Abstract
Artificial cells are best defined as micrometre-sized structures able to mimic many of the morphological and functional characteristics of a living cell. In this mini-review, we describe progress in the application of droplet-based microfluidics for the generation of artificial cells and protocells.
Keywords: artificial cell, microfluidics, droplet-based microfluidics, liposome, polymersome
1. Introduction
The construction of entities that resemble and mimic the basic unit of structure and function in living organisms is an immensely challenging task, but one that offers the possibility of replacing faulty biological components (in individuals affected by pathologies such as diabetes, liver or immune deficiency) and facilitating the understanding of life by bottom-up assembly of functional constituents. Although a strict definition of an artificial cell is still a matter of some debate [1,2], over the last century, the term artificial cell has been commonly applied to any system that, by merging natural and synthetic chemical components, is able to mimic or substitute cellular functions [3,4]. Unsurprisingly, artificial cells have already found applications as blood cell substitutes [5], in gene therapy [6] and in targeted drug delivery [7]. Nevertheless, robust and high-efficiency methods for the routine generation of artificial cells are still a rarity [8].
Recent progress in emerging fields such as synthetic biology and material science is beginning to yield new tools for the design and construction of biological pathways and systems that exist in nature, with microfluidic technologies playing a key role [4,9,10]. Microfluidic systems are well recognized for their ability to efficiently handle, control and process small volumes of fluids on the micrometre scale [11] and more specifically for their facility in producing monodisperse droplets, with volumes ranging from femtolitres to nanolitres, at kilohertz to megahertz rates [12]. Significantly, in the context of the current discussion, the dimensions of these droplets can be made to match those of living cells (1–150 µm) [9]. In this mini-review, we highlight recent progress in the application of droplet-based microfluidic techniques (incorporating both single and double emulsion templates) for artificial cell generation. We also discuss selected methods that to date have only been achieved using conventional technologies, but which could eventually be reproduced using microfluidic components.
2. Droplet-based microfluidics
Since the first report of monodisperse droplet formation in microfluidic channels by Thorsen et al. [13], a diversity of chip- and capillary-based systems have been used to produce, load, process and assay droplet populations for quantitative experimentation in the chemical and biological sciences [12,14]. Beyond the critical advantages of compartmentalization, monodispersity and high-throughput generation, the ability to perform functional operations on droplets (such as reagent mixing, droplet splitting, sampling, fusion, dilution and sorting) has further enlarged the spectrum of possible applications of both passive and active droplet-based microfluidic systems [12,14,15].
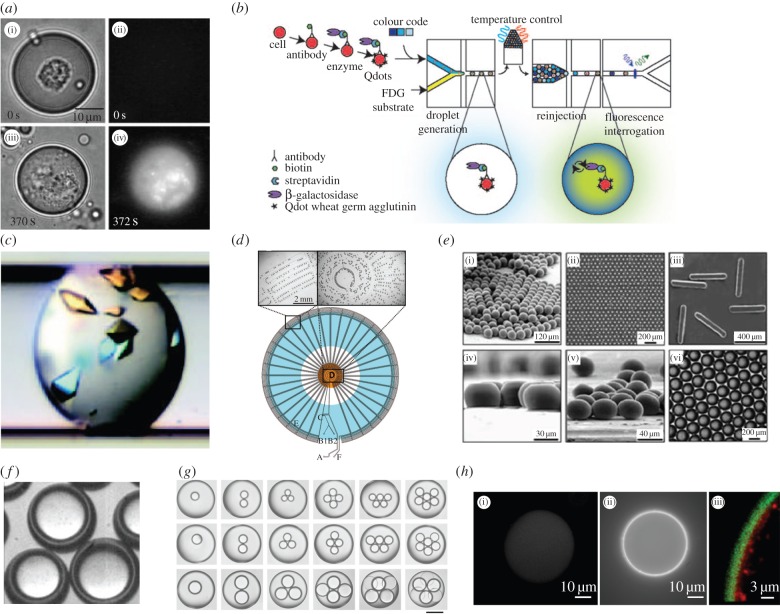
As noted, droplet-based microfluidic systems have been used to perform a wide range of biological and chemical experiments [14,16]. These include high-throughput biochemical assays [17–19], protein crystallization studies [20], cell-based enzymatic assays [21], DNA amplification [22], the synthesis of microspheres [23] and core–shell structures [24–28] and the bespoke synthesis of monodisperse nanomaterials (figure 1a–g). Additionally, drugs, green fuels and high value biomaterials like proteins and antibodies have also been produced using droplet-based reactors [12,14,30] (figure 1h).
Figure 1.
The application of droplet-based microfluidics in chemistry and biology. (a) A cell-based enzymatic assay performed within a droplet containing a single cell. After photolysis, intracellular enzyme is released into the droplet and reacts with a substrate to form a fluorescent product. (b) A droplet-based microfluidic system for high-throughput cell sorting. (c) Photograph of protein crystals formed in a microdroplet after chaotic mixing. (d) Segmented-flow PCR where droplets are motivated through alternating temperature zones to perform DNA denaturation, primer annealing and template extension. (e) Images of polyTPGDA ((i)–(iv)) and agarose discs (vi) produced in droplet-based microfluidic reactors. (f) Core–shell structures, also known as double emulsion, generated using glass capillaries. (g) Multiple emulsions generated using scalable microcapillary devices: scale bar is 200 µm. (h) Cell-free protein expression within a double emulsion: (i) water-soluble protein RFP, ((ii)(iii)) mReB-RFP protein adhering to hydrophobic regions. Images adapted from [20–23,25,28–30].
3. Artificial cells: the basics
Beginning with the idea that an artificial cell is an entity that can mimic some of the functions and structures of a living cell, it is evident that the design of an artificial cell requires a complete understanding of cell biology and the ability to simplify the complexity associated with cellular structure and function [31,32]. In this respect, artificial cells themselves can also be used as platforms to understand cellular mechanisms via bottom-up methods [8,32]. Figure 2a illustrates a simplified representation of a living cell. The cartoon shows a membrane that provides a boundary between the external and internal environments, where cellular processes occur. Significantly, the membrane is an active component, containing membrane receptors that move, communicate and sense their local environment. The inner compartment contains genetic material and enzymes responsible for cellular processes such as protein synthesis, replication and metabolism or growth-related processes. Furthermore, the cell may possess a cytoskeleton, a dynamic structure that gives a shape to the cell, anchors organelles and aids in the movement. Finally, as with all living systems, an artificial cell requires energy to perform its functions and maintain ‘life’. A strategy to unravel cell complexity using a bottom-up approach has been proposed by Brizard et al. [33] and is shown in figure 2b. In their approach, four component steps are used to design an artificial cell. Each defines properties that a cell-like system must possess to guarantee and maintain its vital properties.
Figure 2.
