Abstract
T cell antigen receptor (TCR) signaling drives distinct responses depending upon the differentiation state and context of CD8+ T cells. We hypothesized that access of signal-dependent transcription factors (TFs) to enhancers is dynamically regulated to shape transcriptional responses to TCR signaling. We found that the TF BACH2 restrains terminal differentiation to enable generation of long-lived memory cells and protective immunity following viral infection. BACH2 was recruited to enhancers where it limited expression of TCR-driven genes by attenuating the availability of activator protein 1 (AP-1) sites to Jun family signal-dependent TFs. In naïve cells, this prevented TCR-driven induction of genes associated with terminal differentiation. Upon effector differentiation, reduced expression of BACH2 and its phosphorylation enabled unrestrained induction of TCR-driven effector programs.
Introduction
Following infection or immunization, naïve CD8+ T cells undergo burst-like clonal proliferation and differentiation to generate a population of effector cells reactive against pathogen-associated antigens. Following resolution of infection, the majority of responding cells are eliminated, allowing brisk restoration of immune homeostasis. A fraction of cells escape this fate and persist as memory cells1–6. The presence of greater numbers of antigen-specific memory cells enable more efficient pathogen clearance upon secondary infection. Thus, dynamic regulation of T cell differentiation, proliferation and survival is required to generate and then curtail effector responses while maintaining a subset of pathogen-specific memory cells following withdrawal of antigen.
T cell antigen receptor (TCR) signaling is critical to both initiation and diversification of CD8+ T cell responses. Strong or repeated TCR signaling drives progressive changes in gene expression that result in loss of lymphoid homing potential, acquisition of effector cell functions, and ultimately, terminal effector differentiation and apoptosis7, 8. Conversely, memory cells differentiate in response to weak antigen signals that are insufficient to drive full effector differentiation1, 5, 9. Consequently, memory cells manifest only a subset of transcriptional changes accompanying effector differentiation and their intermediate state of differentiation enables them to remain functionally quiescent, survive and circulate among secondary lymphoid tissues where they can be efficiently recruited into secondary responses10–12. TCR signaling not only plays a role in diversification of CD8+ T cell responses, but induces functionally distinct outcomes within diverse subpopulations of CD8+ T cells. While TCR stimulation of naïve cells predominantly results in proliferation and differentiation, stimulation of effector cells drives rapid induction of effector cytokines and cytotoxic molecules while stimulation of terminally differentiated effector cells induces apoptosis1, 8, 9.
AP-1 family TFs play a central role in transducing TCR-driven effector programs. AP-1 TFs, including Jun (c-Jun, JunD, JunB), Fos (c-Fos, Fosb, Fosl1, Fosl2) and BATF (BATF1, BATF2, BATF3) TFs, contain basic leucine-zipper (bZip) domains that enable them to form heterodimeric complexes at palindromic 12-O-Tetradecanoylphorbol-13-acetate (TPA) response elements (TRE; 5'-TGA(C/G)TCA-3')13, 14. Members of the Jun family of AP-1 TFs are phosphorylated in response to TCR signaling and are recruited to TRE within the enhancers of multiple genes involved in effector differentiation where they predominantly activate gene expression15–20. We hypothesized that modulation of the availability of AP-1 sites to Jun family TFs allows TCR-driven effector programs to be modulated in a stage-specific and contextual manner in CD8+ T cells, allowing for generation of transcriptionally intermediate memory cells.
BACH2 is a 92 kDa transcriptional repressor of the bZip TF family21. We have previously found that BACH2 promotes the differentiation of Foxp3+ regulatory T (Treg) cells and that this function is required under homeostatic conditions to prevent lethal inflammation22. In B cells, BACH2 is critical for somatic hypermutation and class-switch recombination, and its absence leads to impaired generation of class-switched antibody responses23, 24. BACH2, like AP-1 TFs, contains a bZip domain and binds to Maf recognition elements (MARE) which embed a TRE sequence21. Silencing of Bach2 mRNA following activation of CD8+ T cells in vitro results in reduced cellular persistence25. These observations led us to explore whether BACH2 regulates CD8+ T cell differentiation in vivo by controlling access of AP-1 family TFs to the regulatory elements of TCR-induced genes.
Results
BACH2 is required for CD8+ T cell memory
Defective generation of Foxp3+ Treg cells in Bach2–/– mice results in unrestrained effector differentiation among conventional T cells22. To evaluate the cell-intrinsic function of BACH2 in CD8+ T cells, we reconstituted C57BL/6 mice with 1:1 mixtures of congenically distinct CD45.1+ wild-type (WT) and Thy-1.1+ Bach2–/– mature lineage-depleted (Lin–) bone marrow (BM) cells (Supplementary Fig. 1a) and evaluated CD8+ T cells in these animals. We observed diminished frequencies of both effector (CD62L–) and central memory (CD62L+ CD44+) cells within the Bach2–/–compartment, suggesting a cell-intrinsic function of BACH2 in either the generation or maintenance of antigen-primed CD8+ T cell responses (Supplementary Fig. 1b-c). We therefore sought to test whether BACH2 controls responses to defined antigens. We utilized an acute viral infection model wherein recombinant vaccinia virus expressing chicken egg ovalbumin (VV-OVA) is recognized by minimal numbers of adoptively transferred TCR-transgenic CD8+ T cells specific for the OVA257-264 epitope (hereinafter, OT-I cells). First, mice were reconstituted with mixtures of congenically distinct WT CD45.1+ and Bach2–/– Thy-1.1+ OT-I transgenic BM and naïve CD44– CD62L+ OT-I cells of both genotypes were isolated from reconstituted animals. Naïve WT and Bach2–/– cells were co-transferred at a 1:1 ratio into recipient C57BL/6 mice (Fig. 1a and Supplementary Fig. 1d) prior to infection with VV-OVA. Bach2–/– CD8+ T cells exhibited impaired expansion and a near-complete failure to establish long-lived memory responses (Fig. 1b,c). The reduced ratio of KO:WT cells found in spleens of immunized animals was similar to that in the lungs and liver, but there was a further reduction in the frequency of KO cells in lymph nodes (Fig. 1d). Thus, BACH2 is required for maintenance of CD8+ T cell responses following primary infection in vivo.
Figure 1. BACH2 is required for maintenance of primary and secondary CD8+ T cell responses in vivo and establishment of protective immunity.
a, Pre-transfer flow cytometry of WT and KO naïve OT-I cells mixed at ~1:1 ratio. b-c, Kinetic analysis of cells in a following transfer into recipient mice and infection with VV-OVA. Representative flow cytometry (b) and replicate measurements (c) are shown. d, Ratio of WT and KO CD8+ T cells in indicated organs at day 21 following infection normalized to ratios found in the spleen. e, Bacterial titers following LM-OVA infection of animals which were previously administered individual transfers of naïve WT or KO OT-I cells and infected with VV-OVA. Primary infection with VV-OVA occurred 60 days prior to secondary infection with LM-OVA. f-g, Secondary recall responses mediated by WT and KO central memory (f) and effector memory (g) cells isolated at day 14 following primary infection as in c upon ~1:1 mixed transfer into infection-naïve recipients subsequently infected with VV-OVA. Numbers in gates indicate percentages. *P<0.05; **P<0.01; ***P<0.005; ****P<0.001. Data (a-g) are representative of two independent experiments with 4-6 mice per genotype and timepoint (c-g; mean and s.e.m.).
To test the function of BACH2 in mediating secondary protective immunity following primary infection, we infected mice with a recombinant strain of Listeria monocytogenes expressing the OVA257-264 epitope (LM-OVA) 60 days after individual transfer of normalized numbers of naïve WT or Bach2–/– OT-I cells and primary infection with VV-OVA. Consistent with decreased maintenance of CD8+ T cell responses in the absence of BACH2, mice that had initially received Bach2–/– OT-I cells exhibited significant impairment in their ability to clear LM-OVA compared with mice that had received WT cells (Fig. 1e). We also asked whether BACH2 is required for secondary recall responses. We co-transferred 1:1 ratios of WT and Bach2–/– central memory (CM; CD44+ CD62L+) or effector memory (EM; CD44+ CD62L-) cells isolated at day 14 following primary infection into infection-naïve recipients and infected these mice with VV-OVA. Notably, both Bach2–/– CM and EM cells exhibited defective secondary response kinetics, although the defect was more striking among CM than EM cells (Fig. 1f-g). Collectively, these findings indicate that BACH2 plays a non-redundant cell-autonomous role in maintenance of primary and secondary CD8+ T cell responses in vivo and in generation of protective immunity following primary viral infection.
