Abstract
Bile acid CoA:amino acid N-acyltransferase (BAAT) is the terminal enzyme in the synthesis of bile salts from cholesterol and catalyzes the conjugation of taurine or glycine to bile acid CoA thioesters to form bile acid N-acylamidates. BAAT has a dual localization to the cytosol and peroxisomes, possibly due to an inefficient carboxy-terminal peroxisomal targeting signal (PTS), -serine-glutamine-leucine (-SQL). Mutational analysis was used to define the role of the carboxy terminus in peroxisomal localization and kinetic activity. Amidation activity of BAAT and BAAT lacking the final two amino acids (AAs) (BAAT-S) were similar, whereas the activity of BAAT with a canonical PTS sequence (BAAT-SKL) was increased >2.5-fold. Kinetic analysis of BAAT and BAAT-SKL showed that BAAT-SKL had a lower Km for taurine and glycine as well as a greater Vmax. There was no difference in the affinity for cholyl-CoA. In contrast to BAAT, BAAT-SKL forms bile acid N-acylamidates with β-alanine. BAAT-S immunoprecipitated when incubated with peroxisomal biogenesis factor 5 (Pex5) and rabbit anti-Pex5 antibodies; however, deleting the final 12 AAs prevented coimmunoprecipitation with Pex5, indicating the Pex5 interaction involves more than the -SQL sequence. These results indicate that even small changes in the carboxy terminus of BAAT can have significant effects on activity and substrate specificity.
Keywords: bile acid conjugation, bile acid amidation, taurine, glycine, N-acyltransferase, bile acid ligase, peroxisomal biogenesis factor 5, peroxisomal targeting signal
Bile acids are weak organic acids and the end products of cholesterol metabolism. Their role as biological detergents is critical for the digestion and solubilization of dietary fats and their metabolic products. Under normal physiological conditions, over 99% of bile acids in bile are found conjugated to the amino acids (AAs), glycine and taurine (1). Conjugation of bile acids with glycine or taurine lowers their pKa, facilitating excretion into bile and allowing the bile acids to maintain aqueous solubility in the wide range of pH conditions throughout the intestinal tract to ensure the absorption of fat and fat-soluble vitamins.
Bile acid amidation involves the successive action of two enzymes: bile acid CoA ligase and bile acid CoA:amino acid N-acyltransferase (BAAT). Bile acid CoA ligase catalyzes the formation of a bile acid acyl-thioester between CoA and a 27- or 24-carbon bile acid (2). BAAT mediates the conjugation of a CoA-activated bile acid with an AA in a reaction that involves two steps. In the first step, BAAT binds to the bile acid-CoA and hydrolyzes the thioester bond of the CoA moiety, transferring the bile acid linkage to a cysteine (Cys) residue in the enzyme. In the second step, BAAT catalyzes the addition of the AA to the bile acid, forming a bile salt (N-acylamidate) through an amide linkage. BAAT is a multifunctional enzyme that can catalyze reactions as a thioesterase or N-acyltransferase (3). BAAT has been reported to use a catalytic triad of Cys-235, His-362, and Asp-328 (3), similar to the catalytic triads in thioesterase enzymes that use a Ser-His-Asp motif (4). BAAT also has high homology (≥40%) to several peroxisomal, mitochondrial, and cytosolic long-chain acyl-CoA thioesterases or acyl-CoA thioesterases (3, 4).
To date, the investigation of the AA residues affecting BAAT activity has been limited to residues considered as part of the active site located near the interior of the protein (3). The carboxy (C)-termini of N-acetyltransferases have been shown to be critical for activity, and the final three AAs of BAAT are thought to associate with peroxisomal biogenesis factor 5 (Pex5) and facilitate translocation to peroxisomes (5, 6). Approximately 25–30% of BAAT in hepatocytes is localized to peroxisomes; the remainder is in the cytosol (7–9). The majority of proteins targeted to peroxisomes utilize a three AA carboxy-terminal peroxisomal targeting signal (PTS)-1 sequence that interacts with Pex5 (10, 11). Typically, the PTS-1 sequence is -serine/alanine-arginine/lysine-leucine/methionine [S/A][R/K][L/M] located at the C-terminus (12); however, BAAT contains a degenerate PTS-1 sequence, -serine-glutamine-leucine (-SQL) (13). Because approximately 25% of BAAT is localized in peroxisomes (7–9), it was hypothesized that the BAAT-SQL sequence was responsible for a lower level of binding to Pex5 and, thus, a limited targeting of BAAT to peroxisomes. Here, the changes in the activity and properties of BAAT conferred by specific mutations in the C-terminus of the protein are examined. Relatively minor changes in the sequence of BAAT at the C-terminus had significant effects on BAAT activity and substrate selectivity, indicating the importance of this region in the catalytic properties of the enzyme. A point mutation in the penultimate C-terminal residue of BAAT, resulting in formation of a canonical Pex5 binding sequence (BAAT-SKL), increased amidation activity, thioesterase activity, and resistance to thermal inactivation. The PTS sequence was not necessary for interacting with Pex5, as evaluated by immunoprecipitation; however, this sequence plays an important role in the recognition of AAs capable of conjugating the thiol-bile acid intermediate.
MATERIALS AND METHODS
Materials
Our laboratories have previously reported the cloning and expression of human liver BAAT (hBAAT) (13), rat liver BAAT (14), and mouse liver BAAT (15). The goat anti-human Pex5 polyclonal antibody and ExactaCruz C were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The [α-32P]dCTP (3,000 Ci/mmol) and [3H]taurine (31 Ci/mmol) were from Amersham Pharmacia Biotech (Piscataway, NJ). The rabbit anti-mouse BAAT antiserum was generated as described previously (14). Cloned human Pex5S cDNA (AK225126) in a PCMV6-XL4 vector was purchased from Origene (Rockville, MD).
Construction of BAAT mutants
To construct C-terminal mutants, a pKK233-2-hBAAT construct (13) was utilized for PCR mutagenesis. To disrupt the PTS-1 sequence, the Gln417 residue was changed to a termination codon by converting nucleotide 1247 from a C→T using the forward primer 5′-CAGACCATGGTCCAGTTGACAGCTAC-3′ and reverse primer 5′-CCCAAGCTTGGGCTTAGAGTTAACTGGTCACATCTGG-3′. The generated TAA termination codon is in bold. The forward primers contained an NcoI restriction site (underlined) and the reverse primers contained a HindIII restriction site (underlined) to facilitate cloning into pKK233-2 for bacterial expression. To change the wild-type BAAT PTS-1 sequence (-SQL) to a more canonical sequence (-SKL), Gln417 was changed to a Lys by mutating the hBAAT sequence from CAA to AAA using the forward primer 5′-CAGACCATGGTCCAGTTGACAGCTAC-3′ and the reverse primer 5′-CCCAAGCTTTGGGCTTAGAGTTTACTGGTCACATCTGG-3′. The generated Lys codon is in bold. The final 12 AAs were removed from the C terminus of BAAT by using the forward primer 5′-CAGACCATGGTCCAGTTGACAGCTAC-3′ and the reverse primer 5′-CAAGCTTGTTAGAGAAATCTCTGGATCTCCTTCCAAG -3′ to introduce a termination codon at AA position 407 by mutating the nucleotide 1204 from C→A. The termination codon is in bold. The first exon of BAAT (147 AA) was deleted by amplifying the remaining codons using the forward primer 5′-CCATGGGAGCTCTTTCTCCCTCCAG-3′ and the reverse primer 5′-CCCAAGCTTTGGGCTTAGAGTTGACTGGTCACATCTGG-3′. The truncated BAAT sequence was subcloned into pKK233-2 for expression.
