Abstract
The onco-protein epidermal growth factor (EGF) initiates a cascade that includes activation of the ERK and AKT signaling pathways and alters gene expression. We describe a new action of EGF–EGF receptor (EGFR), rapid anticipatory activation of the endoplasmic reticulum stress sensor, the unfolded protein response (UPR). Within 2 min, EGF elicits EGFR dependent activation of phospholipase C γ (PLCγ), producing inositol triphosphate (IP3), which binds to IP3 receptor (IP3R), opening the endoplasmic reticulum IP3R Ca2+ channels, resulting in increased intracellular Ca2+. This calcium release leads to transient and moderate activation of the IRE1α and ATF6α arms of the UPR, resulting in induction of BiP chaperone. Knockdown or inhibition of EGFR, PLCγ or IP3R blocks the increase in intracellular Ca2+. While blocking the increase in intracellular Ca2+ by locking the IP3R calcium channel with 2-APB had no effect on EGF activation of the ERK or AKT signaling pathways, it abolished the rapid EGF-mediated induction and repression of gene expression. Knockdown of ATF6α or XBP1, which regulate UPR-induced chaperone production, inhibited EGF stimulated cell proliferation. Supporting biological relevance, increased levels of EGF receptor during tumor progression were correlated with increased expression of the UPR gene signature. Anticipatory activation of the UPR is a new role for EGF. Since UPR activation occurs in <2 min, it is an initial cell response when EGF binds EGFR.
Keywords: Epidermal growth factor (EGF), Breast cancer, Unfolded protein response (UPR), Calcium, Gene regulation
1. Introduction
Epidermal growth factor (EGF) stimulates cell proliferation and tumor growth by binding to a family of epidermal growth-factor receptors (EGFRs). The EGFR family consists of four members: ErbB1 (EGFR), ErbB2 (HER2/neu), ErbB3, and ErbB4 (Yarden and Pines, 2012). Binding of EGF to EGFR leads to formation of activated EGFR dimer, which exhibits increased autophosphorylation. Phosphorylated EGFR activates several cell signaling pathways, including the ERK and AKT pathways and rapidly alters gene expression. The activated signaling pathways and altered gene expression program promote cell proliferation and invasion, and are anti-apoptotic (Masuda et al., 2012).
EGF receptor is overexpressed in many human cancers including lung, pancreatic, brain, bladder, and breast cancer (Herbst and Langer, 2002; Holbro et al., 2003; Saxena and Dwivedi, 2012). EGFR is expressed in most breast cancers and is overexpressed in ~50% of triple-negative (ERα−, PR−, and HER2/neu−) breast cancers and in highly-aggressive invasive breast cancer (IBC) (Dent et al., 2007; Zell et al., 2009; Reis-Filho et al., 2006). EGFR over-expression is associated with larger tumors, poor differentiation, and poor clinical outcome (Masuda et al., 2012; Sainsbury et al., 1987; Salomon et al., 1995). Activated EGFR reduces sensitivity to anticancer drugs. Although, responses to EGFR inhibitors given alone were modest (Baselga et al., 2005; Dickler et al., 2009; Baselga and Arteaga, 2005), sustained inhibition of EGFR with erlotinib led to increased doxorubicin response and death of triple-negative breast tumors (Lee et al., 2012). This suggests that EGFR inhibitors may enhance chemosensitivity of breast tumors to cytotoxic agents.
The ability of EGF-EGFR to reduce sensitivity to anticancer drugs suggested activated EGFR might alter responses to chemotherapy-induced stress by influencing the activity of stress response pathways. The sensor system for endoplasmic reticulum stress is the unfolded protein response. In the well-studied “reactive” mode of UPR activation, UPR sensors react to an excess of unfolded or misfolded protein, metabolic stress or anticancer drugs by activating signaling pathways that increase protein-folding capacity and reduce protein production (Ron and Walter, 2007; Walter and Ron, 2012). Moderate activation of the UPR is protective. Consistent with the protective role of the UPR, a correlation between UPR activation and resistance to therapy has been described for several cancers (Auf et al., 2010; Crozat et al., 1993; Nawrocki et al., 2005; Shuda et al., 2003; Prabhu et al., 2012). It was widely accepted that UPR activation in tumors arose by clonal evolution and selection of cells that survived in part because they responded to therapy-induced stress by activating the UPR (Clarke et al., 2015; Clarke and Cook, 2015). In contrast to this well-known “reactive” mode of UPR activation, a little studied alternative mode of UPR activation termed “anticipatory” UPR activation occurs in the absence of endoplasmic reticulum stress (Walter and Ron, 2012). Anticipatory UPR activation was initially described in B-cells. Differentiation factors interleukin 4 and lipopolysaccharide induce UPR activation in B lymphocytes prior to increased immunoglobulin synthesis. Very recent studies demonstrate that estrogen in humans (Andruska et al., 2015a), ecdysone in insects (Liu et al., 2014; Dong et al., 2015) and human vascular endothelial growth factor (VEGF) (Karali et al., 2014) elicit biologically significant anticipatory activation of the UPR. Bioinformatic studies indicate that anticipatory estrogen activation of the UPR occurs early in breast cancer development, prior to detection and initiation of therapy (Andruska et al., 2015a).
Many, but not all, mitogenic hormones elicit anticipatory activation of the UPR as an early event in the proliferation program. We therefore tested whether EGF elicits anticipatory activation of the UPR and whether rapid activation of the anticipatory UPR pathway is important for subsequent actions of EGF-EGFR. We demonstrate EGF-EGFR-mediated rapid anticipatory activation of the UPR through activation of phospholipase C γ (PLCγ), resulting in increased production of inositol triphosphate (IP3). The IP3 binds to and opens endoplasmic reticulum IP3 receptors (IP3R) leading to rapid efflux of calcium stored in the lumen of the endoplasmic reticulum into the cytosol. This activates the UPR, resulting in induction of the anti-apoptoptic chaperone BiP/GRP78/HSPA5 (Lee, 2014). Although intracellular calcium was increased in ~1 min, blocking the increase in intracellular calcium did not inhibit subsequent EGF-EGFR activation of the ERK and AKT signaling pathways. However, blocking the increase in intracellular calcium completely inhibited EGF-EGFR induction of pro-proliferation immediate early gene expression and reversed EGF-EGFR mediated down-regulation of pro-apoptotic genes. Moreover, knockdown of the chaperone-inducing arms of the UPR strongly inhibited EGF stimulated cell proliferation. Anticipatory activation of the UPR is a newly revealed early action of EGF-EGFR with very different effects on plasma membrane and nuclear EGF-EGFR-regulated pathways.
2. Experimental procedures
2.1. Cell culture
MDA MB-468 cells were maintained in phenol-red free DMEM/F12, supplemented with 10% fetal bovine serum ([FBS], Atlanta Biological, Atlanta, GA). MCF10A cells were maintained in DMEM/F12, supplemented with 2% charcoal-dextran (cd)-stripped FBS. MCF10A cells were supplemented with 0.5 μg/ml hydrocortisone, 10 μg/ml insulin, 20 ng/ml epidermal growth factor (EGF), and 0.1 μg/ml cholera toxin. HCC1954 cells were maintained in RPMI1640, supplemented with 10% FBS. T47D cells were maintained in MEM, supplemented with 10% FBS. Estrogens were removed from the medium by maintaining T47D cells for at least three days in 10% charcoal dextran treated FBS (cd-FBS), and cells were plated in MEM containing 10% cd-calf serum.
