Abstract
OBJECTIVE
Automated analysis of abdominal CT has advanced markedly over just the last few years. Fully automated assessment of organs, lymph nodes, adipose tissue, muscle, bowel, spine, and tumors are some examples where tremendous progress has been made. Computer-aided detection of lesions has also improved dramatically.
CONCLUSION
This article reviews the progress and provides insights into what is in store in the near future for automated analysis for abdominal CT, ultimately leading to fully automated interpretation.
Keywords: computer-aided detection, image processing, CT, segmentation, volumetrics
Unlike for images of the brain and breast, automated analysis of images of the abdomen has been a relative latecomer to radiology. However, the pace of innovation for automated abdominal image analysis has accelerated in just the last few years. Much of this progress has focused on CT, the workhorse of abdominal diagnosis. For example, in a concerted effort over the last 15 years, great progress was made in image processing of CT colonography, leading to such advances as virtual colonoscopy flythrough and computer-aided detection (CAD) of polyps. Beginning in the same time frame and extending to the present, numerous articles describe methods to automatically detect, quantitate, and classify imaging findings on routine abdominal CT. These advances are moving the field closer to achieving the promise of fully automated image analysis and interpretation [1, 2].
Automated image analysis will be considered here in the broadest sense, including quantitative analyses and CAD and classification of disease. This review will cover the broad swath of applications in the abdomen, including organ, lymph node, adipose tissue, muscle, bowel, spine, and tumor analysis. Some speculations about the future of this dynamic field will conclude the review.
Overview
Radiologists perform numerous high-level tasks when interpreting abdominal CT images. These tasks include assessment of organs and detection, classification, and measurement of lesions. Incidental findings must be considered and accepted or rejected. The findings must be put into the proper clinical context of the particular patient. For example, the knowledge that a patient has cancer influences the classification of a new lesion as metastatic versus infectious or inflammatory.
Each of these tasks is amenable to automation. Organs can be located by the computer using atlas- and landmark-based methods. Organ volume and shape can be assessed by finding the edges of the organs in three dimensions, a process known as segmentation. Lesions can be detected and segmented by assessing the patterns of Hounsfield unit intensities in the organs to identify anomalies. Example patterns include variations in intensities, texture, and shape. The quantitative measurements of these patterns are known as features.
To perform accurate detection and segmentation, organs and lesions must be distinguished from other surrounding tissues to avoid false recognition. To do so, features calculated for organs, lesions, and surrounding tissues are fed into classifiers to teach the computer how to distinguish them. The final outputs include those useful for diagnosis (true-positives) and incorrect ones that are not useful (false-positives). At the present time, for most applications, the computer always produces false-positives, necessitating radiologist review. However, the number of false-positives is steadily decreasing as the computer techniques improve.
There are two approaches to developing accurate automated radiologic image analysis software. In one, the software developer designs (handcrafts) features that best identify a particular disease. For example, curvature and shape are useful features for distinguishing polyps from normal colonic mucosa [3, 4]. In the other, generic features are used and a machine-learning algorithm is taught to distinguish disease from nondisease sites by being trained on labeled cases, without the need for handcrafted features. The latter approach, which is made feasible by recent advances in computer science known colloquially as deep learning, is increasingly being used because it markedly increases the efficiency of image analysis development [5].
To perform fully automated abdominal CT image interpretation at the level of a trained radiologist, the computer must assess all the organs and detect all the abnormalities present in the images. Although this is a seemingly daunting task for the software developer, the numbers of organs and potential abnormalities are finite and can be addressed methodically. This methodical approach has been pursued by numerous investigators in the field and is reviewed in the next sections. Because the literature is vast, representative examples of the various approaches are described.
Organs
The automated localization and volume assessment of the abdominal solid visceral organs is a major area of investigation [6]. The liver, spleen, and kidneys have been the most studied of these organs. A number of different techniques have been used for detection. One is identification of important anatomic landmarks, such as the diaphragm, important vessels, and spinal levels [7]. Another is the registration of images to a labeled anatomic atlas [8, 9]. Advances in organ analysis have been accelerated by open challenges or competitions in which various research groups compete to obtain the most accurate organ segmentations on a common dataset [10, 11].