Artificial cell architecture. (a) Cartoon representation of an artificial cell incorporating a membrane that defines the boundary between the outer and inner environments (where cellular processes occur). The semi-permeable membrane contains receptors, which can sense and interact with the external world. The cell interior is connected to a frame, called the cytoskeleton, which imparts structure to the cell. (b) A proposed strategy to unravel cell complexity via a bottom-up approach involving artificial cells. Image adapted from [33].
4. Artificial cell membranes
All cells possess a cell membrane which defines the boundary between the extracellular environment and the aqueous internal compartment [34]. The membrane itself provides a non-polar environment where anchored or embedded channel proteins, that control the flow of ions across the membrane, reside. Transport of ions or small molecules across the cell membrane is of crucial importance for the majority of cellular processes and, in a living cell, transport across the membrane is performed by a variety of passive and active mechanisms. The former are governed by the permeability of the cellular membrane relying on diffusion, facilitated diffusion or osmosis. Conversely, active mechanisms require an energy input that activates a membrane protein responsible for the transport of a species flowing against a chemical gradient [34].
Over the years, much effort has focused on reproducing artificial membranes that resemble the membrane of a living cell in both function and morphology. Part of this effort has centred on reconstructing the exact composition of natural membranes and accessing the ability to incorporate protein channels, while others have tried to reproduce the ability to respond to external stimuli through synthetically derived materials [1]. In this respect, vesicle-like structures, made from amphiphilic molecules (assembled to form ordered and closed bilayers) have been used as cell membrane mimics [35,36]. In the following sections, we review recent studies involving the use of droplet-based microfluidic techniques for lipid (liposome) and polymeric (polymersome) vesicle formation. It is also important to mention that some researchers have achieved selective permeability and encapsulation of biological components in structures that do not incorporate a membrane. Such structures, known as coacervates, represent powerful models as protocells [37], for mimicking molecular crowding and have also recently been generated using droplet-based microfluidic tools [38].
4.1. Artificial cells based on liposomes
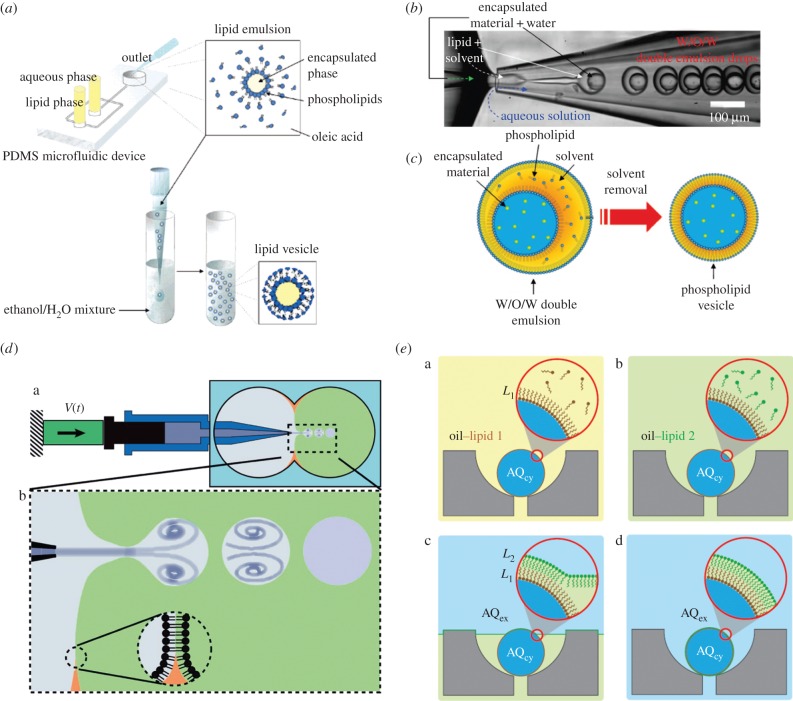
The majority of experimental studies on protocell models incorporate bilayer membranes constructed from one (or occasionally two) lipid species [32,39]. Lipidic vesicles, also known as liposomes, are structures derived from lipid molecule assembly and contain a thin bilayer membrane tens of nanometres thick [40]. Giant liposomes or giant unilamellar vesicles (GUVs; having diameters in excess of 10 µm) are comparable in size to eukaryotic cells, and it has been shown that they can efficiently encapsulate biomaterials such as DNA, RNA and proteins [41]. First synthesized in bulk reactors using conventional methods, such as electroformation [42], freeze-drying [43] and sonication [44], conventional liposome production is time consuming and irreproducible and results in polydisperse vesicle populations containing multilamellar structures [41]. Microfluidic methods have been shown to offer unprecedented control over vesicle size, encapsulation efficiency and membrane homogeneity [40]. The first droplet-based microfluidic system for forming unilamellar liposomes was reported by Tan et al. [45] (figure 3a). This study demonstrated the feasibility of encapsulating proteins, beads and cells in liposomes with diameters ranging from 27 to 55 µm, and facilitating ion exchange between the inner compartment and external environment [45]. Two years later, glass capillary devices, used for the generation of core–shell structures [25], were adopted to form liposomes from double emulsion templates [46]. Using a two-step emulsification process performed entirely on-chip, liposomes with diameters ranging from 20 to 150 µm could be generated in a reproducible fashion (figure 3b,c). Subsequently, a wide variety of droplet-based microfluidic strategies have been employed for liposome generation [9,40,41]. The majority of these reports have demonstrated the possibility of generating liposomes loaded with biologically active compounds (such as DNA and enzymes) and able to host proteins within the lipid membrane. For example, Stachowiak et al. [48] described an elegant pulsed-jetting method [49] to generate monodisperse giant vesicles loaded with α-haemolysin monomers, which spontaneously migrate towards the lipid membrane and form protein pore useful for transporting solutes across the vesicle boundary (figure 3d).
Figure 3.
Liposome generation using droplet-based microfluidics. (a) Droplet emulsion transfer method. (b,c) Formation of a phospholipid-stabilized W/O/W double emulsion in a glass microcapillary device. A toluene and chloroform mixture allows phospholipid dissolution and subsequent solvent evaporation. As the solvent layer becomes thinner during evaporation, phospholipids concentrate forming a bilayer. (d) Schematic of a pulsed jet made from a piezoelectric actuator connected to a pipette and immersed in a two-phase system containing phospholipids. When a pulse is produced, a droplet of one liquid type (grey) is generated within a second phase (green) and a bilayer automatically generated. (e) Asymmetric liposome generation. (i) Cartoon showing a trap containing a droplet of aqueous cytoplasmic material (AQcy, blue) in a mixture of oil–lipid 1 (yellow) that stabilizes the droplet. (ii) Oil–lipid 2 (green) mixture replacing oil–lipid 1. (iii) Extracellular aqueous phase (AQex, blue) replacing oil–lipid 2 and (iv) while the AQex/oil interface traverses the array, it envelopes a second interfacial monolayer composed of oil–lipid 2 that deposits on the trapped droplet. Images adapted from [41,45–47].