2.2. Cell proliferation assays
Cell proliferation assays were carried out as described (Andruska et al., 2015a,b). Briefly, cells were plated in 96-well plates at the following densities: MDA MB-468 (2000 cells/well), T47D (2000 cells/well), MCF10A (1000 cells/well), and HCC-1954 (2000 cells/well). Cell viability was determined using CellTiter 96® Aqueous One Solution Cell Proliferation Assay (MTS, Promega, WI). Cell number was determined from a standard curve of absorbance versus cell number for each cell line.
2.3. Luciferase assay
Luciferase assay were as previously described (Cherian et al., 2012). Hela cells were plated (300,000 cells/well in 1 ml) in 6-well plates in 10% CD-CS 3 days before transfection. On the day of transfection, the medium was changed to 0.2 ml of Opti-MEM (Invitrogen). DNA and Lipofectamine 3000 (Invitrogen) were diluted in Opti-MEM. A total of 2500 ng of DNA (1260 ng of XBP1-Luc, 40 ng Renilla-Luc and 1200 ng IP3 phosphatase or PTZ carrier) was transfected into each well at a DNA:Lipofectamine 3000 ratio of 1:1. 24 h after transfection, 20 ng/ml EGF was added and the cells were incubated for 24 h. Cells were then lysed in 100 μl of passive lysis buffer (Promega), and luciferase activity was determined using a Dual-Luciferase Reporter Assay (Promega #E1910). XBP1-Luc and Renilla-Luc plasmids were constructed and modified by our lab. The IP3 phosphatase plasmid was previously described (Hernandez et al., 2008), and was generously provided by Dr. Mark Shapiro (University of Texas Health Science Center).
2.4. Western blots
Western blotting was carried out as previously described (Masuda et al., 2012; Saxena and Dwivedi, 2012). Briefly, whole cell extracts were prepared using RIPA lysis buffer (Millipore, CA) and mini protease inhibitor mixture (Roche Applied Science, Germany). Bound antibodies were detected using horseradish peroxidase-conjugated secondary antibodies and chemiluminescent immunodetection. The following antibodies from Cell Signaling Technology (MA) were used: Phospho-eIF2α (Ser51) (#3398), eIF2α (#5324), Phospho-AKT (Thr308) (#13038P), AKT (#9272S) Phospho-p44/42 MAPK (#4370), p44/42 MAPK (#4695), BiP (#3177). Other antibodies used: pan-IP3R (sc-28613; Santa Cruz Biotechnology), ATF6α (Imgenex, CA), β-Actin (Sigma, Saint Louis, MO), and α-Tubulin (Sigma, Saint Louis MO).
2.5. qRT-PCR analysis
Cells were seeded into 6-well plates and grown to ~70% confluence. For EGF regulated gene expression assays, cells were washed two times with HEPES buffer (140 mM NaCl, 4.7 mM KCl, 1.13 mM MgCl2, 10 mM HEPES, 10 mM Glucose, pH = 7.4) and then incubated in medium containing 100 nM 2-APB (Sigma, Saint Louis MO) or 10 μM UO126 for 5 min at 37 °C before EGF treatment. Cells were treated with a saturating concentration (20 ng/ml) or a subsaturating concentration (2 ng/ml) of EGF or vehicle-control for the indicated times, and RNA was extracted and purified using the Qiagen RNeasy kit. cDNA was prepared from 1 μg of RNA with M-MuLV reverse transcriptase from New England Biolabs. Diluted cDNA was used to perform quantitative RT-PCR using power SYBR® Green (Life technologies, NY) with actin as the internal control.
2.6. siRNA knockdowns
siRNA knockdowns were performed using DharmaFECT1 Transfection Reagent and 100 nM ON-TARGETplus non-targeting pool or SMARTpools for PLCγ (PLCG1) or pan-IP3R (GE Healthcare, UK). The pan-IP3R SmartPool consisted of three individual Smart-Pools, each at 33 nM, directed against each isoform of the IP3R (ITPR1, ITPR2, and ITPR3).
2.7. Calcium imaging
A calcium-sensitive dye, Fluo-4 AM, was used to measure cytoplasmic Ca2+. Cells were grown on 35 mm fluorodish plates (World Precision Instruments, FL) for two days prior to experiments. Cells were loaded with 5 μM Fluo-4 AM (Life Technologies, NY) in calcium free HEPES buffer for 30 min at 37 °C. Cells were washed three times with the buffer and incubated for 10 min with either 2 mM CaCl2 or without CaCl2. Fluoresence was monitored for 1 min to determine basal fluorescence intensity, and then the appropriate treatment was added. Measurements were taken using a Zeiss LSM 700 confocal microscope with a Plan-Four 20X objective (N.A. = 0.8) and 488-nM laser excitation (7% power). Images were obtained by monitoring fluorescence emission at 525 nM, and data analysis was performed using AxioVision and Zen software (Zeiss, Germany).
2.8. IP3 quantitation
MDA MB-468 cells were incubated for 10 min in 20 ng/ml EGF or vehicle. Intracellular IP3 levels were determined by extracting cell lysate, and determining IP3 levels in an assay based on competitive binding to a recombinant IP3R fragment (Perkin Elmer, MA) that was saturated with 3H-IP3. Unlabeled IP3 was used as a standard for the assays. 1.5 × 106 MDA MB-468 cells were used in this assay and the protocol has been described recently (Andruska et al., 2015b).
2.9. Statistical analysis
All data is reported as mean ± S.E.M. A two-tailed student’s t-test is used for comparisons between groups. One-way ANOVA followed by LSD or Tukey’s post hoc test is used for multiple comparisons with SPSS 13.0 for Windows (SPSS, Chicago, IL, USA). Significance was established when p < 0.05.
3. Results
3.1. EGF activates the UPR in breast cancer cells
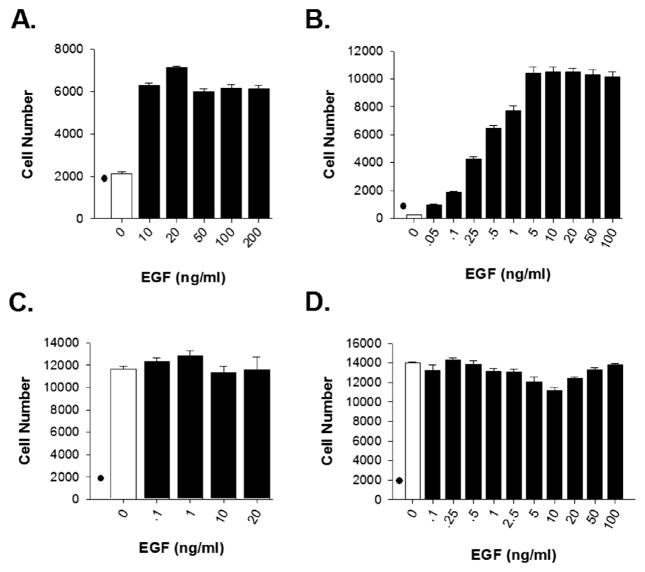
To assess whether EGF rapidly activates the UPR, we selected a diverse set of breast cancer cell lines with different expression levels of EGFR protein and different effects of EGF on cell proliferation. (i) ERα+ T47D breast cancer cells express low levels of EGFR, and are EGF-dependent for cell proliferation (Fig. 1A) (Andruska et al., 2015b; Pattarozzi et al., 2008); (ii) MCF10A pre-tumorigenic breast cells grossly overexpress EGFR, express minimal levels of HER2/neu and are fully dependent on EGF for proliferation (Fig. 1B); (iii) triple-negative MDA MB-468 breast cancer cells, grossly over-express EGFR through gene amplification, and EGF does not stimulate their proliferation (Fig. 1C); and (iv) ERα− HCC-1954 breast cancer cells, which overexpress equal amounts of EGFR and HER2/neu, and do not depend on EGF for cell proliferation (Fig. 1D) (Sahin et al., 2007; Lin et al., 2005).
