Abstract
SM is a fundamental component of mammalian cell membranes that contributes to mechanical stability, signaling, and sorting. Its production involves the transfer of phosphocholine from phosphatidylcholine onto ceramide, a reaction catalyzed by SM synthase (SMS) 1 in the Golgi and SMS2 at the plasma membrane. Mammalian cells also synthesize trace amounts of the SM analog ceramide phosphoethanolamine (CPE), but the physiological relevance of CPE production is unclear. Previous work revealed that SMS2 is a bifunctional enzyme producing both SM and CPE, whereas a closely related enzyme, sphingomyelin synthase-related protein (SMSr)/SAMD8, acts as a monofunctional CPE synthase in the endoplasmatic reticulum. Using domain swapping and site-directed mutagenesis on enzymes expressed in defined lipid environments, we here identified structural determinants that mediate head group selectivity of SMS family members. Notably, a single residue adjacent to the catalytic histidine in the third exoplasmic loop profoundly influenced enzyme specificity, with glutamic acid permitting SMS-catalyzed CPE production and aspartic acid confining the enzyme to produce SM. An exchange of exoplasmic residues with SMSr proved sufficient to convert SMS1 into a bulk CPE synthase. This allowed us to establish mammalian cells that produce CPE rather than SM as the principal phosphosphingolipid and provide a model of the molecular interactions that impart catalytic specificity among SMS enzymes.
Keywords: cell-free expression, ceramide phosphoethanolamine, click chemistry, enzyme mechanisms, Golgi apparatus, lipid biochemistry, lipidomics, model membranes, protein engineering
SM is a major structural component of mammalian cell membranes and one of the end points in sphingolipid biosynthesis. The bulk of SM is produced in the Golgi lumen and delivered by vesicular transport to the plasma membrane, where it accumulates in the exoplasmic leaflet (1). Owing to its unique ability to form extensive hydrogen bonds with other membrane molecules, SM participates in a multitude of cellular processes. SM is the preferred interaction partner of cholesterol, and this interaction has important physiological consequences. Cell surface SM degradation causes cholesterol to redistribute to the endoplasmatic reticulum (ER) (2), leading to downregulation of HMG-CoA reductase, the rate-limiting step in cholesterol biosynthesis (3). Besides directly influencing cellular cholesterol homeostasis, SM (along with cholesterol) likely contributes to the high packing density and thickening of the lipid bilayer of the trans Golgi and plasma membrane, which may influence protein sorting through hydrophobic mismatching of membrane spans (4, 5). Specific interactions between SM and membrane spans have been reported to influence the activity of various integral membrane proteins, including ion channels, receptors, and membrane trafficking machinery (6, 7). SM also serves as receptor for viruses and pore-forming toxins (8, 9). Moreover, the plasma membrane pool of SM acts as a reservoir of lipid signaling molecules, the liberation of which is catalyzed by neutral, alkaline, or acidic SMases in response to diverse stimuli (10). Ceramide generated by this pathway, along with its downstream metabolites sphingosine and sphingosine-1-phosphate, are critical regulators of cell survival, proliferation, and migration (11).
SM synthesis is mediated by a phosphatidylcholine (PC):ceramide choline-phosphotransferase or SM synthase (SMS). This enzyme catalyzes the transfer of phosphocholine from PC onto ceramide, yielding SM and diacylglycerol (DAG). Mammals contain two SMS isoforms, namely SMS1, primarily responsible for de novo SM synthesis in the lumen of the trans Golgi, and SMS2, presumably serving a role in regenerating SM from ceramides liberated by SMases on the cell surface (12–15). Studies in mice revealed a role of SMS1 and SMS2 in inflammation, atherosclerosis, and diabetes (15–17), suggesting that these enzymes represent relevant pharmacological targets. Besides bulk amounts of SM, mammals also produce small quantities of ceramide phosphoethanolamine (CPE), a widespread but poorly studied SM analog. We and others previously showed that SMS2 is a bifunctional enzyme that produces both SM and CPE on the cell surface, whereas a closely related enzyme, sphingomyelin synthase-related protein (SMSr) SAMD8, acts as a monofunctional CPE synthase in the lumen of the ER (18–21). Interestingly, SMSr is by far the best-conserved SMS family member, with homologs in insects and various marine organisms that lack SM (18, 22). While the biological relevance of SMS2-mediated CPE production remains to be established, acute disruption of SMSr catalytic activity in cultured mammalian and insect cells causes an accumulation of ER ceramides, leading to a structural collapse of ER exit sites and induction of mitochondrial apoptosis (18, 23). These phenotypes are suppressed by blocking de novo ceramide synthesis, stimulating ER export of ceramides or targeting SMS1 to the ER, supporting a role of SMSr as critical regulator of ER ceramide levels. However, ubiquitous inactivation of SMSr catalytic activity in mice primarily disrupted CPE biosynthesis in the brain without any obvious impact on steady-state ceramide levels, cell integrity, or survival (20, 21). These contradictory outcomes could be due to compensatory mechanisms that overcome a deregulation of ER ceramides over time. The physiological relevance of SMSr-mediated CPE production and the consequences of its acute disruption in mammals remain to be established.
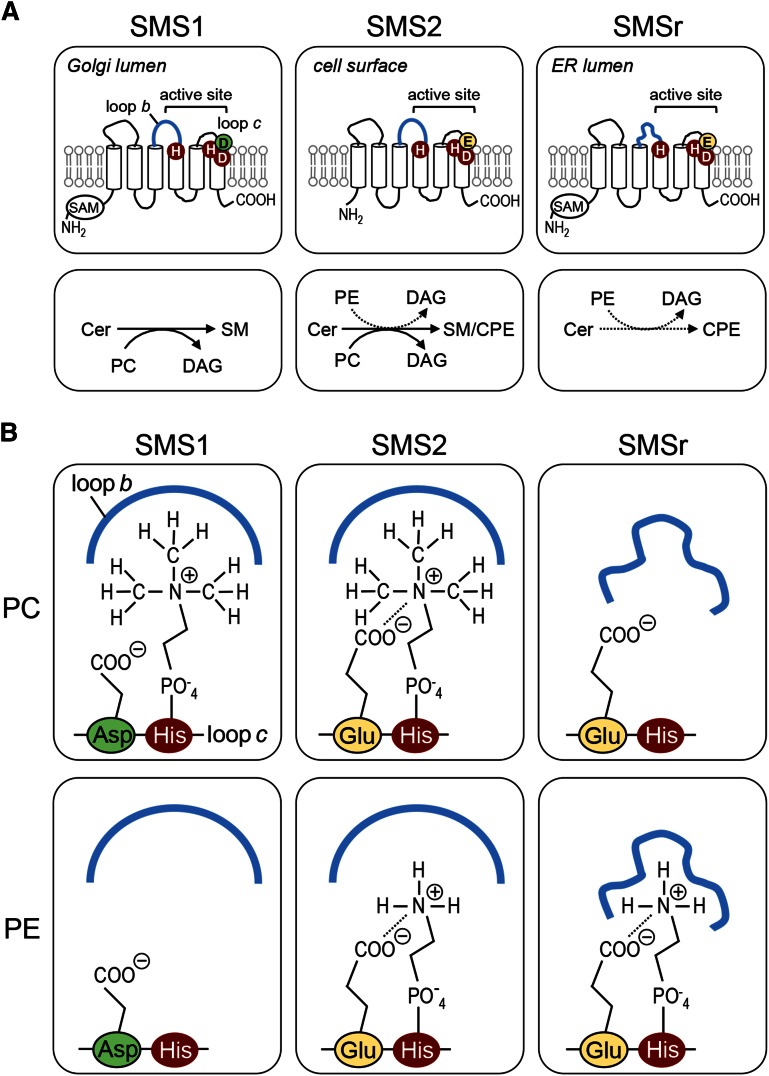
Analogous to members of the lipid phosphate phosphatase (LPP) superfamily, SMS enzymes likely contain a six-times membrane-spanning core domain with an active site comprising a conserved catalytic triad of two histidine (His) residues and one aspartic acid (Asp) residue located in or near the second and third exoplasmic loop (Fig. 1A) (12, 24). This strongly suggests that SMS-catalyzed SM and CPE production follows an LPP-type reaction cycle, which starts when a phospholipid head group donor, PC or phosphatidylethanolamine (PE), enters a single binding site in the enzyme. Next, the conserved His in the third exoplasmic loop carries out a nucleophilic attack on the lipid-phosphate ester bond, assisted by the conserved Asp. After formation of a choline or ethanolamine phospho-His intermediate, DAG is released and replaced by ceramide. The conserved His near the second exoplasmic loop then acts as a nucleophile to attack the primary hydroxyl group of ceramide. This results in formation of SM or CPE, which are released from the active site to allow a next round of catalysis. While the importance of the catalytic triad residues for SMS-catalyzed SM and CPE production has been confirmed by site-directed mutagenesis (18, 25), information on the structural determinants that mediate head group selectivity of SMS enzymes is lacking. As these enzymes use the same reaction chemistry, their specificity for SM and/or CPE production is most likely defined by differences in the geometry of the phospholipid binding site. Identification of residues involved in substrate recognition should not only help to further elucidate the reaction mechanism of this important class of enzymes, but also facilitate the development of specific inhibitors to target the biological role of each enzyme individually.