Microfluidic tools have also been used to fabricate layer-by-layer asymmetric lipid vesicles. In 2003, Pautot et al. developed a method for systematically engineering vesicles with asymmetric bilayers where each leaflet was assembled independently [50]; in 2011, Matosevic et al. presented an assembly-line strategy able to achieve a completely parametrized and reproducible phospholipid vesicle generation [51]. Later on, the same authors [47] demonstrated the formation and subsequent (phospholipid) stabilization of droplets using a flow focusing device. After a defined delay, droplets could be trapped within pockets and the continuous phase gradually exchanged with a secondary phase containing a different type of phospholipid that deposits on the previously formed bilayer. Following this simple strategy, the authors demonstrated the generation of multilamellar asymmetric vesicles (figure 3e). More recently, a microfluidic approach involving on-chip electroformation demonstrated the generation, handling and analysis of interaction dynamics of the pore-forming antimicrobial peptide melittin in trapped GUVs containing two types of lipid molecules [52]. Other examples of artificial cells based on liposomes are presented in the following sections, where examples of lipid vesicles employed to mimic cellular processes are reported.
4.2. Artificial cells based on polymersomes
Owing to their similarity to the natural components contained within cell membranes, phospholipidic vesicles represent a useful method for producing cell mimics [53], despite their susceptibility to breakage and oxidation [35]. Polymeric vesicles, also known as polymersomes, are obtained by self-assembly of amphiphilic block copolymers and represent an interesting liposome proxy due to their enhanced stability and functionality [54]. Depending on the relative length of the hydrophobic and hydrophilic chains, they are able to assemble (in aqueous environments) into a variety of structures such as vesicles and wormlike or spherical micelles [55]. Furthermore, membrane thicknesses (from a few nanometres to a few micrometres), elasticity, permeability and mechanical stability can all be tuned by control of the chemical composition of the chosen polymer [56].
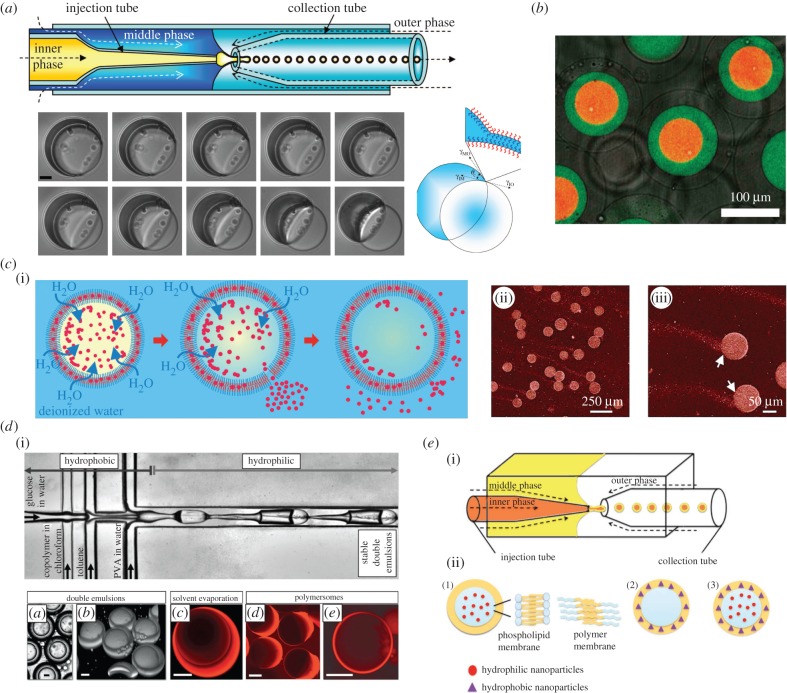
Extensively employed in foods, drugs and cosmetics [57], polymersomes also play a relevant role as cell mimics [58–60]. Again, droplet-based microfluidics has been shown to be an extremely valuable tool in the rapid and reproducible production of such species [61]. For example, Shum et al. [62] used an ‘all-in’ glass capillary format to generate monodisperse vesicle structures of amphiphilic diblock copolymers. The authors elegantly showed the formation of a stable core–shell structure originating from a water–oil–water (W/O/W) double emulsion system containing a poly(ethylene glycol)–poly(lactic acid) (PEG–PLA) diblock after undergoing a dewetting transition (figure 4a). The diblock polymer is dispersed in a mixture of chloroform and toluene, and after double emulsion generation, chloroform evaporates and toluene detaches leaving behind a bilayer shell (figure 4a). In this study, the authors also showed that the shell is able to respond to osmotic shock, a characteristic of fundamental importance in the delivery of encapsulated material. Indeed, the core method has since been used to generate ‘multiple compartment’ polymersomes [63], polymersome-in-polymersome structures [64] (figure 4b) and polymersomes able to express and release proteins in their interiors [59] (figure 4c).
Figure 4.
Polymersome generation using droplet-based microfluidics. (a) Glass capillary device used for PEG-b-PLA polymersome production. Dewetting transition of double emulsion consisting of an aqueous droplet surrounded by a shell of PEG-b-PLA diblock copolymer dissolved in a toluene/chloroform mixture. The diblock polymer is dispersed in a mixture of chloroform and toluene, and after double emulsion generation, chloroform evaporates and toluene detaches leaving behind a bilayer shell. Scale bar, 10 µm. (b) Confocal microscope image showing triple polymersomes. (c) Protein expression and delivery of expressed proteins within polymersomes. Protein release is triggered by osmotic shock, with release tails being clearly visible. This is possible thanks to the semi-permeability of the PEG–PLA bilayer membrane which allows the passage of small molecules like water when the two compartments have different osmolarities. (d) Microfluidic formation of diblock copolymer-stabilized W/O/W double emulsions. Scale bar, 20 µm. (e) Schematic of different types of vesicles containing NPs obtained using capillary device. Vesicles containing hydrophilic NPs in the aqueous core (1), vesicles with hydrophobic NPs in the organic shell (2), vesicles with hydrophilic NPs in the aqueous core and hydrophobic NPs in the organic shell (3). Images adapted from [59,63–66].
In all these approaches, the solvent composition is a critical parameter that cannot be balanced during experiment using capillary-based devices. In this regard, Thiele et al. [65] have demonstrated the ability to tune solvent composition by using a glass-coated polydimethylsiloxane (PDMS) microdevice, and have analysed how solvent composition affects the stability of the emulsion (figure 4d). This strategy also represents an interesting alternative to capillary-based devices not only for the polymersome formation but also for the generation of other double emulsion systems.
In addition to the ability of polymersomes to react to osmotic shock, PEG-b-PLA polymersomes and lipid vesicles have also been enriched with nanoparticles (NPs). For example, hydrophilic and hydrophobic magnetic NPs have been introduced into the shell, the core or both, thereby transferring their magnetic properties to the entire vesicle [66] (figure 4e). Moreover, both synthetic [67] and natural [60] hydrogel materials have been encapsulated in the core, providing an inner solid scaffold within the entire vesicle that can react to external stimuli (figure 5c).