Fig. 1.
Effects of EGF on proliferation of breast cancer cell lines. Effects of EGF on proliferation of (A) T47D breast cancer cells, (B) pre-tumorigenic MCF10A cells, (C) MDA MB-468 breast cancer cells, and (D) HCC1954 breast cancer cells. “●” denotes cell number at day 0. Data is mean ± SEM (n = 6).
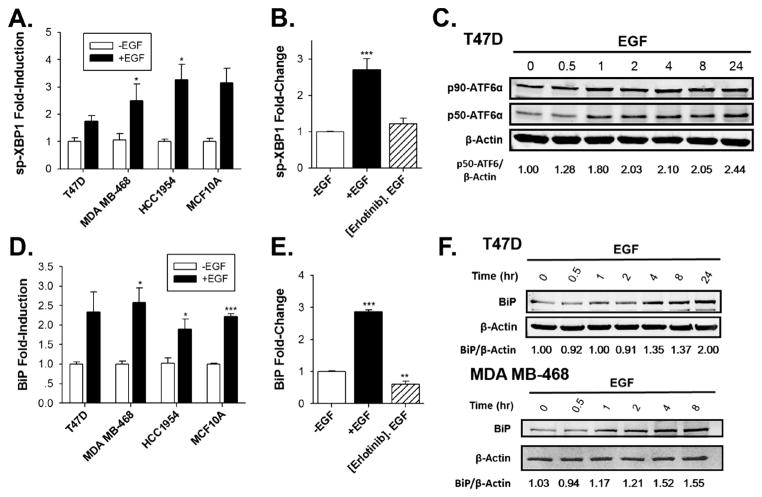
To assess whether EGF activates the UPR, we focused on markers indicative of activation of each arm of the UPR (Supplementary Fig. 1). The UPR regulates protein production by autophosphorylation of the transmembrane kinase, PERK (Walter and Ron, 2012; Ron and Walter, 2007). p-PERK phosphorylates eukaryotic initiation factor 2α (eIF2α), resulting in transient inhibition of protein synthesis. The other UPR arms initiate with activation of the transcription factor ATF6α, leading to increased chaperone production and activation of the ER splicing factor IRE1α, which splices the transcription factor XBP1, leading to production of active spliced-XBP1, increased protein folding capacity and altered mRNA decay and translation (Walter and Ron, 2012; Ron and Walter, 2007). We first assessed whether EGF could activate the IRE1α arm of the UPR. In response to UPR activation, active spliced-XBP1 (sp-XBP1) enters the nucleus and regulates expression of UPR targets (Supplementary Fig. 1A). EGF rapidly activated the IRE1α arm of the UPR, as shown by increased splicing of XBP1 mRNA in T47D, MDA MB-468, HCC-1954 and MCF10A breast cancer cells (Fig. 2A). Pre-treating cells with the EGFR inhibitor, erlotinib, blocked EGF-induction of sp-XBP1 mRNA (Fig. 2B), demonstrating that activation of the IRE1α arm of the UPR is dependent on activation of EGFR. Although our primary focus was on EGF in breast cancer cells, EGF also activated the UPR in HeLa cervical cancer cells as shown by activation of a transfected luciferase reporter activated by sp-XBP1 (Supplementary Fig. 2).
Fig. 2.
EGF activates the IRE1α and ATF6α arms of the UPR and induces production of the chaperone, BiP. (A) qRT-PCR comparing the effect of EGF on sp-XBP1 mRNA in T47D, MDA MB-468, HCC1954 and MCF10A cells after EGF treatment for 2 h (n = 3; -EGF set to 1). (B) qRT-PCR analysis of sp-XBP1 mRNA in MDA MB-468 breast cancer cells after pre-treating MDA MB-468 cells with erlotinib or with DMSO-vehicle for 30 min, following by treatment with EGF for 2 h (n = 3; -EGF set to 1). (C) Western blot analysis showing full-length ATF6α (p90-ATF6α) and cleaved-ATF6α (p50-ATF6α) in EGF-treated T47D breast cancer cells. The numbers below the gel indicate the ratio of p50-ATF6α/β-actin. (D) qRT-PCR analysis of BiP mRNA in T47D, MDA MB-468, HCC1954 and MCF10A breast cell lines after treatment with EGF for 4 h (n = 3; -EGF set to 1). (E) qRT-PCR analysis of BiP mRNA in MDA MB-468 breast cancer cells after pre-treating MDA MB-468 cells with erlotinib or DMSO vehicle for 30 min, following by treatment with EGF for 4 h (n = 3; -EGF set to 1). (F) Western blot analysis of BiP protein levels in T47D, MDA MB-468 cells treated with EGF. The numbers below the gel are the ratio of BiP/β-actin. Data is the mean ± SEM (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001.
We next evaluated whether EGF activates the ATF6α arm of the UPR. ATF6α activation involves release from the ER and transport to the Golgi apparatus where ATF6α undergoes proteolytic cleavage to form active p50-ATF6α (Supplementary Fig. 1B) (Walter and Ron, 2012; Ron and Walter, 2007; Wu et al., 2007). Supporting rapid EGF activation the ATF6α arm of the UPR, EGF modestly increased proteolysis of ATF6α (Fig. 2C, Supplementary Fig. 3B), which was blocked by pre-treatment with erlotinib (Supplementary Fig. 3A).
p50-ATF6α regulates induction of BiP and other UPR-regulated chaperones (Wu et al., 2007; Okada et al., 2002a,b). Inducing BiP increases ER protein folding capacity, contributing to resolution of the stress, and helps reverse UPR activation (Lee, 2007). Over-expression of BiP is linked to a poor prognosis in breast and other cancers (Luo and Lee, 2013). EGF rapidly induced BiP mRNA in T47D, MDA MB-468, HCC-1954 and MCF10A breast cancer cells (Fig. 2D), leading to increases in BiP protein levels (Fig. 2F). Consistent with BiP induction being dependent on activation of EGFR, pre-treating MDA MB-468 cells with erlotinib blocked EGF-induction of BiP mRNA (Fig. 2E). While induction of BiP mRNA led to increases in BiP protein in T47D and MDA MB-468 cells (Fig. 2F), EGF did not increase BiP protein levels in pre-tumorigenic MCF10A cells (Supplementary Fig. 3C). XBP1 can directly interact with and activate estrogen receptor α (Ding et al., 2003). Although the mechanism by which XBP1-ERα interaction might impact translation is unclear, this may provide an explanation for the different levels of BiP expression in estrogen receptor positive T47D cells and estrogen receptor negative MCF10A cells. Alternatively, since BiP expression is elevated in breast cancer cells relative to surrounding normal tissue (Cook et al., 2012), the different stages of tumor development in non-tumorigenic MCF10A cells compared to tumorigenic T47D and MDA-MB-468 cells may play a role.
Inducing BiP helps protect cancer cells against protein-misfolding and other forms of UPR stress (Andruska et al., 2015a). Extensive and sustained activation of the UPR is toxic. This enabled us to test whether prior exposure of MDA MB-468 cells to EGF or to a low concentration of the UPR activator tunicamycin (TUN) could alter the concentration of TUN required to subsequently induce cell death. Pretreating cells with EGF or with TUN increased the concentration of TUN required to induce cell death by ~2 fold (Supplementary Fig. 4).