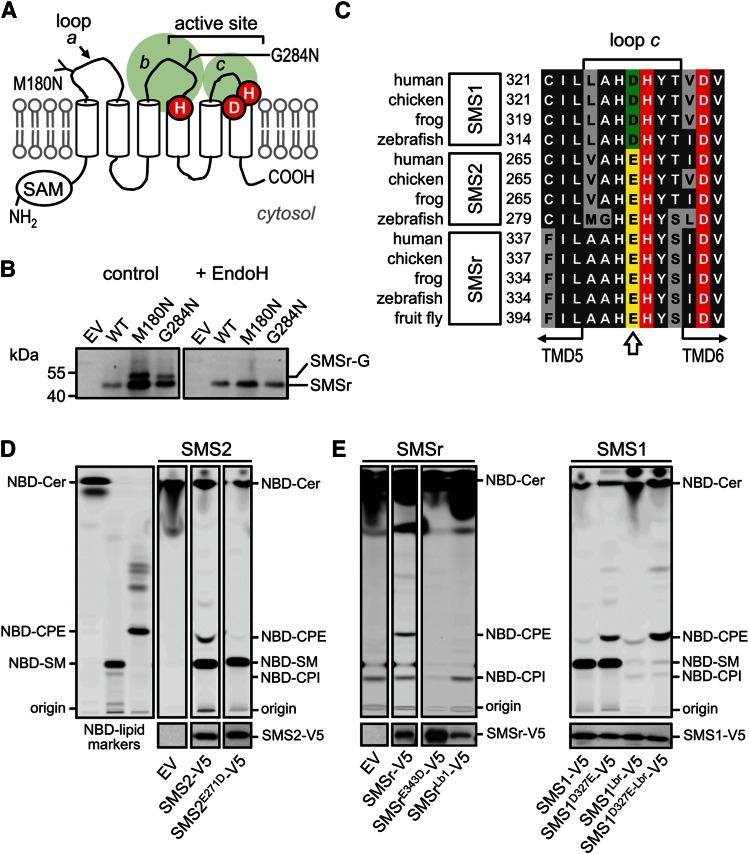
Fig. 1.
The exoplasmic loops of SMSr and SMS1 harbor structural determinants of substrate selectivity. A: Predicted membrane topology of SMSr and SMS1. Active site residues are marked in red. The positions of two N-linked glycosylation sites introduced by site directed mutagenesis in human SMSr are indicated. SAM, sterile α motif. B: Anti-V5 immunoprecipitates prepared from HeLa cells transfected with V5-tagged human SMSr, SMSrM180N, or SMSrG284N constructs were mock treated or treated with EndoH and then subjected to immunoblot analysis with anti-V5 antibody. SMSr-G marks the migration of an N-glycosylated, EndoH-sensitive form of SMSr that is produced exclusively in SMSrM180N- and SMSrG284N-expressing cells. C: Sequence alignment of loop c in SMS family members from vertebrates and fruit fly. The position of a residue critical for discriminating the phospholipid head group donors PC and PE [Asp or glutamic acid (Glu)] is marked by an arrow. Database accession numbers are as follows: human SMS1, BAD16809.1; chicken SMS1, ADY69193.1; frog SMS1, NP_001008197.1; zebrafish SMS1, NP_001071082.1; human SMS2, NP_689834.1; chicken SMS2, XP_420492.1; frog SMS2, AAH88568.1; zebrafish SMS2, zgc:100911; human SMSr, Q96LT4; chicken SMSr, XP_426501.3; frog SMSr, Q28CJ3; zebrafish SMSr, zgc:162183; fruit fly SMSr, CG32380. D: TLC analysis of reaction products formed when lysates of yeast strains expressing V5-tagged human SMS2 or SMS2E271D were incubated with NBD-C6-ceramide (NBD-Cer). EV denotes yeast lysate from strain transfected with empty vector. SMS expression was verified by immunoblotting with anti-V5 antibody (bottom). Note that residue substitution E271D in loop c converts SMS2 into a monofunctional SMS. E: TLC analysis of reaction products formed when lysates of yeast strains expressing V5-tagged human SMSr, SMSrE343D, SMS1, SMS1D327E, or SMS1/r chimera in which loop b was swapped (SMSrLb1, SMS1Lbr) were incubated with NBD-Cer. SMS expression was verified by immunoblotting with anti-V5 antibody (bottom). Note that residue substitution D327E combined with swapping loop b against that of SMSr converts SMS1 into a monofunctional CPE synthase (SMS1D327E-Lbr or SMS1CPE). Data shown in D and E are representative of two independent experiments.
In this work, we use domain swapping and site-directed mutagenesis on enzymes produced in a liposome-coupled cell-free expression system to map structural elements responsible for head group selectivity of SMS family members. Our data reveal that enzyme specificity is strongly influenced by single residues in close proximity of the catalytic triad. We show that exchanging a limited number of exoplasmic residues between SMS1 and SMSr is sufficient to convert a conventional SMS into a monospecific bulk producer of CPE. This enabled the establishment of mammalian cells that produce CPE as their principal phosphosphingolipid, thus creating an opportunity to elucidate the biological significance of this enigmatic membrane component.
MATERIALS AND METHODS
Chemicals and reagents
Wheat germ phosphatidylinositol (PI), egg PC, and egg PE were from Lipid Products UK. d-erythro-sphingosine and 1-palmitoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoethanolamine (NBD-PE) were from Avanti Polar Lipids. NBD-C6-ceramide (NBD-Cer) and NBD-C6-SM (NBD-SM) were from Molecular Probes. NBD-C6-CPE (NBD-CPE) was generously provided by Philippe Devaux (Institut de Biologie Physico-Chimique, Paris). WEPRO2240 wheat germ extract was from Cell-Free Sciences, Accudenz from Accurate Chemicals and [1,2-14C]ethanolamine hydrochloride (100 μCi/ml, 100–115 mCi/mmol) from BioTrend. EndoH was from New England Biolabs.
Synthesis of clickable ceramide analog
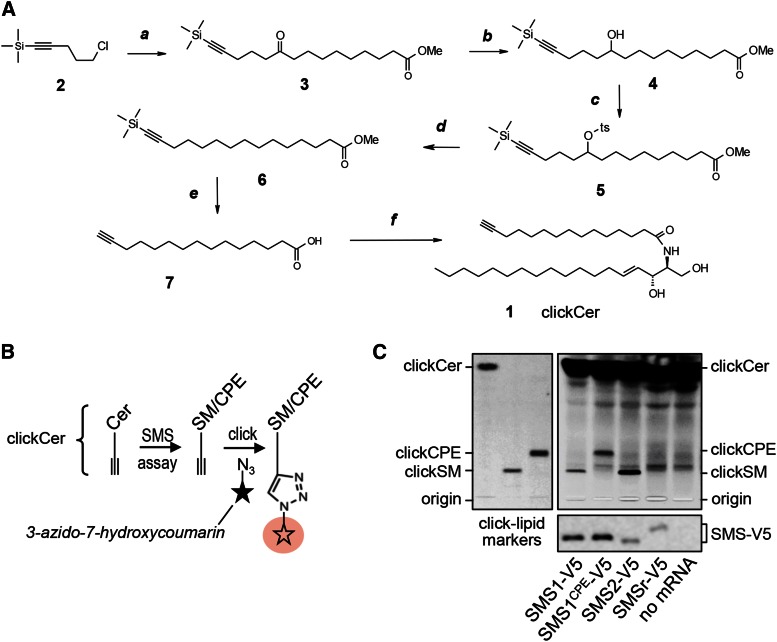
As outlined in Fig. 3A, clickable ceramide (clickCer, 1) was synthesized by the condensation of d-erythro-sphingosine and a terminal acetylenic C15-fatty acid (7). The latter was prepared in five steps starting from the condensation of Grignard reagent, prepared from the commercially available chloride (2), and corresponding decanoyl chloride. The keto-function in the intermediate ketoester (3) was completely reduced using a sequence of three steps, namely, reduction into the alcohol (4), tosylation, and final reduction of the tosylate into the saturated fragment of 6. Simultaneous removal of the TMS-protecting group and hydrolysis of the Me-ester in 6 gave click C15-fatty acid (7) with a yield of 22% over five steps. Structure of clickCer (molecular weight 519.9) was confirmed by NMR spectroscopy (1H, 13C). The synthesis of clickable analogs of SM (clickSM) and CPE (clickCPE) will be described elsewhere (S. Korneev and J. Holthuis, unpublished observations).
Fig. 3.
