Abstract
Hypovascularization in tumors such as liver metastases originating from breast and other organs correlates with poor chemotherapeutic response and higher mortality. Poor prognosis is linked to impaired transport of both low- and high-molecular weight drugs into the lesions and to high washout rate. Nanoparticle albumin-bound-paclitaxel (nAb-PTX) has demonstrated benefits in clinical trials when compared to paclitaxel and docetaxel. However, its therapeutic efficacy for breast cancer liver metastasis is disappointing. As macrophages are the most abundant cells in the liver tumor microenvironment, we design a multistage system employing macrophages to deliver drugs into hypovascularized metastatic lesions, and perform in vitro, in vivo, and in silico evaluation. The system encapsulates nAb-PTX into nanoporous biocompatible and biodegradable multistage vectors (MSV), thus promoting nAb-PTX retention in macrophages. We develop a 3D in-vitro model to simulate clinically observed hypo-perfused tumor lesions surrounded by macrophages. This model enables evaluation of nAb-PTX and MSV-nab PTX efficacy as a function of transport barriers. Addition of macrophages to this system significantly increases MSV-nAb-PTX efficacy, revealing the role of macrophages in drug transport. In the in vivo model, a significant increase in macrophage number, as compared to unaffected liver, is observed in mice, confirming the in vitro findings. Further, a mathematical model linking drug release and retention from macrophages is implemented to project MSV-nAb-PTX efficacy in a clinical setting. Based on macrophage presence detected via liver tumor imaging and biopsy, the proposed experimental/computational approach could enable prediction of MSV-nab PTX performance to treat metastatic cancer in the liver.
1. Introduction
Metastases are associated with poor prognosis and account for most deaths in patients suffering from breast cancer. With more than 234,000 new diagnoses in the United States each year, patients with distant metastasis are characterized by a five-year survival rate of < 22 %. The liver is one of the major organs to which breast cancer disseminates, accounting for approximately half of patients with metastatic disease, with median survival under six months1, 2. Despite recent developments in surgical techniques, radiation, and chemo- and target-specific immune/hormone therapies, liver metastases remain the main cause of mortality for breast cancer patients. Unfortunately, development of more efficacious chemotherapeutic strategies has been hampered by the complexity of the metastatic lesion microenvironment3. The liver has a dense network of capillaries, called sinusoids, that reaches the innermost cells in the organ and efficiently provides oxygen and soluble nutrients. Liver metastases from breast cancer have very distinct structures, forming hypovascularized lesions and receiving nutrients mainly from surrounding vessels. Uniquely, liver metastases preserve the stromal structure of the liver and do not rely on angiogenesis for survival4. This vascularization pattern in which tumor cells primarily use existing vasculature in the surrounding parenchyma is unconventional compared to most solid tumors5. This structure significantly limits diffusive transport and, in particular, availability of oxygen/nutrients/macromolecules to the interior of the lesions; consequently, breast cancer metastases in the liver are characterized by poor permeation of molecules and are clinically observed as hypo-attenuating spots which intravenously injected contrast agents do not permeate6. This unfavourable diffusion pattern is an important factor limiting adequate concentration of therapeutic and could explain why chemotherapy fails to cure unresectable liver lesions. This problem is especially acute with high molecular weight therapeutics, as has been shown for m99Tc microaggregated albumin7.
We hypothesize that to deliver a sufficient concentration of a macromolecule-based drug to liver lesions, innate mechanisms, such as macrophage infiltration, could be leveraged to increase retention and concentration of therapeutics. A major cell population in normal and diseased liver parenchyma is resident liver macrophages that constitute ~10% of all cells in the healthy liver and 80–90% of tissue macrophages in the body8. Studies have shown that liver macrophage density is 2-to-5 times higher in peritumoral regions for both hepatic primary cancers and metastatic lesions9–11. Miyagawa et al. reported doubling in the number of liver macrophages in peritumoral regions as compared to uninvolved tissue (154±49 vs. 74±24 macrophages per field, respectively)11. Previous studies have linked the prognosis of liver metastases to the presence and activation of the macrophages, namely, depletion of liver macrophages leads to larger tumor lesions, while increase in the number of activated macrophages causes tumor burden reduction12. Further reports have shown that tumor-associated-macrophages can be a viable target for accumulating nanotherapeutics13–17. For instance, macrophages from circulating monocytes were able to infiltrate brain metastases through intact blood-brain-barrier and deliver nanoparticles to breast cancer metastasis18.
Previously19, we designed a multistage system in which nanoparticle albumin bound paclitaxel (nAb-PTX) was packaged into solid particles to increase interaction with macrophages. nAb-PTX is clinically employed for therapy of advanced breast tumors20, 21, although its efficacy with liver metastasis is usually lacking and can cause hepatic toxicity22. The mechanism of action of nAb-PTX relies on enhanced transport across the tumor endothelium using albumin transporters23. However, this mechanism cannot yield much benefit with hypovascularized tumors, as is the case with liver metastasis. To enhance nAb-PTX efficacy, we shifted its transport from endothelial cells towards liver macrophages by encapsulating nAb-PTX in a biocompatible and biodegradable multi-stage nanovector (MSV) system24, 25. The newly designed MSV-nAb-PTX was observed to efficiently increase survival in a mouse model of liver metastasis19. Further in vivo studies showed a significantly increased therapeutic response of liver tumors treated with MSV-nAb-PTX compared to nAb-PTX19. In spite of such significant positive results, the in vivo studies did not allow for the drug transport evaluation at the cellular level, required to model the therapeutic response in humans.
Here, we investigate the anti-tumor effects of macromolecule-bound nAb-PTX by associating the formulation with macrophage phagocytosis in the tumor microenvironment. To this end, we employ in vivo, 2D and 3D in vitro tumor models, along with in silico modelling of tumor drug retention, diffusion and cancer cell cytotoxicity. Since nAb-PTX is not natively taken up by macrophages, the drug is packaged into MSV particles. Biodegradation and biocompatibility of MSV have been shown in vitro and in vivo25–29. The role of MSV in this study is to anchor nAb-PTX inside the macrophages located in the peritumoral regions enriched with cells. We hypothesize that by anchoring MSV-nAb-PTX in peritumoral macrophages, the local concentration available for diffusion into hypo-perfused metastatic lesions will increase, overcoming the unfavourable drug transport into the lesions. In the experimental models of breast tumor liver metastasis we evaluate the flow patterns and the kinetics of macromolecule penetration as well as the number of macrophages available on the tumor periphery. As an in vitro model of hypovascularization with under-developed stroma, we use breast cancer cell spheroids to simulate liver metastasis30, 31 in co-culture with macrophages. Based on the data on drug retention and release from macrophages as well as the experimentally measured number of macrophages, we simulate vascularized in silico tumor responses in the clinical setting. The proposed approach may benefit patients with unresectable liver metastasis of breast cancer.
