Abstract
High-affinity–antibody production, T-cell activation, and interferon upregulation all contribute to protective immunity that occurs in humans following influenza immunization. Hematopoietic cell-specific PTPN22 encodes lymphoid phosphatase (Lyp), which regulates lymphocyte antigen receptor and pattern recognition receptor (PRR) signaling. A PTPN22 variant, R620W (LypW), predisposes to autoimmune and infectious diseases and confers altered signaling through antigen receptors and PRRs. We tested the hypothesis that LypW-bearing humans would have diminished immune response to trivalent influenza vaccine (TIV). LypW carriers exhibited decreased induction of influenza virus–specific CD4+ T cells expressing effector cytokines and failed to increase antibody affinity following TIV receipt. No differences between LypW carriers and noncarriers were observed in virus-specific CD8+ T-cell responses, early interferon transcriptional responses, or myeloid antigen-presenting cell costimulatory molecule upregulation. The association of LypW with defects in TIV-induced CD4+ T-cell expansion and antibody affinity maturation suggests that LypW may predispose individuals to have a diminished capacity to generate protective immunity against influenza virus.
Keywords: PTPN22, influenza, vaccine, CD4 T cells, affinity
Influenza virus infection poses a substantial public health burden, resulting in approximately 200 000 hospitalizations and 7505–37 102 deaths annually in the United States [1, 2]. Annual influenza vaccination can provide protection against influenza virus strains circulating during the influenza season, provided there is good matching between viral vaccine strains and circulating strains [3–5].
Protective immunity to influenza virus results from combined humoral and cellular immune responses. Neutralizing antibodies can provide sterilizing immunity to homologous viral strains by preventing viral attachment and entry to host epithelial cells [6]. Antibody neutralizing capacity is heavily dependent upon specificity and epitope binding affinity [7]. However, owing to high mutation rates in influenza virus genes encoding hemagglutinin (HA) and neuraminidase (NA), most neutralizing antibodies elicited by vaccination are strain specific and often do not provide heterosubtypic immunity [8, 9].
T cells are important cellular effectors of antiviral immunity. CD8+ T cells mediate direct lysis of infected epithelial cells [10], and high levels of virus-specific CD8+ T cells correlate with lower viral shedding and lower disease severity in humans [11, 12]. CD4+ T cells contribute to antiviral immunity by providing help to B cells for efficient isotype switching [13] and affinity maturation [14] needed in the generation of virus-specific antibodies [15] and by providing help to CD8+ T cells [16]. CD4+ T cells can also secrete interferon γ (IFN-γ) in the lungs [17] and can directly lyse virus-infected, major histocompatibility complex (MHC) class II–expressing bronchial epithelial cells in a perforin-dependent manner [18]. Both CD8+ and CD4+ T cells are strongly implicated in heterosubtypic immunity to influenza, owing to their recognition of viral proteins that are largely conserved across strains [11, 18–20].
In addition to stimulating adaptive responses, influenza virus infection or vaccination can induce transcription of numerous IFN-regulated genes [21, 22]. An early IFN signature correlates with B-cell responsiveness to vaccination and the development of neutralizing antibodies, suggesting that IFN signaling plays a role in anti–influenza virus antibody production [23]. Indeed, in animals, type I IFNs are critical for the generation of both protective antibody and CD4+ T-cell responses to influenza vaccination [24]. The requirements for type I IFN and innate immunity in vaccine-induced protective immunity among humans remain incompletely defined.
The protein tyrosine phosphatase nonreceptor 22 gene (PTPN22) encodes lymphoid phosphatase (Lyp), a hematopoietic-specific intracellular protein. Lyp functions as a negative regulator of T-cell receptor (TCR) signaling [25] and positively modulates interferogenic Toll-like receptor (TLR) signaling in macrophages and dendritic cells (DCs) [26, 27]. A human PTPN22 coding polymorphism, C1858T, is associated with a significantly increased risk of numerous autoimmune diseases, including rheumatoid arthritis, systemic lupus erythematosus, and type 1 diabetes and with altered susceptibility to select gram-positive bacterial and mycobacterial infections [28–30]. C1858T is carried by 6%–9% of persons of European ancestry [31]. C1858T encodes a single amino acid substitution–bearing (R620W) protein variant, termed LypW, that exhibits altered function in TCR signaling and in TLR-driven type I IFN production.
To date, studies testing LypW function in primary human lymphocyte and innate immune cell responses have used in vitro or ex vivo approaches [32–38]. Information concerning the role of human LypW during immunization responses has not been reported. Since PTPN22 regulates both lymphocyte and innate immune cell signaling and activation, we tested the hypothesis that R620W carriers would mount diminished or defective innate, cellular, and humoral responses to influenza vaccination.
METHODS
Subject Recruitment and Sample Collection
Healthy adult volunteers submitted DNA for PTPN22 rs2476601 (C1858T) genotyping. Eighteen PTPN22 LypW carriers (17 heterozygotes and 1 homozygote) and 17 age- and sex-matched noncarriers (LypR) received intramuscular Fluzone (2013–2014 trivalent inactivated influenza vaccine [TIV]; Sanofi Pasteur). Subjects submitted blood before vaccine receipt and on days 1, 14–15, and 25–28 following vaccination. Institutional review board approval was obtained (University of Minnesota; protocol 1210M21901), and all subjects provided informed written consent.
Genotyping
DNA was extracted from participant blood, using the DNeasy Blood and Tissue kit (Qiagen), and subjects were genotyped for the PTPN22-C1858T single-nucleotide polymorphism by the Taqman assay (Applied Biosystems). HLA-A genotype was determined by BLAST analysis of sequenced HLA-A polymerase chain reaction amplicons [39], using the IMGT/HLA database (available at: https://www.ebi.ac.uk/ipd/imgt/hla/).
Hemagglutination Inhibition Assay (HAI)
Serum was mixed with Aprotinin (Sigma) before freezing. HAIs were performed using the World Health Organization 2013–2014 influenza reagent kit and turkey red blood cells in Alsever solution (Colorado Serum) according to the manufacturer's instructions.