Multiorgan Analysis
A number of investigators have studied methods to identify and measure multiple abdominal organs simultaneously on CT [12–16]. By considering the organs in this way, constraints such as relative organ positions and nonoverlap can be enforced. Accuracy has generally been best for the liver, spleen, and kidneys (> 92%) and lower for more variable organs, such as the gallbladder and pancreas (67–73%) [13]. CAD for simultaneous identification of multiple organs frequently relies on the use of one or more labeled atlases [15]. However, registration of new cases to the different cases in the atlas is usually time consuming, even with modern computer hardware. Solutions to the problems of missing organs in a multiorgan framework, such as nephrectomy or splenectomy, have also been proposed [17].
Liver and Spleen
Manual assessment of organ volumetrics is time consuming and inefficient. Radiologists tend to either over- or underestimate the presence of organomegaly when using longitudinal measurements such as craniocaudal length. Recent advances in image processing have led to fully automated software that can measure organ volumes [18, 19]. The volumetric automated measurements are highly reproducible and accurate and enable more routine organ volumetrics in clinical practice. Nomograms have been developed for liver and spleen volumetry [20, 21]. For example, normal liver volumes averaged 1.51 ± 0.25 L, and normal spleen volumes averaged 0.24 ± 0.08 L. For livers, it was helpful to calculate the ratio of liver volume to body surface area (i.e., the H-score). Using the H-score, mild and massive hepatomegaly were defined as exceeding 0.92 and 1.08 L/m2, respectively. Mild and massive splenomegaly were defined as exceeding 0.31 and 0.43 L, respectively.
Improved liver segmentation on unenhanced CT images has been reported [22]. The segmentation of livers with unusual shapes or with large lesions has been investigated [23]. Liver segmentation can be performed very rapidly using marginal space learning, with detection times less than half a second reported [24].
The biliary tract and hepatic vasculature can be automatically extracted and analyzed [25, 26]. Hepatic vessel analysis has been used for treatment planning, including surgery and radiofrequency ablation [27–29]. Liver volumetrics and vessel analysis are useful for liver transplantation [30].
Kidneys
Automated and semiautomated renal volumetrics have been reported [31–33]. Automated renal cortex segmentation and measurement have been evaluated on healthy kidney donors [34].
Pancreas
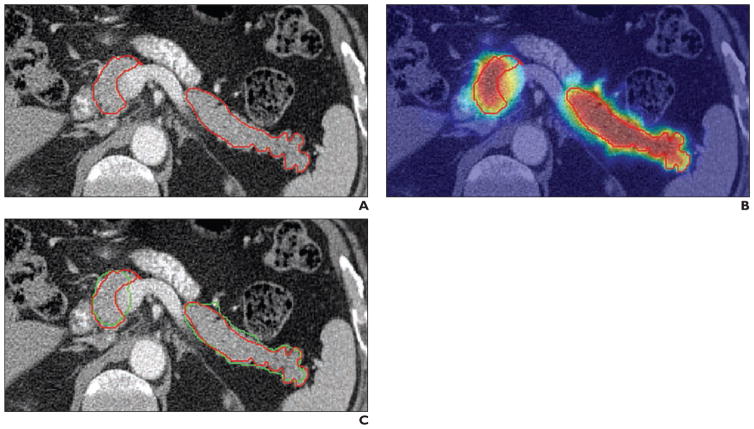
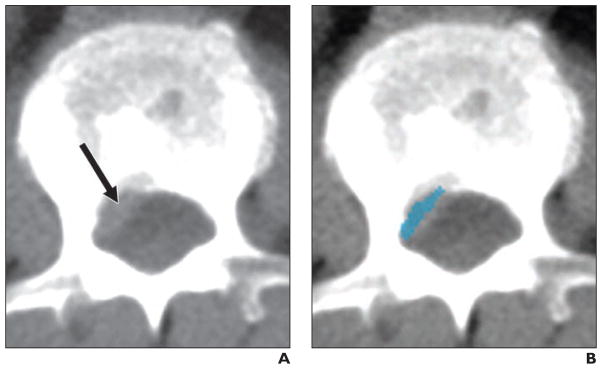
The pancreas is a highly deformable organ that has a shape and location that is greatly influenced by the presence of adjacent structures. This makes automated image analysis of the pancreas extremely challenging. A number of different approaches have been taken to automated pancreas analysis, including the use of anatomic atlases, the location of the splenic and portal veins, and state-of-the-art computer science methods such as deep learning [8, 9, 35–41] (Fig. 1).
Fig. 1.
