale G, Ritter J, Rollin D, Shieh WJ, et al. Notes from the field: Evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses - Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:159–160. doi: 10.15585/mmwr.mm6506e1. [DOI] [PubMed] [Google Scholar]
- 20.PAHO. Implications for public health in the Americas. PAHO/WHO; 2015. Neurological syndrome, congenital malformations, and Zika virus infection. [Google Scholar]
- 21.Calvet G, Aguiar RS, Melo ASO, Sampaio SA, Filippis I, Fabri A, Araujo ESM, Sequeira PC, Mendonca MCL, Oliveira L, Tschoeke DA, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: A case study. Lancet Infect Dis. 2016 doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 22.Besnard M, Lastere S, Teissier A, Cao-Lormeau V, Musso D. Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February 2014. Euro Surveill. 2014;19:20751. doi: 10.2807/1560-7917.ES2014.19.13.20751. [DOI] [PubMed] [Google Scholar]
- 23.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, Christian KM, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Victora CG, Schuler-Faccini L, Matijasevich A, Ribeiro E, Pessoa A, Barros FC. Comment: Microcephaly in Brazil: How to interpret reported numbers. Lancet. 2016;387:621–624. doi: 10.1016/S0140-6736(16)00273-7. [DOI] [PubMed] [Google Scholar]
- 25.WHO Interim Report. Assessment of infants with microcephaly in the context of Zika virus. 2016 [Google Scholar]
- 26.Ventura CV, Maia M, Bravo-Filho V, Góis AL, Belfort R., Jr Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet. 2016;387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 27.Nunes MR, Faria NR, Vasconcelos HB, Medeiros DB, Silva de Lima CP, Carvalho VL, Pinto da Silva EV, Cardoso JF, Sousa EC, Jr, Nunes KN, Rodrigues SG, et al. Phylogeography of dengue virus serotype 4, Brazil, 2010–2011. Emerg Infect Dis. 2012;18:1858–1864. doi: 10.3201/eid1811.120217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pybus OG, Suchard MA, Lemey P, Bernardin FJ, Rambaut A, Crawford FW, Gray RR, Arinaminpathy N, Stramer SL, Busch MP, Delwart EL. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc Natl Acad Sci U S A. 2012;109:15066–15071. doi: 10.1073/pnas.1206598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanluca C, Melo VC, Mosimann AL, Santos GI, Santos CN, Luz K. First report of autochthonous transmission of Zika virus in Brazil. Mem Inst Oswaldo Cruz. 2015;110:569–572. doi: 10.1590/0074-02760150192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer AG, Spielman SJ, Bedford T, Wilke CO. Time dependence of evolutionary metrics during the 2009 pandemic influenza virus outbreak. Virus Evol. 2015;1:vev006. doi: 10.1093/ve/vev006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park DJ, Dudas G, Wohl S, Goba A, Whitmer SL, Andersen KG, Sealfon RS, Ladner JT, Kugelman JR, Matranga CB, Winnicki SM, et al. Ebola Virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell. 2015;161:1516–1526. doi: 10.1016/j.cell.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunes MR, Faria NR, de Vasconcelos JM, Golding N, Kraemer MU, de Oliveira LF, Azevedo RS, da Silva DE, da Silva EV, da Silva SP, Carvalho VL, et al. Vasconcelos, Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015;13:102. doi: 10.1186/s12916-015-0348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunes MR, Palacios G, Faria NR, Sousa EC, Jr, Pantoja JA, Rodrigues SG, Carvalho VL, Medeiros DB, Savji N, Baele G, et al. Air travel is associated with intracontinental spread of dengue virus serotypes 1–3 in Brazil. PLOS Negl Trop Dis. 2014;8:e2769. doi: 10.1371/journal.pntd.0002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musso D. Zika virus transmission from French Polynesia to Brazil. Emerg Infect Dis. 2015;21:1887. doi: 10.3201/eid2110.151125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, Russell CA, Smith DJ, Pybus OG, Brockmann D, Suchard MA. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLOS Pathog. 2014;10:e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pybus OG, Tatem AJ, Lemey P. Virus evolution and transmission in an ever more connected world. Proc Biol Sci. 2015;282:20142878. doi: 10.1098/rspb.2014.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao-Lormeau VM, Blake A, Mons S, Lastóre S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial AL, et al. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet. 2016 doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowd KA, Pierson TC. Antibody-mediated neutralization of flaviviruses: A reductionist view. Virology. 2011;411:306–315. doi: 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res. 2003;59:141–175. doi: 10.1016/S0065-3527(03)59005-4. [DOI] [PubMed] [Google Scholar]
- 40.Gèrardin P, Sampèriz S, Ramful D, Boumahni B, Bintner M, Alessandri JL, Carbonnier M, Tiran-Rajaoefera I, Beullier G, Boya I, Noormahomed T, et al. Neurocognitive outcome of children exposed to perinatal mother-to-child Chikungunya virus infection: The CHIMERE cohort study on Reunion Island. PLOS Negl Trop Dis. 2014;8:e2996. doi: 10.1371/journal.pntd.0002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagbami A, Halstead SB, Marchette N, Larsen K. Heterologous flavivirus infection-enhancing antibodies in sera of Nigerians. Am J Trop Med Hyg. 1988;38:205–207. doi: 10.4269/ajtmh.1988.38.205. [DOI] [PubMed] [Google Scholar]