Tracing catalytic activity of cell-free produced SMS enzymes by click chemistry. A: A clickable ceramide analog, clickCer (1), was synthesized by condensation of d-erythro-sphingosine and a C15-fatty acid carrying a terminal alkyne group (7). The latter was synthesized in five steps, as detailed in Materials and Methods. Reagents and conditions: a, Mg, Et2O, C11H19O3Cl, room temperature; b, NaBH4, methanol-water, room temperature; c, TsCl, Et3N, 4-dimethylaminopyridine, room temperature; d, NaBH4, DMSO, 75°C; e, KOH, methanol, 50°C; f, sphingosine, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, hydroxybenzotriazole, room temperature. B: Schematic outline of the click-chemistry-based SMS assay. ClickCer was incorporated as SMS substrate in liposomes present during the cell-free translation of SMS-V5 mRNA. After lipid extraction, the alkyne moiety was click reacted with the fluorogenic dye 3-azido-7-hydroxycoumarin to yield fluorescently labeled lipids. The scheme was adapted from Gaebler et al. (28). C: V5-tagged SMS1, SMS1CPE, SMS2, and SMSr were produced cell free in the presence of liposomes containing 2 mol% clickCer. After lipid extraction, SMS reaction products were click reacted with 3-azido-7-hydroxycoumarin, separated by TLC, and analyzed by fluorescence detection. SMS expression was verified by immunoblotting using anti-V5 antibody (bottom). Data shown are representative of two independent experiments.
SMS expression constructs
For expression in yeast, open reading frames (ORFs) of human SMS enzymes were PCR amplified and cloned into the pYES2.1/V5-His TOPO vector (Invitrogen) according to the manufacturer’s instructions. To create loop b swap constructs SMSrLb1 and SMS1Lbr, the sequences encoding Gly239-Gly278 in SMS1 and Gly256-Gly294 in SMSr were exchanged by two consecutive fusion PCRs. Single amino acid substitutions were introduced by site-directed mutagenesis using the megaprimer PCR method (26). For expression in human HeLa and KBM7 cells, ORFs of SMS enzymes were PCR amplified in-frame with a C-terminal V5-His or hemagglutinin (HA) epitope and cloned into pcDNA3.1 or retroviral expression vector pLNX2 (Clontech). For cell-free expression, ORFs of SMS enzymes were PCR amplified in-frame with a C-terminal V5 epitope and cloned into the wheat germ pEU-Flexi expression vector (kind gift of Brian G. Fox and James D. Bangs, University of Wisconsin, Madison). All constructs were sequence verified before use.
SMS glycosylation studies
HeLa cells were transfected with SMSr-V5-His, SMSrM180N-V5-His, or SMSrG284N-V5-His constructs using Effectene Transfection Reagent (Qiagen). After 24 h, cells were washed twice with PBS and lysed in Lysis Buffer (1% Triton X-100, 1 mM EDTA pH 8.0, 150 mM NaCl, 20 mM Tris pH 7.5) containing 10 mM N-ethyl maleimide and protease inhibitor cocktail (1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 5 µg/ml antipain, 157 µg/ml benzamidine). Cell lysates were centrifuged at 7,000 g for 10 min, and the supernatants were incubated with Ni2+-NTA beads (Qiagen) for 2 h at 4°C while gently shaken. The beads were washed with Lysis Buffer, resuspended in 40 µl Glycoprotein Denaturing Buffer (0.5% SDS, 40 mM DTT) and incubated for 10 min at 99°C. Next, 40 µl of 50 mM sodium acetate was added, and the beads were split in two equal aliquots. After addition of 500 U of EndoH to one aliquot, the beads were incubated at 37°C for 1 h and subjected to SDS-PAGE and immunoblot analysis using mouse anti-V5 antibody (Invitrogen) and HRP-conjugated goat anti-mouse antibody (ThermoFischer).
Preparation of yeast lysates
Yeast strain IAY11 (MATa, ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-52 ade3-Δ853) was transformed with pYES2.1/SMS-V5-His TOPO expression constructs and then grown in synthetic medium containing 2% (w/v) galactose to early midlogarithmic phase. Cells were collected by centrifugation and washed in ice-cold buffer R (15 mM KCl, 5 mM NaCl, 20 mM HEPES/KOH pH 7.2). The wet cell pellet (2 g) was resuspended in a final volume of 5 ml buffer R containing protease inhibitors (1 μg/ml aprotinin, 1 ug/ml leupeptin, 1 ug/ml pepstatin, 5 ug/ml antipain, 1 mM benzamidine, and 1 mM PMSF). Cells were lysed by vigorous vortexing with 3 g glass beads at 4°C with intermittent cooling on ice. A postnuclear supernatant was prepared by centrifugation at 700 g for 10 min at 4°C. After addition of 0.11 vol of glycerol, lysates were aliquoted, snap-frozen in liquid nitrogen, and stored at −80°C.
Preparation of liposomes
Phospholipid stocks were prepared in chloroform-methanol (9:1, v/v), briefly flushed with nitrogen gas, stored at −20°C in brown glass vials, and periodically checked by TLC and iodine staining. Phospholipid concentrations were determined as described (27). Unilamellar liposomes were prepared from a defined lipid mixture (egg PC/egg PE/wheat germ PI, 2:2:1 mol%) using a miniextruder (Avanti Polar Lipids). In brief, 20 μmol of total lipid was dried under a flow of nitrogen to create a thin film. The film was resuspended in 1 ml lipid rehydration buffer (25 mM HEPES pH 7.5, 100 mM NaCl) by vigorous vortexing to create a 20 mM lipid suspension. After six freeze-thaw cycles using liquid nitrogen and a 40°C water bath, liposomes with an average diameter of 400 nm were obtained by extrusion of the lipid suspension through a 0.4 μm track-etched polycarbonate membrane (Whatman-Nuclepore). To obtain liposomes with an average diameter of 100 nm, the 400 nm liposomes were extruded through a 0.1 μm membrane. Liposomes with a diameter of 30–50 nm were obtained by tip sonication of the 100 nm liposomes on ice until the suspension turned optically clear. The average diameter of liposomes was confirmed by dynamic light scattering. Liposome suspensions were aliquoted, snap frozen in liquid nitrogen, and stored at −80°C.
Cell-free expression of SMS enzymes
pEU-Flexi-SMS expression constructs were treated with Proteinase K to remove trace amounts of RNase, purified by phenol/chloroform extraction and dissolved at 1 μg/μl in water. In vitro transcription was carried out in 50 µl reaction volume containing 5 µg of DNA construct; 2 mM each of ATP, GTP, CTP, and UTP; 20 units of Sp6 RNA polymerase; and 40 units of RNasin in 100 mM HEPES-KOH pH 7.8, 25 mM Mg-acetate, 2 mM spermidine, and 10 mM DTT (28). After incubation at 37°C for 4 h, the reaction mixture was centrifuged at 3400 g for 5 min at room temperature. The supernatant, containing SMS mRNA, was collected to set up a cell-free translation reaction. To this end, 20 µl of mRNA-containing supernatant was used per 100 µl translation reaction containing 0.3 mM each of all 20 amino acids, 2 mM liposomes, 40 µg/ml creatine kinase, 15 optical density (OD)260 WEPRO2240 wheat germ extract, 15 mM HEPES-KOH pH 7.8, 50 mM potassium acetate, 1.25 mM Mg-acetate, 0.2 mM spermidine, 2 mM DTT, 0.6 mM ATP, 125 µM GTP, 8 mM creatine phosphate, and 0.0025% sodium azide. The reaction mixture was incubated for 4 h at 26°C (tube mode) or transferred to a 12 kDa molecular weight cut-off dialysis cup (Novagen) dipped in 5 ml reservoir buffer (same composition as translation reaction but omitting wheat germ extract, mRNA, creatine kinase, and liposomes) and incubated for 16 h at 26°C (dialysis mode). Translation reactions were processed for SMS enzyme activity (see below) or subjected to quantitative immunoblotting using mouse monoclonal anti-V5 antibody (Invitrogen Life Technologies) and known amounts of a V5-tagged 75 kDa reference protein purified from Escherichia coli (MBP-ceramide transfer protein-His6-V5).
Density flotation
Typically, 75 µl of translation reaction was mixed with 75 µl of 80% (w/v) Accudenz prepared in gradient buffer (25 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol) and transferred to the bottom of a 800 µl 5 × 41 mm thinwall Ultra-Clear tube (Beckman Coulter), carefully overlayed with 350 µl of 30% Accudenz and then with 100 µl of gradient buffer. Next, the samples were centrifuged at 100,000 g in a MLS-50 rotor (Beckman Coulter) for 4 h at 4°C. Fractions of 60 µl were collected from the top to the bottom of the gradient and processed for immunoblotting or enzyme assays as indicated below. Gradient fractions were snap frozen in liquid nitrogen and stored at −80°C.
