Abstract
Studies investigating the pathogenic role of the microtubule associated protein tau (MAPT) gene in Parkinson’s disease (PD) have indicated that DNA methylation of the promoter region is aberrant in disease, leading to dysregulated MAPT expression. We examined two potential regulators of MAPT gene expression in respect to PD, a promoter-associated long non-coding RNA MAPT-AS1, and DNA methyltransferases (DNMTs), enzymes responsible for new and maintenance of DNA methylation. We assessed the relationship between expression levels of MAPT and the candidate MAPT-AS1, DNMT1, DNMT3A and DNMT3B transcripts in four brain regions with varying degrees of cell loss and pathology (putamen, anterior cingulate cortex, visual cortex and cerebellum) in N = 10 PD and N = 10 controls. We found a significant decrease in MAPT-AS1 expression in PD (p = 7.154 x 10−6). The transcript levels of both MAPT-AS1 (p = 2.569 x 10−4) and DNMT1 (p = 0.001) correlated with those of MAPT across the four brain regions, but not with each other. Overexpression of MAPT-AS1 decreased MAPT promoter activity by ∼2.2 to 4.3 fold in an in vitro luciferase assay performed in two cell lines (p ≤ 2.678 x 10−4). Knock-down expression of MAPT-AS1 led to a 1.3 to 6.3 fold increase in methylation of the endogenous MAPT promoter (p ≤ 0.011) and a 1.2 to 1.5 fold increased expression of the 4-repeat MAPT isoform transcript (p ≤ 0.013). In conclusion, MAPT-AS1 and DNMT1 have been identified as potential epigenetic regulators of MAPT expression in PD across four different brain regions. Our data also suggest that increased MAPT expression could be associated with disease state, but not with PD neuropathology severity.
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder, characterised by tremor and bradykinesia, that affects 2% of the population over the age of 65 [1]. The neuropathologic hallmarks of PD include loss of dopaminergic neurons in the substantia nigra and the presence of Lewy bodies, cytoplasmic inclusions composed primarily of α-synuclein [2]. Monogenic forms of PD have been linked to mutations in genes including α-synuclein (SNCA) [3], leucine-rich repeat kinase 2 (LRRK2) [4] and parkin (PARK2) [5]. These monogenic forms, however, explain only 6% of PD cases, while the remaining are ‘idiopathic’ or sporadic, as their aetiology is unknown [6].
Multiple susceptibility genes have been identified for sporadic forms of PD [7], with SNCA, LRRK2 and the gene encoding microtubule-associated protein tau (MAPT) being the most consistently replicated loci [8–10]. Microtubule-associated protein tau (tau), a protein that aids in stabilising the axonal cytoskeleton, is particularly interesting as it has been indicated as a susceptibility gene in other neurodegenerative conditions such as progressive supranuclear palsy [11], and MAPT mutations are a cause of monogenic frontotemporal dementia [12]. There are two main MAPT haplotypes, termed H1 and H2, resulting from single nucleotide polymorphisms (SNPs) in a region of absolute linkage disequilibrium that spans the entire MAPT gene [13]. The major allele, H1, has a higher level of expression than the H2 allele and is associated with increased risk of PD [14]. A recent large-scale Quantitative Trait Loci (QTL) study of brain tissues, comprising N = 773 individuals and measuring transcript levels using the Whole Genome DASL assay, further supported the finding that H1 haplotype elevates gene expression in two brain regions [15]. Given that a higher level of MAPT expression may potentially lead to increased neurodegeneration [16] it can be hypothesised that mechanisms regulating the expression of MAPT could contribute to neurodegenerative disease. Previously, dysregulation of MAPT promoter methylation has been associated with PD [17]. Moreover, in leukocyte DNA, MAPT methylation levels also served as a biomarker for age of PD onset in an idiopathic PD cohort [17]. Finally, the MAPT promoter has been shown to be aberrantly methylated in both leukocyte and brain tissue of PD patients [17, 18].
While there are several studies implicating dysregulation of DNA methylation in PD, the mechanisms driving this process are yet to be elucidated. One possible mechanism includes the activity of non-coding RNAs (ncRNAs) which do not code for proteins [19]. One class of ncRNAs are long ncRNAs (lncRNAs) that are diverse in length (≥∼200 bases) and function [20]. An example of the importance of lncRNAs is in the regulation of the neurotrophic and cognition-related gene BDNF [21], whose expression is controlled not only by ncRNAs but also by DNA methylation and histone modifications. Of interest, a similar lncRNA exists at the MAPT locus: MAPT-AS1 is an 840 bp lncRNA transcribed from the anti-sense strand of the MAPT promoter region [22] (Figure 1 in S1 File). Another mechanism includes DNA methyltransferases (DNMTs) which are enzymes carrying a C-terminal catalytic domain that methylates CpG dinucleotides. Catalytically active members of the group include Dnmt1 (gene: DNMT1), responsible for maintenance of methylation by targeting hemimethylated DNA, and Dnmt3a (gene: DNMT3A) and Dnmt3b (gene: DNMT3B), which catalyse de novo methylation and are mainly active during embryonic development and neurogenesis in brain tissue [23]. Aberrant expression of DNMT3A has been linked to atypical DNA methylation levels in frontotemporal dementia [24]. Similarly, alterations to the activity of Dnmt3a and Dnmt1 have been implicated in neuronal cell death pathways in mice and amyotrophic lateral sclerosis in humans [25].
We postulate that dysregulation of MAPT expression in PD is due to altered levels of the lncRNA MAPT-AS1 and/or at least one DNMT gene in a disease-specific manner. In this study we examined the relationship between MAPT, MAPT-AS1, DNMT1, DNMT3A and DNMT3B expression levels in PD and control brain tissue samples across four brain regions. We found disease-specific effects of MAPT-AS1 and DNMT1 on MAPT expression. In vitro cellular assays established that MAPT promoter activity is altered via an epigenetic mechanism when MAPT-AS1 expression is manipulated in vitro.
