Abstract
The study objective was to retrospectively analyze the metabolic variables derived from 18 F-fluorodeoxyglucose (18F-FDG) positron emission tomography/computed tomography (PET/CT) as predictors of progression-free survival (PFS) and overall survival (OS) in advanced lung adenocarcinoma stratified by epidermal growth factor receptor (EGFR) mutation status. A total of 176 patients (91, EGFR mutation; 85, wild-type EGFR) who underwent 18F-FDG PET/CT before treatment were enrolled. The main 18F-FDG PET/CT-derived variables: primary tumor maximum standardized uptake value (SUVmaxT), primary tumor total lesion glycolysis (TLGT), the maximum SUVmax of all selected lesions in whole body determined using the Response Evaluation Criteria In Solid Tumors (RECIST) 1.1 criteria (SUVmaxWBR), and whole-body total TLG determined using the RECIST 1.1 criteria (TLGWBR) were measured. Survival analysis regarding TLGWBR, and other factors in advanced lung adenocarcinoma patients stratified using EGFR mutation status, were evaluated. The results indicated that high TLGWBR (≥259.85), EGFR wild-type, and high serum LDH were independent predictors of worse PFS and OS in all patients with advanced lung adenocarcinoma. Among patients with wild-type EGFR, only TLGWBR retained significance as an independent predictor of both PFS and OS. Among patients with the EGFR mutation, high serum LDH level was an independent predictor of worse PFS and OS, and high TLGWBR (≥259.85) was an independent predictor of worse PFS but not worse OS. In conclusion, TLGWBR is a promising parameter for prognostic stratification of patients with advanced lung adenocarcinoma and EGFR status; however, it cannot be used to further stratify the risk of worse OS for patients with the EGFR mutation. Further prospective studies are needed to validate our findings.
Introduction
Lung cancer is the most common cancer and the leading cause of malignant diseases worldwide. Non-small-cell lung cancer (NSCLC) accounts for 80–85% of lung cancer cases [1]. Adenocarcinoma is the most frequently diagnosed histological subtype of primary lung cancer in most countries, accounting for almost half of all lung cancers. Most patients with lung adenocarcinomas are diagnosed with advanced disease, which is clinically aggressive and has high metastatic potential [2]. Despite remarkable advances in surgical resections and targeted therapies, the prognosis of lung adenocarcinoma patients remains poor [3]. Thus, identifying novel prognostic methods is very important in improving the predictive ability of outcomes for patients with lung adenocarcinoma.
Because of its advantages of noninvasive evaluation and accuracy, positron emission tomography and computed tomography (PET-CT), as assessed using the maximum standardized uptake value (SUVmax), has been increasingly used for the assessment of the initial staging of NSCLC, restaging, recurrence, and monitoring the response to therapy [4–6]. The rationale for using FDG-PET in tumors is its ability to measure increased glucose metabolism in tumor cells. Recent studies have also shown that the degree of tumor FDG uptake (SUVmax) regarding PET was a significant prognostic factor in NSCLC [7–9]. When analyzing the prognostic capability of FDG PET, the most commonly used method for the quantification of FDG uptake is SUVmax [10]. However, there are many disadvantages to the use of SUVmax, particularly the variability introduced by the high statistical noise associated with single voxel analysis [11]. Total lesion glycolysis (TLG) not just denotes the SUVmax but also the tracer uptake of the entire lesion [12]. Despite several advantages of TLG over SUVmax, further study is needed to verify if TLG is a better prognostic predictive factor.
The biological characteristics of patients with the epidermal growth factor receptor (EGFR) mutation differ greatly from those with wild-type EGFR; this leads to the use of different therapies and different clinical outcomes in patients with advanced (≥stage IIIB) lung adenocarcinoma. Consequently, in patients with advanced-stage disease, it is important to study the prognostic value of the EGFR mutation and wild-type EGFR, and to identify a new non-invasive, convenient, and practical predictor of outcome. To our knowledge, no previous studies have demonstrated the value of whole body TLG (lesions were selected in accordance with the Response Evaluation Criteria In Solid Tumors (RECIST) 1.1 criteria) in the prediction of progression-free survival (PFS) in advanced lung adenocarcinoma stratified using EGFR mutation status. Our study was designed to investigate the prognostic value of the 18F-FDG PET-derived parameter TLG per RECIST 1.1 criteria in patients with advanced lung adenocarcinoma stratified using EGFR mutation status.
Materials and Methods
Patient characteristics
All participants were examined at our PET/CT center at least 4 weeks before receiving any therapy. One hundred and seventy-six consecutive nonsurgical patients with histologically proven advanced stage (≥stage IIIB lung adenocarcinoma who underwent a 18F-FDG PET/CT scan before treatment) were included in this retrospective study (from February 2009 to October 2013). Patients with advanced lung adenocarcinoma were identified for inclusion in this study in accordance with the following criteria. (1) No brain metastasis (the characterization of brain metastasis was influenced by high physiological cerebral uptake of 18F-FDG). (2) No other types of concurrent cancer. (3) Known EGFR gene mutation status. (4) Unequivocal primary lung tumors with delineated borders. (5) Treatment according to the institutional guidelines, and clinical follow-up for at least 24 months in our hospital. The study was approved by the Ethics Committee of Harbin Medical University Cancer Hospital. All patients have provided oral consent forms for the use of their medical data. We could not obtain written informed consent from all participants as this was a retrospective study and the majority of the patients had been discharged from hospital at the time of analysis. The oral informed consent was documented in the electronic or paper patient file and approved by the local ethics committee. We collected and analyzed the data anonymously, and no results were ever connected to their identities.
Staging was performed according to the Union for International Cancer Control/the American Joint Committee on Cancer Staging System for NSCLC (UICC/AJCC) [13]. Tissue specimens obtained by conventional bronchoscope or CT-guided percutaneous transthoracic needle biopsy excision were used for EGFR gene detection. EGFR mutations (exons 18, 19, 20, and 21) and v-Ki-ras2 Kirsten rat sarcoma viral oncogene (KRAS) mutations (codons 12, 13, and 61) were analyzed using the amplification refractory mutation system [14]. For analysis of the EML4 (echinodermmicrotubule-associated protein like 4)–ALK (anaplastic lymphoma kinase) gene fusion, we performed fluorescence in situ hybridization analysis for ALK immunohistochemistry-positive cases. According to the National Comprehensive Cancer Network (NCCN) guidelines [15], patients with mutant EGFR receive targeted drugs (gefitinib), while patients with rapidly progressive disease undergo second-line treatment including docetaxel or pemetrexed. For patients with wild-type EGFR, standard first-line treatment usually consists of platinum-based doublet chemotherapy, and second-line treatment options available to patients who experience failure of first-line treatment include additional chemotherapy (docetaxel and pemetrexed). Patients with EML4–ALK rearrangement receive targeted drugs (crizotinib), while patients with rapidly progressive disease undergo second-line treatment consisting of platinum-based doublet chemotherapy. The treatment of patients with mutant KRAS is the same as the treatment of patients with wild-type EGFR.