2. Results and discussion
Intravital microscopy (IVM) evaluation of tracer transport in liver metastases
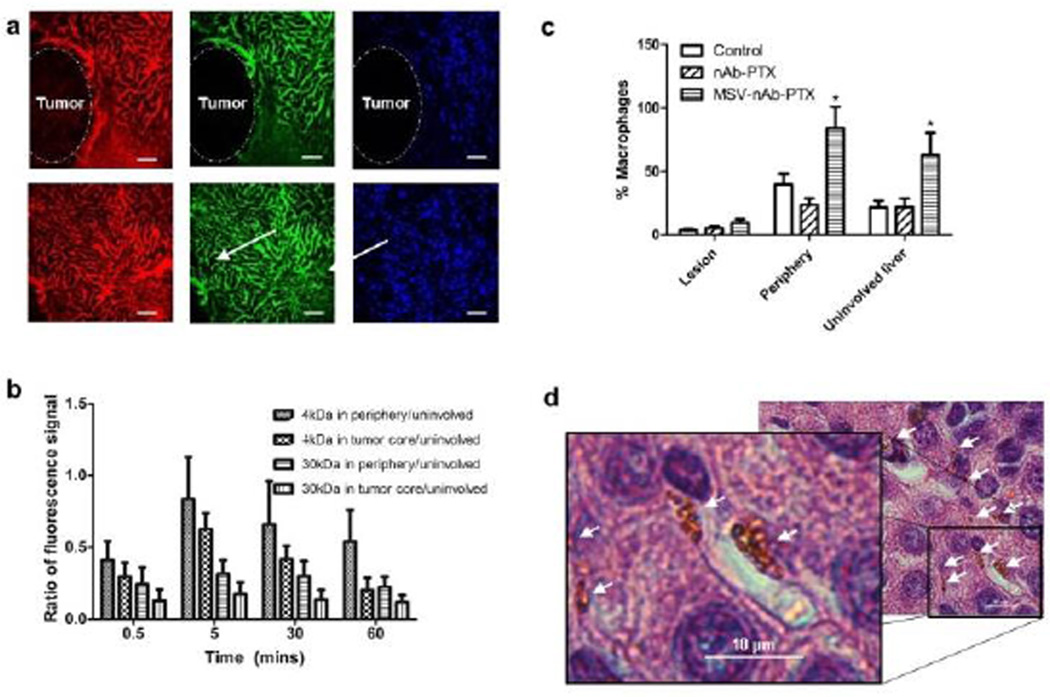
To evaluate the transport patterns in the breast tumor liver metastases, animals injected with tracers were followed by IVM for 60min. Fig 1a shows that while unaffected liver parenchyma appeared as extensively vascularized and permeable for the injected tracers, tumor lesions were impenetrable to 40KDa dextran (hydrodynamic radius ~4.5nm). 3kDa Dextran (<1nm hydrodynamic radius) was detectable in the tumor vasculature but at significantly lower intensity compared to unaffected liver parenchyma. Quantification of the IVM data revealed that in the tumor core (100 µm in depth) only 10–30% of the intensity of liver parenchyma could be detected (Fig 1b). We also observed abundant macrophages surrounding the lesions, but not in the lesions themselves, and which may include both circulating and liver resident macrophages (Kupffer cells). The number of macrophages increased significantly in animals treated with MSV-nAb-PTX (Fig1c and 1d). On average, 9.7±3.2 MSV-nAb-PTX particles per macrophage were found in liver sections in the proximity of the lesions (Fig. 1d). Based on the biodistribution studies19, the number of MSV-nAb-PTX particles expected in the liver is 25%–30% of total injected, which corresponds to 2.5–3×108 particles in the mouse liver (~2g). There are 70–200×106 hepatocytes in gram liver tissue32 and macrophages account for 10–25% of cells in the diseased liver (7–50×106 cells/g, 0.14–1×108 cells/liver). This means that, since hepatocytes do not take up particles, theoretically, each macrophage phagocytoses 2.5–20 particles. This corresponds well with our observations from counting MSV-nAb-PTX particles in mice liver histology slides as exemplified in Fig 1d. Detailed analysis of mice liver has also shown the specific co-localization of MSV with macrophages, confirming that MSV are mostly taken up by macrophages (SI-Fig 1).
Figure 1.
Characterization of breast cancer liver metastasis in mice: a) Intravital microscopy images of tumor lesion (upper panel) and unaffected liver (lower panel) perfused with 3kDA (red), 40kDA (green) dextrans. Scale bar=100µm. b) Quantification of dextran marker penetration within the periphery (up to 100µm depth into the tumor) or the tumor core normalized to the dextran fluorescence signal in uninvolved liver. c) Quantification of macrophages in vivo based on the location within the lesion, on the periphery of the lesion (the area within 50µm border outside the lesion), or in the uninvolved liver. d) Microscope image of particles taken up by macrophages in the liver (arrows). Scale bar=10µm. n=9, mean±SD, *indicates significant difference (p<0.05) as compared to untreated control.
Studies with 2D co-culture of breast cancer cells and macrophages
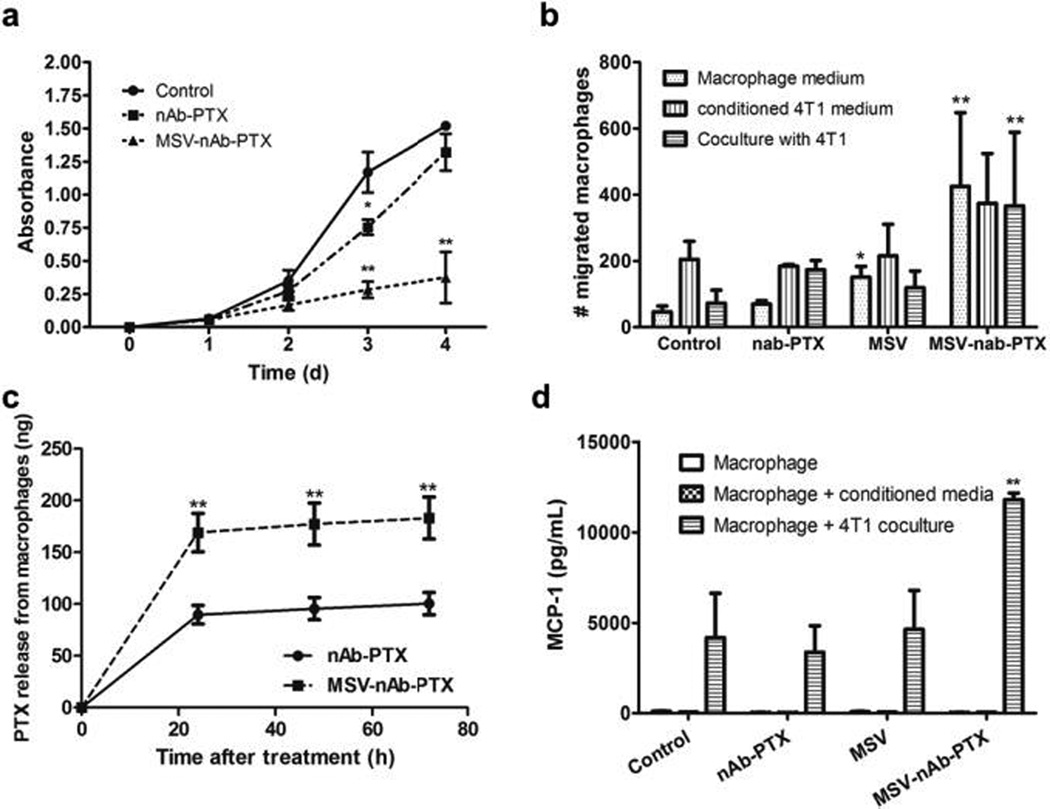
The 2D transwell co-culture model enabled undisrupted evaluation of the direct effect of macrophage pre-treatment with MSV-nAb-PTX. Inhibition of 4T1 breast cancer cell proliferation was more pronounced when macrophages grown on the apical side of the transwell were pre-incubated with MSV-nAb-PTX as compared to nAb-PTX (Fig 2a). Moreover, in co-culture, MSV-nAb-PTX treated macrophages migrated to the basolateral compartment (Fig 2b). To understand the main stimuli for migration, transwell experiments were carried out with macrophages on the apical membrane, and either culture medium from breast cancer cells (static stimulus) or breast cancer cells (dynamic interaction) in basolateral compartment. Fig 2b shows that pre-treatment of macrophages with MSV-nAb-PTX significantly increased their migration capability under all tested conditions. Over 400 macrophages/field of view were found on the basolateral compartment when cells were treated with MSV-nAb-PTX, regardless of the stimuli from basolateral side, while <100 cells per field of view were found for untreated control. A small increase was also observed in untreated macrophages incubated with 4T1 conditioned media.