Materials and Methods
Study cohorts
We examined 80 brain tissue samples from 10 PD and 10 age- and sex-matched controls (Table 1) across four brain regions to assess underlying disease-specific effects that are independent of differences in pathology and cell loss: putamen (affected early in PD and with tissue loss), anterior cingulate cortex (ACC, affected later in PD and with minimal cell loss), visual cortex (VC, no overt pathology but with abnormal lipid metabolism [26]) and cerebellum (not affected by PD) [27, 28]. All PD cases were Braak stage IV or higher. All cases with head injury, brain tumour, infarction or other significant neurodegenerative pathologies were excluded. Controls had no history of neurological or psychiatric symptoms and no significant neuropathology. These samples were all of Caucasian descent and were provided by the Sydney Brain Bank (HC15008 –ethics approval provided by the University of New South Wales Human Research Ethics Committee) and the NSW Tissue Resource Centre (Protocol No X15-0199 & HREC/11/RPAH/147 –ethics approval provided by the NSW Government Health Sydney Local Health District). Ethics approval for the analyses of human tissue for epigenetic modifications (HREC 11312) was provided by the University of New South Wales Human Research Ethics Committee). Informed written consents were obtained from all participants, or their next of kin, or legal guardian, as the participants were recruited, using procedures approved by the ethic committees detailed above.
Table 1. Comparison of demographics and MAPT haplotype frequency in brain tissue cohort.
| Disease status | Sex | Age(y)a | MAPT haplotype | PMD (h)a | Tissue pHa | ||||
|---|---|---|---|---|---|---|---|---|---|
| N | M | F | Mean ± SD | Range | H1 | H2 | Mean ± SD | Mean ± SD | |
| Normal | 10 | 7 (70%) | 3 (30%) | 77.4 ± 8 | 65–88 | 7 (70%) | 3 (30%) | 21.3 ± 15 | 6.5 ± 0.2 |
| PD | 10 | 7 (70%) | 3 (30%) | 79.5 ± 6 | 69–90 | 7 (70%) | 3 (30%) | 19.4 ± 12.9 | 6.5 ± 0.7 |
a There were no significant differences in mean age at death, tissue pH and post-mortem delay (PMD) between cases and controls as assessed by Student’s t-test.
F, female; M, male; MAPT, microtubule-associated protein tau; PD, Parkinson’s disease; PMD, post-mortem delay; SD, standard deviation.
Nucleic acids extraction and genotyping
Genomic DNA and RNA were extracted from brain tissue using DNeasy and RNAQuick kits (QIAGEN, Venlo, Netherlands). A Taqman Probe Genotyping Assay (Life Technologies, CA, USA) for rs1052553 was used to determine the corresponding H1/H2 MAPT diplotype in brain tissue.
Haplotype-specific methylation analysis
Genomic DNA (gDNA) samples (500 ng) were bisulfite converted using the EpiTect96 Bisulfite Kit (QIAGEN, Venlo, Netherlands) according to manufacturer’s specifications. PCR primers were designed from UCSC Feb 2009 (hg19) draft to amplify a 148 bp region of the MAPT promoter region in bisulfite-converted gDNA (GRCh37_Chr 17: 43,971,386–43,971,534). PCR and sequencing primer design was performed using ABI Methylprimer v1.0 (ABI) and PyroMark Assay Design v2.0 software (QIAGEN). Haplotype-specific pyrosequencing was based on the design of haplotype-specific primers which have either a ‘G’ or ‘A’ nucleotide at its 3’ end, corresponding to bisulfite converted sequence of either the H1 or H2 allele of the rs76594404 polymorphism within the PCR amplified region (Figure 1 in S1 File) as described previously [17]. Bisulfite converted DNAs were amplified using MAPT1_pyroF_biotin 5’-GGGAAAGAGATTTTAGTTAGG-3’, MAPT1_pyroR 5’-CCCTTTACTTTCAATCAA-3’, and sequenced using primers specific to either the H1 (5’-gtaaactaccttccacttaac-3’) or H2 (5’-gtaaactaccttccacttaaa-3’) haplotypes (Figure 1 in S1 File). The CpG dinucleotides at position 2 to 6 in the pyrosequenced region showed high intra- and inter- experiment correlation of methylation levels and thus the arithmetic mean of these CpGs was used as a summary score for our analyses.
Relative quantification of gene expression
Approximately 100mg of frozen tissue, from each brain region of each patient, was extracted using the RNAeasy Mini Kit (Qiagen, Valencia, CA, USA). SuperScript III First Strand cDNA synthesis kit (Life Technologies, CA, USA) was used to generate cDNA from brain total mRNA (100 ng) as directed by manufacturer’s instructions. Transcript levels for MAPT (either total using primers that span exons 11 and 12, or 4-repeat MAPT isoform using primers that span exons 9 and 11) and MAPT-AS1 were assayed using relative quantification–polymerase chain reaction (RQ-PCR). Two housekeeping genes, succinate dehydrogenase complex, subunit A (SDHA) [29] and the eukaryotic translation initiation factor 4A2 (EIF4A2) [30] were used to normalize gene expression levels. Primers for the validation of each gene were designed to span exon/exon boundaries to minimise the effects of genomic DNA contamination. The complete set of primers is listed in Table 1 of S1 File. Four experimental replicate PCR reactions were performed for each sample using 1nM concentration of each primer in 1x Platinum SYBR Green qPCR Supermix–UPG [Invitrogen, Carlsbad, CA, USA) and 1ng of reverse transcribed RNA. Serial dilutions of template cDNAs confirmed that each primer pair had amplification efficiency of ∼2. Transcript levels of the DNMT genes were assayed using commercially available pre-designed Taqman Probes [DNMT1 [Hs00945875_m1), DNMT3A [Hs01027166_m1) and DNMT3B [Hs00171876_m1) [Life Technologies)). Four experimental replicate qPCR reactions were performed for each Taqman probe in 1x Taqman Fast Advanced Master Mix [Life Technologies) with 1ng of cDNA. Gene expression levels [2-∆Ct where -∆Ct = —Ct [target)–Ct [geometric mean of house-keeping genes) [29, 31]) were log transformed to -∆Ct values to improve linearity of data in the Repeat Measure Mixed Linear Model Regression analyses, or expressed as 2-∆∆Ct where ∆∆Ct = (Ct(target gene, control cells)−Ct(geometric mean of house-keeping genes, control cells))−(Ct(target gene, treated cells)−Ct(geometric mean of house-keeping genes, treated cells)) when we were comparing the difference in means between control and treated cells within each independent transfection.