Data regarding clinicopathological characteristics, treatment, follow-up, and Eastern Cooperative Oncology Group (ECOG) performance status were recorded according to the patients’ medical records. The clinical data for each patient were discussed and determined by two oncologists. Lesions were selected in accordance with the Response Evaluation Criteria In Solid Tumors (RECIST) 1.1 criteria as previously described [16]. Briefly, pulmonary tumors with the longest diameter ≥10 mm, lymph nodes with the longest diameter ≥15 mm in the short axis, and metastatic solid lesions with the longest diameter ≥10 mm were selected. All measurable lesions up to a maximum of five in a single patient, and two lesions in one organ were recorded in our study.
PET/CT acquisition
All patients fasted for 4–6 h. Blood glucose levels were checked in the peripheral blood (<150 mg/dL was considered normal) before the PET/CT examination. PET/CT images were obtained using an integrated PET/CT scanner (Discovery ST: GE Medical systems, Milwaukee, WI, USA) at 60 min after intravenous administration of 18F-FDG (5.55–7.40 MBq/kg). The scan range started at the mid-thighs and proceeded to the head. A whole-body unenhanced CT scan was performed using the following parameters: 140 kV, 150 mA, 0.8 s per rotation, 22.5 mm/s table speed, and slice thickness of 3.75 mm. Data from the CT scans were reconstructed from a 512 × 512 matrix to a 128 × 128 matrix to satisfactorily match the PET data and allow image fusion. The PET scan was carried out in the same position for all patients and using the two-dimensional imaging mode. PET image datasets were reconstructed using an iterative algorithm (the ordered subsets expectation maximization). The emission scan was obtained at 3 min per bed position, and six to seven bed positions were generally performed for all patients. All of the transaxial, sagittal, and coronal images were displayed and analyzed on a workstation (Xeleris; General Electric Medical Systems,Milwaukee, WI, USA).
All CT, PET, and PET/CT reconstructed images were loaded onto a workstation (Advanced workstation 4.6; GE Medical Systems, Milwaukee, WI, USA). After identification of the tumors, parameters were measured from the attenuation-corrected torso 18F-FDG PET/CT images and calculated semiautomatically using PET VCAR; the PET-based lesion contour was defined using a threshold of 40% of the tumoral SUVmax, and the corresponding parameters were provided. If the defined tumor margin was not appropriate, relative to the fused CT, adjustment of the SUVmax threshold was required until a satisfactory tumoral outline was achieved. The volume boundaries were automatically drawn to incorporate each target lesion in the axial, coronal, and sagittal PET-CT images. In our study, the output results included the SUVmax, metabolic tumor volume (MTV), and TLG of the tumor. 18F-FDG PET/CT images were assessed by two experienced nuclear medicine physicians with PET/CT imaging experience as well as familiarity with PET-VCAR software and our PACS system. These images were reviewed to localize the target lesions that had been confirmed by two nuclear medicine physicians; any discrepancies were resolved by consensus. For each patient, SUVmaxT was the SUVmax of the primary tumor. SUVmaxWBR was the maximum SUVmax of all selected lesions in the whole body determined using the RECIST 1.1 criteria. TLG was the MTV multiplied by the mean SUV of the tumor. TLG of the primary tumor (TLGT) was the MTV multiplied by the mean SUV of the primary lung tumor. TLGWBR was the whole body TLG calculated as the sum of all corresponding TLG values of the lesions selected using the RECIST 1.1 criteria.
Statistical analysis
The results of 18F-PET/CT were displayed as continuous variables. To test the consistency of the measurement of TLGWBR on PET images, TLGWBR were calculated independently twice by two groups of nuclear medicine physicians (any discrepancies were resolved by consensus in each group of two persons) per RECIST 1.1 in all patients. Reliability of TLGWBR was measured using the intraclass correlation coefficient (ICC) generated by a two-way random-effects model with an absolute agreement definition and is reported as a point estimate with a 95% confidence interval (CI). ICC is interpreted as follows: an ICC of 0.00–0.20 indicates slight reproducibility; an ICC of 0.21–0.40, fair reproducibility; an ICC of 0.41–0.60, moderate reproducibility; an ICC of 0.61–0.80, substantial reproducibility; and an ICC of > 0.80, almost perfect reproducibility [17]. The cutoff values for the categorization of low and high TLGWBR, TLGT, SUVmaxT, SUVmaxWBR, and age were proceeded by R (version 3.3.2) with the package of survival ROC (version:1.0.3) (R Development Core Team, Vienna, Austria, http://www.R-project.org). The cutoff values for the categorization of low and high TLGWBR, TLGT, SUVmaxT, SUVmaxWBR, and age were determined using receiver operation characteristic (ROC) curve analyses. The optimal cut-off value was determined using the value representing the maximal area under the curve, and maximal sum of sensitivity and specificity; the same cut-off value for each parameter was applied to compare the PFS and overall survival (OS) in all group analyses. Univariate analysis of prognostic factors for PFS and OS was achieved using the Kaplan–Meier method, and the log-rank test was used to evaluate the significance of the differences between the survival curves, the Cox proportional hazards model that included significant univariate variables was used to determine independent prognostic factors for PFS and OS in multivariate survival analyses. Risk of death was estimated on the basis of hazard ratios and the 95% confidence interval was recorded. Sex, age, performance status, serum LDH level, serum CEA level, EGFR gene status, and 18F-FDG PET/CT-derived parameters were used for univariate and multivariate prediction of OS and PFS. Variables with a P value < 0.05 on univariate analysis were selected for multivariate analysis. To evaluate multi-collinearity between PET/CT parameters and the relationship between serum LDH and metabolic parameters, Spearman’s rank correlation coefficient was calculated. The SPSS 19.0 (SPSS Inc., Chicago, IL, USA) software package was used for the analysis. A two-tailed P value < 0.05 was considered to indicate statistical significance.
Results
Patient characters and EGFR genetic status
The 176 patients with advanced lung adenocarcinoma consisted of 72 men and 104 women with a median age of 61 (range, 30–87) years. A total of 91 (52%) patients with the EGFR mutation were found. Eleven EGFR mutation-negative patients with KRAS mutations and two EGFR mutation-negative patients with EML4–ALK rearrangement were identified. No co-occurrences of driver mutations were found. Patients with wild-type EGFR consisted of 38 men and 47 women with a median age of 61 (range, 30–84) years. Patients with the EGFR mutation consisted of 34 men and 57 women with a median age of 60 (range, 34–87) years. Their characteristics are summarized in Table 1.