Bio-layer Interferometry Assay of Human Serum Samples
Real-time binding assays of antibodies in human sera and purified hemagglutinin proteins were performed using bio-Layer interferometry with an Octet RED system (ForteBio). Human sera were heated at 56° for 30 minutes before the assay. Biotinylated (EZ-link Micro NHS-PEG4-Biotinylation Kit, Thermo Scientific) A/California/04/09 hemagglutinin proteins (20 µg/mL) were immobilized onto streptavidin-coated biosensors (ForteBio) for 5 minutes. After measuring baseline signal in kinetics buffer (1× phosphate-buffered saline, 0.01% bovine serum albumin, and 0.002% Tween-20) for 3 minutes, biosensor tips were immersed into wells containing human sera (with a starting dilution of 1:10 and 3-fold serial dilutions) for 5 minutes to measure association. Biosensors were later immersed into kinetics buffer to measure dissociation. Binding kinetics were calculated using the Octet RED software package (Data Acquisition 8.2), to fit the observed binding curves in a 1:1 binding model to calculate the association and dissociation rate constants. Equilibrium dissociation constants were calculated as the kinetic dissociation rate constant divided by the kinetic association rate constant.
CD4+ T-Cell Assessments
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood and stored at −80°C until batched analysis. Thawed PBMCs were washed with complete prewarmed Roswell Park Memorial Institute 1640 medium. Cells were rested before stimulating 6 hours in the presence of brefeldin A (eBioscience) with either 2013–2014 Fluzone (10 µL), PMA (50 ng/mL) and ionomycin (1 µg/mL; Sigma), or medium alone. Cells were stained with fixable viability dye and surface antibodies (Supplementary Materials) before permeabilization and intracellular staining. Data were acquired on an LSR Fortessa (BD) and were analyzed using FlowJo software (Tree Star). CD4+ T cells were gated as shown in Supplementary Figure 4. The influenza virus–specific CD4+ T-cell fraction was calculated by subtracting numbers of cytokine-positive events observed in no-stimulation control from total cytokine-producing events observed in Fluzone stimulated conditions.
Statistical Analysis
The χ2 test was used to compare categorical variables of antibody seroconversion and seroprotection. The Student t test was used to compare antibody titers, isotype, and affinity; CD4+ and CD8+ T-cell frequency; and costimulatory molecule mean fluorescence intensities between groups. Within-group baseline and postvaccination values were compared using a paired t test. The Mann–Whitney test was also performed as an alternative nonparametric test. For transcriptional analyses, the Mann–Whitney test was performed for each gene to compare the expression levels between groups, followed by a gene-set enrichment association test [40] with 10 000 permutations. Bonferroni correction was used for multiple testing. Statistical analysis was performed using Stata (version 13.1; StataCorp) and GraphPad Prism 5 software.
For additional methods, refer to the Supplementary Materials.
RESULTS
LypW Carriage Associates With Poor Antibody Affinity Maturation After TIV
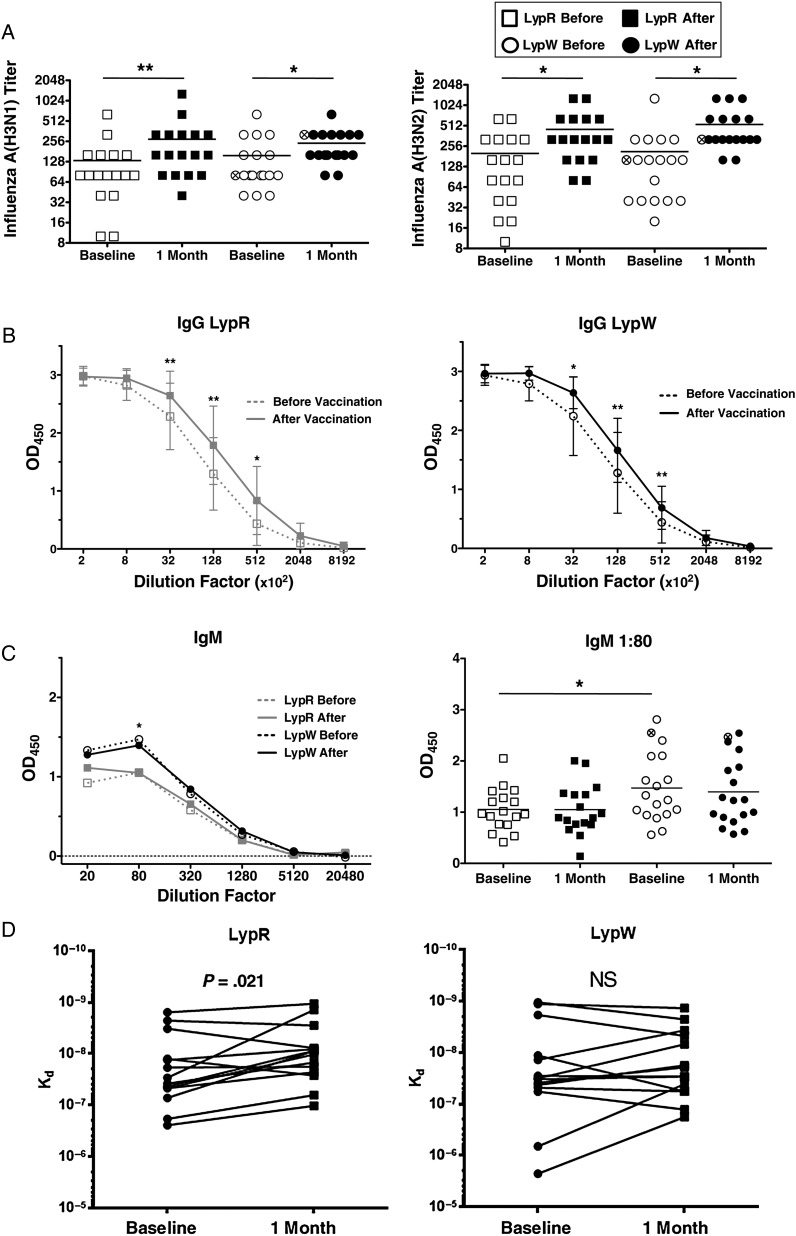
To examine the role of PTPN22 R620W variant (LypW) in human in vivo responses to influenza vaccination, we immunized 18 healthy LypW carriers or 17 noncarriers with TIV (Table 1 shows subject characteristics). First, we compared TIV-induced antibody levels in LypW carriers and noncarriers. We found that prevaccination seroprotection, defined as HAI titer of ≥1:40, was present in 82%–100% of subjects (Table 2). These results suggested high rates of previous vaccination or infection. Twenty-five days after vaccination, the majority of subjects exhibited significant increases in mean neutralizing antibody titers for influenza A(H1N1) and influenza A(H3N2) (Figure 1A). However, we noted no statistically significant differences between LypW carriers and noncarriers (LypR) in average absolute HAI titers, in rates of seroconversion (defined as a fold increase in neutralizing titer of ≥4), or in rates of seroprotection (100%) after TIV receipt (Table 2).
Table 1.