- 42.Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, Tesh RB. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau VM. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Musso D, Nhan T, Robin E, Roche C, Bierlaire D, Zisou K, Shan Yan A, Cao-Lormeau VM, Broult J. Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Euro Surveill. 2014;19:20761. doi: 10.2807/1560-7917.ES2014.19.14.20761. [DOI] [PubMed] [Google Scholar]
- 45.Sorichetta A, Hornby GM, Stevens FR, Gaughan AE, Linard C, Tatem AJ. High-resolution gridded population datasets for Latin America and the Caribbean in 2010, 2015, and 2020. Sci Data. 2015;2:150045. doi: 10.1038/sdata.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zamree I, Drakes N, Rohani A, Lee HL. Sensitivity of Aedes albopictus C6/36 cells line for the detection and infectivity titration of dengue virus. Trop Biomed. 2005;22:217–219. [PubMed] [Google Scholar]
- 47.Lennette EH, Schmidt NJ. In: Diagnostic Procedures for Viral, Rickttsial and Chlamydial Infections. 5th. Lennette EH, Schmidt NJ, editors. American of Public Health Association; Washington: 1974. [Google Scholar]
- 48.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cunha MS, Esposito DL, Rocco IM, Maeda AY, Vasami FG, Nogueira JS, de Souza RP, Suzuki A, Addas-Carvalho M, Barjas-Castro ML, Resende MR, et al. First complete genome sequence of Zika virus (Flaviviridae, Flavivirus) from an autochthonous transmission in Brazil. Genome Announc. 2016;4:e00032–16. doi: 10.1128/genomeA.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.WHO. The WHO Child Growth Standards. Child growth standards. 2016 [Google Scholar]
- 54.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gouy M, Delmotte S. Remote access to ACNUC nucleotide and protein sequence databases at PBIL. Biochimie. 2008;90:555–562. doi: 10.1016/j.biochi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Katoh K, Standley DM. MAFFT: Iterative refinement and additional methods. Methods Mol Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- 57.Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 58.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rambaut A. 2014 available at tree.bio.ed.ac.uk/software/.
- 60.Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, Nasar F, Schuh AJ, Holmes EC, Higgs S, Maharaj PD, et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol. 2010;84:6497–6504. doi: 10.1128/JVI.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLOS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tavarè S. Some probabilistic and statistical problems in the analysis of DNA sequences. In: Waterman MS, editor. Some Mathematical Questions in Biology: DNA Sequence Analysis. American Mathematical Society; Providence, RI: 1986. pp. 57–86. [Google Scholar]
- 65.Ferreira MAR, Suchard MA. Bayesian analysis of elapsed times in continuous-time Markov chains. Can J Stat. 2008;36:355–368. doi: 10.1002/cjs.5550360302. [DOI] [Google Scholar]
- 66.Buathong R, Hermann L, Thaisomboonsuk B, Rutvisuttinunt W, Klungthong C, Chinnawirotpisan P, Manasatienkij W, Nisalak A, Fernandez S, Yoon IK, Akrasewi P, et al. Detection of Zika virus infection in Thailand, 2012–2014. Am J Trop Med Hyg. 2015;93:380–383. doi: 10.4269/ajtmh.15-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roth A, Mercier A, Lepers C, Hoy D, Duituturaga S, Benyon E, Guillaumot L, Souares Y. Concurrent outbreaks of dengue, chikungunya and Zika virus infections - an unprecedented epidemic wave of mosquito-borne viruses in the Pacific 2012–201. Euro Surveill. 2014;19:20929. doi: 10.2807/1560-7917.ES2014.19.41.20929. [DOI] [PubMed] [Google Scholar]
- 68.WHO. Pacific syndromic surveillance report, Week 21, ending 25 May, 2014. WHO Wester Pacific Region. 2014 [Google Scholar]
- 69.Baronti C, Piorkowski G, Charrel RN, Boubis L, Leparc-Goffart I, de Lamballerie X. Complete coding sequence of Zika virus from a French Polynesia outbreak in 2013. Genome Announc. 2014;2:e00500–14. doi: 10.1128/genomeA.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kutsuna S, Kato Y, Takasaki T, Moi M, Kotaki A, Uemura H, Matono T, Fujiya Y, Mawatari M, Takeshita N, Hayakawa K, et al. Two cases of Zika fever imported from French Polynesia to Japan, December 2013 to January 2014. Euro Surveill. 2014;19:20683. doi: 10.2807/1560-7917.ES2014.19.4.20683. [DOI] [PubMed] [Google Scholar]
- 71.Alera MT, Hermann L, Tac-An IA, Klungthong C, Rutvisuttinunt W, Manasatienkij W, Villa D, Thaisomboonsuk B, Velasco JM, Chinnawirotpisan P, Lago CB, et al. Zika virus infection, Philippines, 2012. Emerg Infect Dis. 2015;21:722–724. doi: 10.3201/eid2104.141707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwong JC, Druce JD, Leder K. Zika virus infection acquired during brief travel to Indonesia. Am J Trop Med Hyg. 2013;89:516–517. doi: 10.4269/ajtmh.13-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tappe D, Nachtigall S, Kapaun A, Schnitzler P, Günther S, Schmidt-Chanasit J. Acute Zika virus infection after travel to Malaysian Borneo, September 2014. Emerg Infect Dis. 2015;21:911–913. doi: 10.3201/eid2105.141960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.N.Z.P. Health. New Zealand Public Health Surveillance Report: September 2014. 2014;(3) [Google Scholar]
- 75.Fonseca K, Meatherall B, Zarra D, Drebot M, MacDonald J, Pabbaraju K, Wong S, Webster P, Lindsay R, Tellier R. First case of Zika virus infection in a returning Canadian traveler. Am J Trop Med Hyg. 2014;91:1035–1038. doi: 10.4269/ajtmh.14-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.