Figure 2.
Studies with 2D transwell co-culture of macrophages (apical side) and breast cancer cells (basolateral side). a) Inhibition of 4T1 breast cancer cells proliferation in co-culture with macrophages pretreated with MSV-nAb-PTX, nAb-PTX and PBS (no treatment) as evaluated by NIS Elements image analysis. b) Migration of macrophages from the apical to basolateral compartment of the migration chamber when the tumor cells were incubated in the basolateral compartment. Macrophages were pretreated with nAb-PTX, MSV or MSV-nAb-PTX for 4h prior to seeding on the apical compartment. Migration was analysed 2d after co-culture. c) Amount of active compound PTX released from the nAb-PTX and MSV-nAb-PTX pre-treated macrophages for 24–72h as assessed by LC/MS-MS. d) Monocyte Chemoattractant Protein-1(MCP-1) released by the cancer cells in the co-culture study in transwell setup. n=6 for a, n=3 for b-d, mean±SD, *indicates significant difference (p<0.05), **very significant difference (p<0.01) to untreated control, except c: to nab=PTX.
We further tested PTX concentration released by macrophages following 2 hours pre-incubation with MSV-nAb-PTX and nAb-PTX. After 24 hours of incubation, almost twice as much PTX was released from MSV-nAb-PTX treated macrophages compared to nAb-PTX treated macrophages (169 vs. 89ng, respectively) (Fig 2c). Previously, phagocytic range for diameters 0.3–10 µm33 and maximum phagocytosis for 2–3 µm were reported34. This is more advantageous for the (1µm) MSV-nAb-PTX, compared to the 130nm hydrodynamic diameter of nAb-PTX. These results support the hypothesis that the amount of PTX localized in macrophages is increased when formulated as MSV-nAb-PTX compared to nAb-PTX.
The increased therapeutic concentration prompted the cancer cells to release MCP-1, which may explain the increased macrophage migration (Fig 2d). Higher MCP-1 levels attract macrophages towards the cancer cells. Treatment with PTX was reported to induct MCP-1 and its cognate receptor CCR2 in primary sensory neurons of dorsal root ganglia35. The increase of MCP-1 level was also observed in breast cancer cells treated with PTX (Fig 2). Quantification of the fluorescence signal confirmed the increase of macrophage number within liver metastasis lesions (Fig 1c).
Studies with 3D co-culture of breast cancer cells and macrophages
Monolayer two-dimensional setups lack drug and oxygen diffusion gradients found in live tissue, which can affect therapeutic response36. In order to further study the effects of MSV-nAb-PTX, we, therefore, modified a 3D cancer sphere model we recently described30. Although 3D-culture is a simplification of the in vivo condition, it more appropriately represents cell behavior observed in vivo than cells in (2D) monolayer37–39. In particular, tumor spheroids in 3D cell co-culture with macrophages simulate a representative hypovascularized lesion microenvironment. The results showed that cell proliferation in spheres was not inhibited within the first 5 days (SI-Fig 2), and that diameter and area of the spheres increased with time proportionally to the growth rate.
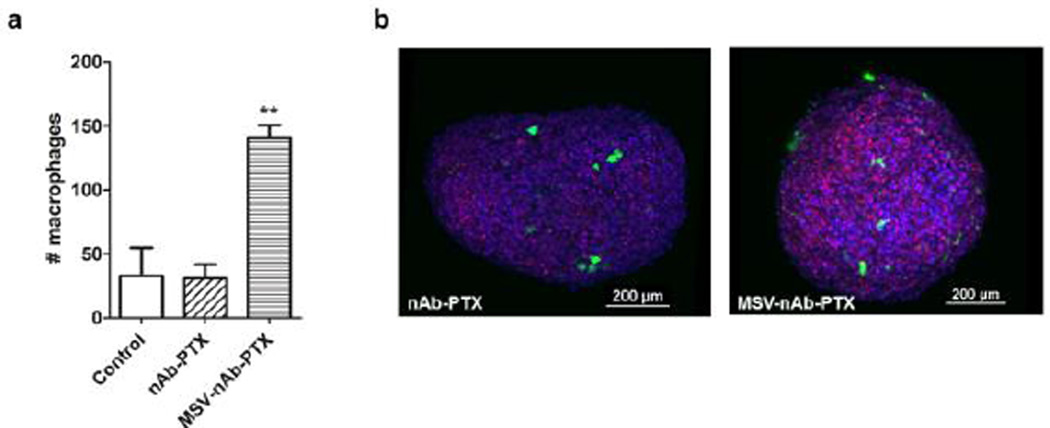
The data from 3D tumor spheres confirmed the results of macrophage migration in the 2D setting. Four to five fold higher number of macrophages pre-treated with MSV-nAb-PTX was found to infiltrate the spheres and, specifically, the core regions (Fig 3). For example, within a sphere with total volume of 0.0342mm3, we found 141 macrophages pre-treated with MSV-nAb-PTX, compared to 31 untreated and 33 nAb-PTX treated macrophages. Additional layer-by–layer gallery of scanned images showing penetration by macrophages can be found in SI-Fig 3.
Figure 3.
Macrophage accumulation and migration into 4T1 tumor spheres: a) Analysis of the total cell numbers associated with the spheroid; b) 3D reconstruction of confocal laser scanning microscopy images of 4T1 spheres (blue) invaded by macrophages (green) and dead cells was imaged by DRAQ7 (red). Macrophages were pre-treated with nAb-PTX or MSV-nAb-PTX.