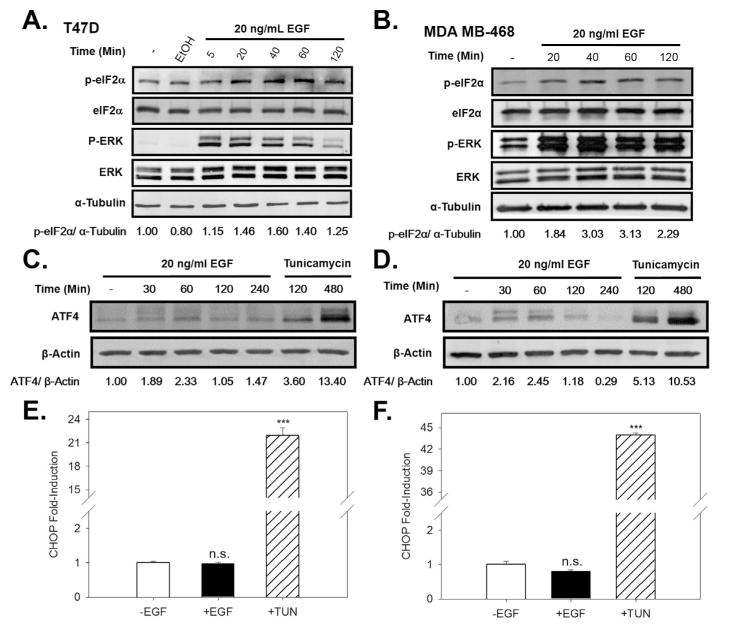
We next assessed whether EGF activates the PERK arm of the UPR. Activated PERK induces increased eIF2α phosphorylation (Supplementary Fig. 1C). EGF induced a rapid increase in phosphorylation of eIF2α (Fig. 3A,B). Phosphorylation of eIF2α leads to preferential translation of ATF4 mRNA, and we observed a moderate and transient increase in ATF4 protein expression. In contrast, the toxic UPR activator, tunicamycin (TUN) robustly induced ATF4 (Fig. 3C,D). Because mild and transient activation of the PERK arm of the UPR does not induce the proapoptotic protein CHOP (Andruska et al., 2015a), CHOP was not induced by the EGF stimulated ATF4 (Fig. 3E,F). This data demonstrates that EGF elicits moderate and transient activation of all three arms of the UPR.
Fig. 3.
EGF activates the PERK arm of the UPR. Western blot analysis showing p-eIF2α levels and total eIF2α levels in ERα+ T47D cells (A) and in MDA MB-468 cells (B) treated with EGF for the indicated times. The numbers below the gel are the ratio of p-eIF2α/α-tubulin ratio. Western blot analysis of ATF4 levels following treatment of T47D cells (C) and MDA MB-468 cells (D) with EGF, or with the UPR activator tunicamycin. The numbers below the gel are the ratio of ATF4/β-actin. qRT-PCR analysis of CHOP mRNA following treatment of T47D cells (E) and MDA MB-468 cells (F) with EGF. Concentrations: EGF, 20 ng/ml, TUN, 10 μg/ml. Data is mean ± SEM (n = 3). ***p < 0.001. n.s., not significant.
3.2. EGF activates the UPR THROUGH a PLCγ-dependent mechanism
Because the pathway is activated in less than 2 min, it was unlikely that accumulation of misfolded protein or other forms of EGF-induced stress triggered UPR activation. We recently identified a different anticipatory pathway of UPR activation in which the estrogen, 17β-estradiol, bound to estrogen receptor α rapidly activates phospholipase C γ (PLCγ). Active phosphorylated plasma membrane PLCγ, hydrolyzes PIP2 to diacylglycerol (DAG) and inositol 1, 4, 5-triphosphate (IP3). The IP3 binds to endoplasmic reticulum IP3 receptors opening the IP3R Ca2+ channels and allowing efflux of Ca2+ stored in the lumen of the endoplasmic reticulum into the cytosol. This activates the UPR. Consistent with a role for Ca2+ efflux in UPR activation, the well-studied UPR activator thapsigargin rapidly activates the UPR by depleting Ca2+ stores in the lumen of the ER and increasing cytosol Ca2+ (Andruska et al., 2015a). Although EGF has been reported to activate PLCγ, the consequences of PLCγ activation were unknown (Margolis et al., 1990). We therefore evaluated the role of Ca2+ release in EGF activation of the UPR.
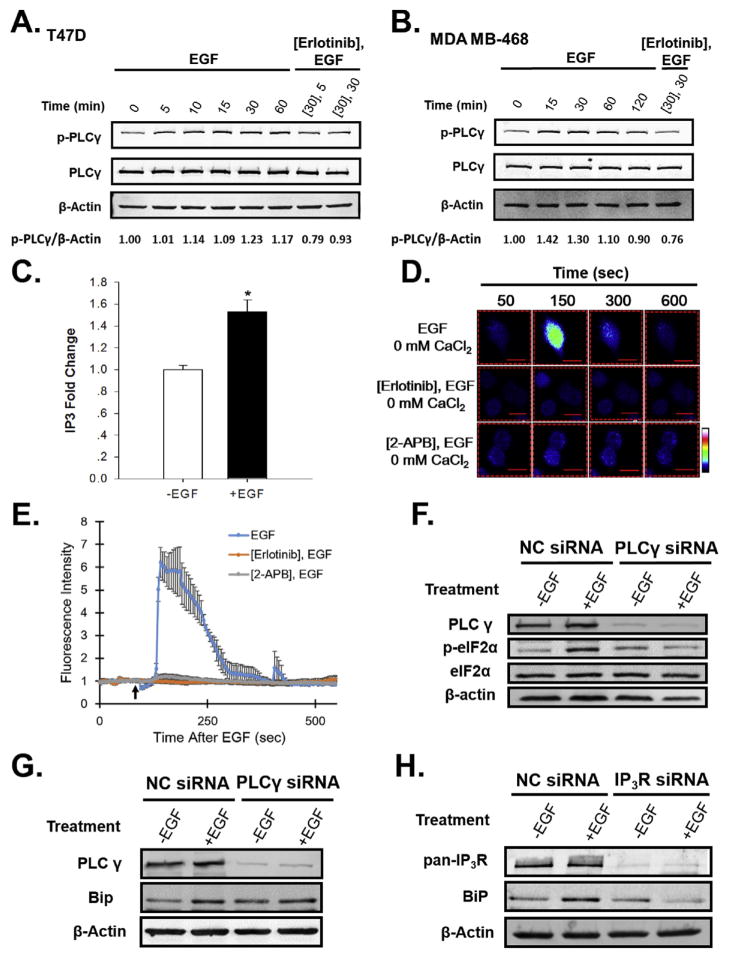
EGF induced rapid phosphorylation and activation of PLCγ in T47D and MDA MB-468 breast cancer cells (Fig. 4A,B). EGF-mediated phosphorylation of PLCγ was blocked by pre-treatment with the EGFR inhibitor, erlotinib (Fig. 4A,B). PLCγ activation rapidly induced IP3 (Fig. 4C).
Fig. 4.