Bacterial expression and purification of hBAAT
To obtain active purified hBAAT protein, an overnight culture of Escherichia coli DH5α containing a pKK-hBAAT plasmid previously constructed by our laboratory (13) was diluted 1:100 in LB broth containing 100 μg/ml ampicillin. This culture was grown to an OD600 = 0.5. IPTG was added to a final concentration of 0.5 mM to induce expression of hBAAT, and the culture incubated an additional 2 h. The culture was centrifuged at 3,000 g for 15 min to pellet the bacteria; then the pellet was resuspended in bacterial lysis buffer [75 mM Tris (pH 8), 250 mM sucrose, 0.25 mM EDTA, 0.02 mg/ml lysozyme] and incubated for 20 min on ice. The lysate was centrifuged at 3,000 g at 4°C for 10 min, resuspended in TEA buffer [10 mM triethanolamine (pH 8), 1.5 mM DTT, 10% glycerol, protease inhibitors (Complete Mini; Roche Biosciences)], and sonicated on ice four times for 10 s each with 30 s in between pulses. The lysed bacteria were centrifuged at 100,000 g for 1 h, and the resulting cytosolic fraction was retained.
For further purification of expressed hBAAT, the bacterial cytosol was loaded onto a 17 × 1.5 cm DEAE-Sepharose Cl-6B column. The column was washed with 30 ml of TEA buffer, and hBAAT was eluted with a 0–200 mM NaCl gradient and collected in 3 ml fractions. Fractions containing hBAAT were identified by BAAT activity assays and immunoblotting. Fractions enriched in BAAT activity were pooled and used for characterization.
Immunoblot analysis
For immunoblot analysis, protein fractions (100 μg) were resolved by SDS-PAGE and electrotransferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat milk followed by incubation with a 1:5,000 dilution of rabbit anti-mouse BAAT antibody, described previously (14), or a commercially available goat anti-human Pex5 antibody (Santa Cruz). Membranes were then incubated with a 1:20,000 dilution of goat anti-rabbit IgG conjugated with horseradish peroxidase (for BAAT antibody) or a 1:20,000 dilution of donkey anti-goat IgG conjugated with horseradish peroxidase (for Pex5 antibody). Immunoconjugates were visualized using the Supersignal West Pico system (Pierce).
Bacterial expression and purification of human Pex5S
Cloned human Pex5S cDNA (AK225126) in a PCMV6-XL4 vector was purchased from Origene (Rockville, MD). For bacterial expression, Pex5 was amplified by PCR using the PCMV6-XL4-Pex5 vector as a template with the forward primer 5′-CCCGGGCCATGGCAATGCGGAG-3′ and the reverse primer 5′-GTCGACTCACTGGGGCAGGCCAAAC-3′. The forward primer contained an XmaI restriction site (underlined) and the reverse primer contained a SalI restriction site (underlined) to facilitate cloning into the mammalian expression vector, pQE-31 (Qiagen, Valencia, CA). PCR products were blunt-end ligated into the pGEM T-Easy vector, and the vector transfected into E. coli DH5α. Bacterial colonies were isolated and sequenced at the University of Alabama at Birmingham Genomics Core Facility to identify Pex5S. The Pex5S cDNA was removed from the pGEM vector by digestion with XmaI and SalI and ligated into pQE-31, which was digested using the same restriction sites. E. coli DH5α was transformed with the Pex5S-pQE vector, and Pex5S was expressed and purified in a similar manner as described for BAAT, with the exception of using a 0–500 mM NaCl gradient to elute the protein. Fractions containing Pex5S were identified via immunoblot analysis using a goat anti-human Pex5 antibody, and fractions containing high levels of Pex5S were pooled and stored at −70°C until used.
Immunoprecipitation
To immunoprecipitate Pex5, recombinant Pex5S protein (alone or in the presence of recombinant BAAT protein) was incubated for 3 h at 4°C with Protein A Sepharose beads bound to a goat anti-human Pex5 antibody, according to the ExactaCruz protocol. After incubation, the beads were washed three times by resuspension in 500 μl 1× PBS followed by centrifugation at 5,000 g before being dissolved in 2× Laemmli sample buffer. The samples were boiled for 5 min before undergoing SDS-PAGE analysis.
BAAT activity radioassay
BAAT amidation activity was determined using the radioassay described by Johnson, Barnes, and Diasio (16), in which [3H]taurine or [3H]glycine are conjugated to unlabeled cholyl-CoA to form 3H-labeled bile acid conjugates. The standard assay mixture contained 100 mM potassium phosphate (pH 8.4), 0.45 mM cholyl-CoA, and the corresponding 3H-labeled AA (0.025 μCi) in a final concentration of 0.250 mM in a total volume of 50 μl. Reactions were initiated by the addition of cholyl-CoA, incubated at 37°C for 15 min, and terminated by addition of 0.5 ml 100 mM sodium phosphate (pH 2.0) containing 1% SDS. Radioactive conjugates were then extracted from unreacted labeled AA with water-saturated n-butanol and quantified by scintillation spectroscopy. Cholyl-CoA was synthesized from cholic acid and CoA by the method of Shah and Staple (17), with modifications as described previously (18).
LC/ESI/MRM for measurement of BAAT activity
Our laboratories have previously described a procedure to simultaneously measure BAAT N-acyltransferase and thioesterase activity (3). Thioesterase activity is measured by the accumulation of free bile acid after BAAT cleaves the CoA moiety from the bile acid-CoA thioester. The N-acyltransferase (amidation) activity is a measurement of the bile acid amidate formed after the conjugation of the bile acid with an AA. A reaction mixture was prepared containing potassium phosphate buffer (pH 8.4), 250 μM taurine, and 60 μM purified cholyl-CoA. The reactions were initiated by the addition of the appropriate BAAT protein and incubated for 15 min at 37°C. The reactions were terminated by addition of 60 μl of 100% methanol per 100 μl reaction volume. Internal standards were then added to aliquots of reaction mixture and stored at −20°C prior to LC/ESI/MRM MS analysis. Because taurine was used as a substrate, the internal standards were taurodeoxycholic acid and chenodeoxycholic acid. Aliquots of the reactions containing internal standards were then injected into a reverse-phase column (RP-300 C8, 4.6 × 100 mm) by an autosampler. Solvent A was 10 mM ammonium acetate in 10% aqueous acetonitrile and solvent B was 10 mM ammonium acetate in 100% acetonitrile. The samples were run on a linear gradient from 100% solvent A to 100% solvent B to elute and introduced into the ESI interface of the API 3 triple quadrupole mass spectrometer (Sciex, Concord, Ontario, Canada) operating in the negative ion mode. The precursor ion/product ion transitions (m/z 407/343 for cholic acid; m/z 391/345 for chenodeoxycholic acid; m/z 514/124 for taurocholic acid; m/z 498/124 for taurodeoxycholic acid) were monitored to detect the formation of cholic acid and taurocholic acid.
To examine the reaction products of the different BAAT forms with nortaurine and homotaurine, the reaction described for the radioassay was used, but taurine was replaced by nortaurine or homotaurine. After incubation for 10 min, enzyme reactions were stopped by addition of 500 μl of methanol, and aliquots of the supernatant infused into the mass spectrometer in 50% methanol-0.1% acetic acid, recording negative mass spectra between m/z 400 and 600.