BACH2 restrains terminal differentiation of CD8+ T cells
Progressive acquisition of effector characteristics is accompanied by loss of lymphoid homing potential and reduction in cellular lifespan1, 7, 11. We asked whether defective maintenance and lymphoid homing of Bach2–/– CD8+ T cells responding to viral infection are accompanied by changes in their differentiation state. Among antigen-experienced CD8+ T cells, CD62L+ central memory cells, CD62L– KLRG1– effector cells and KLRG1+ terminally differentiated effector cells have been described1. We measured expression of CD62L and KLRG1 on adoptively transferred naïve WT and Bach2–/– OT-I CD8+ T cells following viral infection. We observed excessive induction of terminally differentiated KLRG1+ cells and a near-complete loss of CD62L+ Bach2–/– central memory cells as a ratio of surviving cells at late timepoints following infection (Fig. 2a-d). However, the decline in total CD8+ T cell numbers amongst Bach2–/– cells resulted in a decrease in the absolute number of terminally differentiated effector cells (Supplementary Fig. 2a) in addition to memory cells. Consistent with augmented effector differentiation of Bach2–/– cells, we noted higher expression of the effector cytokine interferon-γ (IFN-γ) upon brief restimulation ex vivo (Fig. 2e). Conversely, retroviral overexpression of BACH2 in OT-I CD8+ T cells prevented acquisition of terminally differentiated effector cell characteristics upon transfer into mice subsequently infected with VV-OVA, as indicated by increased expression of CD62L and lower expression of CD44 and KLRG1 (Fig. 2f). Additionally, cells transduced with BACH2 expressed reduced amounts of the effector cytokines IFN-γ and tumor necrosis factor (TNF) (Fig. 2g).
Figure 2. BACH2 prevents terminal differentiation and apoptosis of CD8+ T cells following viral infection.
a-d, Surface phenotype of cells before (a) or at indicated timepoints (b) after mixed transfer of naïve WT and KO OT-I CD8+ T cells into recipient mice and infection with VV-OVA. Representative flow cytometry (a-b) and replicate measurements of CD62L (c) and KLRG1 (d) on the surface of transferred cells are shown. Too few KO cells were detected at day 90 for phenotypic analysis. e, Intracellular cytokine staining of IFN-γ expression by WT and KO CD8+ T cells from spleens of animals 5 days following infection. f, Overexpression of BACH2 prevents terminal effector differentiation following secondary responses to infection in vivo. Expression of CD62L, CD44 and KLRG1 on the surface of CD8+ T cells 5 days following transfer of GFP+ (transduced) OT-I CD8+ T into animals subsequently infected with VV-OVA. g, Expression of IFN-γ and TNF by transferred cells following brief restimulation ex vivo. Numbers in gates indicate percentages. *P<0.05; **P<0.01; ***P<0.005; ****P<0.001. Data (a-g) are representative of two independent experiments with 4-6 mice per genotype and timepoint (c-g; graphs represent mean and s.e.m.).
Terminal differentiation is associated with loss of memory precursor potential amongst effector cells26. Accordingly, we noted a striking loss of CD127+ KLRG1– memory precursor effector cells (MPEC) and an increase in the frequency of CD127– KLRG1+ short-lived effector cells (SLEC) amongst Bach2–/– cells at day 7 following infection (Fig. 3a). We also noted a reduction in the frequency of CD127+ cells within the CD62L– KLRG1– effector population that predominates at day 7 (Supplementary Fig. 2b). We did not, however, observe differences in CD43 and CD27 expression (Supplementary Fig. 2c-d)27–29. Increased acquisition of effector cell characteristics was also observed following stimulation of Bach2–/– CD8+ T cells in vitro and this was antigen-dependent (Supplementary Fig. 3a), proliferation-independent (Supplementary Fig. 3b) and associated with elevated expression of the lymphocyte activation markers CD25 and CD69 (Supplementary Fig. 3c).
Figure 3. BACH2 suppresses differentiation of short-lived effector cells and apoptosis during acute CD8+ T cell responses to viral infection.
a, Expression of CD127 and KLRG1 on the surface of OT-I cells at day 7 following mixed transfer of naïve FACS-sorted WT and KO OT-I CD8+ T cells into recipient mice and infection with VV-OVA. b, BrdU incorporation by WT and Bach2 KO CD8+ T cells at the indicated timepoints following infection. Mice were injected intraperitoneally with BrdU 16 h preceding harvest at each timepoint. *P<0.05, **P<0.005. c, Annexin V and propidium iodide staining of cells at indicated timepoints following infection. Numbers in gates indicate percentages. *P<0.01, **P<0.001. Data are representative of two independent experiments with 4-6 mice per genotype and timepoint (a-c; graphs represent mean and s.e.m.).
Terminal differentiation of CD8+ T cells is accompanied by induction of apoptosis30, 31. We measured rates of proliferation and apoptosis amongst WT and Bach2–/– CD8+ T cells responding to viral infection. While BACH2 was dispensable for sustaining early proliferation (Fig. 3b), we observed substantially increased rates of apoptosis amongst Bach2–/– cells at days 5, 7 and 10 post-infection (Fig. 3c), coinciding with the marked attrition in their cell numbers. Defective survival was not merely attributable to induction of apoptosis at the naïve cell stage, since elevated levels of apoptosis are observed at successive rounds of cell division following stimulation of Bach2–/– naïve CD8+ T cells in vitro (Supplementary Fig. 3d). Terminal differentiation of CD8+ T cells is accompanied by depletion of anti-apoptotic Bcl-2 family members26, 32, 33. We observed decreased expression of the anti-apoptotic Bcl-2 family members Bcl-xL and Mcl1 within Bach2–/– cells activated in vitro (Supplementary Fig. 3e). Corresponding to widespread differences in their phenotype, RNA sequencing (RNA-Seq) of WT and Bach2–/– cells isolated at day 7 following acute infection revealed widespread differences in gene expression (Supplementary Fig. 4a and Supplementary Table 1) and enrichment of transcripts associated with effector differentiation (Supplementary Fig. 4b and Supplementary Table 2). Gene expression differences between WT and Bach2–/– cells persisted, though were substantially mitigated, when fractionated populations of CD62L– KLRG1– effector cells were specifically analyzed (Supplementary Fig. 4c and Supplementary Table 3). Thus, BACH2 prevents terminal effector differentiation and apoptosis, enabling generation of long-lived memory cells following viral infection.
BACH2 controls TCR-driven transcriptional programs
AP-1 TFs bind to genomic TRE and play central roles in transducing TCR-driven effector programs. We hypothesized that BACH2 restrains TCR-driven terminal effector differentiation by binding to TRE and attenuating the activity of AP-1 factors. We mapped genome-wide BACH2 binding-sites using chromatin immunoprecipitation coupled with massively parallel DNA sequencing (ChIP-Seq). To generate sufficient numbers of cells required for ChIP, we stimulated naïve CD8+ T cells isolated by flow cytometric sorting with plate-bound anti-CD3 and anti-CD28 antibodies in the presence of IL-2 for 2 days and harvested cells after 3 further days of culture (hereinafter d5 in vitro activated CD8+ T cells). We observed extensive binding of BACH2 at intergenic and intronic regions (Fig. 4a). Analysis of these sites enabled identification of a BACH2 consensus motif in CD8+ T cells (5'-TGA(C/G)TCAGC-3') which embedded the canonical AP-1 recognition motif (5'-TGA(C/G)TCA34) (Fig. 4b). Importantly, this consensus motif was remarkably similar to a BACH2 recognition motif previously resolved using PCR-assisted binding site selection in vitro21. Given that a majority of BACH2 binding sites were outside annotated promoter regions, we next asked whether BACH2 binds to enhancers. We measured the genome-wide distribution of the coactivator protein p300 and the histone modifications H3K27Ac and H3K4me1, which are characteristically located at enhancers35. We found that a majority of BACH2 binding sites colocalized with one or more of these markers (Fig. 4c). Consistent with similarity in BACH2 and AP-1 binding motifs, BACH2 binding sites were enriched at known genome-wide binding sites of the AP-1 family members JunB, c-Jun and JunD (Fig. 4d-f and Supplementary Tables 4-6)17.
