Abstract
Catalytic enzyme-linked click-chemistry assays (cat-ELCCA) are an emerging class of biochemical assay. Herein we report on expanding the toolkit of cat-ELCCA to include the kinetically superior inverse-electron demand Diels-Alder (IEDDA) reaction. The result is a technology with improved sensitivity and reproducibility, enabling automated high-throughput screening.
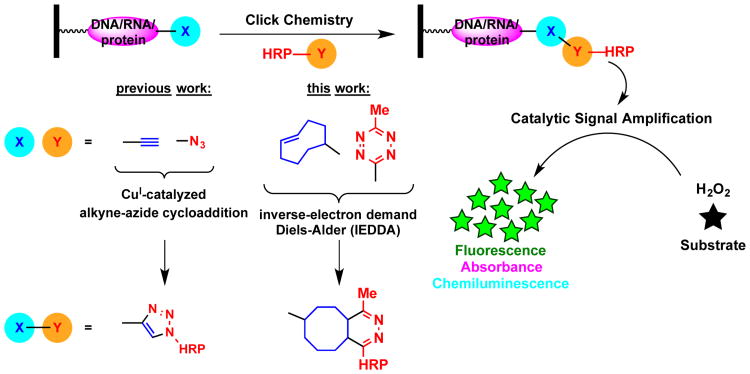
Click chemistry is establishing itself as a critical tool in the development and application of both in vitro and in vivo assays.1-3 Recently, we established a new assay technology, termed catalytic Enzyme-Linked Click Chemistry Assay (cat-ELCCA), as a novel approach for generating click chemistry-based assays armed with catalytic signal amplification.3-5 In cat-ELCCA (Fig. 1), a biotinylated biomolecule is first immobilized in the wells of a streptavidin-coated microtiter plate. This substrate can already contain a click chemistry handle or one can be added via an enzymatic reaction or biomolecular interaction. Detection then occurs via an initial click reaction with a labeled horseradish peroxidase (HRP), followed by addition of a pro-absorbant, -fluorescent or -chemiluminescent HRP substrate. Importantly, this catalytic system, similar to enzyme-linked immunosorbent assays (ELISA), allows for increased sensitivity and a significantly reduced risk of compound interference from screening diverse libraries. Unlike ELISA, however, it eliminates the requirement for enzyme-linked secondary antibodies. To date, we have demonstrated this concept for a post-translation modification3, 4 and microRNA (miRNA) detection.5 Additionally, the first small molecule antagonist for ghrelin O-acyltransferase (GOAT) was identified through a cat-ELCCA-based screen of >4,000 compounds.4
Fig. 1.
cat-ELCCA scheme with modular click chemistry detection.
Over the past 15 years, there has been an explosion in the discovery and application of bioorthogonal reactions.6 While copper-catalyzed azide-alkyne cycloadditions (CuAAC) remain the most common, and the first generations of cat-ELCCA utilized this reaction, some systems may be sensitive to copper or the researcher may have a desire to use other chemistry. Recently, inverse-electron demand Diels-Alder (IEDDA) chemistry has seen increased usage due to its enhanced kinetics and lack of requirement for a catalyst.7 In order to enable ready adaptation of cat-ELCCA to generate reliable, high-throughput screening (HTS)-compatible assays for any biological target, herein we describe our optimization of the click chemistry-based detection step. Using our miRNA maturation assay as a model (Fig. 2), we demonstrate that cat-ELCCA can be employed utilizing IEDDA chemistry to yield an assay with superior reproducibility and high-throughput potential.
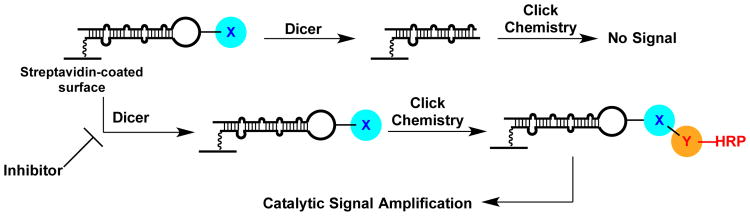
Fig. 2.
cat-ELCCA for Dicer-mediated pre-miRNA maturation.
microRNAs (miRNA) are important post-transcriptional regulators estimated to regulate >60% of mRNA transcripts.8 These small, ∼22 nucleotide (nt) long RNAs are able to use their sequence specificity to bind to the 3′ untranslated region (UTR) of complementary transcripts and cause translational suppression.9 Due to their critical role in gene regulation, dysregulated miRNAs have been implicated in many diseases, including cancer, diabetes, and heart failure.9 To date, targeting miRNAs, and RNA generally, with small molecules has remained a challenging goal.10, 11 Using cat-ELCCA, our goal is to discover new RNA-binding scaffolds that can be used to inhibit pre-miRNA maturation, which is catalyzed by the RNase III enzyme Dicer that digests the precursor into a mature miRNA (Fig. 2),9 in a pre-miRNA-specific manner.