71-year-old man with prostate cancer. Images show examples of automated pancreas detection and segmentation. Reprinted from [36]. Figure is in public domain.
A, Reference standard pancreas segmentation was performed with manual tracing (red).
B, Pancreas probability map was computed using deep learning. Pancreas is outlined in red.
C, Final pancreas segmentation from automated software (green) is superimposed on reference standard (red) for comparison.
Bladder, Uterus, Prostate, and Rectum
Bladder and bladder tumor segmentation and CAD on CT have been reported. For example, bladder cancers were automatically detected during CT urography [42]. At 4.3 false-positives per patient, the sensitivity for detecting bladder cancers was 84.9%. Bladder segmentation was done using a method that separately segments the contrast-enhanced and unenhanced regions of the bladder lumen [43, 44]. For the assessment of pT0 (complete response), the segmentation technique was found to be equivalent to manual 3D segmentation and superior to Response Evaluation Criteria in Solid Tumors (RECIST) and World Health Organization assessments [45].
Segmentation of the bladder, prostate, and rectum were reported [46]. The method was evaluated on CT scans from 188 patients. With an average computation time of under 10 seconds, the average errors were 2.4, 2.8, and 4.2 mm for prostate, bladder, and rectum, respectively.
Pelvic organ segmentations have obvious applications in radiation treatment planning [47–50]. An example is segmentation of the uterus and cervix [51].
Lymph Nodes
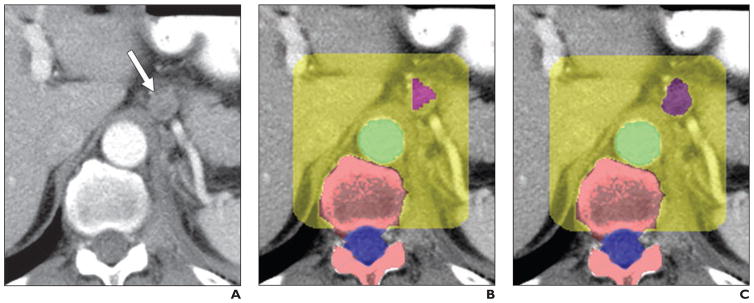
Lymph node analysis is of crucial importance for patients with cancer. In general, the RECIST guidelines are used to evaluate lymph nodes in such patients [52]. A number of investigators have developed automated software for detection and measurement of abdominal adenopathy [53–61] (Fig. 2). Sensitivities for abdominopelvic and retroperitoneal lymphadenopathy are 80–83% at about three false-positives per volume [56, 59].
Fig. 2.
65-year-old man with metastatic prostate cancer. Images show examples of automated lymphadenopathy detection. Reprinted from Hua et al. (presented at 2012 ARRS annual meeting). Figure is in public domain.
A, Original contrast-enhanced abdominal CT image shows centimetric retroperitoneal lymph node (arrow).
B and C, After lymph node detection stage (B) and segmentation stage (C), lymph node (purple), aorta (green), spine (pink), spinal canal (blue), and retroperitoneal lymph node search region (yellow) are shown.
Tumors
Liver Metastases
Liver tumor detection and segmentation have been featured in a number of works. Semiautomated liver tumor segmentation was reported elsewhere [62]. The system could segment the normal liver, enhancing tumor and necrotic tumor with Dice similarity coefficients (a measure of segmentation accuracy) of 93%, 74%, and 72%, respectively. Liver tumor detection and tumor burden—that is, the volumetric fraction of tumor within the liver—were also reported [19]. All of the tumors were detected at a false-positive rate of 2.3 per scan. The tumor burden was estimated with 0.9% error.
A liver tumor segmentation challenge occurred in 2008 [63]. The team with the highest score reported a volumetric overlap error of 31% and an average symmetric absolute surface distance of 1.6 mm [64]. Three other participants in the challenge reported the results of their semiautomated methods [65]. Two of the three evaluated methods were superior but still had high error rates; for example, there were median relative absolute volume differences of about 10% between the semiautomated segmentation and the reference manual segmentation.
Kidney Lesions
Renal lesion volumetrics have been reported [66]. Automated detection of exophytic kidney lesions on CT colonography has been shown [67]. Renal calculi may be accurately detected, with sensitivities exceeding 90% and false-positive rates below one per patient, particularly for larger calculi [68]. Kidney tumor growth rate prediction has been reported [69].
