Abstract
The CRISPR/Cas9 system has emerged as an important tool in biomedical research for a wide range of applications, with significant potential for genome engineering and gene therapy. In order to achieve conditional control of the CRISPR/Cas9 system, a genetically encoded light-activated Cas9 was engineered through the site-specific installation of a caged lysine amino acid. Several potential lysine residues were identified as viable caging sites that can be modified to optically control Cas9 function, as demonstrated through optical activation and deactivation of both exogenous and endogenous gene function.
Many bacteria and archaea utilize an adaptive immune defense based on a system of clustered regularly interspaced short palindromic repeats (CRISPR) that target invasive nucleic acids through the interaction of CRISPR-associated (Cas) proteins and CRISPR arrays, which are transcribed and processed into short CRISPR RNAs (crRNAs).1 The crRNA guides the Cas proteins to sequence-specific duplex cleavage. Type II CRISPR systems employ an additional RNA, the trans-activating crRNA (tracrRNA), which hybridizes with the crRNA.2 These two RNAs can be combined to allow Cas9 targeting with a single guide RNA (gRNA).3 The Cas9 enzyme has been optimized for site-specific DNA cleavage and nicking followed by non-homologous end-joining (NHEJ) or homology-directed repair (HR), enabling gene editing, gene deletion, and gene mutation4 in human cells5 and animal models.6 The ease of customized gRNA design allows for sequence-specific and highly efficient gene targeting without the need for protein engineering.7 In addition, a catalytically inactive Cas9 has been engineered into a transcriptional activator and repressor, expanding the utility of Cas9 as a gene regulatory tool.8
Regulation of protein function with light provides control over biological processes with unprecedented resolution.9 To date, no optical control of Cas9 activity has been reported. Optically regulating Cas9 function enables precise spatial and temporal control of gene editing. Light-activated proteins can be generated in live mammalian cells with an expanded genetic code through the site-specific incorporation of caged amino acids in response to a recoded amber stop codon, TAG.10 In order to develop a system for optochemical control of CRISPR/Cas9 gene editing (Figure 1A), genetic code expansion was used by adding an engineered pyrrolysyl tRNA (PylT)/tRNA synthetase (PCKRS) pair to the translational machinery of human cells to enable the site-specific incorporation of photocaged lysine (PCK, Figure 1C) into proteins.11 Multiple lysines of interest were identified as potential caging sites for the inhibition of CRISPR/Cas9 function (Supporting Figure 1). K76, K163, K510, and K742 are highly conserved across species and, based on recent crystal structures,12 are in close proximity to the gRNA nucleic acid binding sites and thus may be essential for Cas9-gRNA interaction. K866 undergoes a significant conformational change upon binding of the gRNA, orienting the lysine to become surface exposed, which may be necessary to properly position the target DNA strand for cleavage (Figure 1B). However, the exact role of this residue has not been determined.
Figure 1.

(A) Light-activation of caged Cas9 enables optochemical control of gene editing. The caged Cas9 protein contains a site-specifically incorporated photocaged lysine, rendering it inactive until the caging group is removed through light exposure. This generates wild-type Cas9, which induces sequence-specific DNA cleavage followed by subsequent non-homologous end-joining (NHEJ) or homology-directed repair (HR). (B) K866 (red) conformation before (left) and after (right) gRNA binding to Cas9. Renderings are based on PDB 4CMP and 4OO8. (C) Photocaged lysine (PCK), with the photocleavable caging group shown in red. (D) Western blot of PCK-dependent Cas9 K866TAG expression.