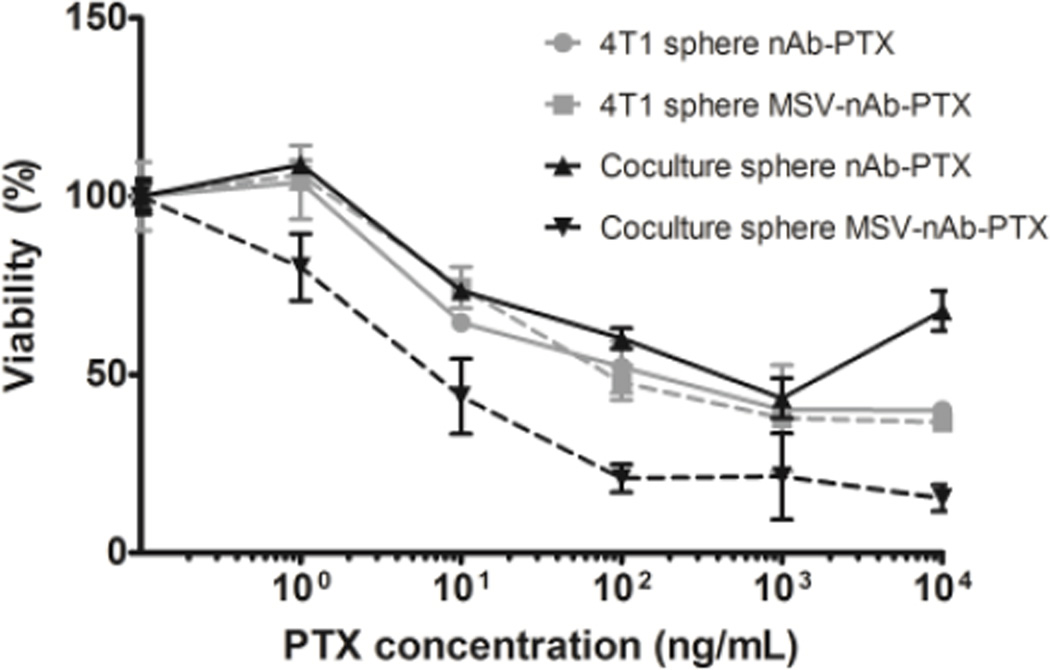
nAb-PTX and MSV-nAb-PTX cytotoxicities were tested using cancer spheres in single culture as well as in co-culture with macrophages with 0–200µg nAb-PTX/mL (Fig 4), exceeding the concentration tested in monolayer. Although the results showed a similar trend of nAb-PTX cytotoxicity in the 3D system compared to 2D, EC50 of nAb-PTX in 2D monolayer was 1 µg/mL, while in 3D spheres it was 30 fold higher (30µg/mL), indicating physical resistance to therapy (Fig 4). The EC50 is the drug concentration required to achieve 50% maximal response. When the drug was instead loaded into macrophages, we observed an increase in nAb-PTX EC50 value compared to drug-only treatment (EC50 of 50 µg/mL). A drastic effect was observed when the MSV-nAb-PTX was loaded into macrophages and co-cultured with the spheres, as the concentration needed to reach EC50 was significantly lower than when treated directly in single culture (Fig 4). In this case, the EC50 value was similar to the mono-culture in monolayer (1 µg/mL), suggesting that macrophages may have a significant role in processing MSV-nAb-PTX, but not nAb-PTX.
Figure 4.
Effects of nAb-PTX and MSV-nAb-PTX on breast cancer cells (4T1) grown in a monoculture in 3D sphere (grey lines) or in co-culture with primary human macrophages (black lines). Solid lines indicate treatment with nAb-PTX and dotted lines with MSV-nAb-PTX. Data were normalized to control untreated cancer cell 3D spheres and cancer cell sphere-macrophage co-culture. n=6, mean±SD.
Simulation of MSV-nAb-PTX vs. nAb-PTX therapeutic efficacy
To explore clinical efficacy, we mathematically modelled the effect of MSV-nAb-PTX on hypo-vascularized liver lesions. The lesion growth was simulated in parallel with the dynamic drug distribution. Given a concentration of 0.0015 ng nAb-PTX per macrophage (which represents 4.167 × 10−5g nAb-PTX/mL, based on our calculation of a minimum 27,778 drug-carrying macrophages/mm3 required to achieve an EC50 of 125ng ABX/ml equivalent to monolayer), we calibrated the tumor regression to match that observed with the 3D cell cultures. This established a baseline value of drug effect. Since the nAb-PTX dose for mice we used was 75mg/kg nAb-PTX (7.5mg/kg PTX)40 one can estimate that for a typical ~23g mouse (1.38mL plasma volume41) the drug concentration in the vasculature reaches 1.25 × 10−4g nAb-PTX/mL, and simulate the corresponding tumor regression via nAb-PTX injection. Using the same nAb-PTX half-life (20 hours42) and assuming similar drug release rates from macrophages and vessels, the tumor regression can be evaluated due to a slower release of drug via macrophages, and compared to bolus injection of nAb-PTX for which a high initial concentration is established followed by rapid washout.
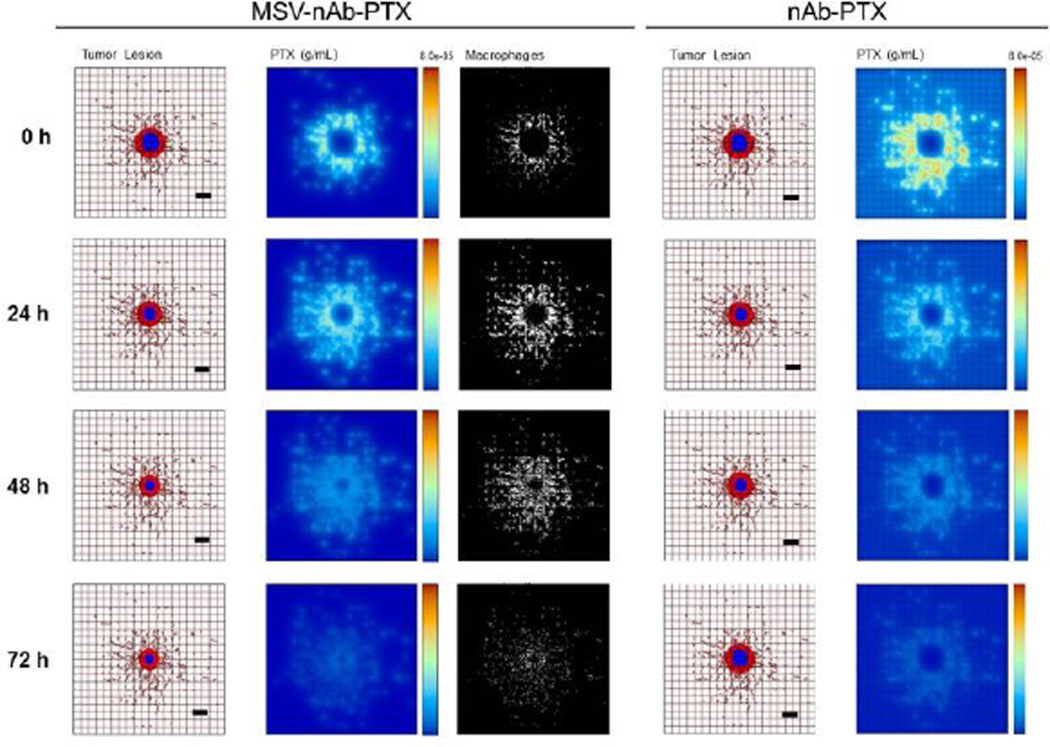
Since the number of cancer cells is a function of tumor lesion size, we estimate that a 1mm3 tumor lesion can contain up to 3×106 cells43, with about 10% of these cells being macrophages. In the simulations, we conservatively assume that the number of macrophages recruited to the lesion is ~25% of that expected in vivo (2.78×104 macrophages/mm3). Fig 5a illustrates the effects of therapy with MSV-nAb-PTX-loaded macrophages compared to bolus nAb-PTX. With MSV-nAb-PTX at 24 hours post treatment initiation, the tumor has slightly shrunk (left panel) compared to the initial lesion (9% radius decrease), while nAb-PTX is being released by macrophages (right), which have extravasated from the surrounding vasculature. Macrophages are recruited to the tumor tissue based on chemoattractants released from hypoxic cells, thus facilitating MSV-nAb-PTX transport in the lesion vicinity. At 72h, the lesion radius has shrunk by 27% of its original size, with most of the drug having been released from the surrounding macrophages.