Automated tumor analysis enabled a radiomics study of hereditary kidney cancer [70]. Different hereditary kidney tumors, such as von Hippel–Lindau syndrome, Birt-Hogg-Dubé syndrome, and hereditary papillary renal carcinoma, were distinguishable to a greater or lesser degree according to lesion contrast enhancement and shape characteristics.
Pancreatic Lesions
Growth modeling of pancreatic neuroendocrine tumors has been reported in patient populations where the tumors are not excised until they reach a certain size [71, 72]. Tumor growth modeling takes into account such factors as tumor blood flow and the effects of adjacent tissues and may one day permit the accurate estimation of when the tumor size will exceed the threshold for surgical intervention.
Peritoneal Ovarian Metastases
CAD of peritoneal metastases from ovarian cancer was the subject of an investigation [73]. The authors showed how perihepatic and perisplenic ovarian cancer metastases could be detected when they caused scalloping of the organ surface. The technique detected such metastases with a sensitivity of 87% at a false-positive rate of two per patient. One significance of this investigation was that it showed that detection of metastases was possible for lesions not just within organs but also those abutting organs. Detection of peritoneal masses that do not abut organs is an area in need of investigation.
Bowel
CT Colonography
Research in CT colonography image analysis underwent intense investigation between about 1996 and 2014. This activity has tapered recently as issues relating to clinical implementation have taken precedence.
A number of CAD systems were reported for colonic polyp and mass detection at CT colonography [3, 4, 74–84]. Review articles cover this topic in more detail [85–87]. Advances in CAD of the colon are leading to improved polyp and mass detection accuracy [88–96]. Different reading paradigms for the use of CAD for polyp detection have been investigated, including the first-, concurrent-, and second-reader paradigms [97, 98].
Electronic cleansing of the colon in the laxative-free or reduced-preparation setting has been proposed with promising results [99–102]. The use of dual-energy CT for electronic cleansing has been proposed [103–105].
Crowdsourcing has been applied to understand perceptual factors important for improving the interpretation of CT colonography using CAD [106, 107]. For example, crowdsourcing experiments showed that video fly-arounds of polyps on the endoluminal view led to significant gains in detection performance.
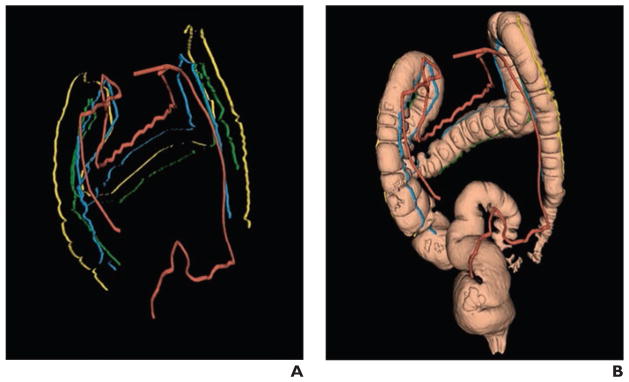
A number of investigations showed how to register the supine and prone CT colonography images longitudinally or circumferentially [108–113]. Approaches to such registration have included using the tenaie coli as fiducial markers and finding ways to register similar haustral folds. One registration technique was evaluated in a radiologist observer trial and for coregistration of polyps at follow-up surveillance [114, 115]. Recent work has shown that the marginal artery of the colon follows the course of the tenia mesocolica and can serve as a fiducial marker for locating the colon in areas of collapse [116, 117] (Fig. 3).
Fig. 3.
51-year-old woman who underwent CT colonography. Reprinted from [117]. Figure is in public domain.
A, Supine 3D reconstruction CT colonography image shows labeled marginal blood vessel (red), detected teniae coli, and segmented colon. Blue indicates tenia mesocolica, yellow indicates tenia omentalis, and green indicates tenia libera.
B, Labeled marginal blood vessel, teniae, and colon are shown together. Image is example of CT colonography image with collapsed segment in sigmoid colon. Marginal blood vessel bridges region of collapse and helps determine colon connectivity there.
It has been shown how to accurately match findings at CT colonography with those of optical colonoscopy [118–120]. This has generally been done by mapping the length of the central path along the colon lumen to the polyp from CT colonography to optical colonoscopy. One such technique refines the prediction of the polyp location by estimating the straightening of the colon that occurs as the colonoscope is inserted into it [121].