EGF activates the UPR through PLCγ-mediated opening of the IP3R Ca2+ channels, leading to the release of Ca2+ from the ER into the cytosol, and UPR activation. Western blot analysis of p-PLCγ and total PLCγ protein levels after treatment of (A) T47D or (B) MDA MB-468 breast cancer cells with EGF or EGF + erlotinib. Cells were treated with either 10 μM erlotinib or vehicle-control, followed by 20 ng/ml EGF for the indicated times. The numbers below the gel are the ratio of p-PLCγ/β-actin. (C) Quantitation of intracellular IP3 levels following treatment of MDA MB-468 cells for 10 min with or without EGF (n = 3). (D) EGF increases intracellular calcium levels in MDA MB-468 breast cancer cells in medium lacking calcium (0 mM CaCl2). Visualization of intracellular Ca2+ using Fluo-4 AM. Color scale from basal Ca2+ to highest Ca2+: blue, green, red, white. (E) Graph depicts quantitation of cytosolic calcium levels in MDA MB-468 breast cancer cells pre-treated with either vehicle-control, the EGFR inhibitor, erlotinib, or the IP3R inhibitor, 2-amino propyl-benzoate (2-APB), followed by treatment with 20 ng/ml EGF in the absence of extracellular calcium (indicated by the black arrow, n = 10 cells). Calcium quantitation data is expressed as mean ± SE (n = 10). (F) Western blotting analysis of PLCγ, p-eIF2α and total eIF2α protein levels after treatment of T47D cells with 100 nM non-coding (NC) or PLCγ siRNA, followed by treatment with EGF (+EGF) or ethanol-vehicle (−EGF) for 30 min. (G) Western blotting analysis of PLCγ and BiP protein levels after treatment of T47D cells with 100 nM non-coding (NC) or PLCγ siRNA, followed by treatment with EGF (+EGF) or ethanol-vehicle (−EGF) for 4 h (H) siRNA knockdown of the three isoforms of the IP3R Ca2+ channel blocks EGF-induction of BiP protein in T47D breast cancer cells. Cells were treated with either 100 nM non-coding (NC) or IP3R siRNA SmartPool, followed by treatment with 20 ng/mL EGF for 4 h. IP3R smartpool contained equal amounts of three individual SmartPools directed against each isoform of IP3R. Data is mean ± SEM (n = 3); *p < 0.05.
To assess changes in the level of intracellular Ca2+, we used the calcium sensitive dye Fluo-4 AM. In the absence of extracellular Ca2+, EGF elicited a transient increase in fluorescence in MDA MB-468 cells (Fig. 4D). Since the increase in cytosol Ca2+ occurs in the absence of extracellular Ca2+, this suggested that the source of intracellular Ca2+ was the Ca2+ stored in the lumen of the endoplasmic reticulum. To identify the Ca2+ channels responsible for promoting the release of Ca2+ from the ER, we assessed whether the EGFR inhibitor erlotinib or 2-APB, which locks the IP3R Ca2+ channel, blocks the EGF-mediated increase in cytosol Ca2+. Pre-treatment of MDA MB-468 cells with erlotinib, or with 2-APB, followed by EGF, blocked the increase in intracellular Ca2+ (Fig. 4D,E). To further confirm that PLCγ activation is required for EGF to activate the UPR, we performed siRNA knockdown of PLCγ, and assessed whether EGF was still capable of activating the UPR and inducing the widely used surrogate marker for UPR activation, the chaperone BiP. Knockdown of PLCγ blocked phosphorylation of eIF2α and induction of BiP (Fig. 4F,G). Similarly, knockdown of IP3Rs blocked EGF-induction of BiP protein (Fig. 4H). These data demonstrate that PLCγ activation, resulting in opening of the ER IP3 receptor, mediates transient EGF activation of the UPR. (Pathway: EGF-EGFR-p (active) → p-PLCγ (active) → IP3↑→ IP3:IP3R (open)→ Ca2+↑→ UPR (activated)→ BiP↑).
3.3. Activation of the anticipatory UPR pathway is required for EGF-EGFR-mediated gene expression and is important for cell proliferation
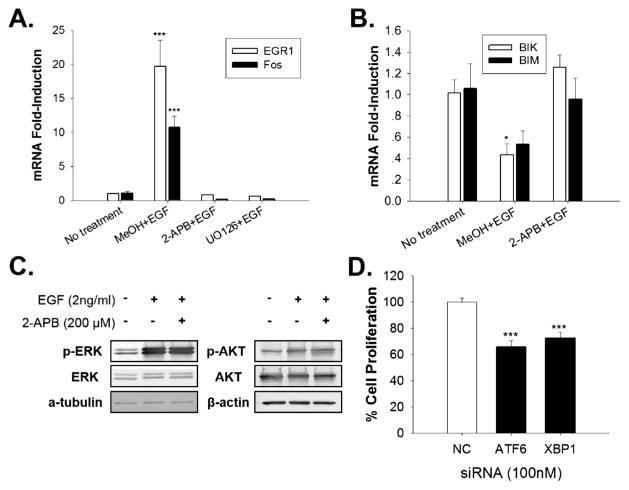
The EGF-EGFR mediated anticipatory pathway of UPR activation can be divided into a rapid early segment that involves activation of PLCγ, increased IP3, opening of the IP3R calcium channel and increased intracellular calcium and a late segment that involves activation of the 3 arms of the UPR and induction of BiP chaperone. Since the EGF induced increase in intracellular Ca2+ occurs in ~1 min (Fig. 4D,E), it may precede other actions of EGF. We therefore tested whether the increase in intracellular calcium is important in subsequent rapid effects of EGF. These rapid pathways include rapid activation of the ERK and AKT signaling pathways and induction of pro-proliferation immediate early genes and rapid repression of pro-apoptotic genes. EGF-EGFR induction of immediate early genes, EGR1 and Fos is thought to play a role in EGF-mediated cell proliferation (Lee, 2007; Murphy et al., 2002). To test whether the extremely rapid increase in intracellular Ca2+ is important for subsequent EGF induction of EGR1 and Fos, we used sub-saturating biologically relevant concentrations of EGF. Fos and EGR1 mRNAs were robustly (>10 fold) induced after only 20 min of EGF treatment. Blocking the EGF mediated increase in intracellular calcium with 2-APB completely blocked EGF induction of EGR1 and Fos mRNAs (Fig. 5A). Since 2-APB also prevents EGF-mediated down-regulation of the pro-apoptotic BIK and BIM mRNAs (Fig. 5B), 2-APB is acting as a selective modulator of EGF-mediated gene expression and not as a general inhibitor of transcription. As expected, inhibition of ERK signaling with the ERK inhibitor UO1026 also blocked EGF-induction of Fos and EGR1 mRNAs (Fig. 5A). To test whether elevated intracellular calcium was stimulating ERK activation (Chao et al., 1992), and activated ERK then induced immediate early gene expression, we examined the effect of 2-APB on rapid EGF activation of the ERK and AKT signaling pathways. Under the same conditions used in the gene expression experiment, 2-APB did not inhibit rapid and robust EGF activation of the ERK and AKT (protein kinase B) signaling pathways (Fig. 5C). Since 2-APB did not block EGF activation of the ERK pathway, the elevated level of intracellular calcium is a previously undescribed independent regulator of EGF-induced gene expression that works together with ERK activation to regulate immediate early gene expression.
Fig. 5.
EGF induced UPR activation is required for EGF regulated gene expression and cell proliferation. (A and B) qRT-PCR of the oncogene mRNAs EGR1 and Fos and the pro-apoptotic mRNAs BIK and BIM in T47D cells pre-treated for 5 min with ethanol, 2-APB or the ERK inhibitor UO126, then treated with 2 ng/ml EGF for 20 min (A), or 2 h (B) (Ca2+-free medium; n = 3, mean ± SEM). (C) Western blot analysis showing p-ERK levels and total ERK and p-AKT and total AKT levels in T47D cells treated with 2 ng/mL EGF with or without 2-APB pretreatment. (D) EGF stimulated proliferation of T47D breast cancer cells treated with 100 nM non-coding (NC), ATF6 or XBP1 siRNA SmartPool (n = 6). Proliferation rates were normalized to cells treated with non-coding (NC) siRNA. *p < 0.05; ***p < 0.001.