We developed a dual reporter assay (based on pIRG13), which switches from expressing DsRed to expressing EGFP in the presence of functional Cas9 and matching gRNAs, and is not responsive to UV exposure in the absence of Cas9 (Supporting Figure 2). Two gRNAs (Supporting Table 1) were designed to target sequences upstream and downstream of the DsRed-terminator cassette. Upon co-expression of Cas9, these gRNAs direct the excision of DsRed, and the plasmid is repaired to allow EGFP expression. This assay was used in an initial alanine scan of K76, K163, K540, K742, and K866. All Cas9 alanine mutants expressed well in HEK293T cells (Supporting Figure 3A), and four of the Cas9 alanine mutants were still active (Supporting Figure 4). However, K866 was identified as being essential for activity, suggesting it as a potential target for the introduction of PCK. Amber stop codon mutations were then introduced at all five lysines of interest, since the K→PCK mutation may induce an additional level of perturbation compared to a K→A mutation, and Western blots confirmed PCK-dependent expression of the caged Cas9 mutants (Figure 1D and Supporting Figure 3B). The function of the caged Cas9 mutants in the presence and absence of UV exposure (365 nm, 2 min) was tested using the dual reporter assay (Figure 2). The incorporation of PCK at K76, K163, and K866 showed full inhibition of Cas9 activity in the absence of UV exposure, while the K742PCK mutant was still functional, similar to wild-type. Additionally, the K510PCK mutant showed a low level of undesired background activity in the absence of UV exposure. After light-activation, the K163PCK, K510PCK, and K866PCK mutants showed successful light-activation of Cas9 as observed through the expression of EGFP, while the K76PCK mutant was surprisingly not activated. In contrast to wild-type Cas9, all cells expressing light-activated Cas9 mutants still showed DsRed fluorescence, since the caged Cas9 activation occurred 24 h after transfection, while the wild-type Cas9 was immediately active once expressed. Thus, in the case of light-activated Cas9, DsRed protein that has already been expressed persists, with a half-life greater than 4 days.14
Figure 2.

Cas9 PCK mutant activity scanning. HEK293T cells were transfected with the caged Cas9 expression system as well as the dual reporter construct and grown in the presence of PCK (2 mM) for 24 h. The cells were kept in the dark or UV irradiated for 120 s and imaged (10×) after 48 h. Scale bar indicates 200 μm.
We successfully identified two lysine sites that are amenable to optically control of Cas9 function, presenting two different pathways for the light-activation of CRISPR/Cas9 gene editing: (1) K163, which might interact with the gRNA, and (2) K866, which may play a role in positioning the non-target DNA strand (Supporting Figure 1). The Cas9 K866PCK mutant showed minimal background activity before irradiation and high activity after light exposure for 2 min, reaching wild-type Cas9 levels (Figure 3A). A UV irradiation time course was performed with the K866-caged Cas9 (Supporting Figure 5), demonstrating that exposure of >2 min did not further enhance activation. In order to show spatial control of CRISPR/Cas9 gene editing in mammalian cells, the activation of K866-caged Cas9 was performed through patterned UV irradiation of a layer of HEK293T cells (Figure 3B). The mechanism of deactivation by the K163PCK and K866PCK mutations was further analyzed through plasmid cleavage assays with purified Cas9 (Supporting Figure 6), showing no DNA cleavage or nicking activity in the absence of UV irradiation. The absence of any catalytic activity of the caged enzyme suggests that interaction with the gRNA and/or target DNA may be inhibited through introduction of PCK, thereby deactivating Cas9 function. Taken together, these experiments successfully demonstrate spatial and temporal control of gene function using a site-specifically caged Cas9 that is genetically encoded in mammalian cells, allowing for conditional light-activation of CRISPR/Cas9 gene editing.
Figure 3.

(A) Analysis of EGFP expression by imaging cytometry. Error bars represent standard deviations from three replicates. (B) Spatial control of CRISPR/Cas9 gene editing through irradiation of a subset of cells defined by a mask. Imaged at 20× magnification with tile stitching (2×3). Scale bar indicates 200 μm.
We then demonstrated silencing of an endogenous gene through light-activated gene editing using the optically activated Cas9. The transmembrane transferrin receptor CD71 (also known as TfR1), associated with leukemia and lymphoma,15 was selected as a target. gRNAs for the 5′UTR8 and several exons within the coding region of CD71 were designed in order to inhibit protein function via disruption of upstream regulatory elements or disruption of the amino acid sequence. First, we confirmed that CD71 knockdown by wild-type CRISPR/Cas9 could be quantified via both qRT-PCR and phenotype analysis via fluorescent antibody staining of HeLa cells. Indeed, we observed repression of CD71 mRNA with the 5′UTR targeting gRNA (∼70%) while the exon-targeting gRNAs showed no effect on mRNA levels. In addition, reduced levels of CD71 protein (∼75%) were observed for all gRNAs, relative to non-treated cells (Supporting Figure 7). Next, CD71 suppression was performed with the K866-caged Cas9 in the absence and presence of light-activation. Quantification of CD71 mRNA revealed a reduction by approximately 60% only in the case of the gRNA targeting the CD71 5′UTR (Figure 4A). This is not surprising, as the introduction of mutations into the coding region of the CD71 gene should not affect transcription. Light-activation of Cas9 function reduced the presence of CD71 protein on the cell surface by approximately 50% (Figure 4B), demonstrating the ability to optically control the silencing of endogenous genes with our caged CRISPR/Cas9 system. The reduced repression observed for the light-activated K866-caged Cas9 compared to wild-type Cas9 (10–25%) may require further optimization for endogenous gene editing applications, although the frequency of indels has not been determined, limiting quantitative assessment on the genomic level. Thus, our optically activated CRISPR/Cas9 system can be applied to the control of endogenous genes on the genomic level.