Figure 4. Enrichment of BACH2 at AP-1 sites within CD8+ T cell genomes.
a, Relationship of genome-wide BACH2 binding sites in d5 in vitro activated CD8+ T cells to annotated genes. b, Consensus motif enriched within BACH2 binding sites in CD8+ T cells (left). AP-1 consensus motif (JASPAR; right). c, Genome-wide co-localization analysis of BACH2 with indicated enhancer associated post-transcriptional histone modifications or p300 within d5 in vitro activated CD8+ T cells. d-f, Co-localization of JunB (d), c-Jun (e) and JunD (f) at genome-wide BACH2 binding sites in CD8+ T cells. Histograms indicate normalized average tag density at indicated distances from BACH2 peak summits. Pie charts indicate proportions of Jun binding sites that directly overlap with BACH2 binding sites (red indicates proportion of Jun sites exhibiting co-localization with BACH2). Binomial tests were used to determine peak significance within ChIP-Seq data and a threshold of p<1 × 10-5 was used for peak calling.
Extensive colocalization of BACH2 at known AP-1 sites in CD8+ T cells led us to enquire whether BACH2 functions to limit TCR-driven gene expression in CD8+ T cells. A prediction of this hypothesis is that a majority of TCR-driven genes it represses will not be differentially expressed prior to stimulation of Bach2–/– compared with WT cells. To test this, we measured global changes in gene expression induced by brief (6h or 18h) ex vivo stimulation of naïve CD44– CD62L+ WT and Bach2–/– CD8+ T cells using RNA-Seq. We identified 192 TCR-driven genes that were upregulated in Bach2–/–compared with WT cells naïve cells at either 0, 6 or 18 h following stimulation (FC>2; p<0.05; Fig. 5a). Of these genes, a substantial majority (163 genes, 84%) lacked significant differences prior to stimulation at 0 h (Fig. 5b-c and Supplementary Table 7). However, as early as 6 hours following stimulation, we observed strikingly increased induction of genes encoding proteins associated with effector differentiation in Bach2–/– cells, including Ifng, Cd44, Ccl1, Ccl9, Icam1, Lif, Lta, Gadd55b, Ttc39c. A majority (72.3%) of excessively induced genes contained mapped BACH2 binding sites within 15 kb upstream and 3 kb downstream of their gene bodies, suggesting that BACH2 may directly repress their TCR-induced expression. The identified genes were highly enriched in transcripts we measured to be induced in terminally differentiated KLRG1+ effector cells responding to VV-OVA infection compared with naïve precursors (Fig. 5d, Supplementary Table 8).
Figure 5. BACH2 represses TCR-driven transcriptional programs in CD8+ T cells.
Naïve WT and KO CD8+ T cells isolated ex vivo by flow cytometric sorting were stimulated briefly with platebound anti-CD3 and anti-CD28. a, Pie chart showing genes upregulated in KO compared with WT cells at 0, 6 or 18h (p<0.05, log2 FC>1). Of these, 192 TCR-induced genes (upregulated by stimulation in WT or KO cells at 6 or 18 h) were identified. b, Hierarchical cluster analysis of TCR-induced genes upregulated in KO cells. FPKM values are normalized to row maxima. TCR-inducibility in WT and KO cells, and BACH2 binding are indicated to right of heatmap. c, Fraction of genes in b differentially expressed in KO compared with WT cells at 0, 6 and 18h (p<0.05, log2 FC>1). d, Enrichment of genes in b with transcriptional differences between terminally differentiated KLRG-1+ OT-I cells and naïve cells. Normalized enrichment score (NES) = 1.9; Q<0.0001. e, Alignments showing mRNA expression in WT and KO naïve CD8+ T cells (top) and mapped BACH2, JunD and H3K4me1 binding sites (bottom). f, Naïve WT and KO CD8+ T cells pretreated for 1 h with JNKi (12.5µM) or vehicle at 37°C were stimulated with platebound anti-CD3 and anti-CD28 and mRNA expression was measured relative to Actb. *P<0.01; **P<0.001. g, mRNA expression at indicated timepoints following anti-CD3 restimulation of GFP+ (transduced) cells infected with indicated retroviruses during primary stimulation. Average FPKM values (b) and analyses of two replicate measurements per condition (a, c-e) are shown. Data (f-g; mean and s.e.m.) are representative of two independent experiments.
BACH2-mediated repression of TCR-driven gene expression is exemplified at the Ifng and Gp49a loci (Fig. 5e), at which colocalization of BACH2 and AP-1 factors is observed at known36 or putative regulatory elements (Fig. 5e). To test whether excessive TCR-induced gene expression in Bach2–/– cells is driven by Jun family AP-1 factors, we utilized a well-characterized pharmacological inhibitor (JNKi) of Jun N-terminal kinase (Jnk)37, which activates Jun family TFs in response to TCR signaling38. Pretreatment with JNKi partially reversed excessive induction of Gp49a and Ifng mRNA following stimulation of Bach2–/– naïve cells (Fig. 5f). Moreover, BACH2-mediated repression of TCR-driven Gp49a and Ifng expression was dependent upon its basic leucine-zipper domain since suppression mRNA expression resulting from BACH2 overexpression was abrogated when the bZip domain was disrupted (Fig. 5g). Collectively, these results indicate that BACH2 restrains TCR-driven transcriptional programs associated with terminal effector differentiation in naïve CD8+ T cells.