RESULTS
hBAAT mutants
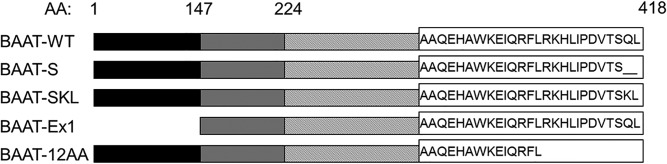
To examine the role of the BAAT-SQL and the adjacent sequence in Pex5 binding and regulation of enzymatic activity, several sequence variants were made to the carboxy terminal of BAAT. A truncated form of hBAAT was generated by removing the terminal Gln417 and Leu418 residues of the PTS sequence, resulting in a carboxy terminus ending in Ser, termed hBAAT-S (Fig. 1). Conversely, the penultimate residue of wild-type BAAT, the Gln of the PTS sequence was changed to a Lys to generate BAAT with a canonical PTS sequence, hBAAT-SKL (Fig. 1). The 12 carboxy-terminal AAs (407-418) were also removed (hBAAT-12AA Trunc) because Brocard and Hartig (19) had reported that a dodecapeptide sequence was necessary for the targeting of certain proteins to peroxisomes instead of a three AA PTS-1 sequence. Additionally, an N-terminal amino (N)-terminal truncation of BAAT (hBAAT-Ex1Del) was made by deleting the AAs encoded by the first exon (147 AAs) to investigate the possibility that a PTS sequence is located near the N terminus (Fig. 1).
Fig. 1.
Schematic representation of BAAT mutants. Wild-type BAAT (BAAT-WT), a two AA C-terminal truncation (BAAT-S), a glutamine (Q) to lysine (K) point mutation in the penultimate residue C-terminal residue (BAAT-SKL), deletion of the first exon (BAAT-Ex1), and a truncation of the final 12 C-terminal AAs (BAAT-12AA) in human BAAT enzyme were generated via PCR mutagenesis as described in the Materials and Methods section. Exon 1 (black bars, AAs 1-147), Exon 2 (gray bars, AAs 148-224), Exon 3 (striped bars, AAs 225-418), and the sequence alignment of the final 26 AAs as single letter codes of wild-type BAAT with the C terminus of each of the mutants are shown.
Mutations in the C-terminus of hBAAT affect enzyme activity
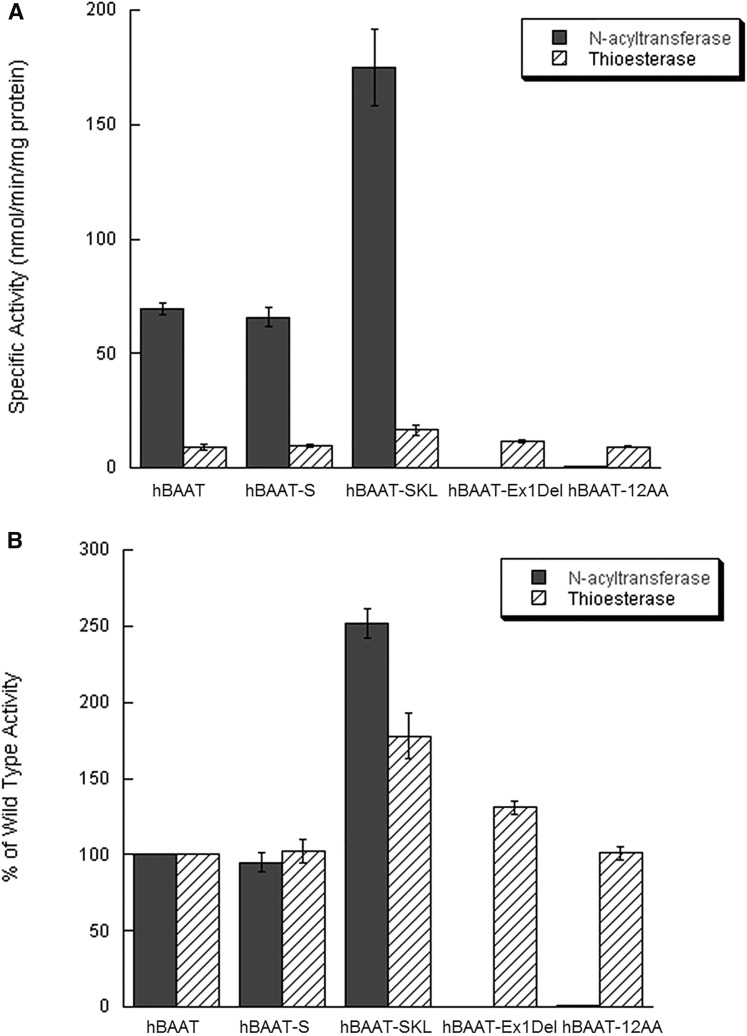
Investigation into the effect of these mutations on enzymatic activity was warranted due to the close proximity of carboxy-terminal mutations to the active site His-362 in BAAT (3). BAAT is capable of catalyzing both N-acyltransferase and thioesterase reactions (3); therefore, simultaneous measurement of both activities was performed using wild-type BAAT, BAAT-S, BAAT-SKL, BAAT-12AA Trunc, and BAAT-Ex1Del. BAAT-SKL was found to have greater than a 2.5-fold increase in N-acyltransferase activity compared with wild-type BAAT using cholyl-CoA and taurine as substrates (Fig. 2A). Deletion of the final two AAs in BAAT-S did not significantly alter N-acyltransferase activity as compared with wild-type BAAT. In contrast, BAAT-12AA Trunc and BAAT-Ex1Del possessed no detectable N-acyltransferase activity, although both enzymes still showed thioesterase activity (Fig. 2A). When thioesterase activities were measured by the production of free cholic acid, BAAT-SKL showed a 78% increase as compared with wild-type BAAT (Fig. 2B). BAAT-S and BAAT-12AA Trunc also had thioesterase activities similar to wild-type BAAT. In contrast, BAAT-Ex1Del showed a 31% increase in thioesterase activity compared with wild-type BAAT (Fig. 2B).
Fig. 2.
Enzymatic activity of BAAT mutants. BAAT-specific N-acyltransferase and thioesterase activities were measured simultaneously for purified recombinant BAAT forms, wild-type, BAAT-S, BAAT-SKL, BAAT-Ex1Del, and BAAT-12AA Trunc, via MS. A: Thioesterase activity (striped bars) was measured by the accumulation of free cholic acid while N-acyltransferase activity was measured by the production of taurocholate (gray bars). B: Specific activity values are expressed as a percentage of wild-type enzyme. Error bars represent the standard error of the mean as calculated for four separate identical experiments.
Kinetic analysis of C-terminal hBAAT mutations
Because mutation of the carboxy-terminus of BAAT resulted in changes in amidation activity, the kinetic parameters of the N-acyltransferase reaction were investigated. The amidase reaction involves two substrates, and changes in activity could be due to an increase in cholyl-CoA binding, an increase in cholyl-CoA turnover, or an increase in the affinity for the AA substrate. The concentration of cholyl-CoA is also important to the reaction, as previous kinetic analysis of the reaction involving the conversion of cholyl-CoA to taurocholate or glycocholate by BAAT suggests a Tetra Uni ping-pong mechanism (20). Therefore, three Michaelis constants (Km) were determined for each form of BAAT (hBAAT, hBAAT-S, and hBAAT-SKL) demonstrating N-acyltransferase activity: one for cholyl-CoA with taurine as substrate, one for glycine, and one for taurine.