Our first step towards transitioning to IEDDA-based cat-ELCCA was to determine the in-solution reactivity of our cat-ELCCA substrates under typical assay conditions (Fig. 3a). We tested conjugation of the highly reactive, albeit larger than our original alkyne substituion,5 trans-cyclooctene (TCO) with mTet,12 and our pre-miR-21 substrate (see the ESI for sequence) was labeled and reacted with either a modified rhodamine or HRP (Fig. 3a). As shown in Fig. 3b (lanes 1–6), successful coupling was observed with both reactants. Importantly, when the pre-miRNA was reacted with its paired HRP (lanes 3,6), a new band appeared indicating formation of the RNA-protein conjugate. This is in stark contrast to the results observed with CuAAC (lanes 7–9), where a weak band was observed only upon overexposure of the gel (Fig. 3b, bottom). Although the rhodamine substrates reacted equally well with both chemistries (qualitative due to overlap of the bands representing pre-miR-21 and pre-miR-21-Rhod), coupling of RNA and protein was significantly enhanced with the IEDDA reaction (reaction efficiencies of 20% and 3% for IEDDA and CuAAC, respectively; see Fig. S1 of the ESI). We attribute this difference to the ≥1,000-fold rate increase reported with IEDDA over CuAAC and the absence of exogenous catalyst,6,13 particularly since this system is untemplated.14-17 This type of proximity effect is more likely to strongly affect CuAAC due to its mechanism, which relies on formation of a metallacycle involving a ligated CuI (one or two molecules), the azide and alkyne.18, 19 Because of the presence of competing copper ligands within biomolecules, this may explain why the rhodamine coupling appears to be equivalent with CuAAC and IEDDA unlike the RNA-HRP reaction. Finally, to verify that these modifications do not interfere with Dicer-mediated pre-miRNA maturation, the TCO- and mTet-labeled pre-miRNAs were subjected to Dicer cleavage. Although more sterically bulky than the alkyne substitution used previously,5 these modifications did not affect Dicer's ability to cleave the substrates (Fig. 3c).
Fig. 3.
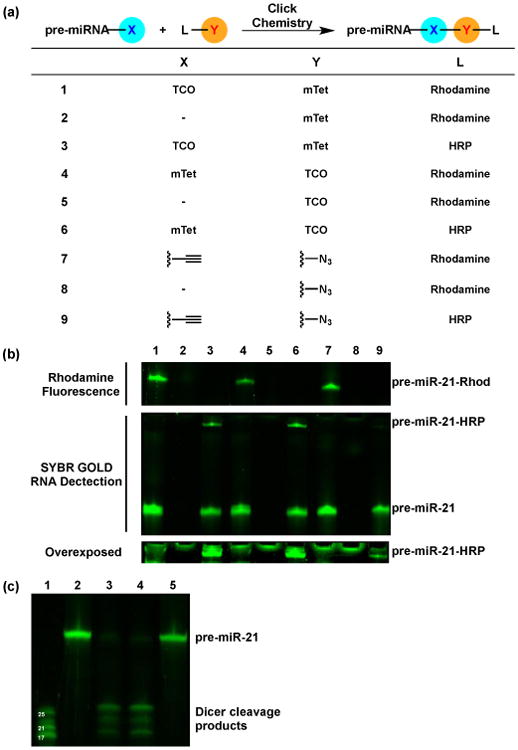
Comparison of in-solution click reactions and Dicer cleavage. (a) General scheme and table of click reactions. - = no pre-miRNA. (b) pre-miRNA IEDDA products detected via fluorescence gel. Click reactions were performed with 2.0 equiv. of L-Y for 2 h at 25 °C. Top = Detection of rhodamine–pre-miRNA conjugate. Rhodamine fluorescence was first detected, followed by addition of SYBR GOLD for 5 min and RNA detection on the same gel. Middle = Detection of HRP–pre-miRNA conjugate using SYBR GOLD. Bottom = Overexposure and zoom of the band representing the HRP–pre-miRNA conjugate. (c) pre-miRNA maturation catalyzed by Dicer. Lane 1 = miRNA ladder; Lane 2 = pre-miRNA-TCO; Lane 3 = pre-miRNA-TCO + Dicer; Lane 4 = pre-miRNA-mTet + Dicer; Lane 5 = pre-miRNA-mTet.