BACH2 limits access of Jun family TFs to its binding sites
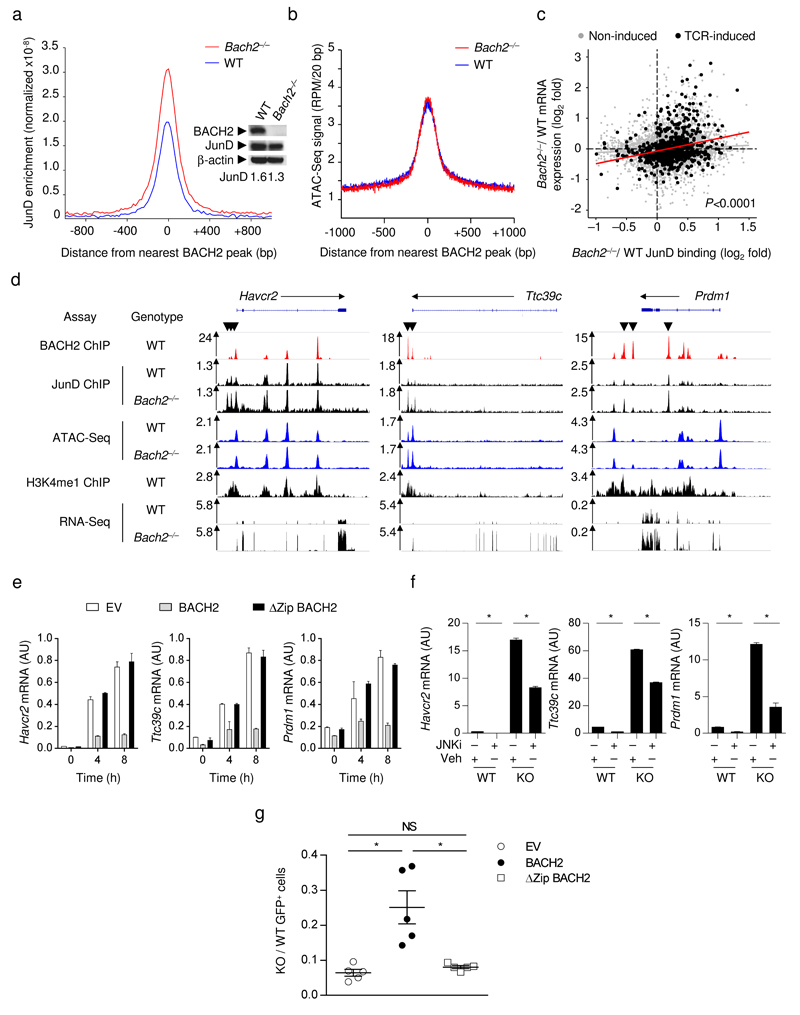
We hypothesized that BACH2 represses TCR-driven gene expression by competing with AP-1 factors for DNA binding. A prediction of this hypothesis is that loss of BACH2 will cause increased binding of Jun family TFs at its binding sites. BACH2 is expressed at readily detectable levels within WT d5 in vitro activated CD8+ T cells (Fig. 6a inset and Supplementary Fig. 5a). WT and Bach2–/– in vitro activated CD8+ T cells were briefly restimulated with platebound anti-CD3 for 2h and subjected to either ChIP-Seq analysis of JunD binding or RNA-Seq analysis. Strikingly, loss of BACH2 resulted in increased global binding of JunD at BACH2 binding sites (Fig. 6a) despite similar levels of total JunD in these cells (Fig. 6a, inset). Increased JunD enrichment was restricted to shared JunD-BACH2 sites since average JunD binding was not significantly increased at JunD sites where BACH2 does not bind (Supplementary Fig. 5b). Moreover, a proportion of JunD binding sites identified in Bach2–/– cells were not detected in WT cells, indicating that BACH2 prevents JunD binding with extremely high efficiency at a subset of loci (Supplementary Fig. 5c-d and Supplementary Table 9). In contrast to increased JunD binding, we observed little difference in chromatin acessibility at BACH2 binding sites in WT and Bach2–/– d5 in vitro activated CD8+ T cells (Fig. 6b). Similarly, we found no correlation between the differences in JunD binding observed at BACH2 binding sites and levels of chromatin accessibility in WT and Bach2–/– cells (Supplementary Fig. 6a). These findings indicate that within in vitro activated CD8+ T cells, BACH2 inhibits JunD binding without affecting chromatin accessibility. This behaviour is consistent with a function of BACH2 as a passive transcriptional repressor39.
Figure 6. BACH2 restricts access of Jun family TFs to regulatory elements of TCR-induced genes.
a, Histogram of JunD enrichment centered around genome-wide BACH2 binding sites in WT and Bach2–/– d5 in vitro activated CD8+ T cells. Western blot shows abundance of indicated total proteins in these cells. b, Average ATAC-Seq signal around BACH2 binding sites in WT and Bach2–/– d5 in vitro activated CD8+ T cells. c, Scatterplot comparing differences in average JunD binding at BACH2 binding sites (x-axis) with differences in mRNA expression of associated genes (y-axis). TCR-induced genes (induced by 2 h anti-CD3 stimulation of d5 in vitro activated CD8+ T cells) and non-induced genes are shown separately. Statistical significance was evaluated using two-sample Kolmogorov-Smirnov test. d, Representative alignments of ChIP-, ATAC- and RNA-Seq measurements in d5 in vitro activated CD8+ T cells at selected loci identified in c. Arrows indicate increased JunD binding at BACH2 binding sites in Bach2–/– cells. e, mRNA expression at specific timepoints following restimulation of transduced (GFP+) d5 in vitro activated CD8+ T cells infected with indicated retroviruses during primary stimulation. f, mRNA expression in WT and KO CD8+ T stimulated in vitro for 5 days in the presence of JNKi (12.5µM) or vehicle. *P<0.001. g, Congenically distinct GFP+ (transduced) WT and KO OT-I cells infected with indicated retroviruses during primary stimulation were co-transferred at ~1:1 ratio into recipient mice. KO/WT ratios within spleens of recipient animals were measured 5 days after infection with VV-OVA. *P<0.01. Data and analyses are based upon one (a), four (b) and two (c) measurements or represent two repeated experiments (e-g; mean and s.e.m.).
We next asked whether differences in JunD binding were associated with differences in gene expression. We compared average differences in JunD binding at BACH2 binding sites with differences in expression of associated genes in WT and Bach2–/– d5 in vitro activated CD8+ T cells. Increased JunD binding in Bach2–/– cells was positively correlated with increased expression of associated genes, especially amongst TCR-induced genes (Fig. 6c and Supplementary Table 10), as exemplified at selected loci identified in the analysis (Figure 6d). We confirmed increased JunD binding by qPCR-based ChIP quantification at selected loci and extended our analysis by measuring occupancy of JunB and c-Jun, in addition to JunD, at these sites (Supplementary Fig. 6b). Increased JunD binding in Bach2–/– cells was observed at all loci examined while increased c-Jun binding was observed at a majority of loci and increased JunB binding was not observed. Loss of BACH2 ChIP signals in Bach2–/– cells provides an important specificity control for the ChIP assay.
Since passive transcriptional repressors mediate their effect through competition with transcriptional activators, their activity is often dependent upon shared DNA-binding domains with the transcriptional activators they antagonize39. BACH2 and AP-1 TFs share possession of DNA-binding bZip domains. Consistent with a requisite role of the bZip domain in repression of TCR-induced genes, BACH2-mediated inhibition of TCR-induced gene expression was abrogated when its bZip domain was disrupted (Fig. 6e). Additionally, similar to our observations in naïve cells, excessive induction of TCR-driven genes in Bach2–/– d5 in vitro activated cells was partially reversed upon pharmacological inhibition of Jun activity using JNKi (Figure 6f). This led us to ask whether maintenance of CD8+ T cell responses to acute infection is BACH2 bZip-dependent. Retroviral overexpression of BACH2 partially rescued the secondary response of d5 in vitro activated Bach2–/– OT-I cells upon transfer and VV-OVA infection in vivo, while overexpression of ΔZip BACH2 failed to do so (Figure 6h). Collectively, these results indicate that BACH2 functions as a passive repressor of TCR-driven transcriptional programs, competing for access with AP-1 factors at a subset of its binding sites.
Stage-specific control of TCR-driven programs by BACH2
Given that effector differentiation proceeds in a subset of cells harboring intact Bach2 loci, we asked whether BACH2 expression is attenuated to permit effector differentiation in a subset of cells. Analyzing known genome-wide distributions of histone modifications in CD8+ T cells40, we noted decreased promoter-bound H3K4me3 at the BACH2 locus in memory and effector cells compared with naïve cells (Fig. 7a-b). Consistently, Bach2 mRNA expression was highest in naïve OT-I CD8+ T cells and was progressively downregulated in central memory (CD62L+ KLRG1–), effector (CD62L– KLRG1–) and terminally differentiated effector (CD62L– KLRG1+) cells 21 days following VV-OVA infection (Fig. 7c). We observed corresponding changes in expression of Bach2 mRNA and the encoded protein in bulk populations of CD8+ T cells at serial timepoints following infection, with early decreased expression at days 5 and 7, when effector cells predominate, followed by increased mRNA expression at day 30, when memory cells recrudesce (Supplementary Fig. 7). Thus, expression of Bach2 mRNA is dynamically regulated in CD8+ T cells at distinct stages of differentiation.