MAPT-AS1 expression constructs and siRNA
A full length 840bp cDNA corresponding to the Homo sapiens MAPT antisense RNA 1 (MAPT-AS1) non coding RNA transcript sequence (NR_024559.1) was commercially synthesized and subcloned into the expression vector pCDNA3.1(+) (GenScript, NJ, USA). Knock-down expression of MAPT-AS1 was achieved using a Custom Silencer® Select siRNA (s237111) (Ambion–Thermofisher Scientific, NY, USA). The sequence of the s237111 siRNA is: Sense 5’-CCACUUCAUGGAUAAGUAAtt-3’ and Antisense 5’-UUACUUAUCCAUGAAGUGGgt-3’. The Silencer® Negative Control No. 2 siRNA (Ambion–Thermofisher Scientific) served as a negative control for non-specific effects.
Cellular assays
The MAPT-promoter luciferase vector, MAPT-pCpG, was as described previously [17]. Two cell lines, the neuroblastoma SK-N-MC (ATTC HTB-10) and the human embryonic kidney HEK293 (ATCC CRL-1573) were co-transfected with either the MAPT-AS1 cDNA expression construct, or siRNAs, and the MAPT-pCpG constructs using Lipofectamine 2000 (Invitrogen, CA, USA). 1.5 x 104 cells/well in a 24 well plate (Nunc Delta Si, Thermofisher Scientific, NY, USA) were transfected with 2μl Lipofectamine 2000, 200ng cDNA expression construct or 2pmol siRNA, 200ng MAPT-pCpG luciferase constructs. An additional 20ng of the pRL-TK renilla luciferase vector (Promega, WI, USA) was added to the transfection mixture for the normalization of transfection efficiencies. We examined whether there was a haplotype-specific effect on promoter activity by co-transfecting MAPT-promoter constructs corresponding to either the H1 or H2 haplotype. Cells were lysed after 48 h and assayed for luciferase activity using the Dual-Luciferase® Reporter assay system (Promega). Results were obtained from 5 independent experiments. No significant differences in raw renilla luciferase values were observed between experimental groups (data not shown).
Statistical analyses
Repeat Measure Mixed Linear Model Regression analysis, with a random intercept model, was used to explore the relationship between transcript levels and disease status across four brain regions. Graphical outputs for these analyses were generated using the alternate form of the repeat measure linear regression model syntax, known as general linear model. Gene expression levels are presented as estimated marginal means because they are adjusted for the effects of significant covariates. The syntax for the statistical analyses are given in full in the Methods of S1 File. Of the cohort demographics (apart from disease status), tissue pH was the only significant predictor for MAPT transcript levels. Thus, relationships between gene expression levels of MAPT, MAPT-AS1, DNMT1, DNMT3A and DNMT3B, were assessed with tissue pH as covariate. Significance of results was adjusted for multiple testing using the Bonferroni correction. Student’s paired samples t-tests were used to assess in vitro promoter luciferase activity, changes in endogenous DNA methylation levels and transcript levels from cell culture experiments. All statistical analyses were performed in SPSS v. 22 (IBM, Armonk, NY).
Results and Discussion
Disease-specific differences in gene expression levels
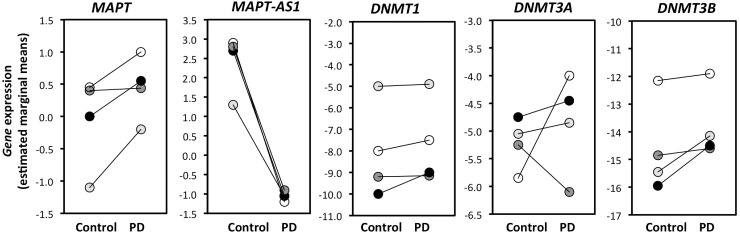
Student’s independent samples t-test was used to examine age, tissue pH, and post-mortem delay between groups, finding no significant difference between PD cases and controls (Table 1). Gender and MAPT haplotype have been previously demonstrated to significantly affect MAPT methylation [17], as such our controls and PD tissue samples were gender-matched and had identical MAPT haplotype frequencies. In addition to the basic demographic factors such as age (Table 2), we specifically examined the effect of disease state on the transcript levels of our five genes of interest, MAPT, MAPT-AS1, DNMT1, DNMT3A and DNMT3B. The Repeat Measure Mixed Linear Model Regression analysis is designed to detect consistent changes across the four brain regions that differ in their level of pathology and cell loss. We speculate that consistent changes in gene expression across four brain regions are more likely to represent factors that are causative of disease, rather than a secondary phenomenon of disease progression. As shown in Table 2, univariate analyses identified that disease state was the strongest predictor for MAPT-AS1 transcript levels (t = -6.287, p = 7.154 x 10−6), followed by DNMT3B (t = 4.044, p = 7.977 x 10−5) and DNMT3A (t = 3.005, p = 0.007). Graphical representations of the change in gene expression with disease state for the five genes are shown in Fig 1.