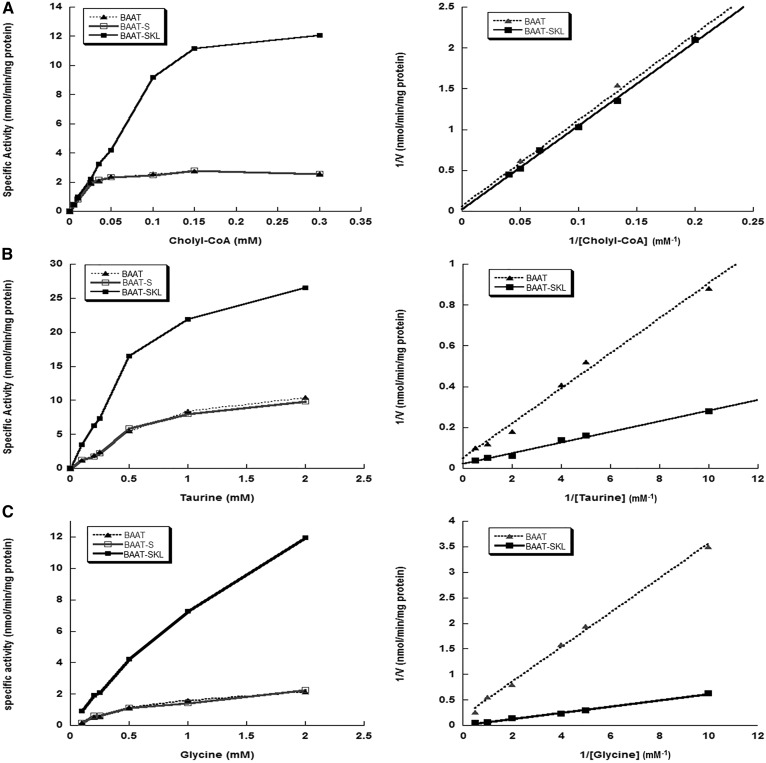
Wild-type BAAT and BAAT-S had similar Km and Vmax values using cholyl-CoA and glycine or taurine as substrates (Fig. 3, Table 1). Wild-type BAAT and BAAT-SKL also had similar Kms for cholyl-CoA; however, the Km values determined for both taurine and glycine with BAAT-SKL were approximately 50% lower (Fig. 3, Table 1). The Vmax for N-acetyltransferase activity catalyzed by BAAT-SKL was about 2.5-fold greater than that for wild-type BAAT. Furthermore, the comparison of specific activity versus cholyl-CoA concentration demonstrated BAAT-SKL activity was not saturated until the cholyl-CoA concentration reached 300 μM, as compared with wild-type BAAT that is saturated at a cholyl-CoA concentration of 50 μM (Fig. 3A).
Fig. 3.
Kinetic analysis of N-acyltransferase activity of BAAT PTS-1 mutants. Concentration curves (left panels) and double reciprocal plots (right panels) of reaction rates involving recombinant purified wild-type BAAT, BAAT-S, and BAAT-SKL with varying substrate concentrations of cholyl-CoA, taurine, and glycine. A: BAAT N-acyltransferase activity of wild-type BAAT (filled triangles), BAAT-S (open squares), and BAAT-SKL (filled squares) was measured via radioassay in the presence of 0.25 mM [3H]taurine and 0, 5, 10, 25, 35, 50, 100, 150, and 300 μM cholyl-CoA. B: BAAT N-acyltransferase activity of wild-type BAAT (filled triangles), BAAT-S (open squares), and BAAT-SKL (filled squares) was measured via radioassay in the presence of 300 μM cholyl-CoA and 0.1, 0.2, 0.25, 0.5, 1.0, and 2.0 mM [3H]taurine. C: BAAT N-acyltransferase activity of wild-type BAAT (filled triangles), BAAT-S (open squares), and BAAT-SKL (filled squares) was measured via radioassay in the presence of 300 μM cholyl-CoA and 0.1, 0.2, 0.25, 0.5, 1.0, and 2.0 mM [3H]glycine. All activities are expressed in units of nanomoles per minute per milligram protein.
TABLE 1.
Michaelis-Menten kinetic parameters based on double reciprocal Lineweaver-Burke plots pictured in Fig. 3
| Taurine | Glycine | Cholyl-CoA | ||||
| Km (mM) | Vmax (nmol/min/mg) | Km (mM) | Vmax (nmol/min/mg) | Km (mM) | Vmax (nmol/min/mg) | |
| BAAT | 1.86 | 21.7 | 3.95 | 8.69 | 0.185 | 17.6 |
| BAAT-S | 1.77 | 22.5 | 4.08 | 8.41 | 0.180 | 17.8 |
| BAAT-SKL | 1.01 | 49.0 | 2.14 | 38.6 | 0.185 | 34.6 |
Wild-type BAAT, BAAT-S, and BAAT-SKL were expressed in E. coli and purified via DEAE column chromatography. The rates of bile acid conjugate formation were measured by radioassay using radioactive AA.
Substrate analysis of BAAT mutants
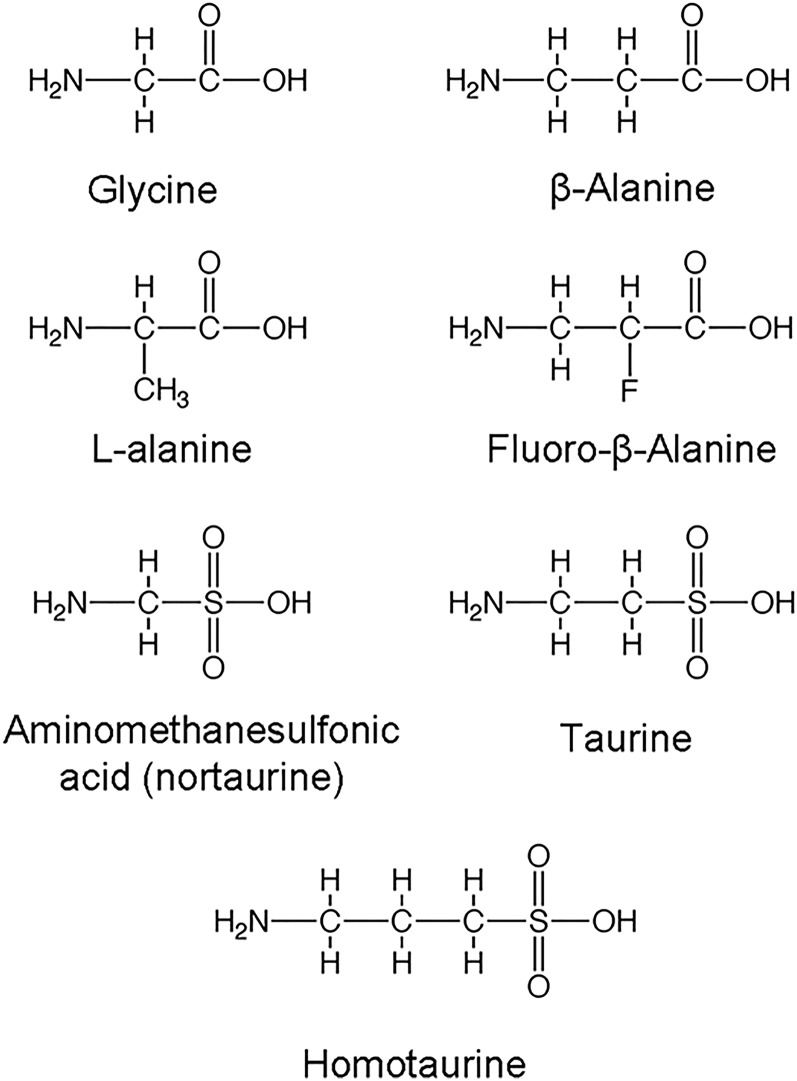
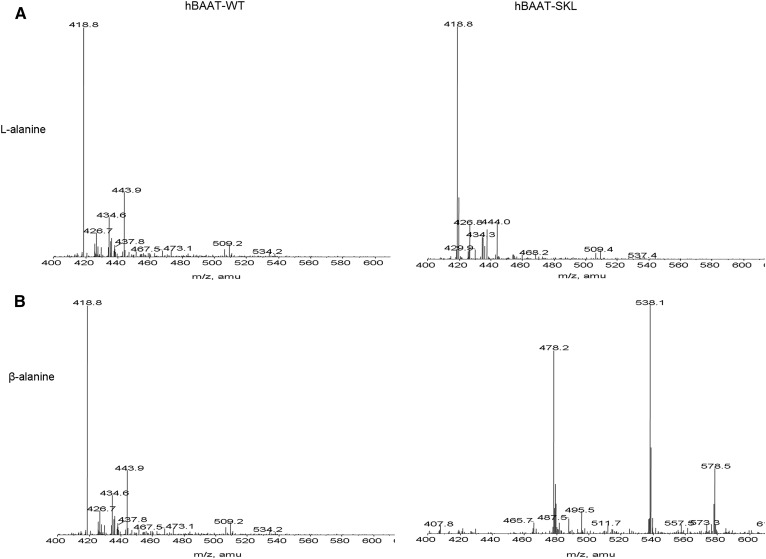
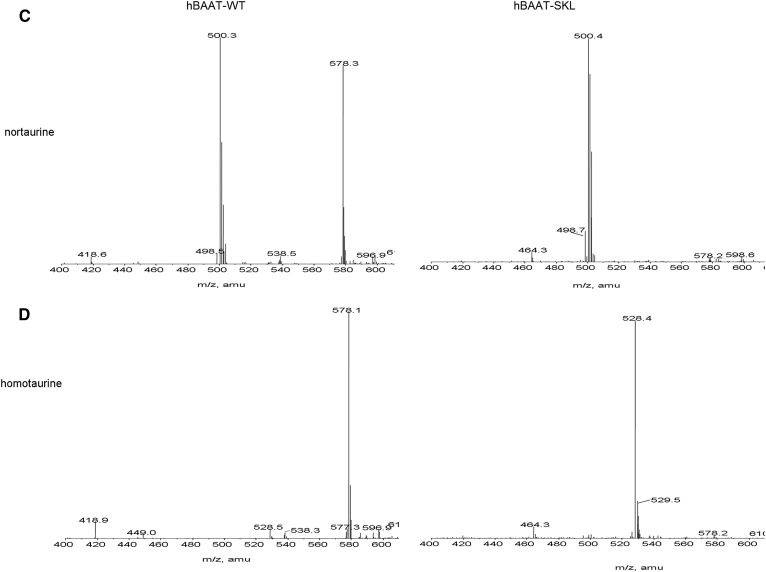