Table 1. General characteristics of patients with advanced lung adenocarcinoma.
| Characteristic | n(%) |
|---|---|
| Age | |
| <64 | 105(60) |
| ≥64 | 71(40) |
| Sex | |
| Male | 72(41) |
| Female | 104(59) |
| History of smoking | |
| + | 103(59) |
| - | 73(41) |
| Mutation(EGRF) | |
| + | 91(52) |
| - | 85(48) |
| ECOG performance status | |
| 0/1 | 122(69) |
| 2/3/4 | 54(31) |
| Serum CEA level | |
| High | 85(48) |
| Normal | 91(52) |
| Serum LDH level | |
| High | 28(16) |
| Normal | 148(84) |
| SUVmaxT | |
| <11.86 | 99(56) |
| ≥11.86 | 77(44) |
| SUVmaxWBR | |
| <16.14 | 99(56) |
| ≥16.14 | 77(44) |
| TLGT | |
| <104.65 | 108(61) |
| ≥104.65 | 68(39) |
| TLGWBR | |
| <259.85 | 99(56) |
| ≥259.85 | 77(44) |
Abbreviations: EGFR: epidermal growth factor receptor; ECOG: Eastern Cooperative Oncology Group; SUVmaxT: primary tumor maximum standardized uptake value (SUVmax); SUVmaxWBR: the maximum SUVmax of all selected lesions in whole body determined using the Response Evaluation Criteria In Solid Tumors (RECIST) 1.1 criteria; TLGT: primary tumor total lesion glycolysis (TLG); TLGWBR: whole-body total TLG determined using the RECIST 1.1 criteria.
18F-FDG PET/CT-derived parameters in advanced lung adenocarcinoma patients
In 176 patients with advanced lung adenocarcinoma, the median TLGWBR was 208.9 (range, 4.09–5038.04), the median TLGT was 62.07 (range, 2.59–4578.27), the median SUVmaxWBR was 13.38 (range, 3.25–50.07), and the median SUVmaxT was 10.58 (range, 1.96–40.44). In 85 patients with wild-type EGFR, the median TLGWBR was 305.59 (range, 4.09–5038.04), the median TLGT was 55.75 (range, 4.09–4578.27), the median SUVmaxWBR was 12.27 (range, 3.81–49.56), and the median SUVmaxT was 9.10 (range, 1.96–40.40). In 91 patients with the EGFR mutation, the median TLGWBR was 182.11 (range, 5.29–2575.26), the median TLGT was 67.30 (range, 2.59–1172.83), the median SUVmaxWBR was 13.47 (range, 3.25–50.07), and the median SUVmaxT was 12.14 (range, 2.03–40.44).
The ICC (the reliability between two group of nuclear medicine physicians with respect to TLGWBR on PET per RECIST 1.1 in all patients) was 0.972 (95% CI: 0.963, 0.979). Because the TLG is calculated by multiplying the MTV and mean SUV, multi-collinearity between MTV and TLG was evaluated. The result of the Spearman’s rank correlation test showed a significant correlation between MTVWBR and TLGWBR (r = 0.898; P<0.0001). Spearman’s rank correlation analyses showed a correlation between serum LDH level and TLGWBR (r = 0.205; P = 0.006). No such correlation was identified between serum LDH level and SUVmaxT, SUVmaxWBR, or TLGT (r = −0.037, P = 0.624; r = 0.079, P = 0.296; and r = −0.004, P = 0.958, respectively). In 91 patients with the EGFR mutation, the LDH level was also correlated with TLGWBR (r = 0.210; P = 0.046); no such correlation was found between serum LDH level and TLGWBR (r = 0.187; P = 0.086) in 85 patients with wild-type EGFR. According to the ROC analysis, the cutoff points for the categorization of low and high SUVmaxT, SUVmaxWBR, TLGT, TLGWBR, and age values were 11.68, 16.14, 104.65, 259.85, and 64, respectively. The ROC curve for identifying the optimal cutoff point of TLGWBR in advanced lung adenocarcinoma patients is shown in Fig 1.
Fig 1. ROC curve for the identification of the optimal cutoff point for whole body total lesion glycolysis (TLGWBR) in advanced lung adenocarcinoma patients.
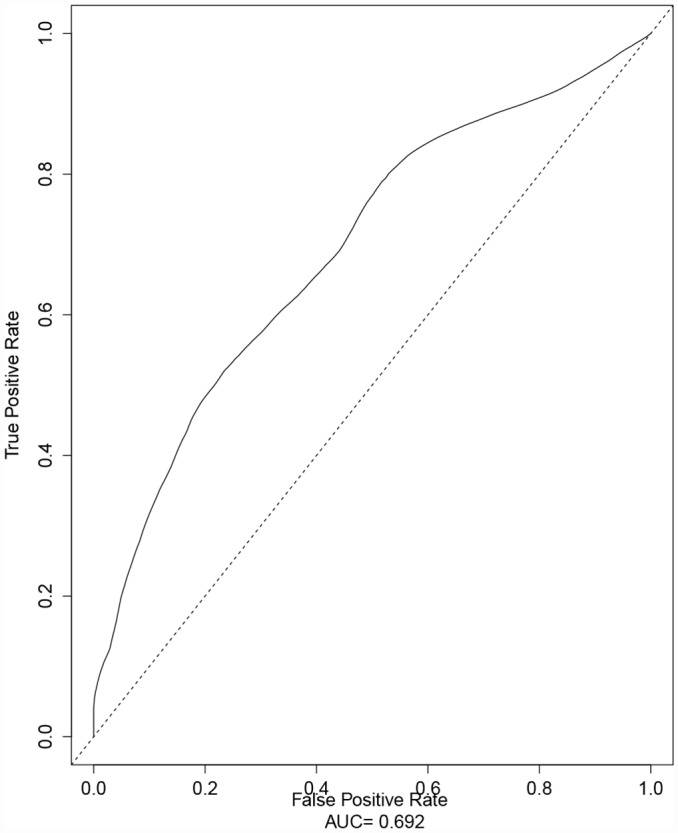
The optimal cutoff value for TLGWBR determined using the Response Evaluation Criteria In Solid Tumors 1.1 criteria was 258.98, and the area under the curve with the maximum summation of sensitivity and specificity was 0.69.