Table 2. Univariate Repeat Measure Mixed Linear Model Regression analyses for predictors of gene expression.
| MAPT | MAPT-AS1 | DNMT1 | DNMT3A | DNMT3B | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Predictor | t-Statistic | p valuea | t-Statistic | p value | t-Statistic | p value | t-Statistic | p value | t-Statistic | p value |
| Age | 0.87 | 0.449 | -0.0776 | 0.447 | 1.821 | 0.085 | 1.02 | 0.32 | -0.152 | 0.88 |
| Gender | 0.811 | 0.474 | -0.298 | 0.769 | -0.563 | 0.597 | -0.378 | 0.709 | 2.271 | 0.042 |
| PMI | -0.504 | 0.655 | 0.103 | 0.919 | -0.385 | 0.718 | -0.551 | 0.588 | 0.32 | 0.753 |
| Tissue pH | -3.212 | 0.004 | -0.594 | 0.559 | 1.022 | 0.378 | -2.039 | 0.057 | -0.048 | 0.963 |
| Disease status | 1.551 | 0.175 | -6.287 | 7.15E-05 | 2.262 | 0.396 | 3.005 | 0.007 | 4.044 | 7.98E-05 |
a significant p values (p < 0.05) are highlighted in bold. Note that the significance levels are not adjusted for multiple testing as this is an exploratory analysis.
Fig 1. Disease-specific expression levels of MAPT, DNMT1, DNMT3A and DNMT3B across four brain regions.
Putamen (black circle), ACC (dark grey circle), visual cortex (light grey circle) and cerebellum (open circle). Data points are derived from estimated marginal means after adjusting for significant demographic predictors apart from disease status.
Predictors of MAPT expression levels
We performed multivariate analyses to explore the relationships between different gene transcript levels and the effect of disease status. We set the threshold p value at < 0.004 for 12 comparisons (4 genes x 3 variables of disease, gene transcript levels, and disease * gene transcript levels). As shown in Table 3, MAPT-AS1 transcript levels was the best predictor of MAPT transcript levels (t = 5.656, p = 2.569 x 10−4) with a significant interaction with disease state (t = -4.286, p = 0.001). DNMT1 transcript levels were also significantly associated with MAPT transcript levels (t = 4.476, p = 0.001), but without significant interaction with disease status after correction for multiple testing. Of interest, DNMT1 transcript levels were not associated with MAPT-AS1 levels (Table 3), suggesting independent effects on MAPT expression. Neither DNMT3A nor DNMT3B were significantly associated with MAPT or MAPT-AS1 transcript levels (Table 3).
Table 3. Multivariate Repeat Measure Mixed Linear Model Regression analyses for the effects of disease status and MAPT transcript levels.
| MAPT | MAPT-AS1 | |||
|---|---|---|---|---|
| Predictor a | t-Statistic | p valueb | t-Statistic | p valueb |
| Disease | -1.456 | 0.179 | - | - |
| MAPT1-AS1 | 5.656 | 2.57E-04 | - | - |
| Disease * MAPT-AS1 | -4.286 | 0.001 | - | - |
| Disease | -2.365 | 0.027 | 0.825 | 0.415 |
| DNMT1 | 4.476 | 0.001 | 0.849 | 0.407 |
| Disease * DNMT1 | -2.991 | 0.006 | -1.014 | 0.318 |
| Disease | -0.551 | 0.583 | 0.696 | 0.49 |
| DNMT3A | 1.512 | 0.135 | 2.62 | 0.013 |
| Disease * DNMT3A | -0.941 | 0.35 | -0.875 | 0.386 |
| Disease | -1.121 | 0.311 | -0.201 | 0.841 |
| DNMT3B | -2.407 | 0.062 | 0.066 | 0.948 |
| Disease * DNMT3B | -0.791 | 0.463 | -0.995 | 0.302 |
a For each gene of interest, demographic covariates were included in the multivariate regression analyses if they were significantly associated in the initial univariate analysis (Table 2). For MAPT and MAPT-AS1: Tissue pH; MAPT and DNMT1: Tissue pH; MAPT and DNMT3A: Tissue PH; MAPT and DNMT3B: Tissue pH and Gender. For MAPT-AS1 and DNMT3B: Gender.
b p values that survived correction for multiple testing are indicated in bold.
Effect of MAPT-AS1 expression on the activity of a MAPT promoter construct
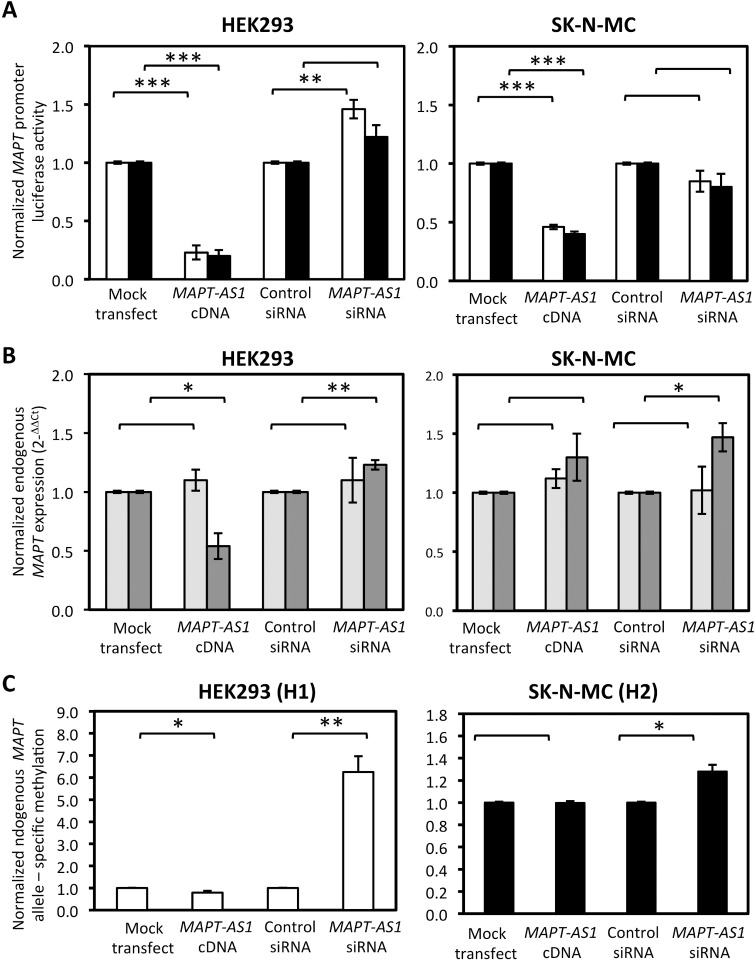
As MAPT-AS1 expression was significantly correlated with MAPT expression, we identified a causative relationship between the two loci in vitro using a sensitive reporter assay in two model cell lines. We co-expressed the MAPT-AS1 cDNA expression construct with MAPT promoter luciferase constructs corresponding to either the H1 or H2 haplotype. In HEK293 cells, for both MAPT haplotypes, there was a significant decrease in luciferase activity when the lncRNA was overexpressed (H1 haplotype, 4.3 fold decrease, p = 2.678 x 10−4; H2 haplotype, 5.1 fold decrease, p = 9.203 x 10−5). A similar effect was observed for SK-N-MC cells (H1 haplotype, 2.2 fold decrease, p = 9.159 x 10−6; H2 haplotype, 2.5 fold decrease, p = 9.855 x 10−6) when compared to mock transfected cells (Fig 2A). Conversely, when the endogenous level of MAPT-AS1 was knocked-down by siRNA specific to the lncRNA, we observed a significant increase in activity from the H1 haplotype reporter construct in HEK293 cells (1.5 fold increase in luciferase activity, p = 0.004), but not the H2 haplotype. No significant effects were observed for either haplotypes in SK-N-MC cells (Fig 2A).