SMS activity assay with NBD-Cer
Catalytic activity of SMS enzymes expressed in yeast were analyzed as described (19). To analyze cell-free produced SMS enzymes for catalytic activity, 20–200 μl of translation reaction mixture was combined with buffer R containing protease inhibitors and 0.5 mM N-ethylmaleinimide to a total volume of 400 μl on ice. NBD-Cer was added from a 2 mM ethanolic stock to a final concentration of 25 μM. Enzyme reactions were incubated at 37°C for 1–2 h with constant shaking. The reaction was stopped by adding 1 ml methanol and 0.5 ml CHCl3. Phase separation was induced by adding 0.5 ml CHCl3 and 0.5 ml of 0.45% (w/v) NaCl. The lower phase was evaporated under a stream of nitrogen, dissolved in 25 μl CHCl3/methanol (2:1), and then spotted at 120 nl/s on NANO-ADAMANT HP-TLC plates (Macherey and Nagel) using a CAMAG Linomat 5 TLC sampler. The TLC was developed first in acetone, dried, and then in CHCl3/methanol/25% NH4OH (50:25:6, v/v/v) using a CAMAG ADC2 TLC developer. Fluorescent lipids were visualized using a Typhoon FLA 9500 Biomolecular Imager (GE Healthcare Life Sciences) operated in Cy2 fluorescence mode with 473 nm excitation laser, BPB1 filter, 50 μm pixel size, and photomultiplier tube (PMT) voltage setting of 290 V. NBD-labeled SMS reaction products were quantified using known amounts of NBD-PE as reference.
SMS activity assay with clickCer
Cell-free expression of SMS enzymes was carried out in the presence of liposomes containing 2 mol% clickCer as above, except that DTT and sodium azide were omitted from the translation-reaction buffer, as their presence was found to interfere with subsequent click reactions. Control experiments showed that removal of these two compounds had no adverse effect on the yield or activity of cell-free produced SMS enzymes. Typically, 200 μl of translation reaction was combined with 200 μl buffer R and incubated for 2 h at 37°C while shaking. Lipids were extracted as above, transferred to Eppendorf Protein LoBind tubes, dried down in a speedvac, and dissolved in 10 μl CHCl3. Lipid extracts were click reacted with the fluorogenic dye 3-azido-7-hydroxycoumarin by addition of 64.5 μl of a freshly prepared click-reaction mix containing 0.45 mM 3-azido-7-hydroxycoumarin and 1.4 mM Cu(I)tetra(acetonitrile) tetrafluoroborate in acetonitrile-ethanol (3:7, v/v) for 2.5 h at 45°C without shaking (29). The reaction was quenched by addition of 150 μl methanol, dried down in a speedvac, dissolved in CHCl3/methanol (2:1, v/v), and applied on a TLC plate. The TLC was developed first in acetone and then in CHCl3/methanol/water/acetic acid (HAc) (65:25:4:1, v/v/v/v). Fluorescent lipids were analyzed using a ChemiDoc XRS+ with UV-trans-illumination (detection settings for ethidium bromide, standard filter) and Quantity One software (Bio-Rad).
Immunofluorescence microscopy
HeLa cells were seeded on glass coverslips and transfected with pLNX2-SMS-HA constructs using Effectene transfection reagent (Qiagen). After 24 h, cells were fixed with 4% (w/v) paraformaldehyde in PBS, quenched in 50 mM ammonium chloride, and permeabilized using PBS containing 0.1% (w/v) saponin and 0.2% (w/v) BSA. Coverslips were incubated with rat anti-HA (Roche) and mouse anti-GM130 (BD Bioscience) antibodies, washed, and then incubated with donkey anti-rat Cy2 and donkey anti-mouse Cy3 (Jackson ImmunoResearch). Coverslips were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) and mounted using Prolong Gold Antifade Reagent (Molecular Probes). Images were captured using a DM5500 B epi-fluorescence microscope (Leica) equipped with a PL-APO 63× (1.40 NA) oil immersion objective and a SPOT Pursuit camera (Leica) with filter sets for differential interference contrast, DAPI, green fluorescent protein, and mCherry (Chroma Technology Corp.).
Retroviral transduction of KBM7 cells
Human myeloid leukemia KBM7-derived SMS1-null cells (30) stably expressing HA-tagged SMS1 or SMS1CPE were created by retroviral transduction. To this end, low-passage human HEK293T cells (ATCC® CRL-3216™) grown in DMEM medium (Life Technologies) supplemented with 10% FBS (Sigma) were cotransfected with pLNX2-SMS-HA expression constructs and packaging vectors (Clontech) using Lipofectamine 2000 (ThermoFisher). The culture medium was changed 6 h post transfection. After 48 h, supernatants were harvested, passed through a 0.45 μm filter, and the virus-containing filtrate was used to transduce KBM7/SMS1-null cells. The cells were grown in Iscove modified Dulbecco medium (IMDM; ThermoFisher) supplemented with 10% FBS and 0.8 mg/ml geneticin (G418; ThermoFisher) for 10 days. Expression of HA-tagged SMS enzymes was confirmed by immunoblotting using rat monoclonal anti-HA antibody.
Metabolic labeling
Insect Sf21 cells (0.5 × 106) were metabolically labeled in 0.5 ml insect X-press medium (Lonza) with 1 μCi of [14C]ethanolamine at 26°C for 24 h. KBM7 cells (2 × 106) were metabolically labeled in 5 ml IMDM medium (Gibco) containing 10% FBS (PAA Laboratories) with 1 μCi of [14C]ethanolamine for 24 h at 37°C and 5% CO2. Lipids were extracted in CHCl3/methanol/10 mM HAc (5:22:1, v/v/v) and then processed according to Bligh and Dyer (31). Half of the extract was subjected to mild alkaline hydrolysis using sodium methoxide. Extracts were applied to TLC and developed in CHCl3/methanol/25% NH4OH (50:25:6, v/v/v). Radiolabeled lipids were detected by exposure to a Storage Phosphor Screen (Fuji Photo Film), which was scanned using a Typhoon FLA 9500 Biomolecular Imager operated at phosphoimaging mode with a 635 nm laser, imaging processing filter, 50 μm pixel size, and PMT setting of 600 V.
Lipid MS
KBM7 cells grown for 48 h in IMDM medium supplemented with 10% delipidated FBS (19) were subjected to total lipid extraction according to Folch et al. (32) with 15:0 SM and 17:0 CPE added as internal standards at the one-phase stage of extraction. After evaporation of the solvent, the lipids were dissolved in methanol/CHCl3 (2:1, v/v), the phosphate contents were determined (27), and the samples were stored at −20°C. To remove glycerolipids, the extract was taken to dryness under a stream of nitrogen, NaOH (0.3 M final concentration) was added, and the sample was incubated overnight at room temperature. After neutralization with 0.3 M HCl, the lipids were extracted as above and dissolved in 40 μl of LC/MS quality methanol. CPE and SM species were separated on Acquity Ultra Performance LC system equipped with an Acquity BEH-C18 1.0 × 150 mm column (Waters Inc.) as in Bickert et al. (20). The column eluent was infused to a Quattro Premier mass spectrometer (Waters) operated in the positive ion mode, and the CPE and SM molecular species were detected using selective reaction monitoring. The individual species were quantified from chromatograms using QuanLynx (Waters Inc.) and Excel (Microsoft) software.
RESULTS AND DISCUSSION
Mapping structural determinants of SMS substrate selectivity
While no crystal structure of SMS enzymes has been determined, protease protection experiments and a comparative hydrophobicity analysis of SMS homologs predict a six-times membrane-spanning core domain with a topology as depicted in Fig. 1A (12, 24). This model positions the catalytic triad of conserved His and Asp residues in or near the second and third predicted exoplasmic loops (loops b and c; Fig. 1A), hence at the side of the membrane where SMS-catalyzed SM and CPE is thought to occur (12, 19). To validate this model, we introduced N-linked glycosylation sites in the first and second putative exoplasmic loops of V5-tagged human SMSr (loops a and b; Fig. 1A), yielding SMSrM180N-V5 and SMSrG284N-V5. Heterologous expression of these constructs in HeLa cells followed by immunoblot analysis in each case yielded a gel-shifted, EndoH-sensitive protein that was absent in SMSr-V5-expressing cells (Fig. 1B). These results indicate that the loops connecting the first with the second and third with the fourth putative membrane span of SMSr face the ER lumen, consistent with the model presented in Fig. 1A.
As SMS-catalyzed SM and CPE production has been reported to take place in the exoplasmic leaflet of the membrane (19), substrate selectivity of SMS enzymes likely involves residues in the exoplasmic loops where interactions with the polar head groups of the phospholipid donors PC and PE are expected to occur. An alignment of SMS sequences revealed an Asp residue immediately proximal to the catalytic His in the third exoplasmic loop (loop c) that is invariably present in monofunctional SMS, SMS1, from diverse species (Fig. 1C). CPE-producing family members SMS2 and SMSr, on the other hand, contain a Glu residue at this position. The influence of this residue on substrate selectivity was investigated by site-directed mutagenesis of V5-tagged human SMS enzymes expressed in budding yeast, an organism lacking endogenous SM and CPE synthase activities. When lysates of yeast cells expressing SMS2 were incubated with the fluorescent ceramide analog NBD-Cer, both NBD-SM and NBD-CPE were formed, in line with previous studies indicating that this enzyme possesses dual SM and CPE synthase activity (Fig. 1D) (19). Substitution of the Glu residue in loop c to Asp selectively abolished SMS2-catalyzed CPE production, thus converting SMS2 into a monofunctional SMS. Exchanging Glu for Asp in loop c of SMSr also blocked SMSr-mediated CPE production (Fig. 1E). Conversely, exchanging Asp for Glu in loop c of SMS1 yielded a bifunctional enzyme with both SM and CPE synthase activity (Fig. 1E). Thus, a Glu residue immediately adjacent to the catalytic His in loop c permits SMS-mediated CPE production while an Asp at this position confines the enzyme to produce only SM.