Survival analysis in 176 patients with advanced lung adenocarcinoma
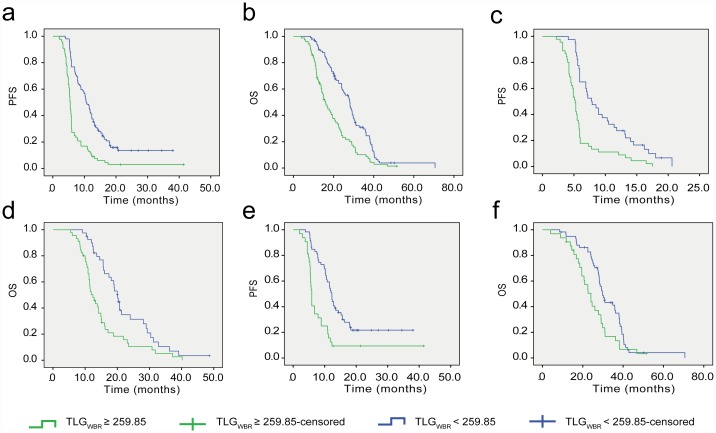
The median PFS and OS for 176 patients in this study were7.4 (range, 2.0–41.4) months and 20.7 (range, 4.0–70.6) months, respectively. Among all patients, univariate survival analysis showed that high TLGWBR (≥259.85), high TLGT (≥104.65), high SUVmaxWBR (≥16.14), EGFR wild-type, high serum LDH level, and poor ECOG performance status were positively correlated with worse PFS. High TLGWBR (≥259.85), high SUVmaxWBR (≥16.14), EGFR wild-type, high serum LDH level, poor ECOG performance status, and male sex were positively correlated with worse OS (Table 2). The Kaplan–Meier survival curves for PFS and OS in 176 patients with advanced lung adenocarcinoma according to TLGWBR are presented in Fig 2a and 2b. Multivariate survival analysis showed that high TLGWBR (≥259.85), presence of wild-type EGFR, and high serum LDH level were independent predictors of worse PFS and OS (Table 3). Representative examples from patients with advanced lung adenocarcinoma who had low and high TLGWBR values are shown in Fig 3.
Table 2. Univariate analysis of progression-free survival and overall survival in 176 patients with advanced lung adenocarcinoma.
| Parameter | No. of patients | Progression-free survival | Overall survival | ||
|---|---|---|---|---|---|
| Mean survival time(months ±SE) | P value | Mean survival time(months ±SE) | P value | ||
| Age | |||||
| <64 | 105 | 10.16±1.07 | 0.311 | 23.06±1.42 | 0.606 |
| ≥64 | 71 | 11.95±1.05 | 24.95±1.25 | ||
| Sex | |||||
| Male | 72 | 10.11±1.08 | 0.192 | 21.30±1.39 | 0.044 |
| Female | 104 | 11.75±1.01 | 26.11±1.23 | ||
| Smoker | |||||
| + | 103 | 9.49±0.71 | 0.057 | 22.52±1.14 | 0.128 |
| - | 73 | 12.87±1.33 | 26.48±1.55 | ||
| ECOG performance status | |||||
| 0/1 | 122 | 12.52±1.02 | 0.003 | 26.18±1.18 | 0.006 |
| 2/3/4 | 54 | 7.83±0.65 | 19.84±1.58 | ||
| Serum CEA level | |||||
| Normal(≤5) | 91 | 11.33±1.12 | 0.972 | 23.94±1.48 | 0.555 |
| High(>5) | 85 | 10.79±0.97 | 24.70±1.25 | ||
| Serum LDH level | |||||
| High | 28 | 6.13±0.65 | 0.000 | 18.34±2.02 | 0.002 |
| Normal | 148 | 12.41±0.94 | 25.58±1.12 | ||
| Mutation | |||||
| EGRF(+) | 91 | 14.64±1.36 | 0.000 | 29.59±1.36 | 0.000 |
| EGRF(-) | 85 | 7.76±0.49 | 18.38±1.06 | ||
| SUVmaxT | |||||
| <11.86 | 99 | 10.49±0.88 | 0.817 | 24.58±1.42 | 0.998 |
| ≥11.86 | 77 | 12.01±1.34 | 23.96±1.30 | ||
| SUVmaxWBR | |||||
| <16.14 | 99 | 12.48±1.00 | 0.002 | 26.79±1.44 | 0.007 |
| ≥16.14 | 77 | 9.61±1.16 | 21.32±1.24 | ||
| TLGT | |||||
| <104.65 | 108 | 12.05±0.99 | 0.026 | 26.21±1.34 | 0.055 |
| ≥104.65 | 68 | 9.79±1.15 | 21.38±1.35 | ||
| TLGWBR | |||||
| <259.85 | 99 | 13.99±1.10 | 0.000 | 28.37±1.35 | 0.000 |
| ≥259.85 | 77 | 7.39±0.79 | 19.11±1.20 | ||
Abbreviations: ECOG: Eastern Cooperative Oncology Group; EGFR: epidermal growth factor receptor; SUVmaxT: primary tumor maximum standardized uptake value (SUVmax); SUVmaxWBR: the maximum SUVmax of all selected lesions in whole body determined using the Response Evaluation Criteria In Solid Tumors (RECIST) 1.1 criteria; TLGT: primary tumor total lesion glycolysis (TLG); TLGWBR: whole-body total TLG determined using the RECIST 1.1 criteria.
Fig 2. Kaplan–Meier survival analysis.
a progression-free survival (PFS) and b overall survival (OS) according to whole-body total lesion glycolysis (TLGWBR) determined using the Response Evaluation Criteria In Solid Tumors 1.1 criteria in 176 patients with advanced lung adenocarcinoma. Kaplan–Meier analysis of c PFS and d OS according to TLGWBR in 85 patients with wild-type EGFR. e Kaplan–Meier analysis of PFS and f OS according to TLGWBR in 91 patients with the EGFR mutation.
Table 3. Multivariate analysis of progression-free survival and overall survival in 176 patients with advanced lung adenocarcinoma.
| Parameter | Progression-free survival | Overall survival | ||||
|---|---|---|---|---|---|---|
| Hazard ratio(exp. B) | 95%CI | P value | Hazard ratio(exp. B) | 95%CI | P value | |
| SUVmaxWBR≥16.14 | 1.466 | 1.043–2.061 | 0.280 | 1.381 | 0.976–1.952 | 0.068 |
| TLGT≥104.65 | 1.001 | 0.706–1.419 | 0.995 | |||
| TLGWBR≥259.85 | 1.896 | 1.296–2.775 | 0.001 | 1.579 | 1.089–2.289 | 0.016 |
| EGFR(-) | 1.763 | 1.262–2.462 | 0.001 | 2.502 | 1.773–3.530 | 0.000 |
| Serum LDH (high) | 2.021 | 1.294–3.156 | 0.002 | 1.620 | 1.007–2.606 | 0.047 |
| ECOG performance status(0/1) | 1.381 | 0.965–1.975 | 0.077 | 1.185 | 0.814–1.727 | 0.375 |
| Sex (male) | 0.722 | 0.516–1.012 | 0.058 | |||
Abbreviations: SUVmaxWBR: the maximum SUVmax of all selected lesions in whole body determined using the Response Evaluation Criteria In Solid Tumors (RECIST) 1.1 criteria; TLGT: primary tumor total lesion glycolysis (TLG); TLGWBR: whole-body total TLG determined using the RECIST 1.1 criteria; EGFR: epidermal growth factor receptor; ECOG: Eastern Cooperative Oncology Group.