Kinetic characterization of BAAT-SKL indicates that small sequence changes in the PTS-1 significantly change the affinity of the enzyme for glycine and taurine, but not cholyl-CoA. Enzymatic studies have demonstrated that few AAs can be used as substrates for BAAT, with the most common in mammals being taurine, glycine, or both, depending on the species (13, 20–22). Our laboratories have previously reported that human BAAT can also use 2-fluoro-β-alanine as a substrate, but not β-alanine (23). If BAAT-SKL has greater affinity for the known substrates glycine and taurine than wild-type BAAT, it is possible that it will accept other potential AAs as substrates. To determine whether mutating the carboxy-terminus of BAAT results in the ability to utilize other substrates, compounds structurally related to taurine and glycine were tested as substrates. The compounds tested were β-alanine, D-alanine, L-alanine, 3-amino-1-propanesulfonic acid (homotaurine), and amino-methanesulfonic acid (nortaurine) (Fig. 4). Reaction products catalyzed by the different BAAT mutants were analyzed qualitatively by LC/ESI/MRM MS to detect formation of cholate conjugates. A positive identification of a substrate for BAAT was made when a peak was apparent at the appropriate molecular mass (atomic mass units) indicating the addition of the compound to cholic acid minus the molecular mass of water. No peak was detected at 478.6 m/z when L-alanine was used in the assay in the presence of cholyl-CoA and either wild-type hBAAT or hBAAT-SKL (Fig. 5A). When β-alanine was incubated with cholyl-CoA in the presence of hBAAT-SKL, however, a peak was detected at 478.6 m/z, indicative of cholic acid conjugated with β-alanine (Fig. 5B). In contrast, no peak was detected when β-alanine was incubated with cholyl-CoA and wild-type BAAT (Fig. 5B). Both wild-type BAAT and BAAT-SKL were capable of conjugating nortaurine to cholic acid, although the activity of BAAT-SKL was apparently greater due to the disappearance of cholyl-CoA (m/z 578.3) (Fig. 5C). For homotaurine, wild-type BAAT had very little conjugating activity with cholyl-CoA (expected ion at m/z 528.3). In contrast, BAAT-SKL completely converted cholyl-CoA to its homotaurine conjugate, as judged by the absence of the m/z 578.3 ion. These results indicate that hBAAT can use nortaurine as a substrate, whereas hBAAT-SKL can use nortaurine and homotaurine, as well as β-alanine, as substrates. Nonetheless, neither wild-type BAAT nor BAAT-SKL could utilize L-alanine as a substrate to conjugate cholyl-CoA.
Fig. 4.
Amino acid substrate candidates for hBAAT mutants. Representative chemical structures for possible AA substrates for wild-type hBAAT, hBAAT-S, and hBAAT-SKL. Glycine, taurine, and fluoro-β-alanine have previously been shown to be substrates for hBAAT (23).
Fig. 5.
MS analysis of BAAT AA substrates. Resulting qualitative mass spectra of reactions of AA conjugates of cholyl-CoA incubated with purified recombinant wild-type hBAAT (left panels) or hBAAT-SKL (right panels) in the presence of L-alanine (A), β-alanine (B), nortaurine (C), or homotaurine (D). Reactions were initiated by the addition of enzyme and stopped with 500 μl of methanol before infusion for MS analysis. The ion at m/z 478.2 is the [M-H]− ion for cholyl-β-alanine. The m/z 538 ion is the [M-H+CH3COOH]− adduct. The m/z 578 ion is doubly charged cholyl-CoA. The ion at m/z 500.3 is the nortaurine conjugate of cholic acid.