Subject Characteristics
| Characteristic | LypW Carriersa (n = 18) | LypR Subjects (n = 17) | Overall (n = 35) |
|---|---|---|---|
| Age, y, median (range) | 21 (18–55) | 21 (18–51) | 21 (18–55) |
| Sex | |||
| Female | 13/18 (72) | 12/17 (71) | 25/35 (71) |
| Ethnicity | |||
| White | 18/18 (100) | 17/17 (100) | 35/35 (100) |
| American Indian | 1/18 (5.5) | 0/17 (0) | 1/35 (2.9) |
| Previous influenza immunization | 17/18 (94) | 16/17 (94) | 33/35 (94) |
Data are proportion (%) of subjects, unless otherwise indicated.
a Seventeen were PTPN22-R620W heterozygous (R/W), and 1 was homozygous (W/W).
Table 2.
Hemagglutination Inhibition Assay (HAI) Seroconversion and Seroprotection
| Serostatus, Strain | LypR Subjects, % | LypW Carriers, % | P Valuea |
|---|---|---|---|
| Seroconversionb | |||
| H1N1 | 23.5 | 27.7 | .774 |
| H3N2 | 35.3 | 44.4 | .581 |
| Seroprotectionc | |||
| Baseline | |||
| Influenza A(H1N1) | 88 | 100 | .134 |
| Influenza A(H3N2) | 82 | 94 | .261 |
| 1 mo | |||
| Influenza A(H1N1) | 100 | 100 | NA |
| Influenza A(H3N2) | 100 | 100 | NA |
Abbreviation: NA, not applicable.
a By the χ2 test.
b Seroconversion is defined as a fold increase in titer of ≥4.
c Seroprotection is defined as an HAI titer of ≥40.
Figure 1.
LypW carriers have reduced affinity maturation after trivalent influenza vaccine (TIV). A, Hemagglutinin (HA) inhibition assays were performed on sera obtained from subjects, to determine influenza A(H1N1) and influenza A(H3N2) titers. B–C, Immunoglobulin isotype enzyme-linked immunosorbent assay for influenza A(H1N1) HA-specific antibodies for immunoglobulin G (IgG) and immunoglobulin M (IgM). B, Anti-HA IgG binding curves for LypR subjects and LypW carriers from serial dilutions of serum specimens obtained at baseline and 1 month after vaccine. Bars represent standard deviations. C, IgM curves and scatterplot for LypR subjects and LypW carriers from serial dilutions of serum specimens obtained at baseline and 1 month after vaccine. D, Dissociation constants from biolayer interferometry analysis of influenza A(H1N1) HA-specific antibodies in sera from 14 LypR subjects and 14 LypW carriers at baseline and 1 month after vaccination. Circle with an x denotes LypW homozygote. *P < .05 and **P < .01, by a paired t test (A, B, and D); *P < .05, by an unpaired t test (C). Abbreviation: NS, not significant.
Antibody isotype switching and affinity maturation are important for development of a neutralizing anti–influenza virus antibody repertoire [7]. To assess immunization-induced isotype switching, we measured serum levels of immunoglobulin M (IgM), immunoglobulin G (IgG), and immunoglobulin A (IgA) specific for influenza A(H1N1) HA. LypW carriers and noncarriers showed equivalent enhancement of IgG and IgA anti-HA in response to vaccination (Figure 1B and Supplementary Figure 1). However, mean anti-HA IgM levels in LypW carriers were significantly higher than levels in LypR subjects at baseline (Figure 1C). We also assessed the role of LypW in antibody affinity maturation during the influenza response. Biolayer interferometry was used to measure the dissociation constants (affinity) of serum antibodies against influenza A(H1N1) HA. While serum from LypR subjects displayed a significant increase in anti-HA binding affinity (mean, 2.6-fold) after vaccination, LypW carrier serum did not (Figure 1D and Supplementary Table 1). Together, these results suggested that LypW carriage associates with increased IgM anti-HA antibodies and with altered capacity to form high-affinity antibody to TIV.
LypW Carriers Show Diminished CD4+ T-Cell Responses to TIV
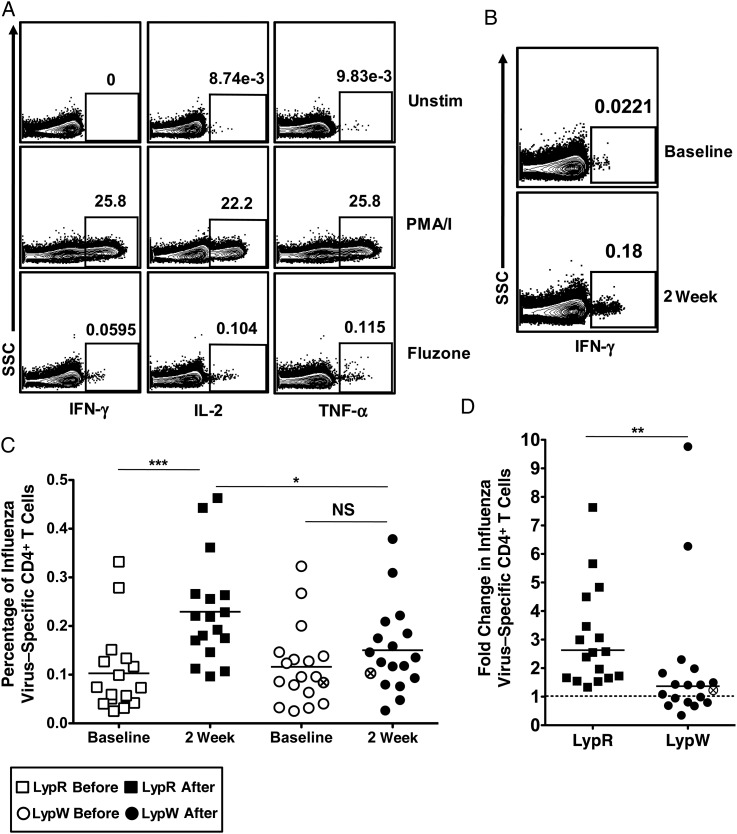
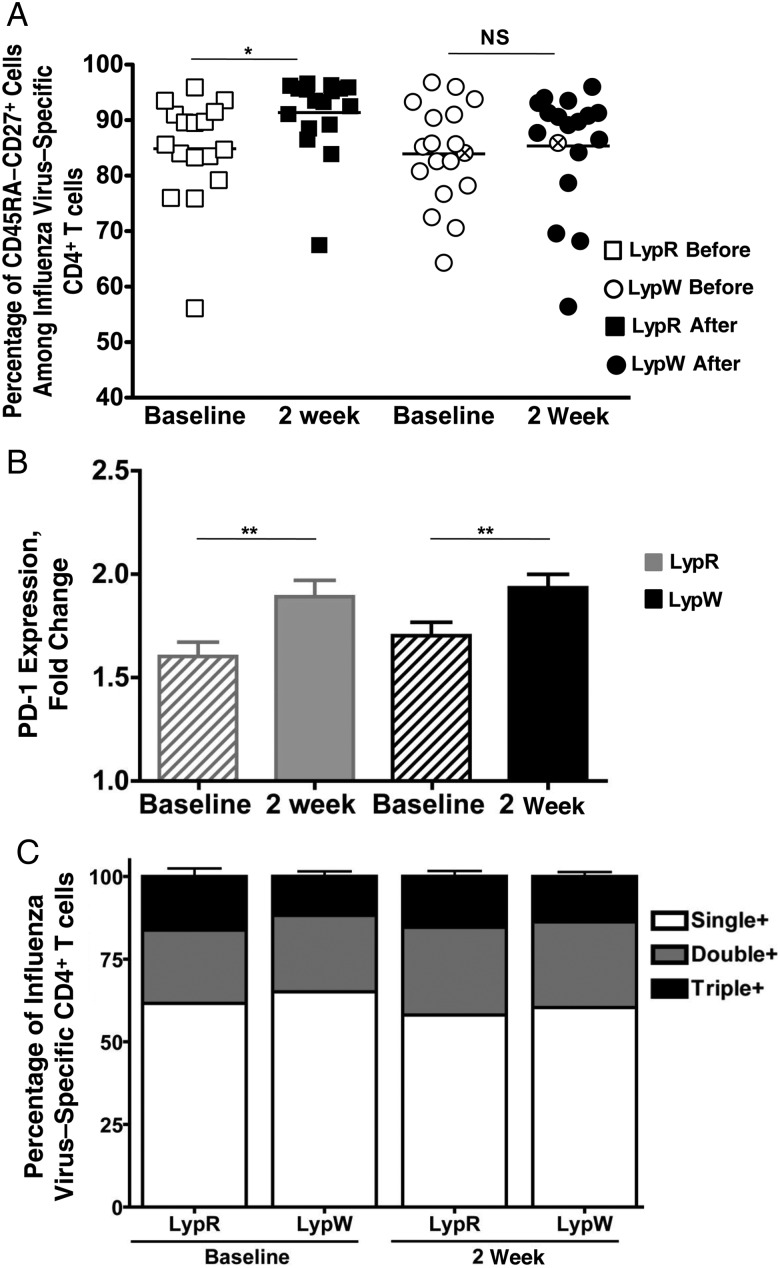