Small- and Large-Bowel Segmentation
The small bowel measures approximately 22 feet in length and is tightly coiled in the abdomen. It has been considered to be a relatively intractable organ for automated image analysis until recently. It is now known that the mesenteric vasculature can be used to accurately locate the small bowel on ultra-high-resolution CT angiography images [122, 123]. This method has potential applications for CAD of small-bowel tumors, obstruction, and inflammatory bowel disease.
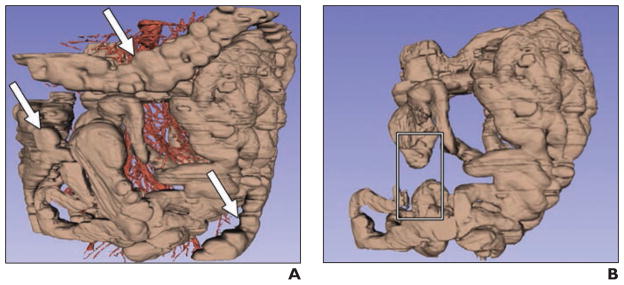
The colon can be distinguished from the small bowel by automated software [124] (Fig. 4). The method relies on detecting the colonic mesenteric vasculature and using a colon location likelihood map taken from CT colonography studies. The technique enabled rejection of most colon false-positives by use of a small-bowel detector. A preliminary investigation has been reported on automated detection of small-bowel strictures on CT enterography [125].
Fig. 4.
51-year-old man with history of midgut carcinoid tumors. Images show example of improved small-bowel segmentation by colon detection and removal. Modified from [124]. Figure is in public domain.
A, Preliminary small-bowel segmentation. Small bowel is shown in brown, and mesenteric and aortoiliac vasculature are shown in red. Arrows indicate false-positives on colon.
B, Improved small-bowel segmentation after colon removal using artery labeling and colon probability map methods. Rectangle indicates missing small bowel, false-negative introduced by method.
Typhlitis or Colitis
Colonic inflammation can be detected by automated software on noncolonographic images by searching for thickened bowel wall. Preliminary investigation shows the feasibility of typhlitis and colitis detection on CT of immunocompromised patients [126]. Using deep learning, 85% of colitis regions were properly detected at a false-positive rate of one per image [127].
Bone
There has been rapid recent progress in automated analysis of bone on CT. Close at hand is complete spine CT image analysis. Much of the research has been conducted on larger-FOV body images in addition to smaller-FOV spine images. Examples include fully automated vertebral level labeling, bone mineral densitometry (i.e., quantitative CT), detection and grading of compression and traumatic vertebral fractures, and detection of metastases, degenerative disease, and epidural masses.
Vertebral Level Labeling
Vertebral level labeling or partitioning is useful for anatomic and lesion localization. It is also required for most other automated spine analysis tools. Automated spinal extraction and partitioning on large-FOV body CT scans have been reported [128–130].
Bone Mineral Densitometry (Quantitative CT)
Quantitative CT is typically done with the use of a densitometry phantom positioned beneath the patient. Quantitative CT can also be performed without a phantom with appropriate calibration of the scanner [131]. Recent work has shown good accuracy and agreement comparing quantitative CT with dual-energy x-ray absorptiometry [132]. This research has opened the possibility of fully automated bone densitometry assessments that are efficient and rapid for all body CT scans, including screening studies such as CT colonography.
Compression Fractures
Vertebral compression fractures are a significant cause of morbidity, particularly in older patients with osteoporosis. Despite their importance, vertebral compression fractures are often overlooked on body CT [133]. Automated software is under development that can accurately detect vertebral compression fractures [134–137]. Future versions of the software will need to both detect and grade the fractures, such as by using a visual semiquantitative scale [138].
Traumatic Fractures
It is crucial to detect vertebral fractures in patients with trauma. Overlooked fractures can lead to significant morbidity. Fractures can occur in the vertebral body and posterior elements. Some current CAD systems treat these two components separately.
Traumatic vertebral body fracture detection software has been reported recently [139]. The software digitally strips the vertebral body cortex and looks for fractures in the cortical shell. The software had a sensitivity of 92% and false-positive rate of 1.6 per patient in 67 test set patients with traumatic vertebral body fractures. The fractures were also classified according to Denis column involvement [140]. Further work is under way to detect posterior element fractures [141]. On a testing dataset, the software achieved an AUC of 0.857 and 81% sensitivity with 10 false-positives per patient. Although this software might be primarily used on dedicated spine CT with a small FOV, it could also be applied to body CT scans with a larger FOV.