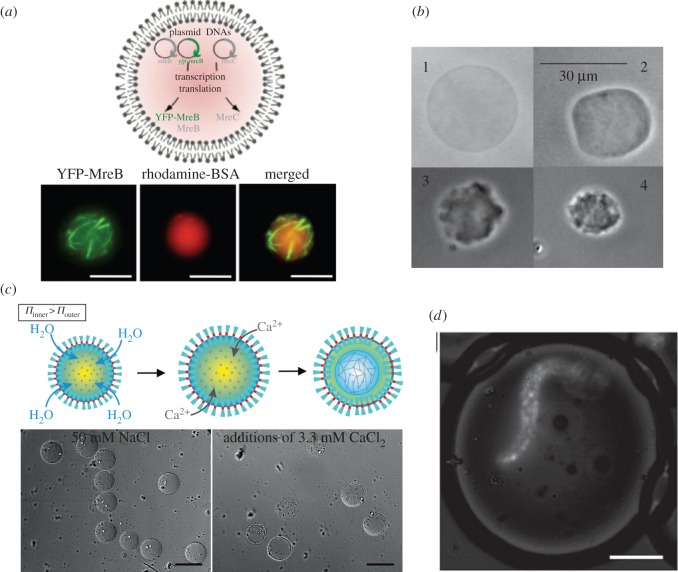
Figure 5.
Artificial cytoskeletons. (a) Cell-free expression of MreB protein in a giant liposome forming filaments (green). The liposome core is filled with bovine serum albumin (red). Scale bar, 10 µm. (b) Agarose-filled lipid vesicles undergoing osmotic shrinkage. (c) Microfluidic generation of polymer vesicles filled with alginate in liquid form. When exposed to calcium ions the semi-permeable membrane expands and allows alginate gelation. Scale bar, 100 µm. (d) Encapsulation of an alginate fibre, loaded with fluorescent beads, within an aqueous droplet. Scale bar, 50 µm. Images adapted from [57,66–68].
5. Artificial cytoskeletons
As previously noted, the cytoskeleton is a central component in both eukaryotic and prokaryotic cells. Some researchers have attempted to mimic this function by directly polymerizing cytoskeleton components encapsulated in lipid vesicles (obtained via conventional approaches; figure 5a) [69–72]. Others have encapsulated hydrogels within lipid vesicles to mimic cytoskeleton deformation and viscoelastic properties [73,74] (figure 5b). Although numerous studies have tried to fill vesicles with hydrogel matrices [33], only a few have used microfluidic tools to exert control over the generation process [60,67,68] (figure 5c,d).
6. Mimics of cellular processes in artificial cells
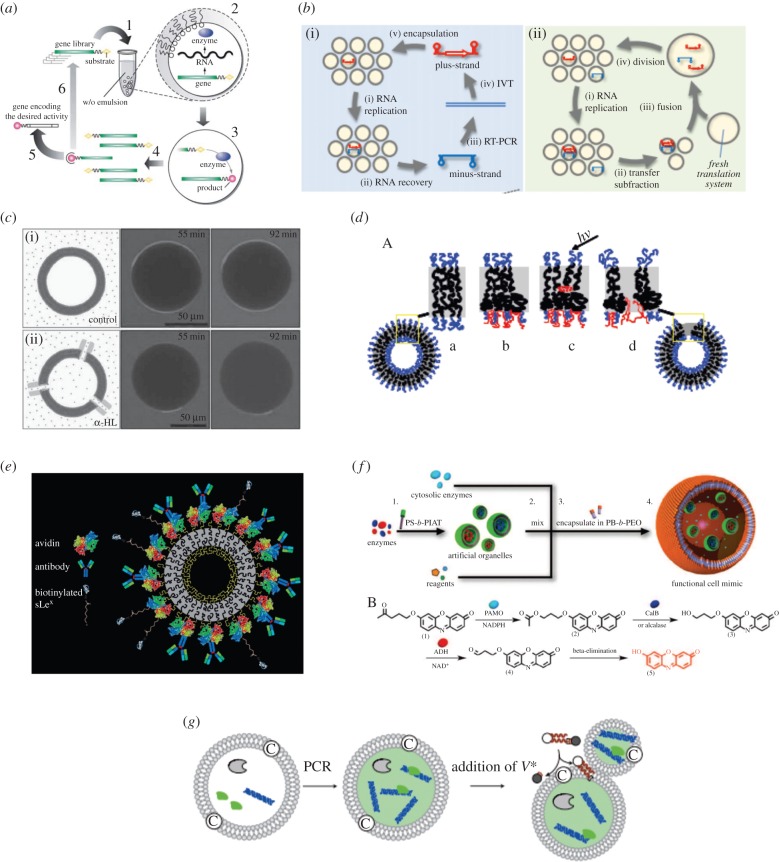
Other essential features of living cells include DNA expression and regulation, self-reproduction, phospholipid and nutrient synthesis and energy generation from carbon sources [4]. Unsurprisingly, it is enormously challenging to mimic even a small part of each of these features, but integration of know-how from different disciplines has made some progress in this respect. For example, in the late 1990s, the compartmentalization of genes and all the basic biologic constituents used in protein expression within small aqueous droplets dispersed in an oil phase was demonstrated for the first time [75]. This technique was named in vitro compartmentalization [76] and elegantly mimicked the generation of proteins confined in tiny, cell-like compartments (figure 6a). Since this time, customized DNA templates and commercially available cell-free protein expression systems have been successfully combined into droplets formed within microfluidic formats, generating water-soluble proteins such as green fluorescent protein (GFP), rsGFP (red-shifted GFP) [77,82], organophosphorus hydrolase enzyme (OpdA) [78] and prokaryotic proteins with membrane affinity expressed both in single [79] and double emulsion formats [30,59,80] (figure 1h). In the same water-in-oil emulsions, Ichihashi et al. [83] demonstrated replication of genomic RNA, involving 600-generation replication experiments in which mutations were spontaneously introduced into RNA by replication error (figure 6b). In 2004, Noireaux et al. demonstrated bacterial cell-free protein expression in liposomes over extended periods. This was made possible by inserting pore channels within the lipid membrane that allowed exchange of material with an external feeding solution [84]. Similarly, in 2014, Arriaga et al. demonstrated the generation of GUVs within a droplet-based microfluidic device and the insertion of an α-haemolysin pore channel within the phospholipid membrane [85] (figure 6c). The ability of polymersomes to respond to external stimuli has also been achieved by inserting light-sensitive components into the polymeric layer, as demonstrated by Kamat and co-workers, who integrated a porphyrin protein into a polyethylene oxide–polybutadiene diblock copolymer membrane [86] (figure 6d). Subsequent addition of antibodies and ligands into the polymeric membrane then confers sensing and targeting properties [87] (figure 6e).
Figure 6.