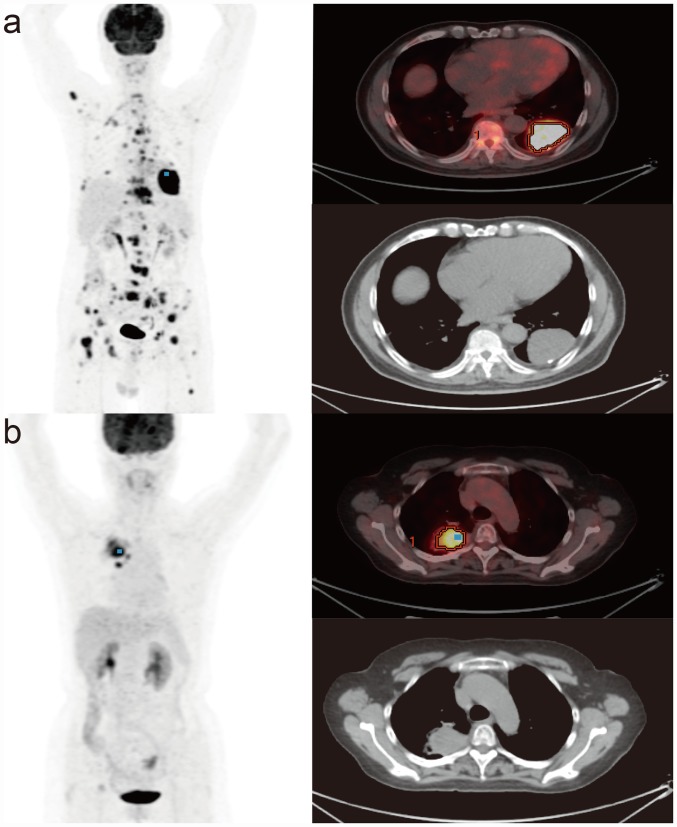
Fig 3. 18F-FDG PET/CT in patients with advanced lung adenocarcinoma.
a A 71-year-old man with high whole-body total lesion glycolysis (TLGWBR) determined using the Response Evaluation Criteria In Solid Tumors 1.1 criteria (7944.00) and primary tumor total lesion glycolysis (TLGT) (1250.46). Progression-free survival (PFS) was 5.9 months and overall survival (OS) was 11.9 months. EGFR mutation status was the wild-type. b A 68-year-old woman with low TLGWBR (179.74) and TLGT (142.41). PFS was 12.3 months and OS was 35.9 months. EGFR mutation status involved the mutation of exon 19 (delE746-A750).
Prognostic value of TLGWBR in 85 patients with wild-type EGFR
Univariate survival analysis indicated a significant association between high TLGWBR (≥259.85), high TLGT (≥104.65), high serum LDH, male sex, and worse PFS and OS (Table 4). The Kaplan–Meier survival curves for PFS and OS of patients with wild-type EGFR according to TLGWBR are shown in Fig 2c and 2d. However, on multivariate analysis only TLGWBR remained significant for both PFS and OS (Table 5).
Table 4. Univariate analysis of progression-free survival and overall survival in patients with wild-type EGFR.
| Parameter | No. of patients | Progression-free survival | Overall survival | ||
|---|---|---|---|---|---|
| Mean survival time(months ±SE) | P value | Mean survival time(months ±SE | P value | ||
| Age | |||||
| <64 | 48 | 7.58±0.73 | 0.973 | 18.16±1.34 | 0.751 |
| ≥64 | 37 | 7.83±0.63 | 18.72±1.76 | ||
| Sex | |||||
| Male | 38 | 6.65±0.67 | 0.038 | 15.46±1.44 | 0.009 |
| Female | 47 | 8.62±0.65 | 20.60±1.42 | ||
| Smoker | |||||
| + | 55 | 7.54±0.64 | 0.606 | 19.30±1.61 | 0.543 |
| - | 30 | 8.21±0.76 | 17.92±1.39 | ||
| ECOG performance status | |||||
| 0/1 | 49 | 8.40±0.69 | 0.147 | 19.20±1.37 | 0.497 |
| 2/3/4 | 36 | 6.94±0.66 | 17.32±1.72 | ||
| Serum CEA level | |||||
| Normal(≤5) | 60 | 7.74±0.51 | 0.948 | 18.17±1.18 | 0.711 |
| High(>5) | 25 | 7.69±1.08 | 18.90±2.31 | ||
| Serum LDH level | |||||
| High | 14 | 5.54±0.95 | 0.007 | 18.34±2.02 | 0.048 |
| Normal | 71 | 8.20±0.54 | 13.78±2.41 | ||
| TLGT | |||||
| <104.65 | 51 | 8.55±0.70 | 0.026 | 20.27±1.54 | 0.024 |
| ≥104.65 | 34 | 6.61±0.59 | 15.73±1.35 | ||
| TLGWBR | |||||
| <259.85 | 40 | 9.74±0.77 | 0.000 | 22.09±1.57 | 0.001 |
| ≥259.85 | 45 | 6.02±0.51 | 15.71±1.26 | ||
Abbreviations: ECOG: Eastern Cooperative Oncology Group; TLGT: primary tumor total lesion glycolysis (TLG); TLGWBR: whole-body total TLG determined using the Response Evaluation Criteria In Solid Tumors 1.1 criteria.
Table 5. Multivariate analysis of progression-free survival and overall survival in patients with wild-type EGFR.
| Parameter | Progression-free survival | Overall survival | ||||
|---|---|---|---|---|---|---|
| Hazard ratio(exp. B) | 95%CI | P value | Hazard ratio(exp. B) | 95%CI | P value | |
| TLGT≥104.65 | 1.229 | 0.766–1.973 | 0.393 | 1.381 | 0.847–2.252 | 0.195 |
| TLGWBR≥259.85 | 2.128 | 1.290–3.509 | 0.003 | 1.881 | 1.127–3.139 | 0.016 |
| Serum LDH (high) | 1.432 | 0.772–2.656 | 0.255 | 1.181 | 0.589–2.369 | 0.640 |
| Sex (male) | 0.701 | 0.444–1.105 | 0.126 | 0.569 | 0.355–0.912 | 0.019 |
Abbreviations: TLGT: primary tumor total lesion glycolysis (TLG); TLGWBR: whole-body total TLG determined using the Response Evaluation Criteria In Solid Tumors 1.1 criteria.
Prognostic values of TLGWBR in 91 patients with the EGFR mutation
Univariate survival analysis revealed that a worse PFS was positively correlated with high TLGWBR (≥259.85), high serum LDH level, and high serum CEA level. Worse OS appeared to be positively correlated with high TLGWBR (≥259.85) and high serum LDH (Table 6). The Kaplan–Meier survival curves for PFS and OS in patients with the EGFR mutation patients according to TLGWBR are shown in Fig 2e and 2f. Multivariate survival analysis revealed that high serum LDH level was an independent predictor of worse PFS and OS, and high TLGWBR (≥259.85) was an independent predictor of worse PFS but not worse OS (Table 7).