Degenerative Changes
Although degenerative changes in the spine are ubiquitous in older patients, in a patient with cancer, it can be challenging to distinguish subtle changes in metastases from extensive degenerative lesions on PET/CT. The cortical shell stripping described already for vertebral body fracture detection can also be applied for degenerative disease detection on body CT [142, 143]. Degenerative disease detection can be particularly valuable for assisting interpretation of sodium fluoride PET/CT in older patients with cancer with osseous metastases [144].
Metastases
Bone metastases are a frequent cause of morbidity in patients with cancer. In the setting of breast and prostate cancer, vertebral lesions may be the first sign of metastatic disease, preceding lymph node and organ involvement. Automated detection of both lytic and sclerotic vertebral metastases on body CT have been reported [145–147]. Sensitivities and false-positive rates for lytic and sclerotic metastases were 94% and 79% and 4.5 and 10.9 false-positives per patient, respectively [145, 146]. Recent improvements led to better performance, with sensitivity and false-positive rates for sclerotic lesions of 70% and three false-positives per patient [59]. Potential future applications include assessment of lesion change and early lesion detection.
Epidural Masses
Epidural masses in patients with cancer are frequently overlooked by radiologists interpreting body CT (Kim LM, et al., presented at the Radiological Society of North America 2015 annual meeting). The authors found that, in 129 patients with epidural masses proven by spine MRI, radiologists did not report the presence of epidural masses in 25% of the 244 CT studies. To address this issue, automated software has been developed that can detect epidural masses with a sensitivity of 82% and false-positive rate of three per patient [148, 149] (Fig. 5).
Fig. 5.
46-year-old man with metastatic melanoma detected on body CT. Reprinted from [149]. Figure is in public domain.
A, Original CT image shows epidural mass (arrow).
B, Computer-aided detection image shows epidural mass (blue).
Adipose Tissue
Visceral fat analysis is routinely used for a number of investigations. The analysis software typically identifies the boundary between the subcutaneous and visceral fat compartments, enabling accurate measurement of both and permitting visceral fat measurement not just at the umbilicus or L4–L5 level used in manual analyses but also whole abdomen visceral fat analysis [150–152]. Approaches have also been developed to choose an optimized level for single slice analysis of adiposity [153].
Visceral fat is known to be associated with metabolic syndrome and colonic polyps [150, 154]. Visceral fat can be assessed on routine abdominal CT and on CT colonography [155–157]. An intriguing association between pericolonic fat and colonic polyps has been recently reported [158].
Muscle
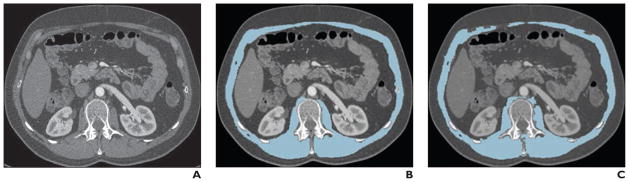
Muscle analysis is important for diseases such as myositis and also for assessment of functional reserve in patients undergoing cancer therapy or surgery [159–163]. Muscle analysis is typically conducted at a single level in the abdomen (L3) using manual or semiautomated tracing of the musculature [164]. Automated image analysis is at a preliminary stage of development but shows promise in enabling whole-body muscle volumetrics [165] (Fig. 6).
Fig. 6.
51-year-old man with history of midgut carcinoid tumors (same patient as Fig. 4). Images are examples of automated muscle segmentation. Reprinted from [165]. Figure is in public domain.
A, Original CT image shows through mid abdomen.
B and C, On reference standard (B) and automated segmentation result (C) images, muscle is indicated in cyan.
Blood Vessels: Abdominal Atherosclerosis and Vessel Mapping
Automated detection of aortoiliac atherosclerosis may enable whole-body calcium scoring and epidemiologic analyses [166]. In a study of 40 CT scans with 249 atherosclerotic calcifications, a sensitivity of detecting calcifications of 83.9% was found, with one false-positive per scan. Patients were accurately grouped into one of four broad categories of aortoiliac plaque burden in 75% of cases and were off by only one category in another 20%.