Mimicking cellular processes in artificial cells. (a) Cell-free protein expression within a water-in-oil emulsion (1). Droplets contain genes attached to substrate molecules and all the material necessary for gene expression (2). When expressed, the genes produce enzymes able to convert the substrate into product (3). Once the emulsion is broken the genes can be collected (4) and, those of interest, isolated (5). (b) RNA replication in a water-in-oil emulsion. A typical cycle consists of replication in droplets and subsequent recovery of new RNA after emulsion breakage (i). In the accumulation method, the emulsion is fused with a fresh translation system, and the cycle repeated up to 600 times (ii). (c) Confocal microscope images of GUVs generated using a microfluidic capillary device and exposed to sulforhodamine in (i) the absence and (ii) the presence of the pore-forming protein α-haemolysin. (d) A light-sensitive membrane incorporating a porphyrin protein within the polymer bilayer. When light illuminates the porphyrin, the receptor modifies its configuration and generates a pore in the polymersome. (e) Insertion of antibodies into a polymersome membrane allows the generation of leucopolymersomes, which mimic the adhesive properties of leucocytes. (f) Mimicking an enzymatic pathway within a multicompartmentalized polymersome. (g) Vesicular self-reproduction induced by the addition of a membrane precursor, V*. This step creates membrane molecules and electrolytes through hydrolysis assisted by an amphiphilic catalyst. Adhesion of the amplified DNA to the inner layer accelerates vesicular growth and division. Images adapted from [77,83–88].
Multistep reactions that in a simple sense resemble natural enzymatic pathways have recently been performed within polymersome-in-polymersome structures generated via emulsion-centrifugation [88]. Specifically, functional eukaryotic cell mimics could be created by loading functional organelle mimics inside polymersomes, with the authors successfully demonstrating an original three-enzyme cascade reaction (figure 6f). Moreover, self-reproduction of supramolecular giant vesicles has also been elegantly demonstrated by Kurihara and co-workers, who initiate the amplification of encapsulated DNA within a self-reproducible cationic giant vesicle [81]. Through addition of a vesicular membrane precursor, growth and spontaneous division of the giant vesicle occurs and is accompanied by distribution of the DNA to the daughter giant vesicle (figure 6g). Finally, Lentini et al. [89] have also demonstrated the generation of artificial cell systems capable of sensing chemical signal and regulating gene expression of living cells like Escherichia coli. The artificial cell is therefore able to expand the senses of E. coli by translating a chemical message, that E. coli cannot sense on its own, to a molecule that activates a natural cellular response.
7. Conclusion
The intimate marriage and cooperation of science and technology are of fundamental importance in better understanding the structure and function of living cells. Artificial cells, minimal cells, protocells and other ‘cell-like’ systems not only provide insights into the origin of life [53,90], but also are beginning to have a dramatic impact in contemporary bioscience as cell and organ replacements [91] and novel therapeutic agents [92]. In this respect, it is clear that over the last decade, droplet-based microfluidic methods have shown much potential for generating and engineering such systems with high precision and in a manner compatible with relevant biological materials. Although technological developments are still in their infancy, the ever-increasing activity within the field suggests that droplet-based microfluidics will soon become an indispensable tool in the generation of sophisticated artificial cells.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Pohorille A, Deamer D. 2002. Artificial cells: prospects for biotechnology. Trends Biotechnol. 20, 123–128. ( 10.1016/S0167-7799(02)01909-1) [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Ruder WC, LeDuc PR. 2008. Artificial cells: building bioinspired systems using small-scale biology. Trends Biotechnol. 26, 14–20. ( 10.1016/j.tibtech.2007.09.006) [DOI] [PubMed] [Google Scholar]
- 3.Hammer DA, Kamat NP. 2012. Towards an artificial cell. FEBS Lett. 586, 2882–2890. ( 10.1016/j.febslet.2012.07.044) [DOI] [PubMed] [Google Scholar]
- 4.Caschera F, Noireaux V. 2014. Integration of biological parts toward the synthesis of a minimal cell. Curr. Opin. Chem. Biol. 22, 85–91. ( 10.1016/j.cbpa.2014.09.028) [DOI] [PubMed] [Google Scholar]
- 5.Chang TMS. 1964. Semipermeable microcapsules. Science 146, 524–525. ( 10.1126/science.146.3643.524) [DOI] [PubMed] [Google Scholar]
- 6.Chang TMS. 1998. Artificial cells with emphasis on cell encapsulation of genetically engineered cells. Artif. Organs 22, 958–965. ( 10.1046/j.1525-1594.1998.06243.x) [DOI] [PubMed] [Google Scholar]
- 7.Swi Chang TM. 1995. Artificial cells with emphasis on bioencapsulation in biotechnology. In Biotechnology annual review, vol. 1 (ed El-Gewely MR.), pp. 267–295. Amsterdam, The Netherlands: Elsevier. [DOI] [PubMed] [Google Scholar]
- 8.Szathmary E. 2005. Life: in search of the simplest cell. Nature 433, 469–470. ( 10.1038/433469a) [DOI] [PubMed] [Google Scholar]
- 9.Takinoue M, Takeuchi S. 2011. Droplet microfluidics for the study of artificial cells. Anal. Bioanal. Chem. 400, 1705–1716. ( 10.1007/s00216-011-4984-5) [DOI] [PubMed] [Google Scholar]
- 10.Jewett MC, Forster AC. 2010. Update on designing and building minimal cells. Curr. Opin. Biotechnol. 21, 697–703. ( 10.1016/j.copbio.2010.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitesides GM. 2006. The origins and the future of microfluidics. Nature 442, 368–373. ( 10.1038/nature05058) [DOI] [PubMed] [Google Scholar]