Figure 5.
Simulation of breast cancer liver metastasis therapy with MSV-nab-PTX (left) and nab-PTX (right): Presentation of tumor lesions, concentration of drug (PTX) released from either macrophages or vessels, and macrophages (only in the case of MSV-nab-PTX). Tumor lesions at the indicated times post treatment initiation. In this simulation, the viable tumor tissue (red) encloses a hypoxic region (blue) without necrosis. The dense capillary network in the liver is modelled by the rectangular grid, with a few irregular sprouts generated through angiogenesis. Size of the lesion is simulated in parallel with drug distribution. PTX represents drug concentration (gm/mL) in the lesion. MSV-nab-PTX are retained in the lesion through the interaction with macrophages while drug is slowly released in the proximity of the tumor cells. Individual macrophages (white) are recruited to vicinity of the lesion based on chemoattraction to hypoxic regions. Bar, 200µm.
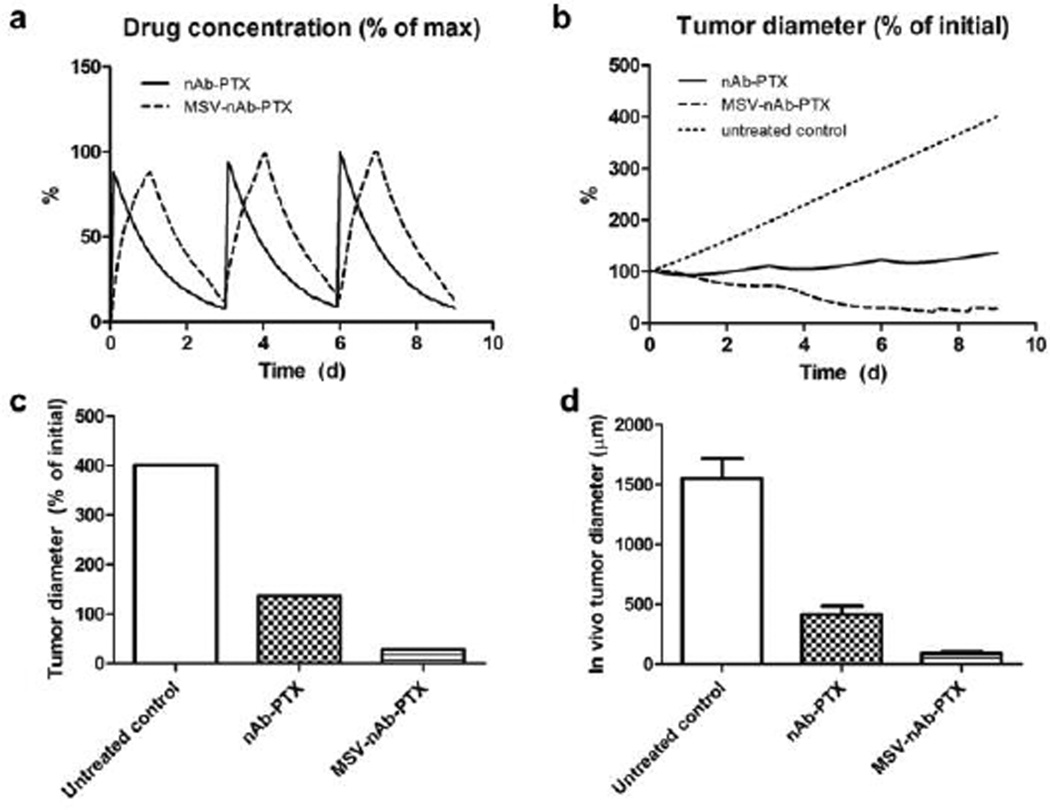
In the case of nAb-PTX bolus injection (Fig 5b), the free drug concentration is initially higher than with macrophages. By 24h, however, most of the drug is washed away, and the lesion resumes growth. The bolus injection is predicted to only achieve a transient 6% radius reduction after a single treatment. We further simulated the response due to repetitive therapy with MSV-nAb-PTX and nAb-PTX. The drugs were administered to the simulated lesions on Days 0, 3, and 6 over the course of 9 days (Fig 6a). The bolus nAb-PTX treatment shows that regression is predicted to be minimal after each treatment, with an overall 37% increase in lesion radius (Fig 6b). In contrast, the MSV-nAb-PTX therapy is projected to achieve a 71% radius decline by treatment completion. The order of magnitude of the endpoints from the in silico results (Fig 6c) are in agreement with a comparable in vivo study (Fig 6d) we have recently reported19. By the end of 9 days, the tumor diameter after in silico treatment with nAb-PTX and MSV-nAb-PTX was 2.9- and 14.1-fold of the untreated control, respectively (Fig 6c), while the in vivo results showed regression of 3.7- and 17.1-fold, respectively (Fig 6d).
Figure 6.
Comparison of simulation results (a–c) to response observed in vivo due to repeated therapy over the course of 9d, showing a) simulated drug (as % of maximum blood levels) and b) simulated tumor effect (as % of initial lesion diameter) after nAb-PTX and MSV-nAb-PTX injection. In all cases, therapy is simulated to begin on Days 0, 3, and 6. c) Simulated tumor diameter after 3 treatments as percentage of initial tumor. d) Comparable results from in vivo tumor after 3 treatments as reported in our recent publication19. The longer-acting and spatially focused drug release with macrophages achieves a more pronounced regression over the course of therapy than with bolus injection.
The simulations assume that about the same number of macrophages delivers drug during each pass. Interestingly, the third pass inflicts minimal shrinkage (7.5% from original size) as the lesion resumes growth, suggesting a diminishing return in therapeutic effect at 3-day intervals due to impaired surrounding vascular access to the shrunken lesion. This is consistent with previous in vivo modelling work44.
3. Experimental
Fabrication, characterization of MSV and loading of nAb-PTX
Discoidal MSV with 1×0.4 µm (dxh) were fabricated in a microelectronics facility via photolithography and electrochemical etching and modified with 3-aminopropyl-triethoxysilane (APTES) as previously described26 (see also Supplementary Information, SI). The particles were lyophilized using Freezone Freeze Dry System (Labconco, Kansas City, MO). nAb-PTX (Abraxane®, Celgene, Summit, NJ) concentrated solution was loaded to MSV particles in aliquots, followed by drying-incubating the particles in a steriflip filtration tube (Millipore Corporation, Billerica, MA) under low pressure45.