Fig 2. Effect of MAPT-AS1 over- and knock-down expression on MAPT expression in HEK293 and SK-N-MC cells.
A) H1 (white columns) and H2 (black columns) haplotype MAPT promoter-driven luciferase activity. Luciferase activity is normalized to each control transfection levels. B) Endogenous transcript levels of either total MAPT (light grey columns) or 4 repeat MAPT transcript (dark grey columns). Transcript levels are normalized to each control transfection levels C) Endogenous haplotype-specific DNA methylation with H1 (white columns) and H2 (black columns) specific data indicated. Methylation levels from MAPT-AS1 over- or under-expression are normalized to control transfection levels. Error bars indicate standard error of the mean from 5 independent experiments. *, p < 0.05; *** p < 0.0001.
MAPT-AS1 expression and effects on endogenous MAPT expression
In order to determine whether increased MAPT-AS1 expression influenced MAPT expression via MAPT promoter methylation we examined endogenous MAPT methylation and expression in vitro using the same cellular models as above, but omitting the co-transfection of the MAPT promoter luciferase constructs. For these experiments, we measured MAPT-AS1 transcript levels in transfected cells by RQ-PCR, and confirmed that transfection with MAPT-AS1 cDNA, compared to mock transfected cells, led to a significant 573- to 2227-fold increase in gene expression for SK-N-MC (p = 0.002) and HEK293 (p = 0.013) cells respectively (Figure 2A in S1 File). Similarly, transfection with the MAPT-AS1 siRNA, compared with the control siRNA, led to a significant 1.7-fold knock-down (or 60% of pre-knock-down levels) of endogenous MAPT-AS1 transcript levels in both SK-N-MC (p = 0.042) and HEK293 cells (p = 0.045) (Figure 2B in S1 File). We also noted that there were no significant differences in the levels of our two house-keeping genes SDHA and EIF4A2 between any experimental groups (data not shown). For endogenous MAPT expression, we measured both total transcript levels as well as the levels of the longer isoform with an additional alternatively spliced exon (4 repeat MAPT isoform). In HEK293 cells, we observed a significant 1.9 fold decrease in transcript levels for the 4 repeat MAPT isoform (p = 0.015), but not for total MAPT transcripts (Fig 2B), in cells over-expressing the MAPT-AS1 cDNA compared with mock transfected cells. No significant effect of MAPT-AS1 over-expression on endogenous MAPT transcript levels was observed in SK-N-MC cells (Fig 2B). Conversely, when we knocked-down expression of MAPT-AS1, we observed a significant 1.2- to 1.5-fold increase in 4 repeat MAPT transcripts for HEK293 cells (p = 0.005) and SK-N-MC cells (p = 0.019) respectively, compared to cells transfected with the control siRNA.
MAPT-AS1 expression and effects on endogenous MAPT promoter methylation
HEK293 and SK-N-MC cells differ in their MAPT haplotypes with HEK293 reported to be H1/H1 homozygotes and SK-N-MC cells reported to be H1/H2 heterozygotes [32]. We performed haplotype-specific pyrosequencing to determine the haplotype-specific effects of MAPT-AS1 expression on DNA methylation of the MAPT promoter. In HEK293 cells, we observed a 1.4 fold decrease in DNA methylation (p = 0.032) of the H1 haplotype when we over-expressed the MAPT-AS1 cDNA compared to mock-transfected cells (Fig 2C). Conversely, we observed a marked 6.3 fold increase in DNA methylation (p = 0.002) on the H1 haplotype when we knocked-down the expression of the lncRNA (Fig 2C). In the SK-N-MC cells, the assay for the H1 haplotype failed. Sanger sequencing across the pyrosequencing primer binding sites from both cell lines confirmed the presence of the appropriate homozygote genotype of rs76594404 in HEK293 and heterozygote genotype for SK-N-MC cells (Figure 3 in S1 File). However, the sequencing also revealed that there was higher level of basal DNA methylation in two CpG sites within the primer binding site for SK-N-MC cells compared with HEK293 cells (Figure 3 in S1 File). This could have affected the binding of the H1 haplotype-specific sequencing primer in SK-N-MC cells. For the H2 haplotype in SK-N-MC cells, we observed a 1.3 fold increase in DNA methylation (p = 0.011) when we knocked-down the expression of the lncRNA.