CD4+ T-cell help is critical for optimal isotype switching and affinity maturation [13, 14]. To study the role of LypW in CD4+ T-cell immunization responses, we exposed PBMCs from immunized subjects to TIV in vitro. Vaccine-stimulated CD4+ T-cell production of any of 3 cytokines—IFN-γ, tumor necrosis factor, or interleukin 2 (IL-2)—was used as an indicator of influenza virus specificity (Figure 2A) [41]. Unstimulated and PMA/ionomycin-stimulated CD4+ T cells were used as negative and positive controls for cytokine production, respectively, and no difference between LypR and LypW subjects was detected in either of these stimulation conditions (Supplementary Table 2). The frequency of influenza virus–specific CD4+ T cells among PBMCs increased in LypR subjects (noncarriers) 2 weeks after vaccination (Figure 2B and 2C). However, LypW carriers showed no vaccination-induced increase in the frequency of influenza virus–specific CD4+ T cells, either by analysis of pooled cytokine producers (Figure 2C) or of CD4+ T cells producing individual cytokines (Supplementary Figure 2). A similar pattern was seen with enumeration of absolute numbers of TIV-specific CD4+ T cells (Supplementary Figure 3). The geometric mean fold change increase in TIV-induced influenza virus–specific CD4+ T cells among noncarriers was 2.6 (95% confidence interval [CI], 2.019–3.426), compared with 1.3 among LypW carriers (95% CI, .926–2.018; Figure 2D). The percentage of TIV-induced influenza virus–specific CD4+ T cells showing evidence of prior activation (CD45RA−CD27+) increased significantly in LypR subjects but not in LypW carriers. These data suggested that LypW carrier CD4+ T cells might be subject to impaired TCR-dependent priming (Figure 3A). We observed no alteration in expression of PD1, a marker for dampened T-cell responsiveness, in either basal or vaccine-induced influenza virus–specific CD4+ T cells between LypR subjects and LypW carriers (Figure 3B). We observed no differences in responder T-cell polyfunctionality [42] between noncarriers and LypW carriers, as the fractions of influenza virus–specific CD4+ T cells expressing single or multiple cytokines were unchanged by vaccination or genotype (Figure 3C). Together, these findings suggest that LypW carriage confers reduced capacity for TIV-induced expansion and activation of CD4+ T cells but that restricted T-cell expansion is not a result of increased PD1 expression in LypW carrier T cells.
Figure 2.
LypW carriers exhibit a reduced CD4+ T-cell response to influenza vaccine. Subject peripheral blood mononuclear cells were stimulated in vitro with the 2013–2014 inactivated influenza vaccine Fluzone and assayed for cytokine production to denote influenza virus–specific CD4+ T cells. A, Representative cytokine staining for CD4+ T cells with indicated stimulus. B, Representative interferon γ (IFN-γ) staining of CD4+ T cells stimulated with Fluzone before and 2 weeks after immunization. C, Total frequency of influenza virus–specific CD4+ T cells. D, Fold change of influenza virus–specific CD4+ T cells in response to vaccination. Circles with an x denotes LypW homozygote. *P < .05 and ***P < .0001, by paired and unpaired t tests (C); **P < .01, by the Mann–Whitney test (D). Abbreviations: IL-2, interleukin 2; NS, not significant; SSC, side scatter; TNF-α, tumor necrosis factor α; Unstim, unstimulated.
Figure 3.
Influenza virus–specific CD4+ T cells from LypW carriers exhibit decreased activation but equal polyfunctionality and PD-1 expression. A, Frequency of influenza virus–specific CD4+ T cells that are CD45RA−CD27+ in LypW carriers and LypR subjects. Circles with an x denotes LypW homozygote. B, Surface PD-1 expression depicted as fold change of influenza virus–specific CD4+ T cells in LypW carriers (black) and LypR subjects (gray) before and after immunization. C, Frequency of influenza-specific CD4+ T cells that produce 1, 2, or 3 cytokines. Error bars represent standard deviation. *P < .05 and **P < .01. Abbreviation: NS, not significant.