Table 6. Univariate analysis of progression-free survival and overall survival in patients with the EGFR mutation.
| Parameter | No. of patients | Progression-free survival | Overall survival | ||
|---|---|---|---|---|---|
| Mean survival time(months ±SE) | P value | Mean survival time(months ±SE) | P value | ||
| Age | |||||
| <64 | 57 | 13.27±2.04 | 0.504 | 30.45±1.65 | 0.503 |
| ≥64 | 34 | 14.77±1.61 | 27.42±1.93 | ||
| Sex | |||||
| Male | 34 | 13.90±1.90 | 0.869 | 27.68±1.85 | 0.494 |
| Female | 57 | 14.05±1.64 | 30.22±1.66 | ||
| Smoker | |||||
| + | 48 | 11.57±1.19 | 0.147 | 27.79±1.52 | 0.344 |
| - | 43 | 16.34±2.09 | 30.95±2.02 | ||
| ECOG performance status | |||||
| 0/1 | 73 | 14.98±1.51 | 0.215 | 30.57±1.49 | 0.236 |
| 2/3/4 | 18 | 9.58±1.35 | 25.20±3.03 | ||
| Serum CEA level | |||||
| Normal(≤5) | 31 | 18.72±2.65 | 0.049 | 33.31±2.30 | 0.078 |
| High(>5) | 60 | 11.75±1.18 | 27.07±1.38 | ||
| Serum LDH level | |||||
| High | 14 | 6.71±0.91 | 0.000 | 21.87±2.81 | 0.002 |
| Normal | 77 | 16.09±1.54 | 31.07±1.48 | ||
| TLGT | |||||
| <104.65 | 57 | 14.91±1.57 | 0.215 | 30.87±1.66 | 0.506 |
| ≥104.65 | 34 | 12.98±2.09 | 26.77±1.95 | ||
| TLGWBR | |||||
| <259.85 | 59 | 16.66±1.57 | 0.000 | 31.83±1.61 | 0.018 |
| ≥259.85 | 32 | 9.84±1.85 | 24.62±1.87 | ||
Abbreviations: ECOG: Eastern Cooperative Oncology Group; TLGT: primary tumor total lesion glycolysis (TLG); TLGWBR: whole-body total TLG determined using the Response Evaluation Criteria In Solid Tumors 1.1 criteria.
Table 7. Multivariate analysis of progression-free survival and overall survival in patients with the EGFR mutation.
| Parameter | Progression-free survival | Overall survival | ||||
|---|---|---|---|---|---|---|
| Hazard ratio(exp. B) | 95%CI | P value | Hazard ratio(exp. B) | 95%CI | P value | |
| TLGWBR≥259.85 | 2.197 | 1.305–3.696 | 0.003 | 1.532 | 0.949–2.472 | 0.086 |
| Serum CEA (high) | 1.355 | 0.806–2.278 | 0.251 | |||
| Serum LDH (high) | 2.215 | 1.177–4.166 | 0.014 | 2.128 | 1.149–3.941 | 0.014 |
Abbreviations: TLGWBR: whole-body total lesion glycolysis determined using the Response Evaluation Criteria In Solid Tumors 1.1 criteria.
Discussion
The variation in the survival of patients with advanced lung adenocarcinoma is associated with multiple factors (EGFR mutation status, ECOG performance status, metabolism variables, serum markers, and sex). 18F-FDG PET/CT is a promising method and provides parameters for the selection of patients who have a better prognosis. In contrast to conventional methods, this approach for selecting patients could reveal the metabolism-specific differences in the prognostic value of PET/CT.
To the best of our knowledge, the current study involves the largest number of cases of any clinical study date, regarding the analysis of the prognostic significance of metabolic and volumetric parameters derived from 18F-FDG PET/CT in advanced lung adenocarcinoma patients with the EGFR mutation. Our study showed that TLGWBR (TLGWBR ≥259.85), EGFR mutation status, and serum LDH level for baseline 18F-FDG PET/CT were significant independent prognostic factors for PFS and OS in 176 patients with advanced lung adenocarcinoma. Other studies have reported that whole-body TLG was an independent prognostic factor in advanced lung adenocarcinoma patients with the EGFR mutation [18–19]. It should be noted that we have used TLGWBR to represent whole-body metabolic tumor burden by selecting tumors in accordance with the RECIST 1.1 criteria. In addition, we analyzed the LDH level, which has not been previously been considered as a prognostic factor for PFS and OS; in our study, high serum LDH level was found to be an independent predictor of worse OS and PFS for all advanced lung adenocarcinoma patients. Furthermore, we found that high LDH serum level was an independent predictor of PFS and OS in patients with the EGFR mutation; this may have been because serum LDH level is related to intratumoral angiogenesis, tumor invasion ability, and resistance to therapy [20–21]. A previous study has reported that LDH levels had a significant effect on PFS in patients treated with erlotinib [22]. The present study also indicated that LDH level exhibited a correlation with TLGWBR in the patients with advanced lung adenocarcinoma. However, no such correlation was found between serum LDH level and other PET/CT parameters (SUVmaxT, SUVmaxWBR, TLGT). Furthermore, this correlation was mainly shown in patients with the EGFR mutation.
Regarding the correlation between the PET/CT metabolic parameters (SUVmax, primary TLG, and whole-body TLG) and survival in patients with advanced NSCLC, there has been some disagreement among researchers. Some studies have reported that whole-body TLG is a better prognostic predictor of worse OS and PFS in patients with advanced NSCLC [23–24], whereas other studies have found no such correlation [25].
SUVmax as a metabolic parameter derived from PET/CT is easily quantified and widely used; nevertheless, SUVmaxT and SUVmaxWBR only denote the highest metabolic activity within the tumor, and do not consider the tumor extent [26–27]. In our study, SUVmaxT and SUVmaxWBR did not show any significant prognostic value as an independent predictor. This finding is consistent with those of recent studies [19, 24, 28]. In contrast, previous studies have revealed that SUVmaxT or SUVmaxWBR were independent predictors of survival in NSCLC [12, 29]. In contrast to SUVmax, TLG incorporates the metabolic burden and disease extent and may provide a higher predictive value. The results of our study revealed that whole-body tumor glycolysis represented by TLGWBR was a strong predictor of survival, which was similar to previously reported findings [18, 23–24]. However, our study indicated that total glycolysis in the primary tumor alone (TLGT) was not an independent predictor of survival, in line with some recent studies [19, 30]. The cause of this may be related to the fact that the TLGT, although accounting for the metabolic burden of the primary tumor, does not take the metabolic burden of the metastatic tumors into account. This finding contradicts previous reports suggesting that FDG uptake (TLGT) in the primary tumor was an independent prognostic factor [24, 31].
To date, only one other study has investigated the predictive value of metabolic and volumetric variables concerning the use of pretreatment 18F-FDG PET/CT in advanced lung adenocarcinoma patients stratified by EGFR mutation status using the RECIST 1.1 criteria [30]. In our study, the result of almost perfect reproducibility between nuclear medicine physicians with respect to TLGWBR on PET per RECIST 1.1 criteria was found. In patients with wild-type EGFR, we found that TLGWBR is an independent predictor of OS, which is consistent with findings of another study involving fewer (56) patients, and only OS as an endpoint [30]. In our study, we evaluated both PFS and OS as endpoints, because in a previous study regarding advanced NSCLC, PFS and OS were not confirmed to be consistently correlated [32]. It was found that TLGWBR was an independent predictor of PFS both in patients with wild-type EGFR and the EGFR mutations. In contrast, TLGWBR had no statistical significance as an independent predictor of OS in patients with the EGFR mutation. These findings will need to be validated in a large-scale cohort prospective study.