The observation that exchanging the Glu and Asp residues in loop c did not cause a complete switch in head group selectivity implied that additional residues in the exoplasmic domains of SMS enzymes contribute to substrate specificity. Sequence alignments of the first and second exoplasmic loops did not reveal any obvious clues on the identity of these residues. However, swapping the second exoplasmic loop (loop b) of SMSr for that of SMS1 abolished SMSr-mediated CPE production (SMSrLb1; Fig. 1E). Strikingly, an SMS1 chimera carrying loop b of SMSr (SMS1Lbr) lost the ability to produce SM but gained CPE synthase activity. Moreover, an exchange of Glu for Asp in loop c of the SMS1Lbr chimera dramatically increased its CPE synthase activity (Fig. 1E). These data suggest that the Asp and Glu residues in loop c along with structural determinants in loop b mediate phospholipid head group donor selectivity of SMS enzymes. The CPE-producing variant of SMS1, SMS1D327E-Lbr, will henceforth be referred to as SMS1CPE.
Functional analysis of SMS enzymes in defined lipid environments
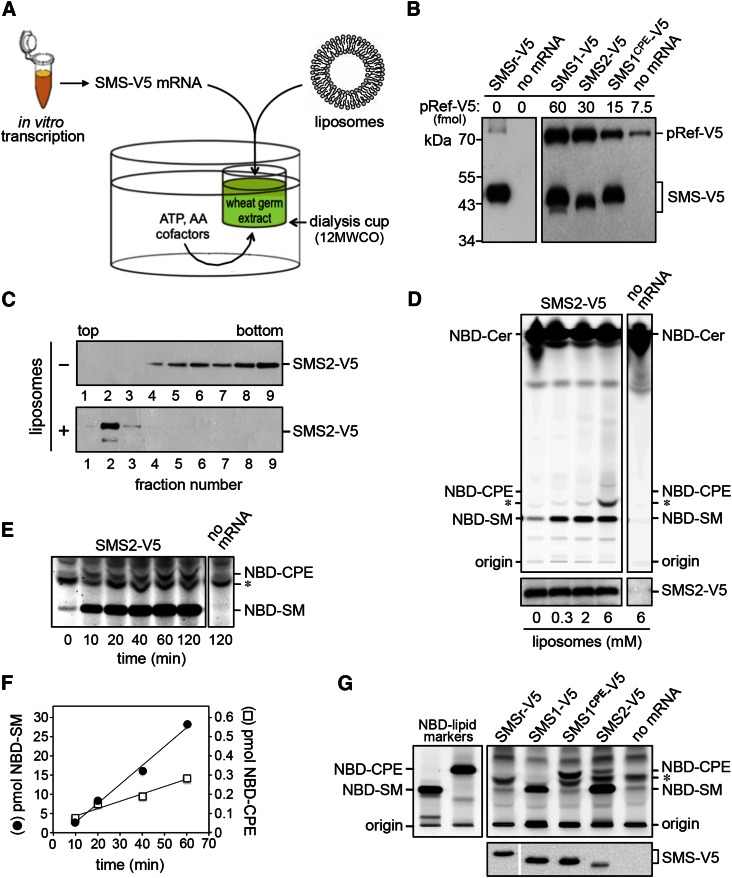
Eukaryotic cells, from mammals to yeast, contain ATP-fueled flippases that translocate PE and PC from the exoplasmic to the cytosolic leaflet of late secretory organelles (33–36). These activities may influence SMS-mediated SM and CPE production by limiting the amount of PC and PE available for consumption by SMS enzymes. For instance, the preferential synthesis of SM over CPE by SMS2 in the plasma membrane could be due to a high PC/PE ratio in the exoplasmic leaflet implemented by aminophospholipid-specific flippases rather than determined by an intrinsic property of the enzyme (19). This led us to evaluate the head group selectivity of native and engineered SMS enzymes in a defined lipid environment. To this end, we adopted a recently developed system for the cell-free production of functional polytopic membrane proteins (28, 37). In this system, SMS mRNA is synthesized by in vitro transcription and used for cell-free translation in a wheat germ extract (Fig. 2A). Translation was performed in the presence of unilamellar liposomes containing equimolar amounts of PC and PE. The liposomes are thought to capture and stabilize the nascent membrane spans of cell-free produced proteins, but the mechanistic principles involved remain to be established. For cell-free expression of human SMSr, SMS1, SMS2, and SMS1CPE, the corresponding cDNAs were cloned downstream of the Sp6 promoter in pEU Flexi-vector pFlx (28). To facilitate detection of the cell-free produced SMS proteins, a V5 epitope was introduced at their C termini. As shown in Fig. 2B, cell-free translation of SMS mRNA in the presence of liposomes in each case yielded a V5-tagged protein of the expected size. No such protein was detected when translation was performed in the absence of mRNA.
Fig. 2.
Cell-free expression and functional analysis of human SMS family members. A: Schematic outline of the wheat germ-based dialysis mode for cell-free translation of SMS-V5 mRNA. Unless indicated otherwise, translation reactions were supplemented with liposomes containing equal amounts of phospholipid head group donors PC and PE. B: Translation reactions with or without SMS-V5 mRNA were subjected to immunoblot analysis using anti-V5 antibody. Known amounts of a 75 kDa V5-tagged reference protein, pRef-V5, were included to allow quantification of cell-free produced SMS-V5 protein. Note that translation reactions with SMS-V5 mRNA in each case yielded an immunoreactive protein of the expected size. C: SMS2-V5 mRNA was translated in the absence or presence of 2 mM liposomes. Translation reactions were subjected to density gradient fractionation and immunoblotting using anti-V5 antibody. D: SMS2-V5 mRNA was translated in wheat germ extract in the presence of the indicated amounts of liposomes. Cell-free produced SMS2-V5 was incubated with NBD-Cer, and reaction products were analyzed by TLC. Expression of SMS2-V5 was verified by immunoblotting using anti-V5 antibody (bottom). E: TLC analysis of reaction products formed when SMS2-V5 produced in the presence of 2 mM liposomes was incubated with NBD-Cer for the indicated period of time. Sensitivity of detection was increased 10-fold compared with D to visualize SMS2-mediated production of NBD-CPE. Migration of an unidentified fluorescent lipid that was also present in reactions lacking SMS2 is marked by an asterisk. F: Quantification of reaction products formed by cell-free produced SMS2-V5 when incubated with NBD-Cer for the indicated period of time. G: Functional analysis of cell-free produced SMS1, SMS1CPE, and SMS2. TLC analysis of reaction products formed when the indicated SMS enzymes produced in the presence of 2 mM liposomes were incubated with NBD-Cer. SMS expression was verified by immunoblotting using anti-V5 antibody (bottom). Data shown are representative of three independent experiments.
To address whether cell-free-produced SMS enzymes are incorporated into liposomes, the translation reactions were loaded at the bottom of an Accudenz step gradient and subjected to high-speed centrifugation. As shown in Fig. 2C, the bulk of cell-free produced SMS2 was recovered from the top fractions of the density gradient, which also contained the bulk of externally added liposomes. In contrast, SMS2 produced in the absence of liposomes was mainly found in the bottom fractions of the gradient. Adding > 2 mM of liposomes to the translation reaction caused a gradual loss of SMS2 flotation (data not shown). In addition, flotation of SMS2 was best supported by liposomes with a diameter of ∼100 nm. Reducing the diameter of liposomes to 30 nm or increasing it to 400 nm in each case diminished the protein’s capacity to float (data not shown). Thus, cell-free produced SMS enzymes appear most efficiently incorporated into 100 nm liposomes.
Cell-free-produced SMS2 displayed dual enzymatic activity analogous to that observed for SMS2 expressed in yeast or human cells, with a strong preference for SM synthesis over CPE synthesis ( ). Thus, even though the enzyme was incorporated into liposomes containing equal amounts of the head group donors PC and PE, its specific activity as SMS was nearly 100-fold higher than that as CPE synthase, namely, 2.0 ± 0.9 E-03 s−1 (n = 5) versus 2.3 ± 1.5 E-05 s−1 (n = 2). Omission of liposomes from the translation reaction did not affect the overall yield of SMS2 but strongly reduced its ability to synthesize sphingolipids (Fig. 2D). The finding that SMS2-mediated sphingolipid production was not completely eliminated in the absence of externally added liposomes suggested that the wheat germ extract contained a residual pool of endogenous phospholipid. Indeed, TLC analysis of the extract revealed the presence of trace amounts of PC (data not shown). From these data, we conclude that the imbalance in SM and CPE production by SMS2 is due to an intrinsic property of the enzyme and that the liposome-coupled wheat germ expression system offers a suitable approach for dissecting the enzymatic characteristics of SMS family members in a controlled lipid environment.