LypW Carriage Does Not Affect TIV Induction of Costimulatory Molecules
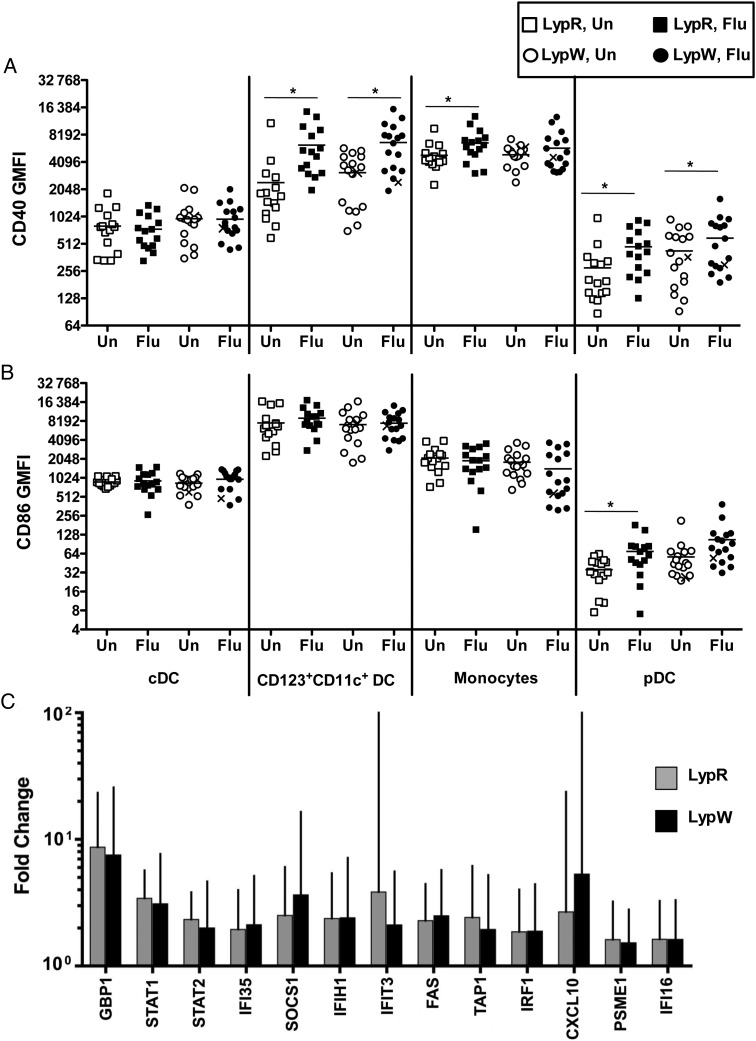
Antigen-presenting cell (APC) activation and IFN signaling represent T-cell–extrinsic factors that enhance CD4+ T-cell responses to immunization [24, 26]. PTPN22 variants differentially modulate these processes in animals [26, 43]; therefore, we investigated potential roles for LypW in human T-cell–extrinsic immunization responses. Optimal T-cell activation and clonal expansion depend upon costimulatory signals delivered through CD86 and CD40 expressed on APCs [44, 45]. In animals, Ptpn22 deficiency results in blunted CD80/CD86 and CD40 upregulation on myeloid APCs after viral infection, and carriage of the LypW allele results in reduced CD80 and CD40 upregulation on macrophages after TLR stimulation [26]. To assess LypW carrier capacity for upregulation of costimulatory molecules, we stimulated PBMCs with Fluzone, medium alone (unstimulated), or lipopolysaccharide (LPS). We monitored CD86 and CD40 expression on myeloid subsets, including monocytes (HLA-DR+CD14+), conventional DCs (cDCs; HLA-DR+CD11c+), CD123+CD11c+ DCs, and plasmacytoid DCs (pDCs; HLA-DR+CD11c−CD123+BDCA-2+; Supplementary Figure 5) [21]. We found that TIV induced significant upregulation of CD40 expression on monocytes, CD123+CD11c+ DCs, and pDCs (Figure 4A) and drove upregulation of CD86 on pDCs (Figure 4B). However, LypW carriers and noncarriers showed no difference in costimulatory molecule upregulation. Additionally, myeloid cell subsets from LypR subjects and LypW carriers showed no difference in upregulation of CD86 (Supplementary Table 3) or CD40 (Supplementary Table 4) in response to LPS. Taken together, these results suggest that LypW carriage does not alter the capacity for APCs to become activated by TIV.
Figure 4.
LypW carriers show no defect in the innate response to influenza vaccine. Geometric mean fluorescence intensity (GMFI) of CD40 (A) and CD86 (B) expression on indicated peripheral blood mononuclear cell subsets from LypR subjects and LypW carriers stimulated with Fluzone (Flu) vaccine in vitro for 18 hours. x's denotes LypW homozygote. C, Fold change in interferon signaling and response genes significantly induced by trivalent influenza vaccine at 1 day for LypR subjects (gray) and LypW carriers (black). Error bars represent standard deviations. *P < .05. Abbreviations: cDC, conventional dendritic cell; pDC, plasmacytoid dendritic cell; Un, unstimulated.
LypW Carriage Does Not Affect IFN Transcriptional Response to TIV
An early transcriptional response dominated by IFN-stimulated genes has been linked with development of adaptive immune responses after TIV [21–23]. LypW carrier myeloid cells show defects in IFN-stimulated gene upregulation after TLR stimuli [26, 27]. We therefore measured transcriptional responses 1 day after TIV receipt, using a limited array enriched for IFN signaling and response genes. Thirteen of 96 analytes were significantly induced 1 day after TIV receipt (Figure 4C). LypW carriers and noncarriers did not differ in either upregulation of individual gene transcripts or in grouped global responses, as assessed by gene-set enrichment association testing [40]. These data suggest that TIV-induced in vivo IFN-induced transcriptional responses are not affected by LypW carriage. Together with vaccine-induced APC activation results (Figure 4A), these findings are consistent with the hypothesis that diminished LypW-associated CD4+ T-cell immunization responses stem from T-cell–intrinsic mechanisms of LypW action.