Figure 7. Dynamic regulation of BACH2 expression enables stage-specific control of TCR-driven gene expression.
a-b, Genomic alignments (a) and quantification (b) of H3K4me3 in naïve, memory and effector CD8+ T cells at the Bach2 gene (UCSC). c, mRNA expression in indicated subsets of OT-I cells sorted 30 days after infection with VV-OVA. *P<0.05, **P<0.01. d, mRNA expression normalized to Actb (left), and protein expression (right) in naïve and CD62L– KLRG1– effector OT-I cells isolated 7 days after infection with VV-OVA (d7 Eff). *P<0.001. e, Comparison of TCR-induced gene expression in KO and WT naïve and d7 Eff cells. Sorted naïve and d7 Eff cells were subjected to brief (6h) stimulation ex vivo with platebound anti-CD3 and anti-CD28. TCR-induced genes (induced by 6 h stimulation of either naïve WT (blue arc) or KO cells (red arc); p<0.05, log2 FC>1) significantly upregulated in KO compared with WT cells at either 0 or 6 h (p<0.05, log2 FC>1) were selected for further analysis. f, Comparison of expression of genes in e in Naïve and d7 Eff cells at 0 or 6 h after stimulation. Color scale of heatmap normalized to row maxima. g, Fraction of genes in f that are significantly differentially expressed between KO and WT cells at either 0 or 6h following stimulation (p<0.05, log2 FC>1). Data representative of two independent experiments (c-d; mean and s.e.m.), or average FPKM values (f) and analyses (e,g) of two replicate measurements per genotype and timepoint.
We asked whether modulation of Bach2 mRNA expression enables stage-specific control of its function. CD62L– KLRG1– cells isolated at day 7 post-infection (d7 Eff) express much lower levels of Bach2 mRNA and the encoded protein than naïve cells (Fig. 7d). Both WT and Bach2–/– d7 Eff cells are present in sufficient numbers for transcriptional analysis. We hypothesized that the effect of Bach2 deficiency on TCR-driven gene expression would be less substantial in d7 Eff cells than in naïve cells, corresponding to its lower expression in these cells. We compared TCR-induced gene expression in WT and Bach2–/– naïve and d7 Eff cells isolated ex vivo using RNA-Seq. We identified TCR-driven genes that were upregulated in Bach2–/– naïve cells compared with WT cells at either 0 or 6 h following stimulation (Fig. 7e). We then compared TCR-induced expression of these genes in WT and Bach2–/– naïve and d7 Eff cells (Fig 7f-g and Supplementary Table 11). Strikingly, a vast majority of genes differentially expressed in naïve cells were not significantly differentially expressed between WT and Bach2–/– cells in d7 Eff cells at either 0h or 6h following stimulation, coinciding with reduced BACH2 expression in this subset (Fig. 7f-g). Thus, dynamic regulation of BACH2 expression allows for stage-specific control of TCR-driven transcriptional programs.
Contextual control of BACH2 function by phosphorylation
Incomplete extinguishment of Bach2 mRNA in KLRG1+ terminally differentiated CD8+ T cells prompted us to explore whether BACH2 is post-translationally modified enabling further contextual control of its function. We noted decreased mobility of BACH2 upon denaturing gel electrophoresis of lysates from cells that were briefly restimulated with anti-CD3 which was reversed upon treatment with lambda phosphatase (Fig. 8a). This suggested that BACH2 is phosphorylated upon TCR stimulation. A highly conserved serine residue at position 520 (Ser520) of murine BACH2 corresponds to a previously reported phosphoresidue of human BACH2 in chronic myeloid leukemia cell lines (Fig. 8b)41. The residues surrounding Ser520 form a consensus motif of the serine/threonine kinase Akt. To test whether BACH2 Ser520 is phosphorylated by Akt following TCR stimulation, we transduced CD8+ T cells with retroviruses encoding WT BACH2 or a substitution mutant BACH2(S520A) following primary stimulation in vitro. Four days post-transduction, cells were restimulated by crosslinking of surface-bound anti-CD3 monoclonal antibodies. Using a previously described Akt-substrate motif-specific antibody which detects RXXp[S], we tested whether inhibition of Akt using a well-characterized and highly-specific allosteric Akt inhibitor (Akti)42 prevents acute phosphorylation of BACH2 Ser520. Indeed, pharmacological inhibition of Akt prevented acute phosphorylation of BACH2 whereas inhibition of downstream mTORC1 activation with rapamycin failed to inhibit this event (Fig. 8c). Moreover, S520A substitution prevented detectable phosphorylation. Thus, Ser520 phosphorylation of BACH2 following stimulation of CD8+ T cells is prevented by pharmacological inhibition of Akt.
Figure 8. Phosphorylation of BACH2 at Ser520 results in functional inactivation.
a, d5 in vitro activated CD8+ T cells were restimulated for 30 minutes with crosslinked anti-CD3 and cellular lysates were treated with or without lambda phosphatase (LP). BACH2 migration was resolved by immunostaining. b, Alignment of BACH2 amino acid sequences in indicated vertebrates. S520A substitution mutation is shown. c, CD8+ T cells were transduced with indicated retroviruses during primary stimulation and restimulated with crosslinked anti-CD3 4 days after transduction, preincubating with indicated pharmacological inhibitors for 1 h. α-Flag-immunoprecipitated proteins or total lysates were subjected to immunostaining. Phospho-BACH2 was detected using an RXXpS phospho-Akt substrate motif-specific antibody. d, Purified Akt1 was combined with ATP and immunoprecipitated Flag-tagged WT or BACH2(S520A). Phospho-BACH2 was detected using an RXXpS phospho-Akt substrate motif-specific antibody. e, CD8+ T cells were transduced with indicated retroviruses during primary stimulation in rhIL-2 and IL-12 prior to restimulation on anti-CD3-coated plates in the presence of Brefeldin A for intracellular cytokine staining. *P<0.05, **P<0.01, ***P<0.005. f-g, CD8+ T cells were transduced with indicated retroviruses and cultured as in e. Cells were briefly restimulated with platebound α-CD3 (2h) and stained for the retroviral transduction marker (Thy-1.1) during the last 30 mins of culture. Fixed cells were stained with DAPI and localization of BACH2-GFP was resolved by imaging flow cytometry. Representative cells (f) and the ratio of nuclear/cytoplasmic mean pixel intensity (MPI) in replicate cells (g) are shown. ***P<0.005. Data representative of two (e, g; mean and s.e.m.) and three (a, c-d) independently repeated experiments.
This observation led us to ask whether BACH2 is a direct substrate of Akt. We performed an in vitro kinase reaction, combining purified Akt1 and BACH2 in the presence of adenosine triphosphate (ATP) in vitro. Addition of purified Akt in the presence of ATP caused phosphorylation of BACH2 in a Ser520-dependent fashion (Fig. 8d). Thus, BACH2 Ser520 is a substrate of Akt kinase in vitro. Since Akt drives effector differentiation in CD8+ T cells43–45 and since effector differentiation is antagonized by its substrate BACH2, we hypothesized that phosphorylation of BACH2 promotes its functional inactivation, enabling effector differentiation to proceed. This was supported by the observation that Akt inhibition suppresses effector differentiation in a partially BACH2-dependent manner (Supplementary Fig. 8). To test whether Ser520 phosphorylation functionally inactivates BACH2, we overexpressed BACH2 or BACH2(S520A) in CD8+ T cells stimulated in vitro in the presence of IL-2 and IL-12. Despite gating on cells expressing similar amounts of the transduction marker Thy-1.1, we noted enhanced suppression of effector cytokines and cell size when BACH2(S520A) was overexpressed (Fig. 8e). Strongly activating in vitro stimulation conditions were used since overexpression of WT BACH2 caused near-complete suppression of effector cytokines following transfer in vivo (Fig. 3b) precluding assessment of a gain-of-function phenotype using this overexpression-based experimental system in vivo.
Phosphorylation of human BACH2 at a homologous residue to murine BACH2 Ser520 in chronic myeloid leukemia cells leads to its functional inactivation through nuclear exclusion41. To test whether phosphorylation of BACH2 causes its nuclear exclusion in T cells, CD8+ T cells were transduced with retroviral vectors expressing BACH2-GFP and BACH2(S520A)-GFP fusion proteins. The subcellular localization of GFP within transduced (Thy-1.1+) cells was resolved following brief restimulation using imaging flow cytometry. This allows for unbiased quantification of nuclear and cytoplasmic localization. WT BACH2 exhibited decreased nuclear localization compared with BACH2(S520A) (Fig. 8f-g) indicating that phosphorylation of BACH2 at Ser520 favors its nuclear exclusion in CD8+ T cells. Thus, Ser-520 phosphorylation provides a mechanism by which the function of BACH2 is modulated by extrinsic signals to enable contextual control of CD8+ T cell differentiation.