Cell culture
4T1 cells (ATCC, Manassas, VA) were cultured in Minimum Essential Medium (MEM) supplemented with 10% FBS, 1% antibiotic-antimycotic mix, 1% GlutaMAX, 1% NEAA, 1% MEM Vitamin and 1% Sodium-Pyruvate in humidified atmosphere at 37°C and 5% CO2. Human macrophages were derived from monocytes isolation with Ficoll gradient centrifugation of buffy coats from healthy donors obtained from blood donation service (IRB (2)1111-0206, Houston Methodist Hospital, Houston, TX). Isolated monocytes were differentiated into macrophages for 7 days with macrophage medium, containing 10% human serum, 1% sodium pyruvate and 1% vitamin in RPMI1640 medium.
In vivo model of breast cancer liver metastasis
Animal studies were performed in accordance with approved protocols by Houston Methodist Research Institute Institutional Animal Care and Use Committee (IACUC). Balb/c mice were purchased from Charles Rivers Laboratories and mouse breast cancer liver metastases xenograft were generated by splenic injection of 105 cells 4T1 tumor cells/100 µL PBS as we previously described19, 40. Splenoctomy was conducted immediately after injection to prevent a primary tumor growth in the spleen and the xenografts were grown for at least 10 days before IVM imaging and therapy.
Intravital microscopy (IVM) analysis
For the evaluation of probe diffusion in the lesion, Alexa fluor 538-labelled 3kDa and Alexa fluor 488-labelled 40kDa dextrans (Molecular Probes, Eugene, Oregon, USA) were injected intravenously into the retro-orbital space of the mice with pre-labelled RBC and macrophages, before IVM recording for visualization of blood flow dynamics as previously described46. Mice were anesthetized with isoflurane and mounted on heated microscope stage during surgery and IVM imaging. To uncover the liver, a midline incision (~1cm) was cut through the abdominal wall, the liver was positioned in the field of view and a cover slip was mounted above the liver with tumor lesions for duration of the experiment. At least 20 tumor lesions were analysed. For analysis of MSV distribution in the liver, macrophages were
In vivo evaluation of macrophage quantity within the microenvironment
For analysis of tumor microenvironment changes in response to therapy in vivo, mice with cancer liver metastasis were randomly divided into 3 groups (n=4): control, nAb-PTX, MSV-nAb-PTX. nAb-PTX and MSV-nAb-PTX containing 75mg/kg nAb-PTX (7.5mg/kg PTX) were injected via tail vein. The treatment was repeated every 3 days and the mice were sacrificed after 3 treatments. Liver was dissected, embedded in OCT compound (Sakura® Finetek USA, Inc., Torrance, CA, USA) and cut in 4µm sections for histological and immunofluorescence analysis. The frozen sections were fixed with ice-cold acetone and stained with Alexa Fluor 488-tagged rat anti mouse F4/80 antibody to detect macrophages and counter stained with DAPI. Particle uptake was quantified in 100 macrophages in the liver section of the MSV-nAb-PTX-treated mice.
Quantification of PTX by LC-MS/MS
To quantify the amount of PTX internalized by macrophages in vitro, human primary macrophages were seeded at the density of 1×106 cells/well in 6-well plates and incubated for 1 or 2 days with culture medium containing one of the following: PBS control; MSV; nAb-PTX or MSV- nAb-PTX (1.5µg/mL PTX). After the incubation, cells were washed thrice with PBS, and either collected and lysed via sonication to assess intracellular levels of PTX, or incubated with a fresh medium for 24h to assess the release of PTX from macrophages. PTX in the cell lysates and media was assessed by LC-MS/MS.
For LC-MS/MS analysis, free PTX (Acros organics, Ceel, Belgium) and D5-PTX (Santa Cruz Biotechnologies, USA) were used as external and internal standard, respectively. PTX was extracted with acetonitrile. LC separation of samples was carried out on a Waters Acquity UPLC system (Waters Corp, Millford MA) using Acquity UPLC 2.1mm × 50mm BEH C-18 column (1.7µm) with guard. MS was carried out on Waters Xevo TQ tandem quadrupole mass spectrometer operated in the SRM/MRM mode under positive ion electrospray conditions. MS and MS/MS conditions were optimized at capillary voltage of 3.5kV, cone voltage 16V, and dwell times of 0.105 sec for each transition. Instrument control and data acquisition was accomplished using MassLynx v4.1 software (Waters Corp.) and sample quantification accomplished by using TargetLynx (v4.1) software (Waters Corp.). Data was quantified relative to the external standard curve and normalized to percent recovery for every sample relative to the internal standard.
Co-culture of breast cancer cells and macrophages in monolayer
4T1 cells were grown on the basolateral compartment of 24-well Transwell® plates for 3 days to obtain ~8,000–10,000 cells/well. Macrophages were pre-treated with nAb-PTX, MSV, and MSV-nAb-PTX for 4 hours before seeded on the apical side of the wells (1,000 cells/well). The co-cultures were analyzed after day 1, 2 and 3. Cell death was analyzed by live/dead assay (Invitrogen, USA) according to the manufacturer’s protocol.
Co-culture of breast cancer spheres and macrophages
Tumor spheres were generated using the Bio-Assembler™ system based on the protocols we recently reported30, 31 (Supplementary information), and grown to ~400µm diameter before cytotoxicity and migration studies. Macrophages were treated with nAb-PTX, MSV or MSV-nAb-PTX for 4h and stained with Vybrant Cell-Labeling Solutions (Molecular Probes, Eugene, Oregon, USA). 1000 of human primary macrophage cells were cultured together with 4T1 spheres in a 96- well plate and kept in incubator. Images were taken by fluorescent microscopy after day 1, 2 and 3, and analyzed with NIS-Elements software.
MTT assay
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide –MTT (Sigma, USA) assay was performed to access cell viability. Either 4T1 in monolayer, 4T1 spheres, or 4T1 spheres in coculture with macrophages were seeded on 96-well plates before treatment. Cells were treated with nAb-PTX and MSV-nAb-PTX in the concentration range from 1 ng to 100µg. After 48 h incubation, the cells were washed twice with PBS and incubated with 0.5 mg/ml MTT reagent for 2 hours. After incubation, cells were washed twice with PBS, dissolved in DMSO, and placed into plate shaker for 30 minutes. The absorbance was determined using a scanning spectrophotometer (Biotek, Minooski, VT) at 570 nm.
Monocyte chemoattractant protein-1 (MCP-1) analysis
MCP-1 chemokine analysis was conducted using Milliplex®MAP Human Cytokine/Chemokine Magnetic Bead Panel Immunoassay (EMD Millipore Corporation, Billerica, MA, USA) according to manufacturer’s protocol and measured by Magpix® System.