TIV Does Not Impact Peripheral Blood CD8+ T-Cell Numbers or Function
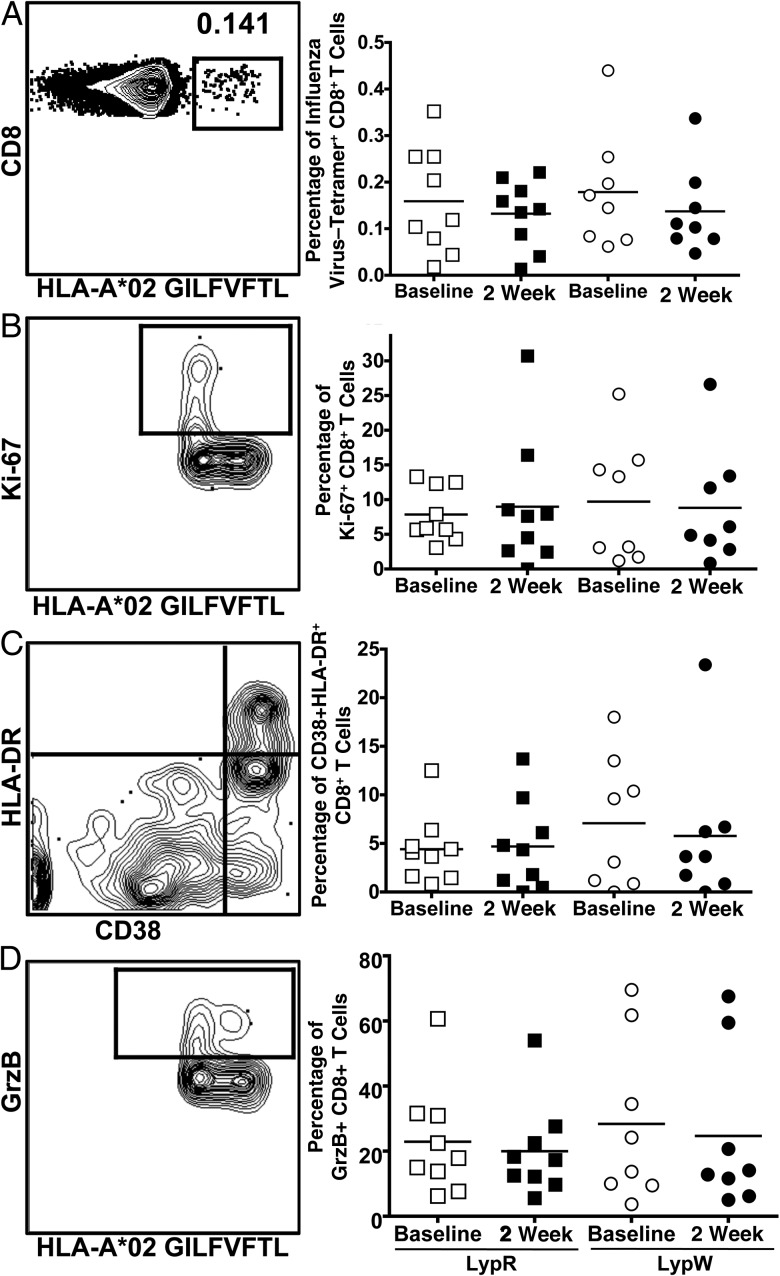
In animals, Ptpn22 deficiency is associated with decreased expansion of virus-specific CD8+ T cells after RNA virus infection [26]. We assessed the role of LypW carriage in expansion of human antigen-specific CD8+ T cells after TIV receipt. We used tetramers to determine the frequency of influenza virus–specific CD8+ T cells among PBMCs in vaccine recipients found to harbor MHC class I HLA-A*02 (9 in the LypR group and 8 in the LypW group). We observed no vaccine-inducible change in the frequency of influenza virus–specific CD8+ T cells 2 weeks after TIV receipt in either LypR or LypW subjects (Figure 5A). Moreover, neither group showed evidence of vaccine-inducible increases in influenza virus–specific CD8+ T-cell proliferation, activation state, or development of effector function, as detected by upregulation of Ki-67 (Figure 5B), by a change in frequency of CD38+HLA-DR+ (Figure 5C), or by expression of granzyme B (Figure 5D), respectively. These data strongly suggest that seasonal TIV does not induce detectable expansion or activation of peripheral blood influenza virus–specific CD8+ T cells at 2 weeks following TIV receipt. The data do not permit conclusions about a potential role for LypW in human CD8+ T-cell responses to vaccine.
Figure 5.
Inactivated influenza vaccine does not induce peripheral CD8+ T-cell response. Peripheral blood mononuclear cells were stained ex vivo with influenza virus peptide–major histocompatibility complex class I tetramers to identify virus-specific CD8+ T cells at baseline and at 2 weeks after immunization. Representative staining and total frequency of tetramer binding CD8+ T cells (A), Ki-67+ influenza virus–specific CD8+ T cells (B), CD38+HLA-DR+ influenza virus–specific CD8+ T cells (C), and GrzB+ influenza virus–specific CD8+ T cells (D).
DISCUSSION
This study addresses PTPN22 variant LypW effects on human immune responses induced in vivo. After TIV vaccination, we found that LypW carriers exhibit markedly impaired influenza virus–specific CD4+ T-cell expansion and show no significant increase in anti–influenza virus antibody affinity, compared with LypR subjects (noncarriers). While both LypR subjects and LypW carriers showed equal induction of isotype-switched antibodies against HA, LypW carriers harbored significantly higher HA-specific IgM titers at baseline and after vaccination. The immunological implication of our findings is that human LypW carriage results in multiple alterations in adaptive immune responses to antiviral immunization. Our results also have potential clinical significance. Since numbers of influenza virus–specific CD4+ T cells correlate with protection against influenza virus infection [18, 46], and since high affinity anti–influenza virus antibodies associate with reduced viral titers after infection [7], the reduced CD4+ T-cell expansion and impaired affinity maturation observed in LypW carriers could translate to increased vulnerability to influenza virus infection following vaccination [18, 46]. One important caveat to our findings is the small sample size of this study. Nevertheless, our data suggest that LypW should be studied further in larger cohorts as a candidate genetic modifier of protective responses to immunization against influenza.
Impaired augmentation of antibody affinity for HA in immunized LypW carriers might stem from altered function in either T-cell or B-cell compartments. Antigen-specific CD4+ T-cell help to B cells is a limiting factor in generating germinal centers and influenza virus–specific antibodies during influenza virus infection [15]. Such CD4+ T-cell help promotes somatic hypermutation that results in affinity maturation [47]. Thus, reduced CD4+ T-cell proliferation in LypW carriers could translate into less help to B cells for initiating immunization-induced germinal center responses. A second possible contributor to blocked immunization-induced anti-HA affinity maturation could be the significantly higher baseline anti-HA IgM levels we observed in LypW carriers. Preexisting anti-HA antibodies can impair vaccine antibody responses [48]. Accordingly, higher prevaccination HA-specific IgM levels in LypW carriers could hamper efficient affinity maturation. Additionally, overall serum HA affinity represents a summation of all affinity-matured immunoglobulin, including IgM, which is less subject than IgG or IgA to hypermutation-mediated affinity boosting. Even if immunization-induced affinity maturation is occurring among IgG subtype antibodies in LypW carriers, affinity modulation may not be detectable because greater amounts of less mutable anti–influenza virus IgM overwhelm the signal arising from hypermutation-matured IgG.