Figure 4.
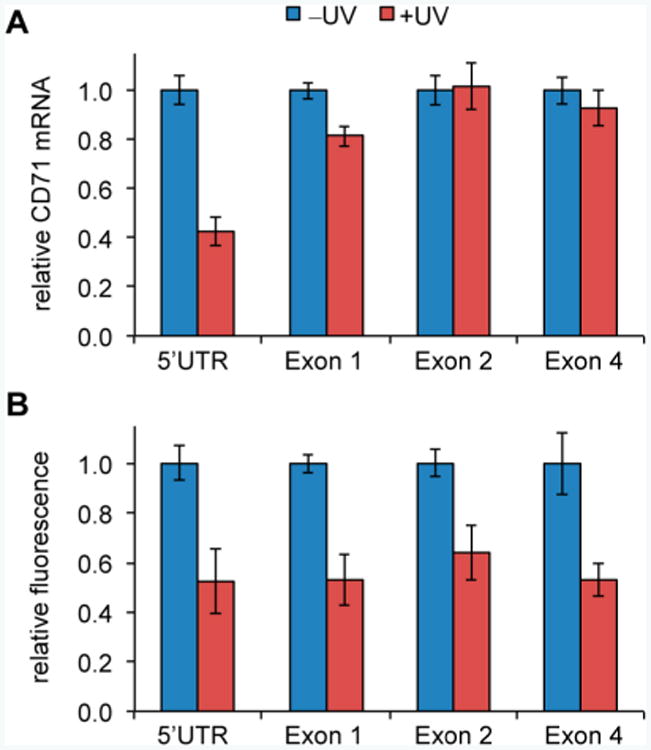
Light-activated CRISPR/Cas9 silencing of CD71 in HeLa cells. (A) Quantitative real-time PCR analysis of CD71 mRNA, normalized to the GAPDH control gene. (B) Fluorescence detection of cell-surface CD71 protein. Data are shown relative to non-irradiated cells for each condition, and error bars represent standard deviations from three replicates.
In summary, we have successfully developed a genetically encoded light-activated CRISPR/Cas9 system for conditional control of gene editing and gene expression. Through both alanine and unnatural amino acid scanning, we identified lysine residues that are important for Cas9 function. Of these sites, K866 was shown to be essential for activity and was utilized in the generation of a light-activated CRISPR/Cas9 system through incorporation of a photocaged lysine at that position. We showed that the genetically encoded caged Cas9 could be applied to gene editing—for activation and deactivation—of both fluorescent reporters and endogenous genes in human cells. Importantly, the caged enzyme was completely inactive before UV illumination, and its activity could be restored up to wild-type levels through a brief 120 s exposure to 365 nm light. In contrast to recently reported light-induced recruitment of a transcriptional activator to a DNA-bound enzymatically inactive dCas9,16 our approach allows for regulation of Cas9 function. Moreover, upon light-induced decaging, wild-type Cas9 is generated. Many cell types and model organisms have been modified by Cas9-mediated gene editing,17 demonstrating the broad applicability of the CRISPR/Cas9 system. Unnatural amino acid mutagenesis based on the pyrrolysine system is expanding into model organisms, such as C. elegans and D. melanogaster,18 providing future opportunities for optical control of CRISPR/Cas9 function beyond cell culture. Light-activation of CRISPR/Cas9 may allow for the study of gene function with high precision, and may reduce toxicity from off-target mutations19 by restricting the function of Cas9 to certain locations or time points.
Supplementary Material
Acknowledgments
This research was supported in part by the NSF (MCB-1330746 to A.D.) and the NIH (R01HL089221 to A.A.). J.H. was supported by an Andrew Mellon Predoctoral Fellowship and E.K.B. was supported by 5T32GM007092.
Footnotes
Supporting Information: Methods and Supporting Figures 1–8 and Supporting Tables 1–5, showing structural analysis of lysines of interest, dual reporter wild-type Cas9 analysis, Western blots, alanine mutation scans, UV irradiation optimization, Cas9 biochemical activity assays, wild-type Cas9 CD71 control experiments, plasmid maps, gRNA sequences, and primer sequence information. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes: The authors declare no competing financial interest.