Mathematical model of tumor response as a function of drug release from MSV-nAb-PTX loaded macrophages
The in silico model46–49 represents viable and necrotic tumor tissue with structure similar to liver metastasis, and with transport of molecules and macrophages through this tissue. Tumor growth is calculated based on the balance of cell proliferation and death. Proliferation depends on adequate cell nutrients and oxygen. Death is induced by levels of oxygen below a certain threshold as well as drug above a certain level of cytotoxicity. Model parameters values are calibrated to experimental data as in46–49. We simulate release of paclitaxel from nAb-PTX carried by MSV-nAb-PTX loaded macrophages infiltrating the tumor tissue. From the experimental data, we know that there are at least 10 MSV per macrophage, and that there is approximately 1.5mg of nAb-PTX per 109 MSV. Since 10% of nAb-PTX consists of Paclitaxel, this implies that the drug typically retained by single macrophages is 0.0015ng PTX. Based on nAb-PTX EC50 of 125ng/mL measured in vitro for 48h with 4T1 cells in monolayer (without diffusion effects), and the observation that 3D tissue (with diffusion gradients) has similar EC50 in the presence of macrophages as the cells in monolayer, we simulate the tumor response at these drug concentrations by varying the numbers of macrophages releasing drug in the tumor tissue. The model and associated parameters are further described in SI.
Statistical analysis
All quantitative parameters are presented as mean values with standard deviation. Statistical analysis was performed by t-test for unpaired samples using Graphpad Prism software, with p-value<0.05 accepted as indicative of significant difference, <0.01 as statistically very significant difference.
4. Conclusions
In the multistage nanovector system described herein, we repurposed and significantly increased the efficacy of nAb-PTX by changing its physical characteristics from a protein aggregate to a solid vector. The system was designed to accumulate in the liver and then be transported to the metastatic lesions. The mathematical modelling enabled assessment of macrophage and MSV-nAb-PTX concentration needed in the lesion microenvironment to achieve inhibition. The results support the notion that MSV association with macrophages can increase drug efficacy compared to bolus injection, and that it is feasible to reach sustained lesion regression with the macromolecule-bound formulation. In the calculation based on the in vitro data, ~280,000 MSV-nAb-PTX particles are needed to achieve 50% cancer cell death per mm3 lesion. Since 1–3×105 macrophages are expected in the lesions (see Results), the numbers show that it is feasible to obtain sufficient EC50 concentration in the target lesions using MSV-nAb-PTX, with macrophages needing to uptake at least three drug-containing particles, which underestimates the in vivo situation. The MSV-nAb-PTX-containing macrophages were also observed to infiltrate the lesions, as shown in vitro and verified in vivo. As a result, a significantly higher number of macrophages were found within lesions in mice treated with MSV-nAb-PTX compared to mice treated with nAb-PTX. The macrophage population in the liver thus acts as a “depot” of drug, ablating the tumor more efficiently than bolus injection. Based on our integrated experimental/computational study, we conclude that the performance of MSV-nAb-PTX is strongly related to macrophage function, and that inadequate drug concentrations to achieve sustained tumor regression associated with conventional drugs may be addressed by leveraging the accumulating effect of MSV-nAb-PTX-carrying macrophages in breast cancer metastases to the liver. Simulation of repeat treatment strategy suggests, as would be expected, that the timing interval is crucial, and that the effect depends on the size and vascularization stage of the lesion. These parameters will be explored in future studies for drug dosage and interval optimization.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge funding from the NCI 1-U54-CA143837 and NIH 1-U54CA151668-01. JL acknowledges the National Institutes for Health through grant P50-GM76516 for a Center of Excellence in Systems Biology at the University of California, Irvine, and P30-CA062203 for the Chao Comprehensive Cancer Center at the University of California, Irvine. JL also acknowledges partial support from the National Science Foundation, Division of Mathematics. HF acknowledges funding from the NCI U54-CA-143907. M.F. and K.Y. would like to acknowledge Ernest Cockrell Jr. Presidential Distinguished Chair.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here].
Notes and references
- 1.Selzner M, Morse MA, Vredenburgh JJ, Meyers WC, Clavien PA. Surgery. 2000;127:383–389. doi: 10.1067/msy.2000.103883. [DOI] [PubMed] [Google Scholar]
- 2.Wyld L, Gutteridge E, Pinder SE, James JJ, Chan SY, Cheung KL, Robertson JFR, Evans AJ. Br. J. Cancer. 2003;89:284–290. doi: 10.1038/sj.bjc.6601038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Eynden GG, Majeed AW, Illemann M, Vermeulen PB, Bird NC, Høyer-Hansen G, Eefsen RL, Reynolds AR, Brodt P. Cancer Res. 2013;73:2031–2043. doi: 10.1158/0008-5472.CAN-12-3931. [DOI] [PubMed] [Google Scholar]
- 4.Stessels F, van den Eynden G, van der Auwera I, Salgado R, van den Heuvel E, Harris AL, Jackson DG, Colpaert CG, van Marck EA, Dirix LY, Vermeulen PB. Br. J. Cancer. 2004;90:1429–1436. doi: 10.1038/sj.bjc.6601727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pezzella F, Pastorino U, Tagliabue E, Andreola S, Sozzi G, Gasparini G, Menard S, Gatter KC, Harris AL, Fox S, Buyse M, Pilotti S, Pierotti M, Rilke F. Am. J. Pathol. 1997;151:1417–1423. [PMC free article] [PubMed] [Google Scholar]
- 6.Liu LX, Zhang WH, Jiang HC. World J Gastroentero. 2003;9:193–200. doi: 10.3748/wjg.v9.i2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daly JM, Butler J, Kemeny N, Yeh SD, Ridge JA, Botet J, Bading JR, DeCosse JJ, Benua RS. Ann. Surg. 1985;202:384–393. doi: 10.1097/00000658-198509000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilzer M, Roggel F, Gerbes AL. Liver Int. 2006;26:1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 9.Ding T, Xu J, Wang F, Shi M, Zhang Y, Li SP, Zheng L. Hum. Pathol. 2009;40:381–389. doi: 10.1016/j.humpath.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Avadanei ER, Wierzbicki PM, Giusca SE, Grigoras A, Amalinei C, Caruntu ID. Folia Histochem. Cytobiol. 2014;52:112–123. doi: 10.5603/FHC.2014.0014. [DOI] [PubMed] [Google Scholar]