Following confirmation of the individual steps of the assay, the IEDDA-enabled pre-miRNA substrates were immobilized in the wells of a 384-well streptavidin-coated plate. After immobilization, the pre-miRNAs were subjected to Dicer cleavage, which removes the chemical handle from the immobilized RNA (Fig. 2). The pre-miRNAs were then reacted with the corresponding HRP conjugate. Subsequently, the wells were washed, treated with SuperSignal West Pico as a HRP substrate, and the resulting chemiluminescence signal was measured. Importantly, regardless of which label was on the RNA, there was a clear difference in signal intensity between no enzyme and Dicer-treated wells (7.3- and 3.9-fold difference for the TCO- and mTet-labeled pre-miRNAs, respectively) (Fig. 4a). Interestingly, the signal intensity that we observed with the IEDDA-based assay was much greater than that observed with the CuAAC variant (25-fold enhancement; Fig. 4b).5 This is likely due to the increased conversion of the IEDDA reaction, as observed by fluorescence gel (Fig. 3b), and demonstrates increased sensitivity with our optimized click detection step.
Fig. 4.
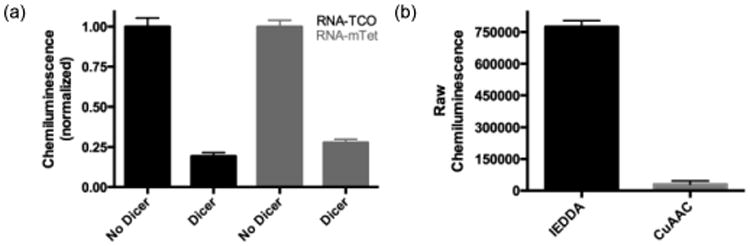
IEDDA-based cat-ELCCA. (a) Comparison of TCO- and mTet-labeled pre-miRNAs. (b) Comparison to CuAAC variant with respect to raw chemiluminescence signal intensity in the absence of Dicer.
Since the pre-miRNA-TCO substrate yielded optimal signal-to-background (S/B), we then further characterizated this second generation cat-ELCCA for Dicer-mediated pre-miRNA maturation. As Fig. 5a–b shows, the assay is dependent on the concentrations of Dicer and immobilized pre-miRNA. Moreover, it is general for other pre-miRNA sequences (Fig. 5c). Finally, to examine how the assay responds to the presence of a known RNA-binding molecule, we tested DAPI (4′,6-diamidino-2-phenylindole; Fig. 5d,e), which acts as an intercalator of RNA.20 High concentrations of DAPI (100 μM) were able to inhibit Dicer activity with statistical significance (p-value <0.0001). Although this is likely through non-specific interactions, it demonstrates that the assay can detect inhibition by small molecules.
Fig. 5.

Characterization of IEDDA-based cat-ELCCA. Dependence on: (a) Dicer; (b) Immobilized pre-miRNA; (c) pre-miRNA substrate; (d) DAPI binding. Significance was determined using a two-tailed, unpaired t-test. p-values of 0.6 and <0.0001 were calculated for 10 μM and 100 μM DAPI, respectively. (e) DAPI structure.
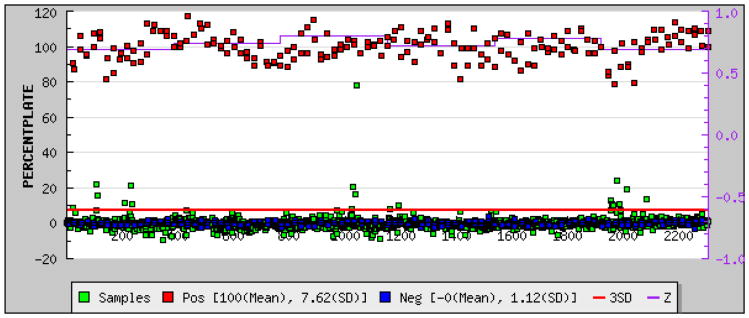
To assess the HTS potential of IEDDA-based cat-ELCCA, we measured the Z′ factor, which is descriptive of dynamic range and data variation associated with signal measurement (see Fig. S2 of the ESI),21 with automated liquid handling. Importantly, we found an average Z′ factor of 0.69, which demonstrates its potential as an excellent assay (Z′ ≥ 0.5) (Fig. 6). This was coupled with excellent S/B and S/N values of 11.5 and >10,000, respectively. As our CuAAC-based cat-ELCCA yielded a Z′ factor of 0.6,5 this represents a significant enhancement with respect to reproducibility in high-throughput over our first generation assay. We then followed this up with a pilot HTS campaign of the commercially available LOPAC library. As shown in Fig. 6, the assay performed as expected even in the presence of a diverse array of compounds. Although we did not find a promising inhibitor scaffold within this library, these results provide the basis for an expanded screening effort in the near future.