In this study we examined the expression of potential epigenetic regulators of gene expression, the lncRNA MAPT-AS1 and the methyltransferase genes DNMT1, DNMT3A and DNMT3B, and their relationship to MAPT expression across four brain regions with differing PD pathology. Of note, MAPT-AS1 expression was highly associated with disease state, and was the strongest predictor of MAPT expression of the four candidates genes. Over-expression of MAPT-AS1 in vitro resulted in decreased MAPT expression in two model cell lines, whilst knock-down expression of the lncRNA had the opposite effect, most clearly observed on the methylation and expression from the endogenous MAPT promoter.
The four brain regions included in this study, the putamen, anterior cingulate and visual cortices and cerebellum, differ in their level of PD neuropathology, as defined by α-synuclein deposition as well as tissue loss. We report for the first time a significant PD-specific decrease in MAPT-AS1 expression across these four brain regions, suggesting that this decrease occurs independently of alpha-synuclein deposition and cell loss. Certain promoter-associated lncRNAs, such as the antisense BDNF-AS, have been shown to act as a repressor of gene expression via epigenetic modifications. This was demonstrated in mice where inhibition of the endogenous Bdnf-AS results in upregulation of the mouse Bdnf expression, changes in histone modifications and increased neuronal growth and differentiation [21]. We observed a similar inhibitory effect on MAPT expression by MAPT-AS1. Future studies will confirm whether the effect of the lncRNA is specific for the MAPT locus, or whether it has other trans-acting effects as well. The role of these promoter-associated lncRNAs are of special interest as we have noted that other neurodegenerative genes, such as the alpha-synuclein (SNCA) [3] and glycogen-synthase kinase 3beta (GSK3B) genes [33], also have similar lncRNAs (SNCA-AS and BC035247 respectively).
MAPT haplotypes are major determinants of MAPT gene expression as well as disease risk [7, 15, 34, 35]. Of note, there are studies that support the role of the H1/H2 haplotypes on differential splicing [32, 36] rather than elevation of total gene expression. Current QTL studies that use gene expression arrays do not effectively capture all alternative splicing information, and there are potential artifacts that can arise from polymorphisms that fall within the binding regions of the probes. Future studies from next generation transcriptomics analyses, which do not rely on hybridization of probes, will provide more accurate information of the expression levels of all splice variants and should provide conclusive evidence on this matter.
Recent studies have suggested that gene transcription and the regulation of alternative splicing are functionally linked [37] and may be due to recruitment of splicing factors by RNAPII, particularly SR proteins such as SRp20 which have been shown to modulate alternative splicing of MAPT [32]. Moreover, epigenetic modifications including DNA methylation also play a crucial role in regulating alternative splicing and have implications for disease process [38]. A DNA-binding protein, CCCTC-binding factor (CTCF), can promote inclusion of alternatively spliced exons by mediating local pausing of RNA polymerase II, and CTCF binding to these exons is inhibited by DNA methylation [39]. In brain tissue of patients with autistic spectrum disorders, altered expression and alternative splicing of SHANK3 gene were observed in samples with increased methylation of CpG islands within SHANK3. Furthermore, a DNA methylation inhibitor modified the methylation of these CpG islands and altered the isoform-specific expression of SHANK3 in cultured cells [40].
Promoter-associated lncRNAs, such as MAPT-AS1, can impact on gene expression in trans, often by direct interaction with key regulators of DNA methylation. Di Ruscio et al. showed that a lncRNA arising from the CEBPA gene locus, termed extra-coding CEBPA, was required for regulation of DNA methylation at this site through the association of the lncRNA with DNMT1 [41]. Similarly, the sustained expression of a central nervous system expressed intergenic lncRNA, Dali, was required for the differentiation of neuroblastoma cells via interactions with DNMT1 and regulates DNA methylation status of multiple CpG island-associated promoters in trans [42]. In our cellular assays, we confirm that the elevation in DNA methylation (in our pyrosequenced region) was significantly associated with an increase in the alternatively spliced 4-repeat MAPT isoform levels, but not total MAPT transcripts. This provides further evidence that epigenetic modifications play a role in determining both gene transcription and alternative splicing.
Certain neuroprotective compounds, such as vitamin E, have been shown to have a haplotype-specific effect on MAPT expression via epigenetic mechanisms as well [17]. Therefore, it was important to demonstrate that MAPT-AS1 was capable of inhibiting the expression of both the H1 and H2 promoter luciferase reporter constructs. We observed a significant effect on each haplotype as assayed in the two cell lines used in this study. The luciferase reporter assays also suggested that MAPT-AS1 can act to repress gene expression in trans.
We noted cell type-specific differences in our two cellular models. Firstly, in control HEK293 cells, endogenous MAPT-AS1 was expressed approximately 30-fold higher than endogenous MAPT (data not shown), whilst in control SK-N-MC cells, MAPT-AS1 was expressed approximately 1.1-fold lower than MAPT (data not shown). Secondly, transfection of the MAPT-AS1 cDNA resulted in a higher level (11-fold) of expression in the HEK cells compared with SK-N-MC cells (Figure 2 in S1 File). The relatively low level of expression of the transfected transgene or transfection efficiency in neuronal cell types has been observed previously [43]. At present, it is unclear whether these differences could fully explain the more consistent and significant effects on luciferase activity, endogenous gene expression and DNA methylation in HEK293 cells compared with SK-N-MC cells. Another aspect we should consider is the difference between over-expression and knock-down expression of MAPT-AS1 and their corresponding effects on the MAPT-promoter luciferase activity (Fig 2A). In the over-expression assays, we observed a consistent effect in both cell lines as we increased the copy numbers of the lncRNA and its putative target on the MAPT promoter by our transfections. Conversely, in the knock-down assays, we only increased the copy number of the exogenous MAPT promoter, whilst knocking down the endogenous lncRNA. This may have led to failure to detect a consistent effect of siRNA knock-down on the luciferase assays.