Thermostability of BAAT in vitro and in vivo
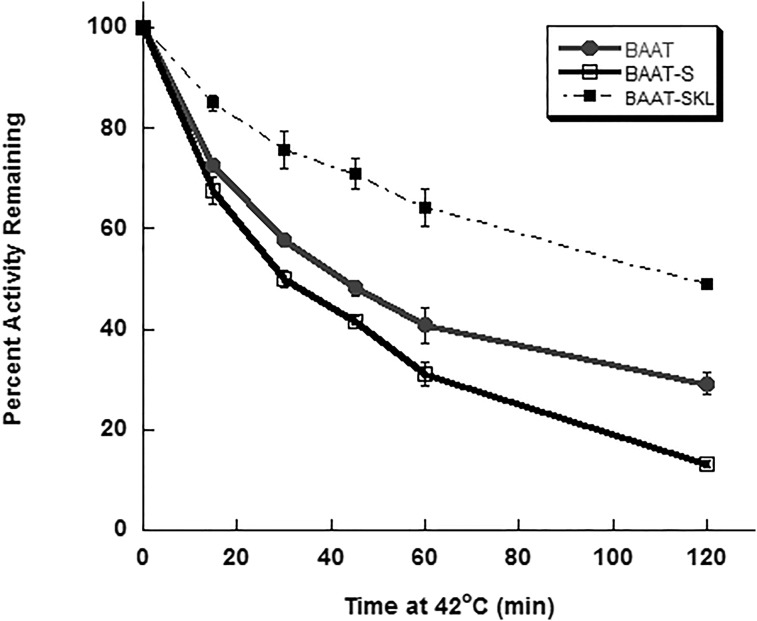
Thermal inactivation of enzymatic activity can reflect changes in the structure of proteins and the stability of the tertiary structure. To discern the role of the carboxy terminus of BAAT in the stability of N-acetyltransferase activity, purified BAAT, BAAT-S, and BAAT-SKL were incubated for increasing lengths of time at 42°C before measuring N-acyltransferase activity. BAAT-S showed a significant increase in the loss of N-acetyltransferase activity as compared with wild-type BAAT (Fig. 6). In contrast, hBAAT-SKL possessed a greater resistance to thermal inactivation than wild-type BAAT (Fig. 6). These results suggest that the C-terminus of BAAT has a significant influence on the thermostability of the enzyme in vitro.
Fig. 6.
In vitro and in vivo thermostability of BAAT N-acyltransferase activity. Purified recombinant wild-type hBAAT (filled circles), hBAAT-S (open squares), and hBAAT-SKL (filled squares) were assayed for BAAT N-acyltransferase activity after incubation at 42°C for 0, 15, 30, 45, 60, or 120 min. Data are expressed as a percentage of control activity (0 h at 42°C) for each BAAT form.
Pex5 interacts with BAAT and inhibits activity
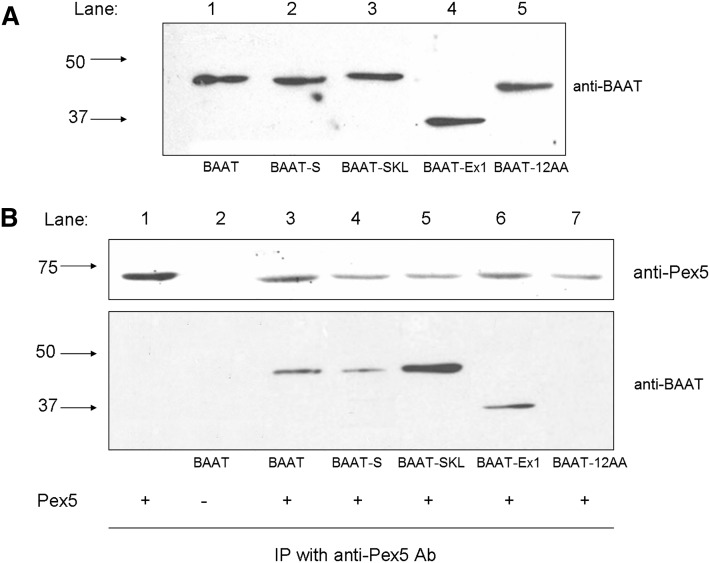
When the expressed BAAT mutants were resolved by SDS-PAGE, BAAT-12AA Trunc and BAAT-Ex1Del exhibited electrophoretic mobilities different from the other forms of BAAT (Fig. 7A). To determine whether Pex5 and the BAAT mutants could interact in vitro, purified wild-type BAAT and the four BAAT mutants were incubated with purified recombinant human Pex5S for 1 h at 37°C, then immunoprecipitated with the goat anti-Pex5 antibody. The precipitates were then immunoblotted with the rabbit anti-BAAT antibody. Wild-type BAAT, BAAT-S, BAAT-SKL, and BAAT-Ex1Del all immunoprecipitated after incubation with Pex5S using the goat anti-Pex5 antibody, whereas hBAAT-12AA Trunc did not (Fig. 7B).
Fig. 7.
Interaction between Pex5 and BAAT mutants. Human wild-type and mutant BAAT forms were expressed in E. coli, purified by DEAE-column chromatography, and analyzed by immunoblot analysis. A: Wild-type BAAT (BAAT), BAAT-S, BAAT-SKL, BAAT-Ex1Del, and BAAT-12AA Trunc were separated via SDS-PAGE analysis before being immunoblotted with a rabbit anti-mouse BAAT antibody (1:5,000). The position of molecular mass standards of 50 kDa and 37 kDa are indicated to the left of the immunoblot. B: Recombinant wild-type BAAT (BAAT), BAAT-S, BAAT-SKL, BAAT-Ex1Del, and BAAT-12AA Trunc protein was incubated with (+) or without (−) an excess of Pex5S purified by DEAE-column chromatography and then immunoprecipitated (IP) with a goat-anti-human Pex5 antibody. The samples were probed with anti-mouse BAAT and anti-Pex5 antibodies as indicated.
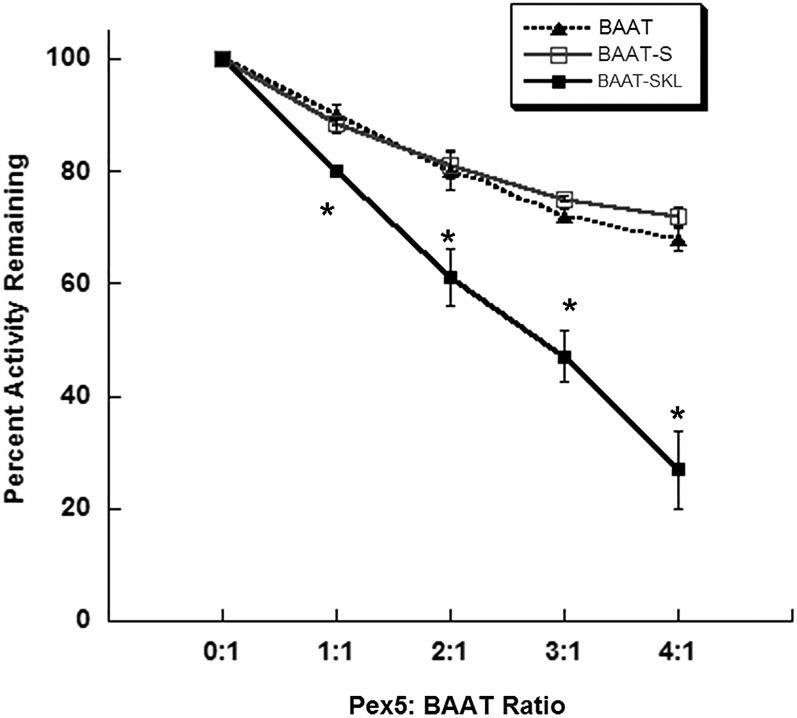
Because Pex5 can bind to BAAT, apparently at the carboxy-terminus, and the last 12 AAs of the carboxy terminus are necessary for N-acyltransferase activity, Pex5 binding was evaluated for a role in the inhibition of N-acyltransferase activity in the BAAT mutants. BAAT N-acyltransferase activity was measured after incubating purified wild-type BAAT, BAAT-S, and BAAT-SKL with varying levels of purified recombinant Pex5S. At equivalent ratios of Pex5 and BAAT, the amidation activity of BAAT-SKL was significantly more inhibited than the activity of wild-type BAAT or BAAT-S (Fig. 8). No significant difference was observed in the changes in amidation activity between wild-type BAAT and BAAT-S. Addition of DEAE-purified bacterial cytosol from cells transformed with an empty pQE31 vector or purified human SULT1E1 to wild-type and mutant forms of BAAT showed no effect on BAAT amidation activity (data not shown).