Discussion
In this study, we have found a cell-autonomous role for BACH2 in regulating CD8+ T cell differentiation and function following primary and secondary infection in vivo. BACH2 restrains terminal effector differentiation and apoptosis within a subset of cells responding to viral infection allowing for generation of long-lived memory cells that mediate secondary protection upon pathogen reencounter. BACH2 binds to AP-1-like sites within enhancers and modulates their availability to Jun family signal-dependent transcription factors. In less differentiated cells, this allows BACH2 to prevent TCR-driven induction of genes associated with terminal differentiation while in effector cells, reduction in Bach2 mRNA expression and phosphorylation of the encoded protein permit TCR driven induction of effector cell transcriptional programs.
The observation that BACH2 modulates binding of Jun factors without affecting chromatin accessibility in activated CD8+ T cells suggests BACH2 engages in passive transcriptional repression by competing with AP-1 factors for DNA binding. This is consistent with previous observations that BACH2 exerts chromatin-independent repression of TPA-induced luciferase reporter gene expression driven by non-chromatinized MARE sequences46. These results do not exclude that BACH2 may modulate chromatin structure in other cell types or in different CD8+ T cell subsets. The localized function of BACH2 in modulating access of AP-1 factors at enhancers is characteristic of short-range repression, whereby local regulation of distinct enhancers permits multiple enhancers to function autonomously within a promoter47, compatible with a requirement for complex regulation of genes involved in lymphocyte differentiation. IRF4 is known to assemble with AP-1 factors at AP-1-IRF composite elements (AICE)48. We did not observe statistically significant differences in Irf4 mRNA expression in WT and Bach2–/– cells (Supplementary Table 7). Since Irf4 is therefore unlikely to be a transcriptional target of BACH2 in CD8+ T cells, we conclude that differences in AP-1 factor binding in Bach2–/– cells are not attributable to an IRF4-dependent mechanism. It is notable that only a subset of TCR-induced gene loci contributed strongly to the correlation between differences in JunD binding and gene expression in WT and Bach2–/– cells. These findings suggest that the extent of BACH2/AP-1 competition or its effect on gene expression may vary in a site-specific fashion. It is an important area of future investigation to dissect the molecular mechanisms and sequence-level determinants for site-specific regulation of BACH2 function.
We found that reduced transcription of the Bach2 gene in effector cells corresponded to a decrease in its ability to restrain TCR-induced gene expression, enabling stage-dependent control of its function. BACH2 was also post-translationally modified to enable further contextual regulation of its function. Our finding that Ser520 phosphorylation of BACH2 results in its functional inactivation are consistent with previous observations in chronic myeloid leukemia cells that phosphorylation at a homologous residue in human BACH2 causes nuclear exclusion and functional inactivation41. Future investigations will be required to test whether modulation of BACH2 expression with progressive differentiation and its nuclear exclusion result in differential BACH2/AP-1 competition. In our analysis, we used TCR stimulation as an experimental system to study BACH2 phosphorylation. However, the finding that BACH2 is phosphorylated by Akt provides a potential mechanism by which a variety of extrinsic cues modulate T cell differentiation. The PI3K/Akt pathway integrates diverse antigenic, inflammatory, costimulatory and metabolic signals and regulate multiple aspects of lymphocyte biology44, 45, 49, 50. It is an important area of future investigation to determine the range of signals that regulate immune function by causing phosphorylation of BACH2. Additionally, it will be important to investigate how phosphorylation affects BACH2 function at endogenous levels of expression.
Collectively, we have found that BACH2 regulates transcriptional responses of CD8+ T cells to TCR signaling, enabling appropriate control of responses to viral infection. Our findings provide a basis for further investigation into how BACH2 controls the differentiation and function of CD8+ T cells and other lymphocyte lineages. Additionally, our results identify a new target for therapeutic manipulation of lymphocyte function in the context of chronic infections, autoimmunity and cancer.
Online methods
Mice and reagents
Experiments were approved by the Institutional Animal Care and Use Committee of the NCI and were performed in accordance with NIH guidelines. C57BL/6J, OT-I transgenic, B6-Ptprca Pepcb/BoyJ (Ly5.1) and B6.PL-Thy1a/CyJ (Thy-1.1) mice were purchased from The Jackson Laboratory. Bach2–/– mice, which have been previously described24, were backcrossed >16 times with C57BL/6 mice. 8-12 week old female mice were used for experiments unless otherwise specified. Congenically distinct WT and Bach2–/– naïve CD62L+ CD44– CD8+ T cells were isolated by flow cytometric sorting from spleens and lymph nodes of mixed BM chimeric animals 6-12 weeks following reconstitution. Naïve CD8+ T cells were activated with plate-bound anti-CD3 (2C11; 1 µg/ml; eBioscience) and soluble anti-CD28 (37.51; 1 µg/ml; eBioscience) in media containing 100 IU recombinant human IL-2 for 5 days. Where indicated, AKTi was added to culture media at a concentration of 1 µM (AKT inhibitor VIII, Calbiochem), JNKi at a concentration of 12.5 µM (JNK Inhibitor I, (L)-Form, Calbiochem) and rapamycin (Sigma) at a concentration of 100ng/ml.
Viral infection and kinetic analysis
For experiments assessing the response of CD8+ T cells to acute viral infection, 2 × 104 congenically distinct WT or Bach2–/– naïve CD44– CD62L+ OT-I CD8+ T cells were transferred into recipient mice. Immediately following transfer, mice were infected with 1 × 106 PFU VV-OVA. Naïve cells for transfer were isolated by flow cytometry from mice reconstituted with 1:1 mixtures of BM from WT CD45.1+ and Bach2–/– Thy-1.1+ OT-I TCR-transgenic mice. Where indicated, WT and Bach2–/– naïve cells were mixed at a 1:1 ratio prior to transfer.
Retroviral transduction
pMIG- and pMIT-BACH2 constructs have been described previously21, 22. S520A substitution mutations were generated by site-directed mutagenesis (Agilent). pMIT-BACH2-GFP and BACH2(S520A)-GFP were generated by cloning synthesized BACH2-GFP and BACH2(S520A)-GFP fusion constructs into the NotI and ClaI restriction sites within pMIT. Platinum-E ecotropic packaging cells (Cell Biolabs) were plated one day prior to transfection on poly-D-lysine coated 10 cm plates (Becton Dickinson) at a concentration of 6 × 106 cells per plate. Packaging cells were transfected with 20 µg of retroviral plasmid DNA along with 6 µg pCL-Eco plasmid DNA using 60 µl Lipofectamine 2000 in 3 ml OptiMEM (Invitrogen) for 8 h in antibiotic-free media. Media was replaced 8 h after transfection and cells were incubated for a further 48 h. Retroviral supernatants were collected and spun at 2000g for 2 h at 32 °C onto 24-well non-tissue culture treated plates coated overnight in Retronectin (20 µg/ml; Takara Bio) and 1 µg/ml anti-CD3 (2C11) and 1 µg/ml anti-CD28 (37.51) (eBioscience). Cells were cultured with 100 IU recombinant human IL-2. For experiments involving functional assessment and subcellular localization of BACH2(S520A), media was additionally supplemented with 0.2-20 ng/ml IL-12 (Peprotech).