- 12.Teh S-Y, Lin R, Hung L-H, Lee AP. 2008. Droplet microfluidics. Lab Chip 8, 198–220. ( 10.1039/b715524g) [DOI] [PubMed] [Google Scholar]
- 13.Thorsen T, Roberts RW, Arnold FH, Quake SR. 2001. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 86, 4163–4166. ( 10.1103/PhysRevLett.86.4163) [DOI] [PubMed] [Google Scholar]
- 14.Huebner A, Sharma S, Srisa-Art M, Hollfelder F, Edel JB, deMello AJ. 2008. Microdroplets: a sea of applications? Lab Chip 8, 1244–1254. ( 10.1039/b806405a) [DOI] [PubMed] [Google Scholar]
- 15.Gulati S, Rouilly V, Niu X, Chappell J, Kitney RI, Edel JB, Freemont PS, deMello AJ. 2009. Opportunities for microfluidic technologies in synthetic biology. J. R. Soc. Interface 6, S493–S506. ( 10.1098/rsif.2009.0083.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon M, Lee A. 2012. Microfluidic droplet manipulations and their applications. In Microdroplet technology: principles and emerging applications in biology and chemistry (eds Day P, Manz A, Zhang Y). Integrated Analytical Systems, pp. 23–50. New York, NY: Springer; ( 10.1007/978-1-4614-3265-4_2) [DOI] [Google Scholar]
- 17.Theberge AB, Courtois F, Schaerli Y, Fischlechner M, Abell C, Hollfelder F, Huck WTS. 2010. Microdroplets in microfluidics: an evolving platform for discoveries in chemistry and biology. Angew. Chem. Int. Ed. 49, 5846–5868. ( 10.1002/anie.200906653) [DOI] [PubMed] [Google Scholar]
- 18.Mark D, Haeberle S, Roth G, von Stetten F, Zengerle R. 2010. Microfluidic lab-on-a-chip platforms: requirements, characteristics and applications. Chem. Soc. Rev. 39, 1153–1182. ( 10.1039/b820557b) [DOI] [PubMed] [Google Scholar]
- 19.Martino C, Zagnoni M, Sandison ME, Chanasakulniyom M, Pitt AR, Cooper JM. 2011. Intracellular protein determination using droplet-based immunoassays. Anal. Chem. 83, 5361–5368. ( 10.1021/ac200876q) [DOI] [PubMed] [Google Scholar]
- 20.Zheng B, Gerdts CJ, Ismagilov RF. 2005. Using nanoliter plugs in microfluidics to facilitate and understand protein crystallization. Curr. Opin. Struc. Biol. 15, 548–555. ( 10.1016/j.sbi.2005.08.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He M, Edgar JS, Jeffries GDM, Lorenz RM, Shelby JP, Chiu DT. 2005. Selective encapsulation of single cells and subcellular organelles into picoliter- and femtoliter-volume droplets. Anal. Chem. 77, 1539–1544. ( 10.1021/ac0480850) [DOI] [PubMed] [Google Scholar]
- 22.Schaerli Y, et al. 2008. Continuous-flow polymerase chain reaction of single-copy DNA in microfluidic microdroplets. Anal. Chem. 81, 302–306. ( 10.1021/ac802038c) [DOI] [PubMed] [Google Scholar]
- 23.Xu SQ, et al. 2005. Generation of monodisperse particles by using microfluidics: control over size, shape, and composition. Angew. Chem. Int. Ed. 44, 724–728. ( 10.1002/anie.200462226) [DOI] [PubMed] [Google Scholar]
- 24.Okushima S, Nisisako T, Torii T, Higuchi T. 2004. Controlled production of monodisperse double emulsions by two-step droplet breakup in microfluidic devices. Langmuir 20, 9905–9908. ( 10.1021/la0480336) [DOI] [PubMed] [Google Scholar]
- 25.Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA. 2005. Monodisperse double emulsions generated from a microcapillary device. Science 308, 537–541. ( 10.1126/science.1109164) [DOI] [PubMed] [Google Scholar]
- 26.Chang FC, Su YC. 2008. Controlled double emulsification utilizing 3D PDMS microchannels. J. Micromech. Microeng. 18, 065018 ( 10.1088/0960-1317/18/6/065018) [DOI] [Google Scholar]
- 27.Bauer WAC, Fischlechner M, Abell C, Huck WTS. 2010. Hydrophilic PDMS microchannels for high-throughput formation of oil-in-water microdroplets and water-in-oil-in-water double emulsions. Lab Chip 10, 1814–1819. ( 10.1039/c004046k) [DOI] [PubMed] [Google Scholar]
- 28.Chu LY, Utada AS, Shah RK, Kim JW, Weitz DA. 2007. Controllable monodisperse multiple emulsions. Angew. Chem. Int. Ed. 46, 8970–8974. ( 10.1002/anie.200701358) [DOI] [PubMed] [Google Scholar]
- 29.Joensson HN, Samuels ML, Brouzes ER, Medkova M, Uhlen M, Link DR, Andersson-Svahn H. 2009. Detection and analysis of low-abundance cell-surface biomarkers using enzymatic amplification in microfluidic droplets. Angew. Chem. Int. Ed. 48, 2518–2521. ( 10.1002/anie.200804326) [DOI] [PubMed] [Google Scholar]
- 30.Martino C, Horsfall L, Chen Y, Chanasakulniyom M, Paterson D, Brunet A, Rosser S, Yuan Y-J, Cooper JM. 2012. Cytoskeletal protein expression and its association within the hydrophobic membrane of artificial cell models. ChemBioChem 13, 792–795. ( 10.1002/cbic.201200038) [DOI] [PubMed] [Google Scholar]
- 31.Walde P. 2010. Building artificial cells and protocell models: experimental approaches with lipid vesicles. BioEssays 32, 296–303. ( 10.1002/bies.200900141) [DOI] [PubMed] [Google Scholar]
- 32.Dzieciol AJ, Mann S. 2012. Designs for life: protocell models in the laboratory. Chem. Soc. Rev. 41, 79–85. ( 10.1039/C1CS15211D) [DOI] [PubMed] [Google Scholar]
- 33.Brizard AM, van Esch JH. 2009. Self-assembly approaches for the construction of cell architecture mimics. Soft Matter 5, 1320–1327. ( 10.1039/b812388h) [DOI] [Google Scholar]
- 34.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. Molecular biology of the cell, 4th edn New York, NY: Garland Science. [Google Scholar]
- 35.Shum H, Thiele J, Kim S-H. 2014. Microfluidic fabrication of vesicles. In Advances in transport phenomena 2011 (ed. Wang L.). Advances in Transport Phenomena, vol. 3, pp. 1–28. Berlin, Germany: Springer International Publishing; ( 10.1007/978-3-319-01793-8_1) [DOI] [Google Scholar]
- 36.Elani Y, Law RV, Ces O. 2014. Vesicle-based artificial cells as chemical microreactors with spatially segregated reaction pathways. Nat. Commun. 5, 5305 ( 10.1038/ncomms6305) [DOI] [PubMed] [Google Scholar]
- 37.Dora Tang TY, Rohaida Che Hak C, Thompson AJ, Kuimova MK, Williams DS, Perriman AW, Mann S. 2014. Fatty acid membrane assembly on coacervate microdroplets as a step towards a hybrid protocell model. Nat. Chem. 6, 527–533. ( 10.1038/nchem.1921) [DOI] [PubMed] [Google Scholar]