SMS1CPE is a monofunctional CPE synthase
As shown in Fig. 2F, cell-free expression of SMS1 and SMS1CPE in the presence of liposomes containing equal amounts of PC and PE yielded monofunctional enzymes with SM and CPE synthase activity, respectively. Moreover, these enzymes displayed similar specific activities (SMS1: 4.2 ± 1.8 E-04 s−1, n = 3; SMS1CPE: 3.3 ± 1.7 E-04 s−1, n = 2). These results corroborate the notion that phospholipid head group selectivity of SMS family members is primarily controlled by residues in the enzyme’s second and third exoplasmic loops (i.e., loops b and c, respectively). Cell-free produced SMSr, on the other hand, displayed hardly any CPE synthase activity. Whether this reflects a low intrinsic specific activity of the enzyme or failure of the wheat germ expression system to generate catalytically active SMSr remains to be established.
So far, catalytic activity of cell-free produced SMS family members was determined by monitoring the enzymatic conversion of the fluorescent short-chain ceramide analog NBD-Cer, which was added to SMS-containing proteoliposomes from an ethanolic stock. To exclude the possibility that the bulky NBD moiety of this analog interferes with SMS substrate specificity and provide further proof that the cell-free produced enzymes are correctly embedded in the lipid bilayer, we synthesized a ceramide analog carrying a C15-acyl chain with a clickable terminal alkyne group that closely mimics natural C16-ceramide (clickCer; Fig. 3A). Liposomes containing 2 mol% clickCer were used as membrane protein acceptor vesicles during cell-free production of SMS enzymes. SMS-mediated conversion of clickCer into clickSM and clickCPE was monitored by TLC analysis after click reacting SMS proteoliposomes with the fluorogenic dye 3-azido-7-hydroxycoumarin [Fig. 3B; Gaebler et al. 2013 (29)]. As shown in Fig. 3C, this approach confirmed that SMS1 and SMS1CPE act as monofunctional SM and CPE synthases, respectively.
Switching head group selectivity in mammalian sphingolipid biosynthesis
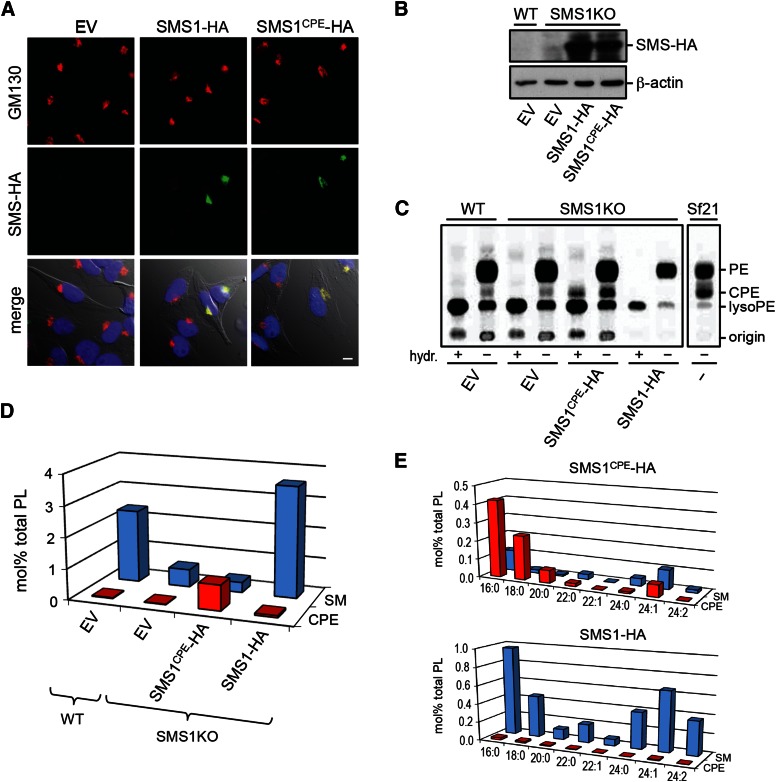
Having demonstrated that swapping exoplasmic residues with SMSr suffices to convert SMS1 into a CPE synthase, we next set out to generate mammalian cells producing CPE rather than SM as bulk phosphosphingolipid. First, we checked whether the exchange of residues necessary for switching head group selectivity of SMS1 had any impact on its subcellular distribution. As shown in Fig. 4A, immunofluorescence microscopy of HeLa cells transfected with HA-tagged versions of SMS1 and SMS1CPE revealed that both enzymes localize to the Golgi complex, where bulk production of SM is known to occur (14). To exchange Golgi-resident SMS1 for SMS1CPE, we used human myeloid leukemia KBM7-derived SMS1-null cells in which the SMS1 gene was inactivated by insertional mutagenesis (30). SMS1-null cells display only residual SMS activity (∼15%) compared with control KBM7 cells (38). This residual activity is likely due to SMS2, which is barely expressed in lymphoid cells (13, 20). SMS1-null cells stably expressing HA-tagged SMS1 or SMS1CPE were created by retroviral transduction and expression of the enzymes was verified by immunoblotting (Fig. 4B). As mammalian cells normally produce only trace amounts of CPE (18), we analyzed KBM7 and SMS1-null cells expressing SMS1 or SMS1CPE for their ability to synthesize CPE by metabolic labeling with [14C]ethanolamine. Labeling of Sf21 insect cells served as control as these cells typically produce bulk amounts of CPE owing to an SMSr-unrelated CPE synthase unique to insects (22). Labeling KBM7, SMS1-null, or SMS1-null cells expressing SMS1 with [14C]ethanolamine for 24 h did not yield any detectable levels of radioactive CPE. In contrast, labeled Sf21 and SMS1CPE-expressing SMS1-null cells contained substantial amounts of radioactive CPE (Fig. 4C), suggesting that SMS1CPE mediates bulk production of CPE.
Fig. 4.
Switching head group selectivity in mammalian sphingolipid biosynthesis. A: HeLa cells were transfected with empty vector (EV), SMS1-HA, or SMS1CPE-HA, fixed and then double labeled with antibodies against the HA-tag (green) and Golgi marker GM130 (red). Note that both SMS enzymes localize to the Golgi. Scale bar, 10 μm. B: Immunoblots of KBM7-derived WT and SMS1-null cells (SMS1KO) transfected with EV, SMS1-HA, or SMS1CPE-HA were stained with antibodies against the HA-tag and β-actin. C: KBM7-derived WT and SMS1KO cells transfected with EV, SMS1-HA, or SMS1CPE-HA were metabolically labeled with [14C]ethanolamine for 24 h and then subjected to lipid extraction, TLC analysis, and autoradiography. In some extracts, glycerolipids were deacylated by mild alkaline hydrolysis (hydr: +) prior to TLC analysis. Note that only SMS1CPE-expressing cells produce a radiolabeled lipid resistant to alkaline hydrolysis and with an Rf of CPE. Metabolic labeling of insect Sf21 cells, which produce bulk amounts of CPE, served as control. D: SM and CPE levels in lipid extracts of KBM7-derived WT and SMS1KO cells expressing SMS1-HA or SMS1CPE-HA were determined by LC/MS/MS and expressed as mol% of total phospholipid analyzed. E: Levels of SM and CPE species in KBM7-SMS1KO cells expressing SMS1-HA or SMS1CPE-HA were determined as in D. Data shown in D and E are representative of two independent experiments.
To assess the relative contribution of SMS1 and SMS1CPE to sphingolipid content at steady state, we next determined the lipid composition of KBM7 and SMS1-null cells expressing SMS1 or SMS1CPE using LC/MS/MS. As reported previously, removal of SMS1 resulted in a 6-fold drop in SM levels (38). Expression of SMS1CPE caused a ∼40-fold increase in CPE levels (43 ± 7, n = 2), making CPE the most abundant phosphosphingolipid in SMS1-null cells (Fig. 4D). This increase concerned both short- and long-chain CPE species, which largely overlapped with SM species synthesized by SMS1-expressing cells in terms of their ceramide backbone structures (Fig. 4E). No such increase in CPE levels was observed in SMSr-overexpressing cells (18 and unpublished observations). Swapping SMS1CPE against SMS1 in KBM7 cells had no significant impact on the levels of other sphingolipid (glucosylceramides, ceramides) or phospholipid classes (PC, PE, PI, phosphatidylserine; unpublished observations). Importantly, these results indicate that SMS1CPE, contrary to SMSr, functions as bona fide bulk CPE synthase. A comprehensive analysis of the consequences of substituting CPE for SM as bulk phosphosphingolipid on mammalian cell physiology will be the subject of a separate study.