Finally, we also examined other candidate trans-acting factors of MAPT expression, specifically the expression profiles of the three DNMTs. In terms of MAPT expression, only DNMT1 transcript levels were significantly associated across the four brain regions, although there does not appear to be a disease-specific effect after adjusting for multiple testing. DNMT1 is required for maintenance of methylation, gene regulation and chromatin stability. DNMT1 is of interest as mutations in this gene have been associated with hereditary sensory neuropathy with dementia and hearing loss [44]. Both DNMT3A and DNMT3B levels were significantly associated with disease state but not with MAPT levels in our cohort of brain samples. This is in contrast to a study by Iwata et al. [45] which demonstrated that DNMT3A over-expression was capable of inhibiting MAPT expression in a cellular model.
Conclusions
The main limitation of our study was the relatively small sample size. Future studies with a larger number of brain samples may also resolve apparent contradictions between tissue and cellular studies for the DNMT3A gene. Moreover, while our pyrosequencing assay focused on 5 CpGs within the CpG island of the MAPT promoter, the examination of additional CpGs in non-proximal promoter regions may identify regions whose methylation levels more accurately reflect MAPT-AS1-mediated MAPT expression changes. In conclusion, we have identified a lncRNA which has an inhibitory effect on the expression of a major neurodegenerative gene, MAPT. The lncRNA may serve as a sensitive biomarker of disease state in PD, as well as a potential therapeutic target for inhibiting the expression of MAPT.
Supporting Information
(PDF)
Acknowledgments
Human brain tissue samples were received from the Sydney Brain Bank at Neuroscience Research Australia and the New South Wales Tissue Resource Centre at the University of Sydney which are supported by the University of New South Wales, Neuroscience Research Australia, Schizophrenia Research Institute and National Institute of Alcohol Abuse and Alcoholism.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Health and Medical Research Council of Australia (NHMRC) seeding fund APP1021269, Project Grant APP1062539 Program Grant 350833 and Capacity Building Grant 568940. GH is a NHMRC senior principal research fellow (grant #1079679). Human brain tissue samples were received from the Sydney Brain Bank at Neuroscience Research Australia and the New South Wales Tissue Resource Centre at the University of Sydney which are supported by the University of New South Wales, Neuroscience Research Australia, Schizophrenia Research Institute and National Institute of Alcohol Abuse and Alcoholism (NIH (NIAAA) R24AA012725). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bertram L, Tanzi RE. The genetic epidemidogy of neurodegenerative disease. Journal of Clinical Investigation. 2005;115(6):1449–57. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickson DW, Braak H, Duda JE, Duyckaerts C, Gasser T, Halliday GM, et al. Neuropathological assessment of Parkinson's disease: refining the diagnostic criteria. Lancet Neurology. 2009;8(12):1150–7. 10.1016/S1474-4422(09)70238-8 [DOI] [PubMed] [Google Scholar]
- 3.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science (New York, N Y). 1997;276(5321):2045–7. Medline:. [DOI] [PubMed] [Google Scholar]
- 4.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–7. Medline:. [DOI] [PubMed] [Google Scholar]
- 5.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–8. 10.1038/33416 . [DOI] [PubMed] [Google Scholar]
- 6.Houlden H, Singleton AB. The genetics and neuropathology of Parkinson's disease. Acta neuropathologica. 2012;124(3):325–38. Medline: 10.1007/s00401-012-1013-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nature genetics. 2014;46(9):989–93. Medline: 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Do CB, Tung JY, Dorfman E, Kiefer AK, Drabant EM, Francke U, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS genetics. 2011;7(6):e1002141 Medline: 10.1371/journal.pgen.1002141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Parkinson Disease Genomics C, Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin U-M, et al. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377(9766):641–9. Medline: 10.1016/S0140-6736(10)62345-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Annals of human genetics. 2010;74(2):97–109. Medline: 10.1111/j.1469-1809.2009.00560.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PMA, Wang L-S, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nature genetics. 2011;43(7):699–705. Medline: 10.1038/ng.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seelaar H, Rohrer JD, Pijnenburg YAL, Fox NC, van Swieten JC. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. Journal of neurology, neurosurgery, and psychiatry. 2011;82(5):476–86. Medline: 10.1136/jnnp.2010.212225 [DOI] [PubMed] [Google Scholar]
- 13.Pittman AM, Myers AJ, Abou-Sleiman P, Fung HC, Kaleem M, Marlowe L, et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. Journal of medical genetics. 2005;42(11):837–46. Medline:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok JBJ, Teber ET, Loy C, Hallupp M, Nicholson G, Mellick GD, et al. Tau haplotypes regulate transcription and are associated with Parkinson's disease. Annals of neurology. 2004;55(3):329–34. Medline:. [DOI] [PubMed] [Google Scholar]
- 15.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS genetics. 2012;8(6):e1002707 Epub 2012/06/12. 10.1371/journal.pgen.1002707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams SJ, Crook RJP, Deture M, Randle SJ, Innes AE, Yu XZ, et al. Overexpression of wild-type murine tau results in progressive tauopathy and neurodegeneration. The American journal of pathology. 2009;175(4):1598–609. 10.2353/ajpath.2009.090462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coupland KG, Mellick GD, Silburn PA, Mather K, Armstrong NJ, Sachdev PS, et al. DNA methylation of the MAPT gene in Parkinson's disease cohorts and modulation by vitamin E In Vitro. Mov Disord. 2013. Epub 2014/01/01. 10.1002/mds.25784 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics. 2013;8(10):1030–8. 10.4161/epi.25865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Souza N. The ENCODE project. Nat Methods. 2012;9(11):1046 Epub 2013/01/03. . [DOI] [PubMed] [Google Scholar]