UPR-induced-chaperones, like BiP/GRP78, are thought to be important for cell proliferation (Dong et al., 2008; Luo et al., 2006). We therefore evaluated the effect of UPR activation in EGF stimulated proliferation of T47D cells. Activation of the ATF6α and IRE1α arms of the UPR induces chaperone production. However, the IRE1α arm of the UPR also play a role in mRNA decay and JNK mediated apoptosis (Chen and Brandizzi, 2013). To dissect out the role of chaperone induction, instead of carrying out an IRE1α knockdown, we knocked down XBP1 which encodes the spliced XBP1 transcription factor when cleaved by the activated IRE1α. Compared to the control siRNA, knockdown of the XBP1 arm of the UPR produced a highly significant 28% decline in EGF stimulated cell proliferation, while ATF6 knockdown produced a 35% decline (Fig. 5D). These data indicate that EGF induced UPR activation and the subsequent chaperone production plays an important role in EGF stimulated cell proliferation.
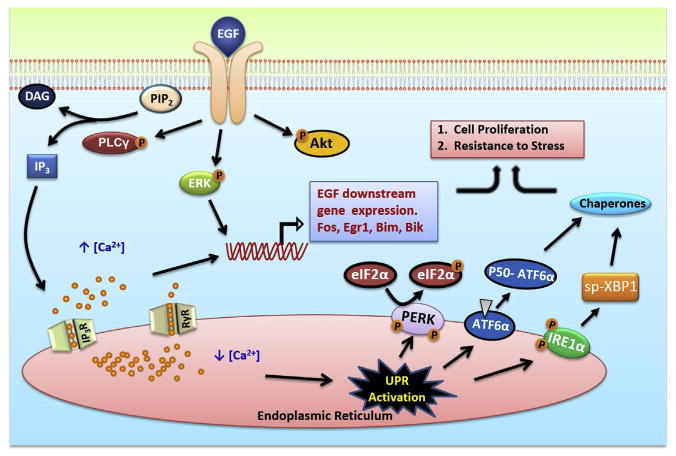
The effects of EGF on the UPR and downstream pathways are depicted in Fig. 6.
Fig. 6.
Schematic of the pathway of EGF induced anticipatory UPR activation.
3.4. Overexpression of EGFR and HER2/neu is correlated with increased UPR activation
The UPR plays an important role in cancer cell migration, metastasis and resistance to therapy-induced apoptosis. Consistent with a role for the anticipatory UPR pathway in the oncogenic actions of EGF, a recent report found that PLCγ signaling was important for EGF to induce an aggressive pro-metastatic phenotype (Dittmar et al., 2002). We therefore evaluated whether over-expression of HER2/neu or EGFR was correlated with increased activation of the UPR in tumors by evaluating a UPR gene signature (Andruska et al., 2015a) in several publically available patient tumor cohorts.
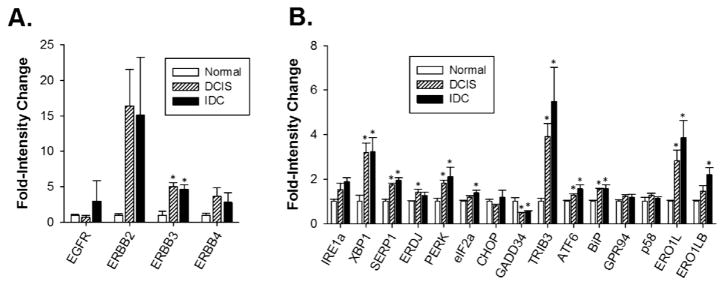
To assess UPR activity early in EGFR+ breast cancer development, we compared EGF receptor family member expression level and UPR pathway activity in samples of histologically normal breast epithelium, ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC). Compared with normal epithelium, DCIS and IDC samples displayed elevated levels of EGF receptor family members mRNA (Fig. 7A). Additionally, DCIS and IDC samples displayed elevated markers of UPR activation. Besides the classic UPR maker genes, additional UPR related genes were correlated with increased expression of EGFR. Specifically, SERP1 mRNA, a marker for IRE1α activation (Lee et al., 2003a,b), TRIB3, which is a marker of PERK activation (Bromati et al., 2011), and BiP chaperone, which is a marker of ATF6α activation (Fig. 7B). These data suggest that UPR activation is correlated with EGF receptor family member expression.
Fig. 7.
Increased expression of EGF receptors is correlated with increased expression of the UPR gene signature. Expression of (A) EGF receptors and (B) UPR sensors and UPR-regulated genes in samples from normal breast epithelial cells (n = 5), ductal carcinoma in situ (n = 9), and invasive ductal carcinoma (n = 5). Normal epithelial samples were set to 1. Data is mean ± SEM, *p < 0.05.
Using data from an independent cohort of 114 ERα− breast cancers, we explored whether the expression of HER2 mRNA correlates with the expression of UPR genes. The expression of several UPR genes displayed highly significant correlation with the expression of HER2 genes (Supplementary Table 1).
4. Discussion
Cancer progression is often correlated with increased protein synthesis and poor glucose and oxygen supply in the microenvironment (Ruggero, 2013; Brown and Wilson, 2004). In response to these different intrinsic and extrinsic stresses, cancer cells activate the UPR pathway. UPR activation has been described in several human tumors including myeloma (Lee et al., 2003a,b), lymphoma (Jenner et al., 2003) and carcinoma of the breast (Lee et al., 2006). The role of UPR activation in resistance of breast cancer and other tumors to tamoxifen and other therapeutics has been extensively studied (Clarke and Cook, 2015). XBP1 is upregulated in antiestrogen resistant breast cancer cells (Gomez et al., 2007). It interacts with and regulates the activity of estrogen receptor (Ding et al., 2003; Fang et al., 2004), NFκB (Gu et al., 2002) and apoptosis related proteins (Moretti et al., 2007); this may contribute to drug resistance in these cells. Induction of BiP and other chaperones may also play a role in this protective UPR activation (Lee et al., 2003a,b).
These studies have focused on the reactive mode of UPR activation in which cells respond to stress by activating the UPR and, if moderate and transient, the UPR is protective (Andruska et al., 2015a; Ma and Hendershot, 2004). In this reactive form of UPR activation, diverse stressors increase the level of unfolded or mis-folded proteins in the lumen of the endoplasmic reticulum. This activates sensors in the endoplasmic reticulum membrane that both sense the stress and activate the 3 major arms of the UPR (Supplementary Fig. 1) (Ron and Walter, 2007; Walter and Ron, 2012; Okada et al., 2002a,b; Novoa et al., 2001).
Here we describe a fundamentally different type of very rapid UPR activation that anticipates future needs and occurs in the absence of cell stress or accumulation of unfolded proteins. In less than 2 min EGF triggers PLCγ-mediated opening of endoplasmic reticulum IP3R calcium channels and release of Ca2+ into the cytosol. This increase in cytosol Ca2+ stimulates activation of all three arms of the UPR. Anticipatory activation of the UPR is a newly identified common pathway shared by several mitogenic hormones. We suggest this newly unveiled pathway is used by many, but not all, mitogenic hormones to prepare cells for the increased protein folding load that will occur during subsequent hormone-stimulated cell proliferation. Although this pathway is conserved from peptide hormones to steroid hormones, there are important differences in the activation mechanisms. VEGF induced UPR activation is not inhibited by blocking the ER calcium signal (Karali et al., 2014). However, EGF mediated activation of the UPR is totally dependent on IP3 mediated calcium release from the ER.
Supporting biological relevance of the UPR pathway, EGFR levels and expression levels of UPR related genes were strongly correlated in samples from 114 ERα negative breast cancers. Moreover, there was a parallel increase in EGFR content and UPR gene index components as cells progress from normal mammary epithelial cells to DCIS and then to IDC. Thus the EGF induced anticipatory UPR pathway not only facilitates tumor cell proliferation but likely also helps protect cancer cells against subsequent apoptosis induced by hypoxia, nutrient deprivation and therapy. Since hypoxia, nutrient deprivation and therapy can all stimulate reactive UPR activation, the anticipatory and reactive modes of UPR activation lead to UPR engagement throughout the entire cycle of tumor development and therapy.