Fig. 8.
Inhibition of BAAT activity in the presence of Pex5. Purified recombinant wild-type BAAT (filled triangles), mutant BAAT-S (open squares), and BAAT-SKL (filled squares) protein were incubated separately with increasing amounts of purified recombinant human Pex5S protein at 37°C for 1 h before measuring BAAT amidation activity using 0.25 mM taurine as a substrate. Activity assays were run for 15 min at 37°C. Error bars represent the standard error of the mean as calculated for three separate, identical experiments. A significant difference (P < 0.05, Student’s t-test) from wild-type enzyme incubated with the same amount of Pex5 is indicated by an asterisk (*).
DISCUSSION
BAAT has the important function of forming bile acid N-acylamidates of both newly formed bile acids as well as those being returned to the liver after reabsorption from the gastrointestinal tract. To aid in this function, BAAT is localized both in peroxisomes and in the cytosol of hepatocytes (7–9, 14). The final steps of bile acid synthesis occur in peroxisomes and approximately 25% of BAAT is present in peroxisomes to amidate the newly formed bile acids. Bile acids that have been deamidated by bacteria in the gastrointestinal tract before reabsorption are efficiently removed by hepatocytes from portal blood and amidated in the cytosol prior to being secreted into the biliary system (1).
Interaction of proteins with a PTS-1 sequence with Pex5 is involved in the translocation of many proteins into peroxisomes (24). The typical high activity PTS-1 sequence is -SKL. However, BAAT possesses a degenerate PTS-1 sequence at its carboxy terminus that may be involved with the differential distribution of BAAT between the cytosolic and peroxisomal compartments. A lower affinity for Pex5 binding may allow BAAT to be out-competed for Pex5 binding by proteins with canonical PTS-1 sequences. This lower affinity for Pex5 is consistent with the greater ability of the BAAT-SKL mutant to inhibit BAAT activity, indicating a greater affinity.
Interaction of the BAAT-SKL mutant with Pex-5 demonstrated the most inhibition of BAAT activity. The inhibition of wild-type BAAT and BAAT-S by Pex5 were similar, indicating that both proteins were interacting with Pex5 in an equivalent manner, even though the PST-1 sequence was deleted in BAAT-S. The physical interaction of BAAT, BAAT-SKL, and BAAT-S with Pex5 is supported by the observation that all three proteins were immunoprecipitated with Pex5 using the rabbit anti-BAAT antibody, and both wild-type and BAAT-S activity were inhibited by Pex5S. Because BAAT-12AA and Pex5 were not immunoprecipitated together, a relatively strong interaction is apparently occurring between Pex5 and the last 12 AAs of BAAT that does not require a PTS-1 sequence. Whether this interaction is sufficient to allow peroxisomal localization in cells remains unknown.
The carboxy-terminus of BAAT also has significant effects on the kinetic and substrate recognition properties of BAAT. The conversion of BAAT to BAAT-SKL resulted in a 2.5-fold increase in specific activity associated with a decreased Km for both taurine and glycine as substrates (Table 1). There was no effect on the affinity for cholyl-CoA. In contrast, the deletion of the final two AAs in BAAT-S did not alter the affinity for either glycine or taurine, indicating that the insertion of the Lys in the penultimate position was responsible for the increased amidation activity. The presence of the Lys also increased resistance of amidation activity to thermal inactivation, as compared with wild-type BAAT.
BAAT is capable of hydrolyzing the thioester linkage between cholate and the active site Cys (3). The alterations in the carboxy-terminus were also associated with changes in thioesterase activity. BAAT-SKL showed a 75% increase in thioesterase activity as compared with wild-type BAAT. The final 12 AAs of BAAT were required for amidase activity; however, BAAT-12AA Trunc that lacked amidase activity still possessed more thioesterase activity than wild-type BAAT. BAAT-ex1, which lacked 150 AAs at the amino terminus, was not capable of catalyzing amidase activity, but still possessed the same thioesterase activity as wild-type BAAT. The ability of BAAT to retain thioesterase activity while losing amidase activity may allow for mutations resulting in loss of bile acid amidate formation, while still catalyzing bile acid CoA hydrolysis.
Human BAAT has been reported to use a catalytic triad of Asp, His, and Cys to form bile acid amidates with the Cys being involved in the bile acid thioester linkage that is attacked by glycine or taurine to form the amidate (3). The sequence of the carboxy-terminal PTS-1 sequence had an unexpected effect on selectivity of the AA substrates. BAAT and BAAT-SKL were both capable of conjugating cholic acid with nortaurine and homotaurine; however, BAAT-SKL was also capable of using β-alanine in the conjugation reaction. This indicates that the presence of the Lys in the SKL PTS-1 sequence increases accessibility to the cholic acid thioester linkage to substrates as well as water. However, the binding of cholyl-CoA and formation of thioester is apparently more robust than AA binding.
Human BAAT essentially forms only taurine and glycine amidates in vivo (25). Rat BAAT also forms taurine and glycine conjugates, whereas mouse BAAT only forms taurine conjugates (14, 15). The carboxy-terminus of BAAT apparently has an important role in taurine and glycine recognition because the BAAT-12AA Trunc lacks amidation activity and BAAT-SKL allowed the use of β-alanine as a substrate. In addition, the single AA residue modification (Q417K) of BAAT to BAAT-SKL also allowed substantial conjugating activity with both nortaurine and homotaurine. These results indicate that the carboxy-terminus forms part of the highly selective AA recognition site without significantly altering the access of the bile acid thioester linkage to water that is responsible for the thioesterase activity. Unfortunately, the structure of BAAT has not been resolved, so the orientation of the PTS-1 sequence to the active site of BAAT has not been established.
None of the mutations examined in this study have been detected in human populations. A nonsynonomous SNP in BAAT (A226G, Met76Val) was associated with familial hypercholanemia in an Amish population (26). Two nonsynonomous SNPs have been reported in a Japanese population, G59A (Arg20Gln) and G602C (Arg201Pro), with allelic frequencies of 0.50 and 0.095, respectively (27). The G602C SNP was also detected in a Caucasian population with an allelic frequency of 0.195 (26). The effect of these variants on the activity or expression of BAAT has not been reported.
The initial hypothesis was that the sequence of the PTS-1 sequence in human BAAT would essentially be responsible for binding Pex5 and transport into the peroxisome. The presence of the -SQL PTS-1 sequence in BAAT would be associated with the low levels of BAAT inserted in the peroxisome due to a lesser affinity for Pex5. Although the results are consistent with this idea, the carboxy-terminus of BAAT also had effects on substrate reactivity, substrate selectivity, thermal stability, and activity. The structure and interactions of the carboxy-terminus of BAAT require analysis to understand the roles of this region in subcellular localization, amidation activity, and thioesterase activity.