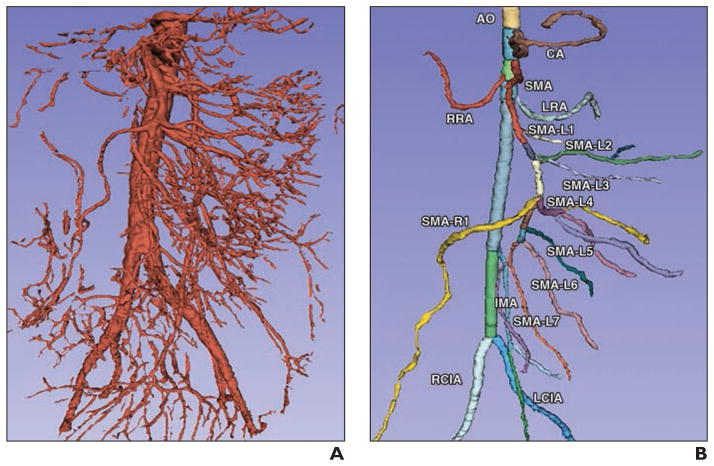
Automated mapping of abdominal vessels has been proposed [26, 167, 168] (Fig. 7). The labeling provided by the mapping may be useful for surgical navigation [169]. Semiautomated volumetry of abdominal aortic aneurysms on CT has also been reported [170].
Fig. 7.
51-year-old man with history of midgut carcinoid tumors (same patient as Fig. 4). Reprinted from [167]. Figure is in public domain.
A, Three-dimensional mesenteric vasculature segmentation was derived from contrast-enhanced abdominal CT angiography.
B, Automated anatomic labeling result is shown. AO = abdominal aorta, CA = celiac axis, SMA = superior mesenteric artery, RRA = right renal artery, LRA = left renal artery, IMA = inferior mesenteric artery, RCIA = right common iliac artery, LCIA = left common iliac artery.
The Future
Advances are likely in several areas pertaining to automated abdominal CT image interpretation. These areas include machine learning, big data, automated report generation, multimodality image analysis, publicly available datasets and competitive challenges, investigation of other organs and diseases, and new applications. If automated interpretation is widely realized, there will be effects on radiologists that will need to be considered.
Machine Learning
Machine learning, a computer science discipline, has had a tremendous effect on the advancements in radiology CAD [86]. A recent advance in computer science is the refinement of neural networks, a type of machine learning classifier used to make decisions from data. This refinement, known generically as deep learning but more specifically as convolutional neural networks, has shown dramatic improvements in automated intelligence applications. Initially drawing attention for impressive improvements in speech recognition and natural image interpretation, deep learning is now being applied to medical images, as described already in the sections on the pancreas and colitis. The results have been particularly promising for reduction of false-positives.
Deep learning led to dramatic improvements in the performance of three different body CT CAD systems [59]. The authors improved existing CAD systems for sclerotic spine metastases, lymphadenopathy, and colonic polyps [75, 146, 171, 172]. Sensitivities improved from 57% to 70%, from 43% to 77%, and from 58% to 75%, respectively, with three false-positives per patient.
There has been a recent explosion of research activity on the use of deep learning for radiologic image analysis that is likely to continue for at least the next few years. Examples for abdominal CT analysis include pancreas and kidney imaging [36, 173].
Big Data and Automated Radiology Reporting
CT scans can be analyzed in combination with their accompanying reports to enable automated report generation and efficient linking of report findings with image findings. In a study of 216,000 radiology key images identified during routine clinical interpretation of scans from 62,000 unique patients, including many from abdominal CT scans, the rate of predicted disease-related words matching the actual words in the report sentences was 56% [174]. Automated hyperlinking of report sentences mentioning specific organs to the proper CT images, along with choice of window width and level settings appropriate to the organ, has been shown [175].
Multimodality Image Analysis
CT images of a lesion can be combined with other modalities such as PET and MRI to improve segmentation. For example, assessment of lesions in the liver and pelvis was improved using cosegmentation from multimodality PET/CT and PET/MRI [176]. This topic demands further exploration.
Publicly Available Datasets and Competitive Challenges
Greater availability of CT datasets is needed to advance the field. Abdominal CT datasets are slowly becoming available, through such sites as The Cancer Imaging Archive [177] and the Visceral project [178]. For example, a dataset of 595 annotated abdominal lymph nodes in CT scans of 86 patients is available online [179]. Some datasets are released as part of various challenges and workshops, such as the Medical Image Computing and Computer-Assisted Intervention Society’s Beyond the Cranial Vault workshop [180]. However, much larger datasets may be necessary to enable the computer to perform as accurately as a trained radiologist [181]. A number of companies have recently sought to collect huge proprietary radiology datasets for this purpose, including IBM, Zebra Medical Vision, and Enlitic.