- 11.Miyagawa S, Miwa S, Soeda J, Kobayashi A, Kawasaki S. Clin. Exp. Metastasis. 2002;19:119–125. doi: 10.1023/a:1014571013978. [DOI] [PubMed] [Google Scholar]
- 12.Pearson HJ, Anderson J, Chamberlain J, Bell PRF. Cancer Immunol., Immunother. 1986;23:214–216. doi: 10.1007/BF00205652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi J, Kim HY, Ju EJ, Jung J, Park J, Chung HK, Lee JS, Lee JS, Park HJ, Song SY, Jeong SY, Choi EK. Biomaterials. 2012;33:4195–4203. doi: 10.1016/j.biomaterials.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Satoh T, Saika T, Ebara S, Kusaka N, Timme TL, Yang G, Wang J, Mouraviev V, Cao G, Fattah EMA, Thompson TC. Cancer Res. 2003;63:7853–7860. [PubMed] [Google Scholar]
- 15.Lei H, Ju DW, Yu Y, Tao Q, Chen G, Gu S, Hamada H, Cao X. Gene Ther. 2000;7:707–713. doi: 10.1038/sj.gt.3301162. [DOI] [PubMed] [Google Scholar]
- 16.De Palma M, Mazzieri R, Politi LS, Pucci F, Zonari E, Sitia G, Mazzoleni S, Moi D, Venneri MA, Indraccolo S, Falini A, Guidotti LG, Galli R, Naldini L. Cancer Cell. 2008;14:299–311. doi: 10.1016/j.ccr.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Fidler IJ, Barnes Z, Fogler WE, Kirsh R, Bugelski P, Poste G. Cancer Res. 1982;42:496–501. [PubMed] [Google Scholar]
- 18.Choi MR, Bardhan R, Stanton-Maxey KJ, Badve S, Nakshatri H, Stantz KM, Cao N, Halas NJ, Clare SE. Cancer Nanotechnol. 2012;3:47–54. doi: 10.1007/s12645-012-0029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanei T, Leonard F, Liu X, Alexander JF, Saito Y, Ferrari M, Godin B, Yokoi K. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-15-1576. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nugent LJ, Jain RK. Cancer Res. 1984;44:238–244. [PubMed] [Google Scholar]
- 21.Ibrahim NK, Samuels B, Page R, Doval D, Patel KM, Rao SC, Nair MK, Bhar P, Desai N, Hortobagyi GN. J. Clin. Oncol. 2005;23:6019–6026. doi: 10.1200/JCO.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Villano JL, Mehta D, Radhakrishnan L. Invest. New Drugs. 2006;24:455–456. doi: 10.1007/s10637-006-6214-0. [DOI] [PubMed] [Google Scholar]
- 23.Nyman DW, Campbell KJ, Hersh E, Long K, Richardson K, Trieu V, Desai N, Hawkins MJ, Von Hoff DD. J. Clin. Oncol. 2005;23:7785–7793. doi: 10.1200/JCO.2004.00.6148. [DOI] [PubMed] [Google Scholar]
- 24.Tasciotti E, Liu X, Bhavane R, Plant K, Leonard AD, Price BK, Cheng MMC, Decuzzi P, Tour JM, Robertson F, Ferrari M. Nat. Nanotechnol. 2008;3:151–157. doi: 10.1038/nnano.2008.34. [DOI] [PubMed] [Google Scholar]
- 25.Godin B, Tasciotti E, Liu X, Serda RE, Ferrari M. Acc. Chem. Res. 2011;44:979–989. doi: 10.1021/ar200077p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Godin B, Chiappini C, Srinivasan S, Alexander JF, Yokoi K, Ferrari M, Decuzzi P, Liu X. Adv. Funct. Mater. 2012;22:4225–4235. doi: 10.1002/adfm.201200869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godin B, Gu J, Serda RE, Bhavane R, Tasciotti E, Chiappini C, Liu X, Tanaka T, Decuzzi P, Ferrari M. J. Biomed. Mater. Res. A. 2010;94:1236–1243. doi: 10.1002/jbm.a.32807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka T, Godin B, Bhavane R, Nieves-Alicea R, Gu J, Liu X, Chiappini C, Fakhoury JR, Amra S, Ewing A, Li Q, Fidler IJ, Ferrari M. Int. J. Pharm. 2010;402:190–197. doi: 10.1016/j.ijpharm.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokoi K, Godin B, Oborn CJ, Alexander JF, Liu X, Fidler IJ, Ferrari M. Cancer Lett. 2013;334:319–327. doi: 10.1016/j.canlet.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaganathan H, Gage J, Leonard F, Srinivasan S, Souza GR, Dave B, Godin B. Sci. Rep. 2014;4:6468. doi: 10.1038/srep06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonard F, Godin B. In: Breast Cancer: Methods and Protocols. Cao J, editor. Springer; 2015. [Google Scholar]
- 32.Wilson ZE, Rostami-Hodjegan A, Burn JL, Tooley A, Boyle J, Ellis SW, Tucker GT. Br. J. Clin. Pharmacol. 2003;56:433–440. doi: 10.1046/j.1365-2125.2003.01881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green TR, Fisher J, Stone M, Wroblewski BM, Ingham E. Biomaterials. 1998;19:2297–2302. doi: 10.1016/s0142-9612(98)00140-9. [DOI] [PubMed] [Google Scholar]
- 34.Champion JA, Walker A, Mitragotri S. Pharm. Res. 2008;25:1815–1821. doi: 10.1007/s11095-008-9562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Boyette-Davis JA, Kosturakis AK, Li Y, Yoon SY, Walters ET, Dougherty PM. J Pain. 2013;14:1031–1044. doi: 10.1016/j.jpain.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Lilly D, Doty C, Podsiadlo P, Kotov N. Small. 2009;5:1213–1221. doi: 10.1002/smll.200801788. [DOI] [PubMed] [Google Scholar]
- 37.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Nat. Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 38.Yamada KM, Cukierman E. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Pickl M, Ries CH. Oncogene. 2009;28:461–468. doi: 10.1038/onc.2008.394. [DOI] [PubMed] [Google Scholar]
- 40.Desai N, Trieu V, Yao Z, Louie L, Ci S, Yang A, Tao C, De T, Beals B, Dykes D, Noker P, Yao R, Labao E, Hawkins M, Soon-Shiong P. Clin. Cancer. Res. 2006;12:1317–1324. doi: 10.1158/1078-0432.CCR-05-1634. [DOI] [PubMed] [Google Scholar]
- 41.Riches AC, Sharp JG, Thomas DB, Smith SV. J. Physiol. (Lond) 1973;228:279–284. doi: 10.1113/jphysiol.1973.sp010086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sparreboom A, Scripture CD, Trieu V, Williams PJ, De T, Yang A, Beals B, Figg WD, Hawkins M, Desai N. Clin. Cancer. Res. 2005;11:4136–4143. doi: 10.1158/1078-0432.CCR-04-2291. [DOI] [PubMed] [Google Scholar]
- 43.Spinney L. Nature. 2006;442:736–738. doi: 10.1038/442736a. [DOI] [PubMed] [Google Scholar]
- 44.Owen MR, Stamper IJ, Muthana M, Richardson GW, Dobson J, Lewis CE, Byrne HM. Cancer Res. 2011;71:2826–2837. doi: 10.1158/0008-5472.CAN-10-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leonard F, Margulis-Goshen K, Liu X, Srinivasan S, Magdassi S, Godin B. Mesoporous biomaterials. 2014;1 doi: 10.2478/mesbi-2014-0002. 10.2478/mesbi-2014-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M, Frieboes HB, Chaplain MAJ, McDougall SR, Cristini V, Lowengrub JS. J. Theor. Biol. 2014;355:194–207. doi: 10.1016/j.jtbi.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu M, Frieboes HB, McDougall SR, Chaplain MAJ, Cristini V, Lowengrub J. J. Theor. Biol. 2013;320:131–151. doi: 10.1016/j.jtbi.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Ven AL, Wu M, Lowengrub J, McDougall SR, Chaplain MAJ, Cristini V, Ferrari M, Frieboes HB. AIP Advances. 2012;2 doi: 10.1063/1.3699060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macklin P, McDougall S, Anderson ARA, Chaplain MAJ, Cristini V, Lowengrub J. J. Math. Biol. 2009;58:765–798. doi: 10.1007/s00285-008-0216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.