The underlying cause of elevated HA-specific IgM levels in immunized LypW carriers is not clear. Studies of B-cell function suggested a role for LypW in altering B-cell receptor and CD40 signaling [32–34], both of which are involved in isotype switching [48]. Further quantifying frequency and phenotype of IgM+ B cells generated in LypW carriers after TIV receipt will likely provide insight into how LypW contributes to elevated IgM levels in carriers.
Animal studies have explored roles of PTPN22 variants in immunization responses. Early observations in Ptpn22-deficient mice suggested a T cell–intrinsic effect of Ptpn22 in restraining T-cell expansion to peptide immunization [36]. Two groups have studied animals bearing knock-ins for Ptpn22-R619W, a murine LypW ortholog [49, 50]. R619W-expressing T cells produce more antigen-stimulated IL-2 following OVA/adjuvant immunization [49]. Further, R619W knock-in mice show increased antibody titers against T-dependent antigens after adjuvanted peptide immunization [49, 50]. Together, these data are evidence that, in mice, Ptpn22 functions to repress immunization-induced T-cell expansion/activation and that R619W is a loss-of-function variant in T-cell responses to vaccination.
Multiple factors could contribute to apparent discordance between the above-described animal study results and our finding of diminished CD4+ T-cell expansion in TIV-exposed human LypW carriers. First, T-cell responses in naive, syngeneic mice may differ from those elicited in antigen-experienced, outbred humans. Second, the present study documents human responses to a nonadjuvanted “split” viral protein preparation, whereas animal immunizations were conducted with adjuvanted neo-antigen. Third, functional differences between murine 619W and human 620W in T-cell signaling could contribute to divergent outcomes of immunization observed in human and animal systems.
Controversy exists regarding the net effect of LypW on TCR signaling in human cells. Some in vitro studies suggest that LypW carriage confers increased TCR signal strength and increased cytokine responsiveness among both naive and memory human CD4+ T cells [37, 50]. These data suggest that LypW acts as a loss-of-function negative regulator. In contrast, other studies suggest that, in antigen-experienced human T cells, LypW exerts gain of function as a TCR signaling suppressor, perhaps because of higher phosphatase enzymatic activity [38, 51, 52]. Such gain-of-function behaviors, which would be consistent with the T-cell expansion defects described here, have not been reported for murine 619W. In the face of uncertainty about the T-cell–intrinsic function of LypW, our findings of diminished CD4+ T-cell expansion in immunized LypW carriers will require validation in larger studies of genotyped humans. Further studies should also address possibilities that LypW carriage could modulate secretion capacity for cytokines such as IL-10 [51] and IL-2 [53] by immunization-induced CD4+ T cells. Discrepancy in data concerning in vivo LypW function might also be approached through the study of immunization responses by LypW-expressing memory T cells in murine models and by studies of adjuvanted neo-antigen immunizations in human LypW carriers. Finally, previous studies raise the possibility that effector T-cell–extrinsic mechanisms such as altered regulatory T cell (Treg) frequency or function could contribute to the decreased CD4+ T-cell response to TIV in LypW carriers [37, 49, 54]. Whether the size or function of the human Treg compartment is altered by LypW carriage after vaccination requires further investigation.
Reports that LypW acts as a loss-of-function variant in myeloid cell pattern recognition receptor signaling [26] lead us to hypothesize that LypW could also differentially regulate vaccine-induced CD4+ T-cell expansion by altering innate immune activation. Vaccine-induced, IFN-driven transcriptional responses and costimulatory molecule upregulation on APC subsets have been linked to pattern recognition receptor signaling [24]. Therefore, we were surprised that we detected no LypW-dependent alterations in APC activation after TIV exposure in the present study (Figure 4). However, interpretation of these results is complicated by the fact that LypW-bearing influenza virus–specific T cells present in PBMCs could be modulating APC responses via cytokine secretion or contact-dependent processes. In addition, O′Gorman et al found that TIV could engage human innate immune cells through Fc receptors, independently of TLRs [55]. In the current study, we observed no effect of LypW carriage on innate immune cell–produced cytokines (eg, CXCL10; data not shown) or transcriptional responses to IFN-γ, both of which are reportedly induced by TIV–immune complex stimulation of Fc receptors [55]. Thus, the degree to which diminished CD4+ T-cell expansion to TIV in LypW carriers reflects differential modulation of innate immune responses remains to be determined.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank Ryan Langlois for critical reading of the manuscript. Study data were collected and managed using REDCap electronic data capture tools hosted at the University of Minnesota.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Center for Advancing Translational Sciences, National Institutes of Health (NIH; award UL1TR000114) to E. J. P.; the University of Minnesota Immunology Training Grant, via the NIH (T32AI007313) to J. N. C.; and the Canadian Institutes of Health Research (operating grant to M. S. M.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Thompson WW, Shay DK, Weintraub E et al. Influenza-associated hospitalizations in the United States. JAMA 2004; 292:1333–40. [DOI] [PubMed] [Google Scholar]
- 2.Foppa IM, Cheng P-Y, Reynolds SB et al. Deaths averted by influenza vaccination in the U.S. during the seasons 2005/06 through 2013/14. Vaccine 2015; 33:3003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis 2012; 12:36–44. [DOI] [PubMed] [Google Scholar]
- 4.Fiore AE, Uyeki TM, Broder K et al. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recomm Rep Morb Mortal Wkly Rep Recomm Rep Cent Dis Control 2010; 59(RR-8):1–62. [PubMed] [Google Scholar]
- 5.Skowronski DM, Janjua NZ, De Serres G et al. Effectiveness of AS03 adjuvanted pandemic H1N1 vaccine: case-control evaluation based on sentinel surveillance system in Canada, autumn 2009. BMJ 2011; 342:c7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couch RB. An overview of serum antibody responses to influenza virus antigens. Dev Biol 2003; 115:25–30. [PubMed] [Google Scholar]
- 7.Verma S, Dimitrova M, Munjal A et al. Oligomeric recombinant H5 HA1 vaccine produced in bacteria protects ferrets from homologous and heterologous wild-type H5N1 influenza challenge and controls viral loads better than subunit H5N1 vaccine by eliciting high-affinity antibodies. J Virol 2012; 86:12283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DJ, Lapedes AS, de Jong JC et al. Mapping the antigenic and genetic evolution of influenza virus. Science 2004; 305:371–6. [DOI] [PubMed] [Google Scholar]
- 9.Westgeest KB, Russell CA, Lin X et al. Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011. J Virol 2014; 88:2844–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Topham DJ, Tripp RA, Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol Baltim Md 1950 1997; 159:5197–200. [PubMed] [Google Scholar]