Structural model of SMS substrate selectivity
In SMS-catalyzed SM/CPE production, transfer of the PC/PE head group onto ceramide is thought to proceed via formation of a choline/ethanolamine phospho-His intermediate (24). Here, we defined two critical determinants of head group selectivity among SMS family members, namely, i) an Asp/Glu residue at the -1 position of the catalytic His that binds the phospholipid head group in the third exoplasmic loop, loop c; and ii) the second exoplasmic loop, loop b, proximal to the other catalytic His (Fig. 5A). As outlined in the model depicted in Fig. 5B, we envision that loop b in SMS1 and SMS2 has a spacious fold that allows accommodation of a bulky phosphocholine head group in the active site, presumably via hydrophobic interactions with the methyl groups. A phosphoethanolamine head group lacks this possibility of interaction with loop b, which would explain why SMS2 preferentially synthesizes SM over CPE even when the enzyme has direct access to equal amounts of the phospholipid head group donors PC and PE. In contrast, we suggest that loop b of SMSr is more tightly folded over the active site, thus excluding entrance of a phosphocholine head group. This idea is supported by our finding that exchanging loop b between SMS1 and SMSr abolishes SMS1-mediated SM production (Fig. 1D). Our model also predicts that the Glu residue just upstream of the catalytic His in loop c of SMSr and SMS2 has a critical role in stabilizing a phosphoethanolamine head group in the active site, quite independently of the proposed loop b interactions. Indeed, exchanging this Glu residue for Asp blocked SMSr-mediated CPE production while exchanging Asp for Glu at the corresponding position in SMS1 yielded an SMS2-like enzyme with both SM and CPE synthase activity (Fig. 1D).
Fig. 5.
Structural model of SMS head group selectivity. A: Membrane topology and structural elements that contribute to substrate specificity of SMS enzymes. The invariant His and Asp residues that form the catalytic triad are marked in red. Head group selectivity of SMS enzymes is determined by a single residue adjacent to the catalytic His in exoplasmic loop c, that is, Asp in SMS1 (marked in green) or Glu in SMS2 and SMSr (marked in yellow), along with structural information in loop b (marked in blue). SAM, sterile α motif. B: Model explaining how structural elements in loops b and c of SMS enzymes cooperate to determine head group selectivity. See text for details.
To our knowledge, the only proposed structure for an SMS enzyme is based on a human SMS1 homology model created by Zhang et al. (39). In their model, loop b is folded on top of part of the active site, compatible with our model. The model by Zhang et al. also shows additional interactions of Phe173 and Phe177 from loop a with the phosphocholine head group of an SM molecule, which are suggested to contribute to SM binding. This leaves the alternative possibility that loop b of SMSr in the context of an SMS1 enzyme blocks the interaction with the two Phe residues, thereby abolishing SM synthesis. However, Phe173 is conserved in SMS1, SMS2, and SMSr ,whereas Phe177 only occurs in SMS1. Therefore, these Phe residues unlikely contribute to the head group selectivity of SMS enzymes as Phe173 is conserved in SMSr (which does not produce SM) while Phe177 is not strictly necessary for binding of phosphocholine head group as SMS2 is a good SM producer without it.
In our model, the point on the reaction coordinate at which the head group “selectivity filter” is applied is not defined. The way the model is drawn, for reasons of simplicity, assumes the coordination/exclusion of a choline/ethanolamine phospho-His intermediate. This would imply that PC and PE are constantly probing the active site, but that the intermediate is formed only when the appropriate phospholipid head group donor has entered. Alternatively, exclusion already takes place at the level of phospholipid entry, that is, depending on the protein’s selectivity, the appropriate phospholipid gains access to the active site. The difference between binding energies of SMS1 to PC and PE calculated by Zhang et al. (90 kJ/mol) suggests that the latter mechanism applies (39). However, photoaffinity labeling experiments with diazirine-containing phospholipid analogs (40) combined with a thorough analysis of enzyme kinetics will be necessary to distinguish the above two mechanisms. The latter will be a challenging task [see e.g., Mina et al. (41)], because for a bifunctional enzyme like SMS2, it involves systematic variation of the concentrations of three different substrates (i.e., ceramide, PC, and PE) when only the forward reactions are considered.
It is of interest to note that in a study on the substrate selectivity of an SMS-related ceramide phosphoinositol (CPI) synthase family from trypanosomes (42), the residue at the -1 position of the catalytic His that binds the phospholipid head group also had a marked effect on the various products formed by these enzymes (CPI, CPE, SM). The residues found at this position are more divergent (Ser, Phe) than those in SMS enzymes (Asp, Glu), in agreement with the increased divergence of the products (the head group of CPI has a net negative charge and is bulkier than that of SM and CPE). However, the authors did not wish to speculate on the specific molecular interactions that could cause the change in selectivity.
CONCLUDING REMARKS
In this work, we defined structural elements near the active site of SMS enzymes that impart catalytic specificity. Residues in the second and third exoplasmic loops proximal to the conserved catalytic triad turned out to be key determinants of head group selectivity. Swapping these residues with SMSr allowed conversion of the Golgi-resident SMS, SMS1, into a bulk producer of CPE and the establishment of a mammalian cell line that synthesizes CPE rather than SM as the dominant phosphosphingolipid. CPE normally represents only a minor fraction of the mammalian sphingolipid pool, with steady state levels ∼300- to 1,500-fold below those of SM (20). In contrast, CPE is the dominant phosphosphingolipid in most insects, including Drosophila (22, 43). Unlike SM, CPE does not interact favorably with cholesterol despite of its hydrogen-bonding properties (44). This may explain why insects are auxotroph for sterols and have a more relaxed structural requirement for sterols in their membranes than mammals, which appear highly adapted to the use of cholesterol (45, 46). Substitution of SM for CPE may therefore have a profound impact on cholesterol homeostasis in mammalian cells. Our current work provides an ideal basis to test this prediction.
At present, the physiological relevance of CPE biosynthesis in mammals is unclear (20, 21). We previously showed that acute disruption of SMSr-catalyzed CPE production in cultured cells causes a deregulation of ER ceramides and induction of mitochondrial apoptosis (18, 23). Whether CPE acts as a signaling lipid in SMSr-mediated ceramide homeostasis remains to be established. To address this possibility, our current efforts focus on manipulating ER-resident CPE pools by targeting active-site-engineered SMS1CPE to the ER. The establishment of SMS variants with altered head group selectivity not only provides fresh insight into the reaction mechanism underlying SM and CPE biosynthesis, but should also facilitate the development of isoenzyme-specific inhibitors.
Acknowledgments
The authors thank Tarja Grundström for technical assistance.
Footnotes
Abbreviations:
- Asp
- aspartic acid
- CPE
- ceramide phosphoethanolamine
- CPI
- ceramide phosphoinositol
- ER
- endoplasmatic reticulum
- Glu
- glutamic acid
- HA
- hemagglutinin
- His
- histidine
- IMDM
- Iscove modified Dulbecco medium
- NBD
- nitrobenzooxadiazole
- NBD-Cer
- NBD-C6-ceramide
- NBD-CPE
- NBD-C6-CPE
- ORF
- open reading frame
- PC
- phosphatidylcholine, PE, phosphatidylethanolamine
- PI
- phosphatidylinositiol
- SMS
- sphingomyelin synthase
- SMSr
- sphingomyelin synthase-related protein
This work was supported by a Marie Curie Intra-European Fellowship (J.G.M.M.), the European Union Seventh Framework Programme Marie-Curie ITN ‘Sphingonet’ Grant (289278), the Deutsche Forschungsgemeinschaft Sonderforschungsbereich/SFB944-P14 (J.C.M.H.), and an Incentive Award of the Faculty of Biology/Chemistry from the University of Osnabrück (M.K.). The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Slotte J. P. 2013. Biological functions of sphingomyelins. Prog. Lipid Res. 52: 424–437. [DOI] [PubMed] [Google Scholar]
- 2.Slotte J. P., and Bierman E. L.. 1988. Depletion of plasma-membrane sphingomyelin rapidly alters the distribution of cholesterol between plasma membranes and intracellular cholesterol pools in cultured fibroblasts. Biochem. J. 250: 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta A. K., and Rudney H.. 1991. Plasma membrane sphingomyelin and the regulation of HMG-CoA reductase activity and cholesterol biosynthesis in cell cultures. J. Lipid Res. 32: 125–136. [PubMed] [Google Scholar]
- 4.Sharpe H. J., Stevens T. J., and Munro S.. 2010. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 142: 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quiroga R., Trenchi A., González Montoro A., Valdez Taubas J., and Maccioni H. J. F.. 2013. Short transmembrane domains with high-volume exoplasmic halves determine retention of type II membrane proteins in the Golgi complex. J. Cell Sci. 126: 5344–5349. [DOI] [PubMed] [Google Scholar]