Antibodies and flow cytometry
The following fluorochrome-conjugated antibodies against surface and intracellular antigens were used at 0.25-1×10–3 mg/ml: anti-Thy-1.1 (OX-7), anti-CD45.1 (A20), anti-KLRG1 (2F1), anti-CD25 (PC61), anti-CD62L (MEL-14), anti-IFN-γ (XMG1.2), anti-TNF (MP6-XT22), anti-CD44 (IM7), and anti-CD8α clone 53-6.7 (BD Biosciences). Cells were incubated with specific antibodies for 30 min on ice in the presence of 2.4G2 mAb to block FcγR binding. All samples were acquired with a FACS Canto II flow cytometer (Becton Dickinson) and analyzed using FlowJo software (TreeStar). To determine cytokine expression, cellular suspensions containing T cells were stimulated with phorbol 12-myristate 13-acetate (PMA), ionomycin and brefeldin-A (Leukocyte activation cocktail with Golgiplug; BD Biosciences) for 4 h. After stimulation, cells were stained an amine-reactive exclusion-based viability dye (Invitrogen) and antibodies against cell-surface antigens, fixed and permeabilized and intracellularly stained with specific anti-cytokine antibodies. Intranuclear staining for cytokines was carried out using the Foxp3 staining kit (eBioscience). Countbright beads (Invitrogen) were added for the flow cytometric quantification of absolute cell number.
Imaging flow cytometric detection of BACH2 subcellular localization
Total CD8+ T cells were isolated by negative magnetic selection (Miltenyi), stimulated using plates coated overnight with 1 µg/ml anti-CD3 (2C11) and 1 µg/ml anti-CD28 (37.51) in the presence of 100 IU IL-2 and 0.2-20 ng/ml IL-12 and transduced as described above with either pMIT-BACH2-GFP or pMIT-BACH2(S520A)-GFP. Four days post-transduction, cells were briefly restimulated using plates coated with 1 µg/ml anti-CD3, stained with Thy-1.1-APC during the last 30 min of culture, fixed using 2% PFA in PBS, stained with 4',6-diamidino-2-phenylindole (DAPI) and analyzed using an Amnis ImageStreamX MKII imaging flow cytometer. Data was analyzed using Amnis IDEAS. Briefly, single cells were selected based on cell size and focus attributes, and Thy-1.1, GFP and DAPI positivity. Cellular masks were defined as bounded by the Thy-1.1-APC cytoplasmic membrane signal, nuclear masks were defined using the DAPI signal, and a cytoplasmic mask was defined by subtracting the nuclear mask from the cytoplasmic mask. Mean pixel intensity of the BACH2-GFP signal was computed within the nuclear and cytoplasmic masks separately, and the ratio of the two values were analyzed to assess nuclear localization.
Immunoblotting and immunoprecipitation
CD8+ T cells and 293T cells were lysed in IP lysis buffer (Pierce). For immunoprecipitation experiments Flag-BACH2/BACH2(S520A) was immunoprecipitated following overnight incubation with M-2 anti-Flag monoclonal antibodies (Sigma) using protein A/G magnetic beads (Dynal). After several washes, immunoprecipitated proteins were released from beads using competitive elution with Flag peptide according to manufacturer’s instructions (Sigma) or by boiling in 1x Laemmli buffer. Proteins similarly immunoprecipitated from 293T cells transfected with WT or BACH2(S520A) were subjected to kinase assays while bound to beads and in the presence of ATP (Sigma) and kinase reaction buffer according to manufacturer’s instructions (Cell Signaling).
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Cells were isolated by flow cytometric sorting or harvested from cell cultures and transferred into RNALater solution (Ambion) for storage at -80 °C. Total RNA from pelleted cells was isolated using the RNAeasy mini kit (Qiagen). First-strand cDNA synthesis was performed using the high-capacity cDNA synthesis kit (Applied Biosystems) in the presence of SuperaseIn RNase inhibitor (Ambion). cDNA was used as a template for quantitative PCR reactions using Taqman primer-probes against specified mRNA transcripts (Applied Biosystems). Reactions were performed using Fast Universal PCR Mastermix (Applied Biosystems) and thermocycled in quadruplicate 10 µl reactions in 384-well plates. Signals in the FAM channel were normalized to ROX intensity, and ct values were calculated using automatically determined threshold values using SDS software (Applied Biosystems).
Bone marrow chimera experiments and measurement of anti-viral response kinetics
For BM reconstitution experiments, C57BL/6 mice were administered 1000 Gy total-body γ-radiation from a 137Cs source prior to intravenous injection of BM cells with or without depletion of mature lineages from single-cell BM preparations from 6-10 week-old mice using antibody-coupled magnetic beads (Miltenyi Biotec). To measure antiviral responses, 2 × 104 OT-I cells were transferred i.v. into recipients prior to infection with 1 × 106 plaque-forming units VV-OVA. Where indicated, WT and Bach2 KO naïve cells were sorted from mixed bone marrow chimeric mice and mixed at a 1:1 ratio prior to transfer.
In vivo BrdU labeling
For analysis of cellular proliferation in vivo, mice were administered 2 mg BrdU suspended in 500 µl PBS at 16 h (anti-viral kinetic experiment) or every day for 5 d (steady state proliferation in mixed BM chimeras) prior to analysis. Incorporated BrdU was detected using the APC BrdU staining kit (BD Biosciences).
RNA Sequencing and analysis
RNA-Seq analyses were performed using ≥2 biological replicates. Data from replicate measurements are publically available from the GEO repository (see Accession Codes). RNA sequencing was performed and analyzed as described previously22. Briefly, total RNA was prepared from cells using the RNeasy Plus Mini kit (Qiagen). 200 ng of total RNA was subsequently used to prepare RNA-Seq library by using TruSeq RNA sample prep kit (FC-122-1001, Illumina) according to manufacturer’s instructions. Sequenced reads were aligned to the mouse genome (NCBI37/mm9) with Tophat 2.0.1151 and uniquely mapped reads were used to calculate gene expression. RefSeq gene database (mm9) was downloaded from the UCSC genome browser for RNA-Seq analysis. Raw counts that fell on exons of each gene were calculated and differentially expressed genes were identified with the statistical R package edgeR52 or Cuffdiff53. Fisher’s exact test or the t-test were used to evaluate significance with indicated P-value and fold-change thresholds. The expression heat maps were generated with the R package “pheatmap”.
Chromatin immunoprecipitation and accessibility assays
T cells were chemically crosslinked and sonicated cells to generate fragmented genomic DNA. Chromatin immunoprecipitation was performed using the following antibodies: anti-BACH2 (N-2; Tohoku University), anti-JunD (sc-74), anti-c-Jun (sc-45), anti-JunB (sc-73) and anti-p300 (sc-586; Santa Cruz) anti-H3K4me1 (ab8895) and anti-H3K27Ac (ab4729; Abcam). For PCR-based confirmation of BACH2 binding, chromatin immunoprecipitation was performed as described above, and qPCR reactions were carried out on input and immunoprecipitated DNA using the Power SYBR Green kit (Applied Biosystems). Genome-wide measurement of chromatin accessibility and computational alignment of generated data were performed using ATAC-Seq on 50,000 d5 in vitro activated CD8+ T cells as previously described54. Subsequent bioinformatic analyses were performed as described below.
ChIP-Seq analysis
For sequencing of immunoprecipitated DNA, DNA fragments were blunt-end ligated to Illumina adaptors, amplified, and sequenced by using the HiSeq 2000 platform (Illumina). Sequenced reads of 50–125bp were obtained by using the Illumina Pipeline. Sequenced reads were aligned to the mouse genome (NCBI37/mm9) with Bowtie 0.12.955; only uniquely mapped reads were retained. The output of Bowtie was converted to .bam files, which represent the genomic coordinates of each read. Reads were mapped into non-overlapping 10-bp windows, and the location of reads on positive (negative) strand was shifted ± 75 bp from its 5′ start to determine the approximate center of the DNA fragment associated with the reads. Genomic graphs were generated and viewed using the IGV (Integrative Genomics Viewer)56, and we aligned the BACH2, JunD, cJun, p300, H3K4me1, H3K27Ac and input control in CD8+ T cells.