Footnotes
Abbreviations:
- AA
- amino acid
- BAAT
- bile acid CoA:amino acid N-acyltransferase
- BAAT-S
- bile acid CoA:amino acid N-acyltransferase lacking the final two amino acids
- BAAT-SKL
- bile acid CoA:amino acid N-acyltransferase with a canonical peroxisomal targeting signal sequence
- Cys
- cysteine
- hBAAT
- human liver bile acid CoA:amino acid N-acyltransferase
- Pex5
- peroxisomal biogenesis factor 5
- PTS
- peroxisomal targeting signal
- -SQL
- -serine-glutamine-leucine
This research was supported in part by Foundation for the National Institutes of Health Grant DKO46390 to S.B. The mass spectrometers were purchased following Shared Instrumentation Awards from the National Center for Research Resources (S10 RR06487) and support from the University of Alabama at Birmingham School of Medicine. Funds for the operation of the University of Alabama at Birmingham Comprehensive Cancer Center Mass Spectrometry Shared Facility were provided by a Division of Cancer Prevention, National Cancer Institute Core Support Grant (P30 CA13148-34; E. Partridge, Principal Investigator). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Hofmann A. F. 1989. Enterohepatic circulation of bile acids. In Handbook of Physiology - The Gastrointestinal System III. S. G. Shultz, editor. American Physiological Society, Bethesda, MD. 567–596. [Google Scholar]
- 2.Falany C. N., Xie X., Wheeler J. B., Wang J., Smith M., He D., and Barnes S.. 2002. Molecular cloning and expression of rat liver bile acid CoA ligase. J. Lipid Res. 43: 2062–2071. [DOI] [PubMed] [Google Scholar]
- 3.Sfakianos M. K., Wilson L., Sakalian M., Falany C. N., and Barnes S.. 2002. Conserved residues in the putative catalytic triad of human bile acid coenzyme A:amino acid N-acyltransferase. J. Biol. Chem. 277: 47270–47275. [DOI] [PubMed] [Google Scholar]
- 4.Huhtinen K., O’Byrne J., Lindquist P. J., Contreras J. A., and Alexson S. E.. 2002. The peroxisome proliferator-induced cytosolic type I acyl-CoA thioesterase (CTE-I) is a serine-histidine-aspartic acid alpha/beta hydrolase. J. Biol. Chem. 277: 3424–3432. [DOI] [PubMed] [Google Scholar]
- 5.Craft C. M., and Zhan-Poe X.. 2000. Identification of specific histidine residues and the carboxyl terminus are essential for serotonin N-acetyltransferase enzymatic activity. Brain Res. Mol. Brain Res. 75: 198–207. [DOI] [PubMed] [Google Scholar]
- 6.Mushtaq A., Payton M., and Sim E.. 2002. The COOH terminus of arylamine N-acetyltransferase from Salmonella typhimurium controls enzymic activity. J. Biol. Chem. 277: 12175–12181. [DOI] [PubMed] [Google Scholar]
- 7.Solaas K., Kase B. F., Pham V., Bamberg K., Hunt M. C., and Alexson S. E.. 2004. Differential regulation of cytosolic and peroxisomal bile acid amidation by PPAR alpha activation favors the formation of unconjugated bile acids. J. Lipid Res. 45: 1051–1060. [DOI] [PubMed] [Google Scholar]
- 8.Solaas K., Ulvestad A. , O. Söreide, and Kase B. F.. 2000. Subcellular organization of bile acid amidation in human liver: a key issue in regulating the biosynthesis of bile salts. J. Lipid Res. 41: 1154–1162. [PubMed] [Google Scholar]
- 9.Styles N. A., Falany J. L., Barnes S., and Falany C. N.. 2007. Quantification and regulation of the subcellular distribution of bile acid coenzyme A:amino acid N-acyltransferase activity in rat liver. J. Lipid Res. 48: 1305–1315. [DOI] [PubMed] [Google Scholar]
- 10.Gould S. G., Keller G. A., and Subramani S.. 1987. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J. Cell Biol. 105: 2923–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holroyd C., and Erdmann R.. 2001. Protein translocation machineries of peroxisomes. FEBS Lett. 501: 6–10. [DOI] [PubMed] [Google Scholar]
- 12.Gould S. J., Keller G. A., and Subramani S.. 1988. Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins. J. Cell Biol. 107: 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falany C. N., Johnson M. R., Barnes S., and Diasio R. B.. 1994. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA:amino acid N-acyltransferase. J. Biol. Chem. 269: 19375–19379. [PubMed] [Google Scholar]
- 14.He D., Barnes S., and Falany C. N.. 2003. Rat liver bile acid CoA:amino acid N-acyltransferase: expression, characterization, and peroxisomal localization. J. Lipid Res. 44: 2242–2249. [DOI] [PubMed] [Google Scholar]
- 15.Falany C. N., Fortinberry H., Leiter E. H., and Barnes S.. 1997. Cloning, expression, and chromosomal localization of mouse liver bile acid CoA:amino acid N-acyltransferase. J. Lipid Res. 38: 1139–1148. [PubMed] [Google Scholar]
- 16.Johnson M. R., Barnes S., and Diasio R. B.. 1989. Radioassay of bile acid coenzyme A:glycine/taurine: N-acyltransferase using an n-butanol solvent extraction procedure. Anal. Biochem. 182: 360–365. [DOI] [PubMed] [Google Scholar]
- 17.Shah P. P., and Staple E.. 1968. Synthesis of coenzyme A esters of some bile acids. Steroids. 12: 571–576. [DOI] [PubMed] [Google Scholar]
- 18.Johnson M. R., Barnes S., Kwakye J. B., and Diasio R. B.. 1991. Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from human liver. J. Biol. Chem. 266: 10227–10233. [PubMed] [Google Scholar]
- 19.Brocard C., and Hartig A.. 2006. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta. 1763: 1565–1573. [DOI] [PubMed] [Google Scholar]
- 20.Czuba B., and Vessey D. A.. 1980. Kinetic characterization of cholyl-CoA glycine-taurine N-acyltransferase from bovine liver. J. Biol. Chem. 255: 5296–5299. [PubMed] [Google Scholar]
- 21.Czuba B., and Vessey D. A.. 1981. Identification of a unique mammalian species of cholyl-CoA: amino acid N-acyltransferase. Biochim. Biophys. Acta. 665: 612–614. [DOI] [PubMed] [Google Scholar]
- 22.Killenberg P. G., and Jordan J. T.. 1978. Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from rat liver. J. Biol. Chem. 253: 1005–1010. [PubMed] [Google Scholar]
- 23.Johnson M. R., Barnes S., Sweeny D. J., and Diasio R. B.. 1990. 2-Fluoro-beta-alanine, a previously unrecognized substrate for bile acid coenzyme A:amino acid:N-acyltransferase from human liver. Biochem. Pharmacol. 40: 1241–1246. [DOI] [PubMed] [Google Scholar]
- 24.Neuberger G., Maurer-Stroh S., Eisenhaber B., Hartig A., and Eisenhaber F.. 2003. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 328: 581–592. [DOI] [PubMed] [Google Scholar]
- 25.Miyazaki S., Tanaka H., Horikawa R., Tsuchiya H., and Imai K.. 1980. Rapid determination of glycine- and taurine-conjugated bile acids in human bile by high-performance liquid chromatography. J. Chromatogr. 181: 177–185. [DOI] [PubMed] [Google Scholar]
- 26.Carlton V. E., Harris B. Z., Puffenberger E. G., Batta A. K., Knisely A. S., Robinson D. L., Strauss K. A., Shneider B. L., Lim W. A., Salen G., et al. . 2003. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat. Genet. 34: 91–96. [DOI] [PubMed] [Google Scholar]
- 27.Tougou K., Fukuda T., Ito T., Yamazaki H., Fujio Y., and Azuma J.. 2007. Genetic polymorphism of bile acid CoA: amino acid N-acyltransferase in Japanese individuals. Drug Metab. Pharmacokinet. 22: 125–128. [DOI] [PubMed] [Google Scholar]