References
- 1.Wiedenheft B, Sternberg SH, Doudna JA. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Garneau JE, Dupuis M, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]; Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. Genetics. 2013;194(4):1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauβ A, Lahaye T. Mol Plant. 2013;6(5):1384–1387. doi: 10.1093/mp/sst075. [DOI] [PubMed] [Google Scholar]; Niu J, Zhang B, Chen H. Mol Biotechnol. 2014;56(8):681–688. doi: 10.1007/s12033-014-9771-z. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HM, Larson DR, Lawrence DS. ACS Chem Biol. 2009;4(6):409–427. doi: 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]; Riggsbee CW, Deiters A. Trends Biotechnol. 2010;28(9):468–475. doi: 10.1016/j.tibtech.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Baker AS, Deiters A. ACS Chem Biol. 2014;9(7):1398–1407. doi: 10.1021/cb500176x. [DOI] [PubMed] [Google Scholar]; Gautier A, Gauron C, Volovitch M, Bensimon D, Jullien L, Vriz S. Nat Chem Biol. 2014;10(7):533–541. doi: 10.1038/nchembio.1534. [DOI] [PubMed] [Google Scholar]
- 10.Edwards WF, Young DD, Deiters A. ACS Chem Biol. 2009;4(6):441–445. doi: 10.1021/cb900041s. [DOI] [PubMed] [Google Scholar]; Groff D, Chen PR, Peters FB, Schultz PG. ChemBioChem. 2010;11(8):1066–1068. doi: 10.1002/cbic.200900690. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gautier A, Deiters A, Chin JW. J Am Chem Soc. 2011;133(7):2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chou C, Deiters A. Angew Chem, Int Ed. 2011;50(30):6839–6842. doi: 10.1002/anie.201101157. [DOI] [PMC free article] [PubMed] [Google Scholar]; Arbely E, Torres-Kolbus J, Deiters A, Chin JW. J Am Chem Soc. 2012;134(29):11912–11915. doi: 10.1021/ja3046958. [DOI] [PubMed] [Google Scholar]; Hemphill J, Chou C, Chin JW, Deiters A. J Am Chem Soc. 2013;135(36):13433–13439. doi: 10.1021/ja4051026. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kang JY, Kawaguchi D, Coin I, Xiang Z, O'Leary DD, Slesinger PA, Wang L. Neuron. 2013;80(2):358–370. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Uprety R, Luo J, Liu J, Naro Y, Samanta S, Deiters A. ChemBioChem. 2014;15(12):1793–1799. doi: 10.1002/cbic.201400073. [DOI] [PubMed] [Google Scholar]; Luo J, Uprety R, Naro Y, Chou C, Nguyen DP, Chin JW, Deiters A. J Am Chem Soc. 2014;136(44):15551–15558. doi: 10.1021/ja5055862. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ren W, Ji A, Ai HW. J Am Chem Soc. 2015;137(6):2155–2158. doi: 10.1021/ja508597d. [DOI] [PubMed] [Google Scholar]
- 11.Gautier A, Nguyen DP, Lusic H, An W, Deiters A, Chin JW. J Am Chem Soc. 2010;132(12):4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- 12.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Science. 2014;343(6176) doi: 10.1126/science.1247997. No. 1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Gasperi R, Rocher AB, Sosa MA, Wearne SL, Perez GM, Friedrich VL, Hof PR, Elder GA. Genesis. 2008;46(6):308–317. doi: 10.1002/dvg.20400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verkhusha VV, Kuznetsova IM, Stepanenko OV, Zaraisky AG, Shavlovsky MM, Turoverov KK, Uversky VN. Biochemistry. 2003;42(26):7879–7884. doi: 10.1021/bi034555t. [DOI] [PubMed] [Google Scholar]
- 15.Parenti R, Salvatorelli L, Magro G. Int J Endocrinol. 2014;2014 doi: 10.1155/2014/685396. No. 685396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. Chem Biol. 2015;22(2):169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]; Polstein LR, Gersbach CA. Nat Chem Biol. 2015;11(3):198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sander JD, Joung JK. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greiss S, Chin JW. J Am Chem Soc. 2011;133(36):14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. Nat Chem Biol. 2012;8(9):748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]
- 19.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, Bao G. Nucleic Acids Res. 2014;42(11):7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