We have used the TLGWBR of selected tumors (selected using the RECIST 1.1 criteria) as a substitute for summing up the TLG values for whole-body tumors [30]. In contrast to previous studies, whole-body TLG for TLGWBR was strictly selected using RECIST 1.1 criteria, and the ICC indicated almost perfect reproducibility, as measured by the two groups of nuclear medicine physicians with respect to TLGWBR, although not all tumoral lesions could be included. The uptake of 18F-FDG is falsely influenced by inflammatory lesions and other lesions with obscure boundaries on images, making it difficult to distinguish between tumors and other nontumoral tissues; using our approach, this problem was relatively diminished and thus the outcomes were relatively credible. TLGWBR could be applied in some cases with fewer limitations in a clinical setting. Conversely, the clinical application of the PERCIST 1.0 criteria may be influenced by a high incidence of hepatitis in China; this is because its reference tissue value relies on FDG uptake by the liver [33]. In addition, according to NCCN guidelines [16], KRAS mutation testing is not conventionally recommended, and targeted therapy is not currently available for patients with mutant KRAS, thus most patients did not undergo KRAS status tests in our study. The prognostic influence of KRAS mutations could not be analyzed for either PFS or OS in this study. In a previous study, no (0.0% (0/945)) lung adenocarcinoma patients had KRAS mutations with EGFR mutations, while one patient (0.1% (1/897)) had EML4–ALK and EGFR mutations in Asian populations [34], thus the very low proportion (nearly zero) (co-occurrences of driver mutations) almost could not influence the prognosis of patients receiving TKIs in this study. Furthermore, NCCN guidelines indicate that KRAS mutation does not appear to affect chemotherapeutic efficacy. The number of positive cases was too small to enable the analysis of prognosis of patients with EML4–ALK rearrangement. Therefore, our results could not be potentially influenced by KRAS mutation and EML4–ALK rearrangement. Further study is needed to analyze the influence of KRAS mutations or EML4–ALK rearrangement on prognosis.
Our study had a number of limitations. First, it was a single-center retrospective study, which could have led to various biases. In most patients, 18F-FDG PET/CT scans were only performed once, and the treatment-related response changes with TLGWBR values were not evaluated. Second, not all the lymph node or distant metastases histopathology was evaluated. Third, all of the malignant lesions could not be included within the measurement of the PET workstation; thus, it was possible to underestimate the whole tumor burden using the imaging method. However, we selected the largest tumor metabolism burden per RESIST 1.1 as possible to avoid large inter-observer variation. Fourth, because there is no uniform or optimal threshold to delineate an accurate tumor boundary [18], a commonly adopted threshold (40% of the maximum SUVmax) was used to determine tumor volume; this has been used in most of the previous studies [19, 27]. These limitations notwithstanding, our current data revealed that the TLGWBR for selected tumors (using the RESIST 1.1 criteria) may be a promising parameter in the evaluation of the prognosis for advanced lung adenocarcinoma patients.
Conclusion
TLGWBR provides a strong prognostic indicator and could be an important guide for making treatment decisions in advanced lung adenocarcinoma patients with EGFR mutation status; however, it cannot be used to further stratify the risk of worse OS for patients with the EGFR mutation.
Acknowledgments
We thank our colleagues from PET/CT and medical oncology at Harbin Medical University Cancer Hospital who contributed PET/CT scans and images analysis to this work.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by Project Research (81171405) from the Natural Science Foundation of China. http://www.nsfc.gov.cn/; Lijuan Yu. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015; 65: 5–29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 2.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007; 450: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conde E, Angulo B, Izquierdo E, Paz-Ares L, Belda-Iniesta C, Hidalgo M, et al. Lung adenocarcinoma in the era of targeted therapies: histological classification, sample prioritization, and predictive biomarkers. Clin Transl Oncol. 2013; 15: 503–508. 10.1007/s12094-012-0983-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicks RJ, Kalff V, MacManus MP, Ware RE, Hogg A, McKenzie AF, et al. (18)F-FDG PET provides high-impact and powerful prognostic stratification in staging newly diagnosed non-small cell lung cancer. J Nucl Med. 2001; 42: 1596–1604. [PubMed] [Google Scholar]
- 5.Hicks RJ, Kalff V, MacManus MP, Ware RE, McKenzie AF, Matthews JP, et al. The utility of (18)F-FDG PET for suspected recurrent non-small cell lung cancer after potentially curative therapy: impact on management and prognostic stratification. J Nucl Med. 2001; 42: 1605–1613. [PubMed] [Google Scholar]
- 6.Hoekstra CJ, Stroobants SG, Hoekstra OS, Vansteenkiste J, Biesma B, Schramel FJ, et al. The value of [18F]fluoro-2-deoxy-D-glucose positron emission tomography in the selection of patients with stage IIIA-N2 non-small cell lung cancer for combined modality treatment. Lung Cancer. 2003; 39: 151–157. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki R, Komaki R, Macapinlac H, Erasmus J, Allen P, Forster K, et al. [18F]fluorodeoxyglucose uptake by positron emission tomography predicts outcome of non-small-cell lung cancer. J Clin Oncol. 2005; 23: 1136–1143. [DOI] [PubMed] [Google Scholar]
- 8.Kaira K, Serizawa M, Koh Y, Takahashi T, Yamaguchi A, Hanaoka H, et al. Biological significance of 18F-FDG uptake on PET in patients with non-small-cell lung cancer. Lung Cancer. 2014; 83: 197–204. 10.1016/j.lungcan.2013.11.025 [DOI] [PubMed] [Google Scholar]
- 9.Hoang JK, Hoagland LF, Coleman RE, Coan AD, Herndon JE 2nd, Patz EF Jr. Prognostic value of fluorine-18 fluorodeoxyglucose positron emission tomography imaging in patients with advanced-stage non-small-cell lung carcinoma. J Clin Oncol. 2008; 26: 1459–1464. 10.1200/JCO.2007.14.3628 [DOI] [PubMed] [Google Scholar]