- 38.van Swaay D, Tang TYD, Mann S, de Mello A. 2015. Microfluidic formation of membrane-free aqueous coacervate droplets in water. Angew. Chem. Int. Ed. 54, 8398–8401. ( 10.1002/anie.201502886) [DOI] [PubMed] [Google Scholar]
- 39.Walde P, Cosentino K, Engel H, Stano P. 2010. Giant vesicles: preparations and applications. ChemBioChem 11, 848–865. ( 10.1002/cbic.201000010) [DOI] [PubMed] [Google Scholar]
- 40.van Swaay D, deMello A. 2013. Microfluidic methods for forming liposomes. Lab Chip 13, 752–767. ( 10.1039/c2lc41121k) [DOI] [PubMed] [Google Scholar]
- 41.Yu B, Lee RJ, Lee LJ. 2009. Microfluidic methods for production of liposomes. Methods Enzymol. 465, 129–141. ( 10.1016/S0076-6879(09)65007-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Angelova MI, Dimitrov DS. 1986. Liposome electroformation. Faraday Disc. Chem. Soc. 81, 303–311. ( 10.1039/dc9868100303) [DOI] [Google Scholar]
- 43.Li C, Deng Y. 2004. A novel method for the preparation of liposomes: freeze drying of monophase solutions. J. Pharmaceut. Sci. 93, 1403–1414. ( 10.1002/jps.20055) [DOI] [PubMed] [Google Scholar]
- 44.Maulucci G, De Spirito M, Arcovito G, Boffi F, Castellano AC, Briganti G. 2005. Particle size distribution in DMPC vesicles solutions undergoing different sonication times. Biophys. J. 88, 3545–3550. ( 10.1529/biophysj.104.048876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan Y-C, Hettiarachchi K, Siu M, Pan Y-R, Lee AP. 2006. Controlled microfluidic encapsulation of cells, proteins, and microbeads in lipid vesicles. J. Am. Chem. Soc. 128, 5656–5658. ( 10.1021/ja056641h) [DOI] [PubMed] [Google Scholar]
- 46.Shum HC, Lee D, Yoon I, Kodger T, Weitz DA. 2008. Double emulsion templated monodisperse phospholipid vesicles. Langmuir 24, 7651–7653. ( 10.1021/la801833a) [DOI] [PubMed] [Google Scholar]
- 47.Matosevic S, Paegel BM. 2013. Layer-by-layer cell membrane assembly. Nat. Chem. 5, 958–963. ( 10.1038/nchem.1765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH, Fletcher DA. 2008. Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proc. Natl Acad. Sci. USA 105, 4697–4702. ( 10.1073/pnas.0710875105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Funakoshi K, Suzuki H, Takeuchi S. 2006. Lipid bilayer formation by contacting monolayers in a microfluidic device for membrane protein analysis. Anal. Chem. 78, 8169–8174. ( 10.1021/ac0613479) [DOI] [PubMed] [Google Scholar]
- 50.Pautot S, Frisken BJ, Weitz DA. 2003. Engineering asymmetric vesicles. Proc. Natl Acad. Sci. USA 100, 10 718–10 721. ( 10.1073/pnas.1931005100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matosevic S, Paegel BM. 2011. Stepwise synthesis of giant unilamellar vesicles on a microfluidic assembly line. J. Am. Chem. Soc. 133, 2798–2800. ( 10.1021/ja109137s) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paterson DJ, Reboud J, Wilson R, Tassieri M, Cooper JM. 2014. Integrating microfluidic generation, handling and analysis of biomimetic giant unilamellar vesicles. Lab Chip 14, 1806–1810. ( 10.1039/c4lc00199k) [DOI] [PubMed] [Google Scholar]
- 53.Luisi PL, Stano P. 2011. Synthetic biology: minimal cell mimicry. Nat. Chem. 3, 755–756. ( 10.1038/nchem.1156) [DOI] [PubMed] [Google Scholar]
- 54.Discher BM, Won Y-Y, Ege DS, Lee JC-M, Bates FS, Discher DE, Hammer DA. 1999. Polymersomes: tough vesicles made from diblock copolymers. Science 284, 1143–1146. ( 10.1126/science.284.5417.1143) [DOI] [PubMed] [Google Scholar]
- 55.Mecke A, Dittrich C, Meier W. 2006. Biomimetic membranes designed from amphiphilic block copolymers. Soft Matter 2, 751–759. ( 10.1039/b605165k) [DOI] [PubMed] [Google Scholar]
- 56.Lee JCM, Bermudez H, Discher BM, Sheehan MA, Won Y-Y, Bates FS, Discher DE. 2001. Preparation, stability, and in vitro performance of vesicles made with diblock copolymers. Biotechnol. Bioeng. 73, 135–145. ( 10.1002/bit.1045) [DOI] [PubMed] [Google Scholar]
- 57.Kamat NP, Katz JS, Hammer DA. 2011. Engineering polymersome protocells. J. Phys. Chem. Lett. 2, 1612–1623. ( 10.1021/jz200640x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meng F, Engbers GHM, Feijen J. 2005. Biodegradable polymersomes as a basis for artificial cells: encapsulation, release and targeting. J. Controlled Release 101, 187–198. ( 10.1016/j.jconrel.2004.09.026) [DOI] [PubMed] [Google Scholar]
- 59.Martino C, Kim S-H, Horsfall L, Abbaspourrad A, Rosser SJ, Cooper J, Weitz DA. 2012. Protein expression, aggregation, and triggered release from polymersomes as artificial cell-like structures. Angew. Chem. Int. Ed. 51, 6416–6420. ( 10.1002/anie.201201443) [DOI] [PubMed] [Google Scholar]
- 60.Martino C, Lee TY, Kim S-H, deMello AJ. 2015. Microfluidic generation of PEG-b-PLA polymersomes containing alginate-based core hydrogel. Biomicrofluidics 9, 024101 ( 10.1063/1.4914112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guan L, Rizzello L, Battaglia G. 2015. Polymersomes and their applications in cancer delivery and therapy. Nanomedicine 10, 2757–2780. ( 10.2217/nnm.15.110) [DOI] [PubMed] [Google Scholar]
- 62.Shum HC, Kim J-W, Weitz DA. 2008. Microfluidic fabrication of monodisperse biocompatible and biodegradable polymersomes with controlled permeability. J. Am. Chem. Soc. 130, 9543–9549. ( 10.1021/ja802157y) [DOI] [PubMed] [Google Scholar]
- 63.Shum HC, Zhao Y-J, Kim S-H, Weitz DA. 2011. Multicompartment polymersomes from double emulsions. Angew. Chem. 123, 1686–1689. ( 10.1002/ange.201006023) [DOI] [PubMed] [Google Scholar]
- 64.Kim S-H, Shum HC, Kim JW, Cho J-C, Weitz DA. 2011. Multiple polymersomes for programmed release of multiple components. J. Am. Chem. Soc. 133, 15 165–15 171. ( 10.1021/ja205687k) [DOI] [PubMed] [Google Scholar]
- 65.Thiele J, Abate AR, Shum HC, Bachtler S, Förster S, Weitz DA. 2010. Fabrication of polymersomes using double-emulsion templates in glass-coated stamped microfluidic devices. Small 6, 1723–1727. ( 10.1002/smll.201000798) [DOI] [PubMed] [Google Scholar]