Since a key feature of rapid anticipatory activation of the UPR is an increase in cytosolic calcium, we explored the effect of blocking this increase on well-known actions of EGF-EGFR. The mitogenic action of EGF is mediated in part by regulation of immediate early gene expression. Two major EGF-induced immediate early genes, c-fos and egr1 encode transcription factors important for cell cycle progression (Zwang et al., 2012; Brown et al., 1998). Rapid (5–30 min) EGF-EGFR activation of the ERK signaling pathway is essential for early gene expression (Figs. 5A and 6) (Murphy et al., 2002). Cytosolic calcium levels play an important role in regulating ERK activation (Schmitt et al., 2004; Agell et al., 2002). Moreover a massive increase in cytosol calcium due to strong and sustained cytotoxic UPR activation is sufficient to activate the ERK pathway (Chao et al., 1992). However, blocking the transient and moderate increase in cytosol Ca2+ induced by EGF-EGFR activation of the protective anticipatory UPR pathway did not inhibit EGF-EGFR activation of the ERK or AKT signaling pathways (Fig. 5 and 6). Since blocking the EGF-induced increase in cytosol Ca2+ abolished induction and repression of gene expression by EGF, the anticipatory UPR pathway is not regulating immediate early gene expression by controlling ERK activation. ERK activation and the elevated calcium resulting from activation of the early stages of the anticipatory UPR pathway are independent EGF activated pathways that converge at the level of immediate early gene expression. Since 2-APB blocks EGF-induced immediate early gene expression without affecting EGF activation of the ERK and AKT signaling pathways, 2-APB represents a useful new probe for dissecting the roles of immediate early gene expression and ERK and AKT activation in downstream actions of EGF. In contrast to 2-APB, the equally useful simultaneous RNAi knockdown of all 3 functionally overlapping, but non-homologous, endoplasmic reticulum IP3R channels (Fig. 4H) is technically challenging and has rarely been reported.
Accumulating evidence suggests a role for UPR chaperones in regulation of cell proliferation. One of the most abundant and well characterized UPR-induced chaperones, GRP78/BiP, influences proliferation of embryonic cells (Luo et al., 2006). In a GRP78 heterozygous mice model where the level of BiP was reduced by about half, growth of breast tumors was significantly reduced (Dong et al., 2008). These results suggest UPR chaperones have functions other than facilitating protein folding within the ER. Using siRNA, we knocked down the major UPR chaperone producing pathways, the XBP1 and ATF6 arms (Supplementary Figs. 1 and 5), and significantly inhibited EGF stimulated cancer cell proliferation (Fig. 5D).
These results indicate that the EGF induced anticipatory UPR pathway facilitates EGF stimulated cell proliferation in at least two ways. First, it releases calcium from endoplasmic reticulum stores and cooperates with the ERK signaling pathway to regulate immediate early gene expression. Second, it increases UPR chaperone production, which facilitates EGF stimulated cell proliferation.
Our studies add a new dimension to the cascade of events that occur when a cell is exposed to EGF.
Supplementary Material
Acknowledgments
We thank members of our laboratory for critical reading and discussion of the manuscript.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.mce.2015.11.005.
Footnotes
This research was supported by NIH grant DK 071909 and DOD Breast Cancer Research Program Grant BC131871.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Agell N, Bachs O, Rocamora N, Villalonga P. Modulation of the Ras/Raf/MEK/ERK pathway by Ca 2+, and calmodulin. Cell Signal. 2002;14:649–654. doi: 10.1016/s0898-6568(02)00007-4. [DOI] [PubMed] [Google Scholar]
- Andruska N, Zheng X, Yang Y, Helferich WG, Shapiro DJ. Anticipatory estrogen activation of the unfolded protein response is linked to cell proliferation and poor survival in estrogen receptor alpha-positive breast cancer. Oncogene. 2015a;34:3760–3769. doi: 10.1038/onc.2014.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andruska ND, Zheng X, Yang X, Mao C, Cherian MM, Mahapatra L, Helferich WG, Shapiro DJ. Estrogen receptor α inhibitor activates the unfolded protein response, blocks protein synthesis, and induces tumor regression. Proc Natl Acad Sci. 2015b;112:4737–4742. doi: 10.1073/pnas.1403685112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auf G, Jabouille A, Guérit S, Pineau R, Delugin M, Bouchecareilh M, Magnin N, Favereaux A, Maitre M, Gaiser T. Inositol-requiring enzyme 1α is a key regulator of angiogenesis and invasion in malignant glioma. Proc Natl Acad Sci. 2010;107:15553–15558. doi: 10.1073/pnas.0914072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2005;23:2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- Baselga J, Albanell J, Ruiz A, Lluch A, Gascon P, Guillem V, Gonzalez S, Sauleda S, Marimon I, Tabernero JM, Koehler MT, Rojo F. Phase II and tumor pharmacodynamic study of gefitinib in patients with advanced breast cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2005;23:5323–5333. doi: 10.1200/JCO.2005.08.326. [DOI] [PubMed] [Google Scholar]
- Bromati CR, Lellis-Santos C, Yamanaka TS, Nogueira TC, Leonelli M, Caperuto LC, Gorjao R, Leite AR, Anhe GF, Bordin S. UPR induces transient burst of apoptosis in islets of early lactating rats through reduced AKT phosphorylation via ATF4/CHOP stimulation of TRB3 expression. Am J Physiol Regul Integr Comp Physiol. 2011;300:R92–R100. doi: 10.1152/ajpregu.00169.2010. [DOI] [PubMed] [Google Scholar]
- Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- Brown JR, Nigh E, Lee RJ, Ye H, Thompson MA, Saudou F, Pestell RG, Greenberg ME. Fos family members induce cell cycle entry by activating cyclin D1. Mol Cell Biol. 1998;18:5609–5619. doi: 10.1128/mcb.18.9.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T, Byron K, Lee K, Villereal M, Rosner MR. Activation of MAP kinases by calcium-dependent and calcium-independent pathways. Stimulation by thapsigargin and epidermal growth factor. J Biol Chem. 1992;267:19876–19883. [PubMed] [Google Scholar]
- Chen Y, Brandizzi F. IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 2013;23:547–555. doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian MT, Wilson EM, Shapiro DJ. A competitive inhibitor that reduces recruitment of androgen receptor to androgen-responsive genes. J Biol Chem. 2012;287:23368–23380. doi: 10.1074/jbc.M112.344671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Cook KL. Unfolding the role of stress response signaling in endocrine resistant breast cancers. Front Oncol. 2015:5. doi: 10.3389/fonc.2015.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer–an overview and update. Mol Cell Endocrinol. 2015;418(3):220–234. doi: 10.1016/j.mce.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KL, Shajahan AN, Wärri A, Jin L, Hilakivi-Clarke LA, Clarke R. Glucose-regulated protein 78 controls cross-talk between apoptosis and autophagy to determine antiestrogen responsiveness. Cancer Res. 2012;72:3337–3349. doi: 10.1158/0008-5472.CAN-12-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Åman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res Off J Am Assoc Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Dickler MN, Cobleigh MA, Miller KD, Klein PM, Winer EP. Efficacy and safety of erlotinib in patients with locally advanced or metastatic breast cancer. Breast cancer Res Treat. 2009;115:115–121. doi: 10.1007/s10549-008-0055-9. [DOI] [PubMed] [Google Scholar]