- 20.Mather KA, Kwok JB, Armstrong N, Sachdev PS. The role of epigenetics in cognitive ageing. Int J Geriatr Psychiatry. 2014;29(11):1162–71. Epub 2014/08/08. 10.1002/gps.4183 . [DOI] [PubMed] [Google Scholar]
- 21.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30(5):453–9. Epub 2012/03/27. 10.1038/nbt.2158 ; PubMed Central PMCID: PMCPmc4144683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speir ML, Zweig AS, Rosenbloom KR, Raney BJ, Paten B, Nejad P, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2015. Epub 2015/11/22. 10.1093/nar/gkv1275 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng Y-L, An R, Shin J, Song H, Ming G-l. DNA modifications and neurological disorders. Neurotherapeutics: the journal of the American Society for Experimental NeuroTherapeutics. 2013;10(4):556–67. Medline:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banzhaf-Strathmann J, Claus R, Mucke O, Rentzsch K, van der Zee J, Engelborghs S, et al. Promoter DNA methylation regulates progranulin expression and is altered in FTLD. Acta neuropathologica communications. 2013;1(1):16 Medline:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31(46):16619–36. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng D, Jenner AM, Shui G, Cheong WF, Mitchell TW, Nealon JR, et al. Lipid pathway alterations in Parkinson's disease primary visual cortex. PLoS One. 2011;6(2):e17299 Epub 2011/03/10. 10.1371/journal.pone.0017299 ; PubMed Central PMCID: PMCPmc3046155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halliday GM, McCann H. The progression of pathology in Parkinson's disease. Annals of the New York Academy of Sciences. 2010;1184:188–95. Medline: 10.1111/j.1749-6632.2009.05118.x [DOI] [PubMed] [Google Scholar]
- 28.Braak H, Ghebremedhin E, Rub U, Bratzke H, Del Tredici K. Stages in the development of Parkinson's disease-related pathology. Cell and tissue research. 2004;318(1):121–34. Medline:. [DOI] [PubMed] [Google Scholar]
- 29.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):Research0034 Epub 2002/08/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penna I, Vella S, Gigoni A, Russo C, Cancedda R, Pagano A. Selection of candidate housekeeping genes for normalization in human postmortem brain samples. International journal of molecular sciences. 2011;12(9):5461–70. Epub 2011/10/22. 10.3390/ijms12095461 ; PubMed Central PMCID: PMCPmc3189726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25(4):402–8. Medline:. [DOI] [PubMed] [Google Scholar]
- 32.Caffrey TM, Joachim C, Paracchini S, Esiri MM, Wade-Martins R. Haplotype-specific expression of exon 10 at the human MAPT locus. Human molecular genetics. 2006;15(24):3529–37. Epub 2006/11/07. 10.1093/hmg/ddl429 . [DOI] [PubMed] [Google Scholar]
- 33.Kwok JB, Hallupp M, Loy CT, Chan DK, Woo J, Mellick GD, et al. GSK3B polymorphisms alter transcription and splicing in Parkinson's disease. Ann Neurol. 2005;58(6):829–39. Epub 2005/11/30. 10.1002/ana.20691 . [DOI] [PubMed] [Google Scholar]
- 34.Kwok JB, Teber ET, Loy C, Hallupp M, Nicholson G, Mellick GD, et al. Tau haplotypes regulate transcription and are associated with Parkinson's disease. Ann Neurol. 2004;55(3):329–34. Epub 2004/03/03. 10.1002/ana.10826 . [DOI] [PubMed] [Google Scholar]
- 35.Caillet-Boudin ML, Buee L, Sergeant N, Lefebvre B. Regulation of human MAPT gene expression. Mol Neurodegener. 2015;10:28 Epub 2015/07/15. 10.1186/s13024-015-0025-8 ; PubMed Central PMCID: PMCPmc4499907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C, et al. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Human molecular genetics. 2012;21(18):4094–103. Epub 2012/06/23. 10.1093/hmg/dds238 ; PubMed Central PMCID: PMCPmc3428157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nature reviews Molecular cell biology. 2013;14(3):153–65. Epub 2013/02/07. 10.1038/nrm3525 . [DOI] [PubMed] [Google Scholar]
- 38.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144(1):16–26. Epub 2011/01/11. 10.1016/j.cell.2010.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–9. Epub 2011/10/04. 10.1038/nature10442 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu L, Wang X, Li XL, Towers A, Cao X, Wang P, et al. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Human molecular genetics. 2014;23(6):1563–78. Epub 2013/11/05. 10.1093/hmg/ddt547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–6. Epub 2013/10/11. 10.1038/nature12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, et al. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife. 2014;3:e04530 Epub 2014/11/22. 10.7554/eLife.04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jordan ET, Collins M, Terefe J, Ugozzoli L, Rubio T. Optimizing electroporation conditions in primary and other difficult-to-transfect cells. Journal of biomolecular techniques: JBT. 2008;19(5):328–34. Epub 2009/02/03. [PMC free article] [PubMed] [Google Scholar]
- 44.Klein CJ, Bird T, Ertekin-Taner N, Lincoln S, Hjorth R, Wu Y, et al. DNMT1 mutation hot spot causes varied phenotypes of HSAN1 with dementia and hearing loss. Neurology. 2013;80(9):824–8. Epub 2013/02/01. 10.1212/WNL.0b013e318284076d ; PubMed Central PMCID: PMCPmc3598458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwata A, Nagata K, Hatsuta H, Takuma H, Bundo M, Iwamoto K, et al. Altered CpG methylation in sporadic Alzheimer's disease is associated with APP and MAPT dysregulation. Human molecular genetics. 2014;23(3):648–56. Epub 2013/10/09. 10.1093/hmg/ddt451 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.