- 66.Seth A, Béalle G, Santanach-Carreras E, Abou-Hassan A, Ménager C. 2012. Design of vesicles using capillary microfluidic devices: from magnetic to multifunctional vesicles. Adv. Mater. 24, 3544–3548. ( 10.1002/adma.201200757) [DOI] [PubMed] [Google Scholar]
- 67.Kim S-H, Kim JW, Kim D-H, Han S-H, Weitz DA. 2013. Polymersomes containing a hydrogel network for high stability and controlled release. Small 9, 124–131. ( 10.1002/smll.201201709) [DOI] [PubMed] [Google Scholar]
- 68.Martino C, Statzer C, Vigolo D, deMello A. 2015. Controllable generation and encapsulation of alginate fibers using droplet-based microfluidics. Lab Chip 16, 59–64. ( 10.1039/C5LC01150G) [DOI] [PubMed] [Google Scholar]
- 69.Merkle D, Kahya N, Schwille P. 2008. Reconstitution and anchoring of cytoskeleton inside giant unilamellar vesicles. ChemBioChem 9, 2673–2681. ( 10.1002/cbic.200800340) [DOI] [PubMed] [Google Scholar]
- 70.Maeda YT, Nakadai T, Shin J, Uryu K, Noireaux V, Libchaber A. 2012. Assembly of MreB filaments on liposome membranes: a synthetic biology approach. ACS Synth. Biol. 1, 53–59. ( 10.1021/sb200003v) [DOI] [PubMed] [Google Scholar]
- 71.Tsai F-C, Stuhrmann B, Koenderink GH. 2011. Encapsulation of active cytoskeletal protein networks in cell-sized liposomes. Langmuir 27, 10 061–10 071. ( 10.1021/la201604z) [DOI] [PubMed] [Google Scholar]
- 72.Pontani L-L, van der Gucht J, Salbreux G, Heuvingh J, Joanny J-F, Sykes C. 2009. Reconstitution of an actin cortex inside a liposome. Biophys. J. 96, 192–198. ( 10.1016/j.bpj.2008.09.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Viallat A, Dalous J, Abkarian M. 2004. Giant lipid vesicles filled with a gel: shape instability induced by osmotic shrinkage. Biophys. J. 86, 2179–2187. ( 10.1016/S0006-3495(04)74277-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Campillo C, Pepin-Donat B, Viallat A. 2007. Responsive viscoelastic giant lipid vesicles filled with a poly(N-isopropylacrylamide) artificial cytoskeleton. Soft Matter 3, 1421–1427. ( 10.1039/b710474j) [DOI] [PubMed] [Google Scholar]
- 75.Tawfik DS, Griffiths AD. 1998. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 16, 652–656. ( 10.1038/nbt0798-652) [DOI] [PubMed] [Google Scholar]
- 76.Kelly BT, Baret J-C, Taly V, Griffiths AD. 2007. Miniaturizing chemistry and biology in microdroplets. Chem. Commun. 1773–1788. ( 10.1039/B616252E) [DOI] [PubMed] [Google Scholar]
- 77.Courtois F, Olguin LF, Whyte G, Bratton D, Huck WTS, Abell C, Hollfelder F. 2008. An integrated device for monitoring time-dependent in vitro expression from single genes in picolitre droplets. ChemBioChem 9, 439–446. ( 10.1002/cbic.200700536) [DOI] [PubMed] [Google Scholar]
- 78.Zhu YG, Wu N, Courtois F, Surjadi R, Oakeshott J, Peat TS, Easton CJ, Abell C, Zhu Y. 2011. Enzyme synthesis and activity assay in microfluidic droplets on a chip. Eng. Life Sci. 11, 157–164. ( 10.1002/elsc.201000043) [DOI] [Google Scholar]
- 79.Chanasakulniyom M, Martino C, Paterson D, Horsfall L, Rosser S, Cooper JM. 2012. Expression of membrane-associated proteins within single emulsion cell facsimiles. Analyst 137, 2939–2943. ( 10.1039/c2an35047e) [DOI] [PubMed] [Google Scholar]
- 80.Ho KKY, Murray VL, Liu AP. 2015. Engineering artificial cells by combining HeLa-based cell-free expression and ultrathin double emulsion template. In Methods in cell biology, vol. 128 (eds Jennifer R, Wallace FM), pp. 303–318. New York, NY: Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurihara K, Tamura M, Shohda K-I, Toyota T, Suzuki K, Sugawara T. 2011. Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem. 3, 775–781. ( 10.1038/nchem.1127) [DOI] [PubMed] [Google Scholar]
- 82.Dittrich PS, Jahnz M, Schwille P. 2005. A new embedded process for compartmentalized cell-free protein expression and on-line detection in microfluidic devices. ChemBioChem 6, 811–814. ( 10.1002/cbic.200400321) [DOI] [PubMed] [Google Scholar]
- 83.Ichihashi N, Usui K, Kazuta Y, Sunami T, Matsuura T, Yomo T. 2013. Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat. Commun. 4, 2494 ( 10.1038/ncomms3494) [DOI] [PubMed] [Google Scholar]
- 84.Noireaux V, Libchaber A, Chaikin PM. 2004. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl Acad. Sci. USA 101, 17 669–17 674. ( 10.1073/pnas.0408236101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arriaga LR, Datta SS, Kim S-H, Amstad E, Kodger TE, Monroy F, Weitz DA. 2014. Ultrathin shell double emulsion templated giant unilamellar lipid vesicles with controlled microdomain formation. Small 10, 950–956. ( 10.1002/smll.201301904) [DOI] [PubMed] [Google Scholar]
- 86.Kamat NP, Robbins GP, Rawson JS, Therien MJ, Dmochowski IJ, Hammer DA. 2010. A generalized system for photo-responsive membrane rupture in polymersomes. Adv. Funct. Mater. 20, 2588–2596. ( 10.1002/adfm.201000659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robbins GP, Saunders RL, Haun JB, Rawson J, Therien MJ, Hammer DA. 2010. Tunable leuko-polymersomes that adhere specifically to inflammatory markers. Langmuir 26, 14 089–14 096. ( 10.1021/la1017032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peters RJRW, Marguet M, Marais S, Fraaije MW, van Hest JCM, Lecommandoux S. 2014. Cascade reactions in multicompartmentalized polymersomes. Angew. Chem. Int. Ed. 53, 146–150. ( 10.1002/anie.201308141) [DOI] [PubMed] [Google Scholar]
- 89.Lentini R, et al. 2014. Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat. Commun. 5, 4012 ( 10.1038/ncomms5012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sutherland JD. 2016. The origin of life—out of the blue. Angew. Chem. Int. Ed. 55, 104–121. ( 10.1002/anie.201506585) [DOI] [PubMed] [Google Scholar]
- 91.Chang TMS. 2004. Artificial cells for cell and organ replacements. Artif. Organs 28, 265–270. ( 10.1111/j.1525-1594.2004.47343.x) [DOI] [PubMed] [Google Scholar]
- 92.Chang TMS. 2005. Therapeutic applications of polymeric artificial cells. Nat. Rev. Drug Discov. 4, 221–235. ( 10.1038/nrd1659) [DOI] [PubMed] [Google Scholar]