- Ding L, Yan J, Zhu J, Zhong H, Lu Q, Wang Z, Huang C, Ye Q. Ligand-independent activation of estrogen receptor α by XBP-1. Nucleic Acids Res. 2003;31:5266–5274. doi: 10.1093/nar/gkg731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar T, Husemann A, Schewe Y, Nofer JR, Niggemann B, Zanker KS, Brandt BH. Induction of cancer cell migration by epidermal growth factor is initiated by specific phosphorylation of tyrosine 1248 of c-erbB-2 receptor via EGFR. FASEB J Off Publ Fed Am Soc Exp Biol. 2002;16:1823–1825. doi: 10.1096/fj.02-0096fje. [DOI] [PubMed] [Google Scholar]
- Dong D, Ni M, Li J, Xiong S, Ye W, Virrey JJ, Mao C, Ye R, Wang M, Pen L. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68:498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- Dong D-J, Jing Y-P, Liu W, Wang J-X, Zhao X-F. The steroid hormone 20-hydroxyecdysone upregulates Ste-20 family serine/threonine kinase Hippo to induce programmed cell death. J Biol Chem. 2015 doi: 10.1074/jbc.M115.643783. jbc. M115. 643783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Yan J, Ding L, Liu Y, Zhu J, Huang C, Zhao H, Lu Q, Zhang X, Yang X. XBP-1 increases ERα transcriptional activity through regulation of large-scale chromatin unfolding. Biochem Biophys Res Commun. 2004;323:269–274. doi: 10.1016/j.bbrc.2004.08.100. [DOI] [PubMed] [Google Scholar]
- Gomez BP, Riggins RB, Shajahan AN, Klimach U, Wang A, Crawford AC, Zhu Y, Zwart A, Wang M, Clarke R. Human X-box binding protein-1 confers both estrogen independence and antiestrogen resistance in breast cancer cell lines. FASEB J. 2007;21:4013–4027. doi: 10.1096/fj.06-7990com. [DOI] [PubMed] [Google Scholar]
- Gu Z, Lee RY, Skaar TC, Bouker KB, Welch JN, Lu J, Liu A, Zhu Y, Davis N, Leonessa F. Association of interferon regulatory factor-1, nucleophosmin, nuclear factor-κB, and cyclic AMP response element binding with acquired resistance to Faslodex (ICI 182,780) Cancer Res. 2002;62:3428–3437. [PubMed] [Google Scholar]
- Herbst RS, Langer CJ. Seminars in Oncology. Elsevier; 2002. Epidermal growth factor receptors as a target for cancer treatment: the emerging role of IMC-C225 in the treatment of lung and head and neck cancers. [DOI] [PubMed] [Google Scholar]
- Hernandez CC, Zaika O, Tolstykh GP, Shapiro MS. Regulation of neural KCNQ channels: signalling pathways, structural motifs and functional implications. J Physiol. 2008;586:1811–1821. doi: 10.1113/jphysiol.2007.148304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Maillard K, Cattini N, Weiss RA, Boshoff C, Wooster R, Kellam P. Kaposi’s sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc Natl Acad Sci. 2003;100:10399–10404. doi: 10.1073/pnas.1630810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali E, Bellou S, Stellas D, Klinakis A, Murphy C, Fotsis T. VEGF signals through ATF6 and PERK to promote endothelial cell survival and angiogenesis in the absence of ER stress. Mol Cell. 2014;54:559–572. doi: 10.1016/j.molcel.2014.03.022. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003a;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci. 2003b;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Nichols P, Spicer D, Groshen S, Mimi CY, Lee AS. GRP78 as a novel predictor of responsiveness to chemotherapy in breast cancer. Cancer Res. 2006;66:7849–7853. doi: 10.1158/0008-5472.CAN-06-1660. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14:263–276. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Hsieh F, Song H, Lin J. Elevated phosphorylation and activation of PDK-1/AKT pathway in human breast cancer. Br J Cancer. 2005;93:1372–1381. doi: 10.1038/sj.bjc.6602862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Cai MJ, Zheng CC, Wang JX, Zhao XF. Phospholipase Cγ1 connects the cell membrane pathway to the nuclear receptor pathway in insect steroid hormone signaling. J Biol Chem. 2014;289:13026–13041. doi: 10.1074/jbc.M113.547018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4:966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- Margolis B, Bellot F, Honegger A, Ullrich A, Schlessinger J, Zilberstein A. Tyrosine kinase activity is essential for the association of phospholipase C-gamma with the epidermal growth factor receptor. Mol Cell Biol. 1990;10:435–441. doi: 10.1128/mcb.10.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti L, Cha YI, Niermann KJ, Lu B. Switch between apoptosis and autophagy: radiation-induced endoplasmic reticulum stress? Cell Cycle. 2007;6:793–798. doi: 10.4161/cc.6.7.4036. [DOI] [PubMed] [Google Scholar]
- Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- Nawrocki ST, Carew JS, Dunner K, Boise LH, Chiao PJ, Huang P, Abbruzzese JL, McConkey DJ. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 2005;65:11510–11519. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002a;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002b;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattarozzi A, Gatti M, Barbieri F, Würth R, Porcile C, Lunardi G, Ratto A, Favoni R, Bajetto A, Ferrari A. 17β-Estradiol promotes breast cancer cell proliferation-inducing stromal cell-derived factor-1-mediated epidermal growth factor receptor transactivation: reversal by gefitinib pretreatment. Mol Pharmacol. 2008;73:191–202. doi: 10.1124/mol.107.039974. [DOI] [PubMed] [Google Scholar]
- Prabhu A, Sarcar B, Kahali S, Shan Y, Chinnaiyan P. Targeting the unfolded protein response in glioblastoma cells with the fusion protein EGF-SubA. PloS One. 2012;7:e52265. doi: 10.1371/journal.pone.0052265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho JS, Pinheiro C, Lambros MB, Milanezi F, Carvalho S, Savage K, Simpson PT, Jones C, Swift S, Mackay A, Reis RM, Hornick JL, Pereira EM, Baltazar F, Fletcher CD, Ashworth A, Lakhani SR, Schmitt FC. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006;209:445–453. doi: 10.1002/path.2004. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007a;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007b;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013;5:a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin Ö, Löbke C, Korf U, Appelhans H, Sültmann H, Poustka A, Wiemann S, Arlt D. Combinatorial RNAi for quantitative protein network analysis. Proc Natl Acad Sci. 2007;104:6579–6584. doi: 10.1073/pnas.0606827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury JR, Farndon JR, Needham GK, Malcolm AJ, Harris AL. Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. Lancet. 1987;1:1398–1402. doi: 10.1016/s0140-6736(87)90593-9. [DOI] [PubMed] [Google Scholar]
- Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol/Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- Saxena R, Dwivedi A. ErbB family receptor inhibitors as therapeutic agents in breast cancer: current status and future clinical perspective. Med Res Rev. 2012;32:166–215. doi: 10.1002/med.20209. [DOI] [PubMed] [Google Scholar]
- Schmitt JM, Wayman GA, Nozaki N, Soderling TR. Calcium activation of ERK mediated by calmodulin kinase I. J Biol Chem. 2004;279:24064–24072. doi: 10.1074/jbc.M401501200. [DOI] [PubMed] [Google Scholar]
- Shuda M, Kondoh N, Imazeki N, Tanaka K, Okada T, Mori K, Hada A, Arai M, Wakatsuki T, Matsubara O. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605–614. doi: 10.1016/s0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2012;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- Zell JA, Tsang WY, Taylor TH, Mehta RS, Anton-Culver H. Prognostic impact of human epidermal growth factor-like receptor 2 and hormone receptor status in inflammatory breast cancer (IBC): analysis of 2,014 IBC patient cases from the California Cancer Registry. Breast Cancer Res BCR. 2009;11:R9. doi: 10.1186/bcr2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwang Y, Oren M, Yarden Y. Consistency test of the cell cycle: roles for p53 and EGR1. Cancer Res. 2012;72:1051–1054. doi: 10.1158/0008-5472.CAN-11-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.