- 10.Paesmans M, Berghmans T, Dusart M, Garcia C, Hossein-Foucher C, Lafitte JJ, et al. Primary tumor standardized uptake value measured on fluorodeoxyglucose positron emission tomography is of prognostic value for survival in non-small cell lung cancer: update of a systematic review and meta-analysis by the European Lung Cancer Working Party for the International Association for the Study of Lung Cancer Staging Project. J Thorac Oncol. 2010; 5: 612–619. 10.1097/JTO.0b013e3181d0a4f5 [DOI] [PubMed] [Google Scholar]
- 11.de Langen AJ, Vincent A, Velasquez LM, van Tinteren H, Boellaard R, Shankar LK, et al. Repeatability of 18F-FDG uptake measurements in tumors: a metaanalysis. J Nucl Med. 2012; 53: 701–708. 10.2967/jnumed.111.095299 [DOI] [PubMed] [Google Scholar]
- 12.Larson SM, Erdi Y, Akhurst T, Mazumdar M, Macapinlac HA, Finn RD, et al. Tumor Treatment Response Based on Visual and Quantitative Changes in Global Tumor Glycolysis Using PET-FDG Imaging. The Visual Response Score and the Change in Total Lesion Glycolysis. Clin Positron Imaging. 1999; 2: 159–171. [DOI] [PubMed] [Google Scholar]
- 13.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007; 2: 706–714. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Wang G, Hao Y, Xu Y, Zhang L. A comparison of ARMS and mutation specific IHC for common activating EGFR mutations analysis in small biopsy and cytology specimens of advanced non small cell lung cancer. Int J Clin Exp Pathol. 2014; 7: 4310–4316. [PMC free article] [PubMed] [Google Scholar]
- 15.Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, et al. Non-Small Cell Lung Cancer, Version 6.2015. J Natl Compr Canc Netw. 2015; 13: 515–524. [DOI] [PubMed] [Google Scholar]
- 16.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009; 45: 228–247. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 17.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979; 86: 420–428. [DOI] [PubMed] [Google Scholar]
- 18.Chen HH, Chiu NT, Su WC, Guo HR, Lee BF. Prognostic value of whole-body total lesion glycolysis at pretreatment FDG PET/CT in non-small cell lung cancer. Radiology. 2012; 264: 559–566. 10.1148/radiol.12111148 [DOI] [PubMed] [Google Scholar]
- 19.Chung HW, Lee KY, Kim HJ, Kim WS, So Y. FDG PET/CT metabolic tumor volume and total lesion glycolysis predict prognosis in patients with advanced lung adenocarcinoma. J Cancer Res Clin Oncol. 2014; 140: 89–98. 10.1007/s00432-013-1545-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulas A, Turkoz FP, Silay K, Tokluoglu S, Avci N, Oksuzoglu B, et al. A laboratory prognostic index model for patients with advanced non-small cell lung cancer. PLOS One. 2014; 9: e114471 10.1371/journal.pone.0114471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koukourakis MI, Giatromanolaki A, Sivridis E, Bougioukas G, Didilis V, Gatter KC, et al. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br J Cancer. 2003; 89: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krawczyk P, Kowalski DM, Krawczyk KW, Szczyrek M, Mlak R, Rolski A, et al. Predictive and prognostic factors in second- and third-line erlotinib treatment in NSCLC patients with known status of the EGFR gene. Oncol Rep. 2013; 30: 1463–1472. 10.3892/or.2013.2553 [DOI] [PubMed] [Google Scholar]
- 23.Olivier A, Petyt G, Cortot A, Scherpereel A, Hossein-Foucher C. Higher predictive value of tumour and node [18F]-FDG PET metabolic volume and TLG in advanced lung cancer under chemotherapy. Nucl Med Commun. 2014; 35: 908–915. 10.1097/MNM.0000000000000145 [DOI] [PubMed] [Google Scholar]
- 24.Liao S, Penney BC, Wroblewski K, Zhang H, Simon CA, Kampalath R, et al. Prognostic value of metabolic tumor burden on 18F-FDG PET in nonsurgical patients with non-small cell lung cancer. Eur J Nucl Med Mol Imaging. 2012; 39: 27–38. 10.1007/s00259-011-1934-6 [DOI] [PubMed] [Google Scholar]
- 25.Yoo SW, Kim J, Chong A, Kwon SY, Min JJ, Song HC, et al. Metabolic Tumor Volume Measured by F-18 FDG PET/CT can Further Stratify the Prognosis of Patients with Stage IV Non-Small Cell Lung Cancer. Nucl Med Mol Imaging. 2012; 46: 286–293. 10.1007/s13139-012-0165-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung HH, Kwon HW, Kang KW, Park NH, Song YS, Chung JK, et al. Prognostic value of preoperative metabolic tumor volume and total lesion glycolysis in patients with epithelial ovarian cancer. Ann Surg Oncol. 2012; 19: 1966–1972. 10.1245/s10434-011-2153-x [DOI] [PubMed] [Google Scholar]
- 27.Fonti R, Larobina M, Del Vecchio S, De Luca S, Fabbricini R, Catalano L, et al. Metabolic tumor volume assessed by 18F-FDG PET/CT for the prediction of outcome in patients with multiple myeloma. J Nucl Med. 2012; 53: 1829–1835. 10.2967/jnumed.112.106500 [DOI] [PubMed] [Google Scholar]
- 28.Hyun SH, Ahn HK, Kim H, Ahn MJ, Park K, Ahn YC, et al. Volume-based assessment by (18)F-FDG PET/CT predicts survival in patients with stage III non-small-cell lung cancer. Eur J Nucl Med Mol Imaging. 2014; 41: 50–58. 10.1007/s00259-013-2530-8 [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Wroblewski K, Liao S, Kampalath R, Penney BC, Zhang Y, et al. Prognostic value of metabolic tumor burden from (18)F-FDG PET in surgical patients with non-small-cell lung cancer. Acad Radiol. 2013; 20: 32–40. 10.1016/j.acra.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 30.Ho TY, Chou PC, Yang CT, Tsang NM, Yen TC. Total lesion glycolysis determined per RECIST 1.1 criteria predicts survival in EGFR mutation-negative patients with advanced lung adenocarcinoma. Clin Nucl Med. 2015; 40: e295–299. 10.1097/RLU.0000000000000774 [DOI] [PubMed] [Google Scholar]
- 31.Liao S, Penney BC, Zhang H, Suzuki K, Pu Y. Prognostic value of the quantitative metabolic volumetric measurement on 18F-FDG PET/CT in Stage IV nonsurgical small-cell lung cancer. Acad Radiol. 2012; 19: 69–77. 10.1016/j.acra.2011.08.020 [DOI] [PubMed] [Google Scholar]
- 32.Maroun JA. The significance of progression-free survival as an endpoint in evaluating the therapeutic value of antineoplastic agents. Curr Oncol. 2011; 18 Suppl 2: S3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong Y, Qiu C, Xia X, Wang J, Zhang H, Zhang X, et al. Hepatitis B virus and hepatitis C virus infection among HIV-1-infected injection drug users in Dali, China: prevalence and infection status in a cross-sectional study. Arch Virol. 2015; 160: 929–936. 10.1007/s00705-014-2311-0 [DOI] [PubMed] [Google Scholar]
- 34.Dearden S, Stevens J, Wu YL, Blowers D. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol. 2013; 24: 2371–2376. 10.1093/annonc/mdt205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.