Fig. 6.
HTS data with IEDDA-based cat-ELCCA.
In conclusion, we have adapted cat-ELCCA to include IEDDA reactions, providing enhanced signal intensity and a higher Z′ score indicative of a more suitable high-throughput screening assay. By demonstrating the versatility of cat-ELCCA, we hope to promote the application of this easily adaptable method to other biological systems and targets. As cat-ELCCA is amenable to both CuAAC and IEDDA click reactions, by combining these or other bioorthogonal reactions, such as strain-promoted alkyne-azide cycloadditions, it should be possible to follow multiple modifications allowing for multiplexing of more complex systems. Future efforts beyond the scope of this report will be dedicated to this goal.
Supplementary Material
Acknowledgments
This work was supported through a generous start-up package from the University of Michigan College of Pharmacy, a pilot grant from the University of Michigan Center for the Discovery of New Medicines, and the NIH (R01 GM118329 to A.L.G. and P30 CA046592 to the University of Michigan Comprehensive Cancer Center). D.A.L. is grateful for a fellowship from the Graduate Assistance in Areas of National Need (GAANN) program. We thank Martha Larsen and Steve Vander Roest for assistance with HTS equipment set-up.
Footnotes
Electronic Supplementary Information (ESI) available: General methods and materials, assay protocols, and supplemental figures.
Notes and references
- 1.Salic A, Mitchison TJ. Proc Natl Acad Sci, U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu X, LaBaer J. Nat Protoc. 2015;10:756–767. doi: 10.1038/nprot.2015.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garner AL, Janda KD. Angew Chem, Int Ed. 2010;49:9630–9634. doi: 10.1002/anie.201003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garner AL, Janda KD. Chem Commun. 2011;47:7512–7514. doi: 10.1039/c1cc11817j. [DOI] [PubMed] [Google Scholar]
- 5.Lorenz DA, Song JM, Garner AL. Bioconj Chem. 2015;26:19–23. doi: 10.1021/bc500544v. [DOI] [PubMed] [Google Scholar]
- 6.Lang K, Chin JW. ACS Chem Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 7.Devaraj NK, Weissleder R. Acc Chem Res. 2011;44:816–827. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman RC, Kai-How Farh K, Burge CB, Bartel DP. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Rana TM. Nat Rev Drug Disc. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 10.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 11.Guan L, Disney MD. ACS Chem Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 12.We also examined the reaction of a methylcyclopropene (J. Am. Chem. Soc., 2012, 134, 18638-18643) with mTet; however, even upon mixing up to 50 mM of the unconjugated reactants, only trace product could be detected (data not shown).
- 13.A similar result was observed with conjugation of HRP to a protein (data not shown), providing additional evidence for the superiority of IEDDA chemistry in coupling biomolecules.
- 14.Paredes E, Das SR. ChemBioChem. 2011;12:125–131. doi: 10.1002/cbic.201000466. [DOI] [PubMed] [Google Scholar]
- 15.Seckute J, Yang J, Devaraj NK. Nucleic Acids Res. 2013;41:e148. doi: 10.1093/nar/gkt540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winz ML, Linder EC, Andre T, Becker J, Jaschke A. Nucleic Acids Res. 2015;43:e110. doi: 10.1093/nar/gkv544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ameta S, Becker J, Jaschke A. Org Biomol Chem. 2014;12:4701–4707. doi: 10.1039/c4ob00076e. [DOI] [PubMed] [Google Scholar]
- 18.Worrell BT, Malik JA, Fokin VV. Science. 2013;340:457–460. doi: 10.1126/science.1229506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin L, Tolentino DR, Melaimi M, Bertrand G. Sci Adv. 2015;1:e1500304. doi: 10.1126/sciadv.1500304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanious FA, Veal JM, Buczak H, Ratmeyer LS, Wilson WD. Biochemistry. 1992;31:3103–3112. doi: 10.1021/bi00127a010. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TDY, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.