Peak calling, motif discovery and genome-wide distribution analysis
We used MACS 1.4.257 to call binding sites (peaks) relative to input control libraries. The binomial distribution test was used to assess peak significance and a P-value threshold of 1 × 10-5 was used unless otherwise specified. For motif discovery, the most significant 1,000 peaks ranked by lowest P-values were used and 100 bp DNA sequences centered on each peak summit were analyzed to using MEME to characterize the BACH2 consensus binding motif in mouse CD8+ T cells. The discovered motifs were compared with the JASPAR motif database to evaluate motif similarities. 5′ UTR, 3′ UTR, introns, exons and intergenic regions were defined according to the RefSeq database. Peaks were identified to be associated with genes if they were found within a region 15□kb upstream of the transcription start site to 3 kb downstream of the transcription end site of that gene. The distance between summits of two peaks was calculated and plotted as histograms using the R package.
Genome-wide binding profiles
Genome-wide binding profiles were established by counting the reads into binned windows (20 bp/bin) relative to peak summits. Normalized read densities were plotted in the range [-1 kb, 1 kb] centered on peak summits. Kolmogorov–Smirnov test was used to evaluate the significance between different distributions.
Statistical testing
Unless otherwise specified, statistical tests were performed using unpaired two-tailed Student’s t-tests. Where necessary, the Shapiro-Wilk test was used to test for normality of the underlying sample distribution. No blinding was necessary since objective quantitative assays, such as flow cytometry, were used. Experimental sample sizes were chosen using Power calculations using preliminary experiments or were based on previous experience of variability in similar experiments. Samples which had undergone technical failure during processing were excluded from analyses. Where relevant, recipient mice were randomized prior to adoptive transfer.
Supplementary Material
Acknowledgements
The research was supported by the Intramural Research Programs of the NCI and NHLBI, the Wellcome Trust / Royal Society grant 105663/Z/14/Z (R.R.) and the UK Biotechnology and Biological Sciences Research Council grant BB/N007794/1 (R.R. and K.O.). We thank S.A. Rosenberg, K. Hanada, K. Hirahara, K. Mousavi, H. Zare, V. Sartorelli, N. Van Panhuys, S. Kerkar and A. Restifo for ideas and discussion, A. Mixon and S. Farid for expertise with cell sorting, members of the NHLBI sequencing core facility for help with sequencing, L. Samsel for help with Imagestream imaging flow cytometry and G. McMullen for expertise with mouse handling.
Footnotes
Author Contributions
R.R., D.C., and N.P.R. wrote the manuscript and designed experiments. R.R., D.C., K.M.Q., Y.J., Z.Y., J.H.P., Y.K., Y.W. L.G. and G.F. performed experiments. P.L. and R.R analyzed bioinformatic data. C.A.K., D.C.P., D.C.M., M.S., S.J.P., H.-Y.S., R.S., A.M., L.G., R.L.E., J.Z., K.O., J.J.O’S., K.I. and W.J.L. edited the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Accession codes
Data are publicly available under NCBI Gene Expression Omnibus (GEO) accession number GSE77857.
References
- 1.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belz GT, Kallies A. Effector and memory CD8+ T cell differentiation: toward a molecular understanding of fate determination. Curr Opin Immunol. 2010;22:279–285. doi: 10.1016/j.coi.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 3.D’Cruz LM, Rubinstein MP, Goldrath AW. Surviving the crash: transitioning from effector to memory CD8+ T cell. Semin Immunol. 2009;21:92–98. doi: 10.1016/j.smim.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 5.Restifo NP, Gattinoni L. Lineage relationship of effector and memory T cells. Curr Opin Immunol. 2013;25:556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15:1104–1115. doi: 10.1038/ni.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teixeiro E, et al. Different T cell receptor signals determine CD8+ memory versus effector development. Science. 2009;323:502–505. doi: 10.1126/science.1163612. [DOI] [PubMed] [Google Scholar]
- 8.Wirth TC, et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 2010;33:128–140. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 10.Roychoudhuri R, et al. Transcriptional profiles reveal a stepwise developmental program of memory CD8(+) T cell differentiation. Vaccine. 2015;33:914–923. doi: 10.1016/j.vaccine.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005;175:5895–5903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 12.Sarkar S, et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner R, Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989;243:1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- 14.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 15.Rincon M, Flavell RA. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 1994;13:4370–4381. doi: 10.1002/j.1460-2075.1994.tb06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 17.Kurachi M, et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol. 2014;15:373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cippitelli M, et al. Negative transcriptional regulation of the interferon-gamma promoter by glucocorticoids and dominant negative mutants of c-Jun. J Biol Chem. 1995;270:12548–12556. doi: 10.1074/jbc.270.21.12548. [DOI] [PubMed] [Google Scholar]
- 20.Falvo JV, et al. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol Cell Biol. 2000;20:2239–2247. doi: 10.1128/mcb.20.6.2239-2247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oyake T, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roychoudhuri R, et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature. 2013;498:506–510. doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muto A, et al. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muto A, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 25.Hu G, Chen J. A genome-wide regulatory network identifies key transcription factors for memory CD8(+) T-cell development. Nat Commun. 2013;4:2830. doi: 10.1038/ncomms3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaech SM, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 27.Hikono H, et al. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med. 2007;204:1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyman O, Cho JH, Tan JT, Surh CD, Sprent J. A major histocompatibility complex class I-dependent subset of memory phenotype CD8+ cells. J Exp Med. 2006;203:1817–1825. doi: 10.1084/jem.20052495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendriks J, Xiao Y, Borst J. CD27 promotes survival of activated T cells and complements CD28 in generation and establishment of the effector T cell pool. J Exp Med. 2003;198:1369–1380. doi: 10.1084/jem.20030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev. 2010;236:151–166. doi: 10.1111/j.1600-065X.2010.00926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opferman JT, et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 34.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shnyreva M, et al. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barr RK, Kendrick TS, Bogoyevitch MA. Identification of the critical features of a small peptide inhibitor of JNK activity. J Biol Chem. 2002;277:10987–10997. doi: 10.1074/jbc.M107565200. [DOI] [PubMed] [Google Scholar]
- 38.Dong C, et al. JNK is required for effector T-cell function but not for T-cell activation. Nature. 2000;405:91–94. doi: 10.1038/35011091. [DOI] [PubMed] [Google Scholar]
- 39.Thiel G, Lietz M, Hohl M. How mammalian transcriptional repressors work. Eur J Biochem. 2004;271:2855–2862. doi: 10.1111/j.1432-1033.2004.04174.x. [DOI] [PubMed] [Google Scholar]
- 40.Russ BE, et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 2014;41:853–865. doi: 10.1016/j.immuni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida C, et al. Bcr-Abl signaling through the PI-3/S6 kinase pathway inhibits nuclear translocation of the transcription factor Bach2, which represses the antiapoptotic factor heme oxygenase-1. Blood. 2007;109:1211–1219. doi: 10.1182/blood-2005-12-040972. [DOI] [PubMed] [Google Scholar]
- 42.Calleja V, Laguerre M, Parker PJ, Larijani B. Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol. 2009;7:e17. doi: 10.1371/journal.pbio.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macintyre AN, et al. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim EH, et al. Signal integration by Akt regulates CD8 T cell effector and memory differentiation. J Immunol. 2012;188:4305–4314. doi: 10.4049/jimmunol.1103568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crompton JG, et al. Akt inhibition enhances expansion of potent tumor-specific lymphocytes with memory cell characteristics. Cancer Res. 2015;75:296–305. doi: 10.1158/0008-5472.CAN-14-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muto A, et al. Activation of Maf/AP-1 repressor Bach2 by oxidative stress promotes apoptosis and its interaction with promyelocytic leukemia nuclear bodies. J Biol Chem. 2002;277:20724–20733. doi: 10.1074/jbc.M112003200. [DOI] [PubMed] [Google Scholar]
- 47.Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994;8:1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- 48.Glasmacher E, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cantrell D. Protein kinase B (Akt) regulation and function in T lymphocytes. Semin Immunol. 2002;14:19–26. doi: 10.1006/smim.2001.0338. [DOI] [PubMed] [Google Scholar]
- 51.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.