- 6.Milescu M., Bosmans F., Lee S., Alabi A. A., Kim J. I., and Swartz K. J.. 2009. Interactions between lipids and voltage sensor paddles detected with tarantula toxins. Nat. Struct. Mol. Biol. 16: 1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contreras F-X., Ernst A. M., Haberkant P., Björkholm P., Lindahl E., Gönen B., Tischer C., Elofsson A., von Heijne G., Thiele C., et al. 2012. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature. 481: 525–529. [DOI] [PubMed] [Google Scholar]
- 8.Miller M. E., Adhikary S., Kolokoltsov A. A., and Davey R. A.. 2012. Ebolavirus requires acid sphingomyelinase activity and plasma membrane sphingomyelin for infection. J. Virol. 86: 7473–7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaji-Hasegawa A., Hullin-Matsuda F., Greimel P., and Kobayashi T.. 2016. Pore-forming toxins: properties, diversity, and uses as tools to image sphingomyelin and ceramide phosphoethanolamine. Biochim. Biophys. Acta.. [DOI] [PubMed] [Google Scholar]
- 10.Adada M., Luberto C., and Canals D.. 2016. Inhibitors of the sphingomyelin cycle: sphingomyelin synthases and sphingomyelinases. Chem. Phys. Lipids. [DOI] [PubMed] [Google Scholar]
- 11.Maceyka M., and Spiegel S.. 2014. Sphingolipid metabolites in inflammatory disease. Nature. 510: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huitema K., van den Dikkenberg J., Brouwers J. F. H. M., and Holthuis J. C. M.. 2004. Identification of a family of animal sphingomyelin synthases. EMBO J. 23: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaoka S., Miyaji M., Kitano T., Umehara H., and Okazaki T.. 2004. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279: 18688–18693. [DOI] [PubMed] [Google Scholar]
- 14.Tafesse F. G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., and Holthuis J. C. M.. 2007. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J. Biol. Chem. 282: 17537–17547. [DOI] [PubMed] [Google Scholar]
- 15.Mitsutake S., Zama K., Yokota H., Yoshida T., Tanaka M., Mitsui M., Ikawa M., Okabe M., Tanaka Y., Yamashita T., et al. 2011. Dynamic modification of sphingomyelin in lipid microdomains controls development of obesity, fatty liver, and type 2 diabetes. J. Biol. Chem. 286: 28544–28555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J., Huan C., Chakraborty M., Zhang H., Lu D., Kuo M-S., Cao G., and Jiang X-C.. 2009. Macrophage sphingomyelin synthase 2 deficiency decreases atherosclerosis in mice. Circ. Res. 105: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z., Zhang H., Liu J., Liang C-P., Li Y., Teitelman G., Beyer T., Bui H. H., Peake D. A., Zhang Y., et al. 2011. Reducing plasma membrane sphingomyelin increases insulin sensitivity. Mol. Cell. Biol. 31: 4205–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vacaru A. M., Tafesse F. G., Ternes P., Kondylis V., Hermansson M., Brouwers J. F. H. M., Somerharju P., Rabouille C., and Holthuis J. C. M.. 2009. Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J. Cell Biol. 185: 1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ternes P., Brouwers J. F. H. M., van den Dikkenberg J., and Holthuis J. C. M.. 2009. Sphingomyelin synthase SMS2 displays dual activity as ceramide phosphoethanolamine synthase. J. Lipid Res. 50: 2270–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bickert A., Ginkel C., Kol M., vom Dorp K., Jastrow H., Degen J., Jacobs R. L., Vance D. E., Winterhager E., Jiang X-C., et al. 2015. Functional characterization of enzymes catalyzing ceramide phosphoethanolamine biosynthesis in mice. J. Lipid Res. 56: 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding T., Kabir I., Li Y., Lou C., Yazdanyar A., Xu J., Dong J., Zhou H., Park T., Boutjdir M., et al. 2015. All members in the sphingomyelin synthase gene family have ceramide phosphoethanolamine synthase activity. J. Lipid Res. 56: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vacaru A. M., van den Dikkenberg J., Ternes P., and Holthuis J. C. M.. 2013. Ceramide phosphoethanolamine biosynthesis in Drosophila is mediated by a unique ethanolamine phosphotransferase in the Golgi lumen. J. Biol. Chem. 288: 11520–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tafesse F. G., Vacaru A. M., Bosma E. F., Hermansson M., Jain A., Hilderink A., Somerharju P., and Holthuis J. C. M.. 2014. Sphingomyelin synthase-related protein SMSr is a suppressor of ceramide-induced mitochondrial apoptosis. J. Cell Sci. 127: 445–454. [DOI] [PubMed] [Google Scholar]
- 24.Tafesse F. G., Ternes P., and Holthuis J. C. M.. 2006. The multigenic sphingomyelin synthase family. J. Biol. Chem. 281: 29421–29425. [DOI] [PubMed] [Google Scholar]
- 25.Yeang C., Varshney S., Wang R., Zhang Y., Ye D., and Jiang X-C.. 2008. The domain responsible for sphingomyelin synthase (SMS) activity. Biochim. Biophys. Acta. 1781: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orr-Weaver T. L., Szostak J. W., and Rothstein R. J.. 1983. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 101: 228–245. [DOI] [PubMed] [Google Scholar]
- 27.Bartlett E. M., and Lewis D. H.. 1970. Spectrophotometric determination of phosphate esters in the presence and absence of orthophosphate. Anal. Biochem. 36: 159–167. [DOI] [PubMed] [Google Scholar]
- 28.Goren M. A., Nozawa A., Makino S-I., Wrobel R. L., and Fox B. G.. 2009. Cell-free translation of integral membrane proteins into unilamelar liposomes. Methods Enzymol. 463: 647–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaebler A., Milan R., Straub L., Hoelper D., Kuerschner L., and Thiele C.. 2013. Alkyne lipids as substrates for click chemistry-based in vitro enzymatic assays. J. Lipid Res. 54: 2282–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carette J. E., Guimaraes C. P., Varadarajan M., Park A. S., Wuethrich I., Godarova A., Kotecki M., Cochran B. H., Spooner E., Ploegh H. L., et al. 2009. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 326: 1231–1235. [DOI] [PubMed] [Google Scholar]
- 31.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 32.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 33.Pomorski T., Lombardi R., Riezman H., Devaux P. F., van Meer G., and Holthuis J. C. M.. 2003. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell. 14: 1240–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alder-Baerens N., Lisman Q., Luong L., Pomorski T., and Holthuis J. C. M.. 2006. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell. 17: 1632–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takatsu H., Tanaka G., Segawa K., Suzuki J., Nagata S., Nakayama K., and Shin H-W.. 2014. Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J. Biol. Chem. 289: 33543–33556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panatala R., Hennrich H., and Holthuis J. C. M.. 2015. Inner workings and biological impact of phospholipid flippases. J. Cell Sci. 128: 2021–2032. [DOI] [PubMed] [Google Scholar]
- 37.Sevova E. S., Goren M. A., Schwartz K. J., Hsu F-F., Turk J., Fox B. G., and Bangs J. D.. 2010. Cell-free synthesis and functional characterization of sphingolipid synthases from parasitic trypanosomatid protozoa. J. Biol. Chem. 285: 20580–20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tafesse F. G., Sanyal S., Ashour J., Guimaraes C. P., Hermansson M., Somerharju P., and Ploegh H. L.. 2013. Intact sphingomyelin biosynthetic pathway is essential for intracellular transport of influenza virus glycoproteins. Proc. Natl. Acad. Sci. USA. 110: 6406–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Lin F., Deng X., Wang R., and Ye D.. 2011. Molecular modeling of three-dimensional structure of human sphingomyelin synthase. Chin. J. Chem. 29: 1567–1575. [Google Scholar]
- 40.Haberkant P., Raijmakers R., Wildwater M., Sachsenheimer T., Brügger B., Maeda K., Houweling M., Gavin A-C., Schultz C., van Meer G., et al. 2013. In vivo profiling and visualization of cellular protein-lipid interactions using bifunctional fatty acids. Angew. Chem. Int. Ed. Engl. 52: 4033–4038. [DOI] [PubMed] [Google Scholar]
- 41.Mina J. G., Mosely J. A., Ali H. Z., Shams-Eldin H., Schwartz R. T., Steel P. G., and Denny P. W.. 2010. A plate-based assay system for analyses and screening of the Leishmania major inositol phosphorylceramide synthase. Int. J. Biochem. Cell Biol. 42: 1553–1561. [DOI] [PubMed] [Google Scholar]
- 42.Goren M. A., Fox B. G., and Bangs J. D.. 2011. Amino acid determinants of substrate selectivity in the Trypanosoma brucei sphingolipid synthase family. Biochemistry. 50: 8853–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho M., Sampaio J. L., Palm W., Brankatschk M., Eaton S., and Shevchenko A.. 2012. Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 8: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Térová B., Heczko R., and Slotte J. P.. 2005. On the importance of the phosphocholine methyl groups for sphingomyelin/cholesterol interactions in membranes: a study with ceramide phosphoethanolamine. Biophys. J. 88: 2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu L., von Bergmann K., Lutjohann D., Hobbs H. H., and Cohen J. C.. 2004. Selective sterol accumulation in ABCG5/ABCG8-deficient mice. J. Lipid Res. 45: 301–307. [DOI] [PubMed] [Google Scholar]
- 46.Carvalho M., Schwudke D., Sampaio J. L., Palm W., Riezman I., Dey G., Gupta G. D., Mayor S., Riezman H., Shevchenko A., et al. 2010. Survival strategies of a sterol auxotroph. Development. 137: 3675–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]