- 11.McMichael AJ, Gotch FM, Noble GR, Beare PA. Cytotoxic T-cell immunity to influenza. N Engl J Med 1983; 309:13–7. [DOI] [PubMed] [Google Scholar]
- 12.Sridhar S, Begom S, Bermingham A et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 2013; 19:1305–12. [DOI] [PubMed] [Google Scholar]
- 13.Parker DC. T cell-dependent B cell activation. Annu Rev Immunol 1993; 11:331–60. [DOI] [PubMed] [Google Scholar]
- 14.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014; 41:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam S, Knowlden ZAG, Sangster MY, Sant AJ. CD4 T cell help is limiting and selective during the primary B cell response to influenza virus infection. J Virol 2014; 88:314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riberdy JM, Christensen JP, Branum K, Doherty PC. Diminished primary and secondary influenza virus-specific CD8(+) T-cell responses in CD4-depleted Ig(-/-) mice. J Virol 2000; 74:9762–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefrançois L, Farber DL. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol Baltim Md 1950 2011; 187:5510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson TM, Li CKF, Chui CSC et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 2012; 18:274–80. [DOI] [PubMed] [Google Scholar]
- 19.Kreijtz JHCM, de Mutsert G, van Baalen CA, Fouchier RAM, Osterhaus ADME, Rimmelzwaan GF. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J Virol 2008; 82:5161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yewdell JW, Bennink JR, Smith GL, Moss B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc Natl Acad Sci USA 1985; 82:1785–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obermoser G, Presnell S, Domico K et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity 2013; 38:831–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsang JS, Schwartzberg PL, Kotliarov Y et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 2014; 157:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao RG, Suarez NM, Obermoser G et al. Differences in antibody responses between trivalent inactivated influenza vaccine and live attenuated influenza vaccine correlate with the kinetics and magnitude of interferon signaling in children. J Infect Dis 2014; 210:224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koyama S, Aoshi T, Tanimoto T et al. Plasmacytoid dendritic cells delineate immunogenicity of influenza vaccine subtypes. Sci Transl Med 2010; 2:25ra24. [DOI] [PubMed] [Google Scholar]
- 25.Bottini N, Peterson EJ. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu Rev Immunol 2014; 32:83–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Shaked I, Stanford SM et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity 2013; 39:111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Ewart D, Crabtree JN et al. PTPN22 Variant R620W Is Associated With Reduced Toll-like Receptor 7-Induced Type I Interferon in Systemic Lupus Erythematosus. Arthritis Rheumatol 2015; 67:2403–14. [DOI] [PubMed] [Google Scholar]
- 28.Stanford SM, Bottini N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol 2014; 10:602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boechat AL, Ogusku MM, Sadahiro A, dos Santos MC. Association between the PTPN22 1858C/T gene polymorphism and tuberculosis resistance. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis 2013; 16:310–3. [DOI] [PubMed] [Google Scholar]
- 30.Chapman SJ, Khor CC, Vannberg FO et al. PTPN22 and invasive bacterial disease. Nat Genet 2006; 38:499–500. [DOI] [PubMed] [Google Scholar]
- 31.Burn GL, Svensson L, Sanchez-Blanco C, Saini M, Cope AP. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? FEBS Lett 2011; 585:3689–98. [DOI] [PubMed] [Google Scholar]
- 32.Arechiga AF, Habib T, He Y et al. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol Baltim Md 1950 2009; 182:3343–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habib T, Funk A, Rieck M et al. Altered B cell homeostasis is associated with type I diabetes and carriers of the PTPN22 allelic variant. J Immunol Baltim Md 1950 2012; 188:487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menard L, Saadoun D, Isnardi I et al. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J Clin Invest 2011; 121:3635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gianchecchi E, Crinò A, Giorda E et al. Altered B cell homeostasis and toll-like receptor 9-driven response in type 1 diabetes carriers of the C1858T PTPN22 allelic variant: implications in the disease pathogenesis. PloS One 2014; 9:e110755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 2004; 303:685–9. [DOI] [PubMed] [Google Scholar]
- 37.Vang T, Landskron J, Viken MK et al. The autoimmune-predisposing variant of lymphoid tyrosine phosphatase favors T helper 1 responses. Hum Immunol 2013; 74:574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottini N, Musumeci L, Alonso A et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 2004; 36:337–8. [DOI] [PubMed] [Google Scholar]
- 39.Bugawan TL, Apple R, Erlich HA. A method for typing polymorphism at the HLA-A locus using PCR amplification and immobilized oligonucleotide probes. Tissue Antigens 1994; 44:137–47. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian A, Tamayo P, Mootha VK et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolfi DV, Mansfield KD, Kurupati RK et al. Vaccine-induced boosting of influenza virus-specific CD4 T cells in younger and aged humans. PloS One 2013; 8:e77164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Precopio ML, Betts MR, Parrino J et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med 2007; 204:1405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes DA, Suto E, Lee WP et al. Autoimmunity-associated protein tyrosine phosphatase PEP negatively regulates IFN-α receptor signaling. J Exp Med 2015; 212:1081–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Gool SW, Vandenberghe P, de Boer M, Ceuppens JL. CD80, CD86 and CD40 provide accessory signals in a multiple-step T-cell activation model. Immunol Rev 1996; 153:47–83. [DOI] [PubMed] [Google Scholar]
- 45.Cella M, Scheidegger D, Palmer-Lehmann K, Lane P, Lanzavecchia A, Alber G. Ligation of CD40 on dendritic cells triggers production of high levels of interleukin-12 and enhances T cell stimulatory capacity: T-T help via APC activation. J Exp Med 1996; 184:747–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayward AC, Wang L, Goonetilleke N et al. Natural T Cell-mediated Protection against Seasonal and Pandemic Influenza. Results of the Flu Watch Cohort Study. Am J Respir Crit Care Med 2015; 191:1422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol 2005; 86:43–112. [DOI] [PubMed] [Google Scholar]
- 48.Andrews SF, Huang Y, Kaur K et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 2015; 7:316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai X, James RG, Habib T et al. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest 2013; 123:2024–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Zahir N, Jiang Q et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet 2011; 43:902–7. [DOI] [PubMed] [Google Scholar]
- 51–55.These references are available in the Supplementary Appendix. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.