Investigation of Other Organs and Diseases
Despite the tremendous progress described already, there are still some understudied organs and diseases. The gallbladder, adrenal glands, and ovaries are difficult to analyze because of their small size. Uterine anomalies and fibroids have not been the subject of fully automated image analysis. Automated quantification of ascites and the detection of pneumoperitoneum would be useful applications. Automated detection of vascular thrombosis, either bland or malignant, and collateral vessels would have clinical value. Hydronephrosis detection and grading would be useful for scan triage for immediate reading and for standardization of severity assessment.
New Applications to Improve Patient Care
An area of tremendous potential is the use of fully automated image interpretation in underserved populations to meet global health needs. Billions of patients live in countries with limited access to trained radiologists [182]. Transport of images via teleradiology from underserved communities to trained radiologists may be insufficient to meet these patients’ needs if the pace of imaging utilization accelerates. Automated analyses may be a more sustainable solution.
Another underutilized approach is the use of automated image analysis running in the background to triage patients with potentially life-threatening conditions, to reduce common interpretative errors, to perform large-scale epidemiologic studies, and to coordinate and interpret large volumes of clinical, genomic, and imaging data. As radiology practices consolidate into larger hospital-led groups, it will be more feasible to implement such systems. Such big data analyses could uncover previously unrecognized associations among imaging findings, drug treatments, and other data in the clinical record. These applications promise improvements in patient care.
Effect on the Radiologist
Autopilots for airplanes changed the role of the pilot. Self-driving cars will change the role of the driver. In both cases, the human is still ultimately responsible for the safety of the passengers. Similarly, fully automated abdominal CT image interpretation is likely to change the role of radiologists, but they will still be responsible for taking care of the patient and making the final diagnosis. The automated report could improve reading efficiency, but radiologists will need to be vigilant to avoid placing too much trust in the computer. Further research on human factors, such as visual perception, and on quality assurance and promotion of a safety culture will be required to understand potential failures of the technology when used in the clinical setting [183–185].
The use of automated image interpretation by nonradiologists will need to be considered. Such users might include radiology technologists, radiologist assistants, and nonradiologist clinicians. The technology could lead to further commoditization of radiology services.
In conclusion, advances in abdominal CT automated image interpretation are occurring at a rapid pace. In the not too distant future, these advances may enable fully automated image interpretation. Similar advances may occur in other body regions and with other imaging modalities. Risks and benefits are difficult to foresee but may include increased pressures for commoditization, better reading efficiency, fewer interpretive errors, and a more quantitative radiology report. The primary focus must ultimately be on improved patient care.
Acknowledgments
Supported by the Intramural Research Program of the National Institutes of Health, Clinical Center.
I thank Andrew Dwyer for critical review of the manuscript.
Footnotes
No NIH endorsement of any product or company mentioned in this manuscript should be inferred. The opinions expressed herein are the author’s and do not necessarily represent those of NIH.
Based on presentations at the Radiological Society of North America 2014 and 2015 annual meetings, Chicago, IL.
R. M. Summers receives patent royalties and has received research support from iCAD Medical.
References
- 1.van Ginneken B, Schaefer-Prokop CM, Prokop M. Computer-aided diagnosis: how to move from the laboratory to the clinic. Radiology. 2011;261:719–732. doi: 10.1148/radiol.11091710. [DOI] [PubMed] [Google Scholar]
- 2.Summers RM. Road maps for advancement of radiologic computer-aided detection in the 21st century. Radiology. 2003;229:11–13. doi: 10.1148/radiol.2291030010. [DOI] [PubMed] [Google Scholar]
- 3.Summers RM, Beaulieu CF, Pusanik LM, et al. Automated polyp detector for CT colonography: feasibility study. Radiology. 2000;216:284–290. doi: 10.1148/radiology.216.1.r00jl43284. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida H, Nappi J. Three-dimensional computer-aided diagnosis scheme for detection of colonic polyps. IEEE Trans Med Imaging. 2001;20:1261–1274. doi: 10.1109/42.974921. [DOI] [PubMed] [Google Scholar]
- 5.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 6.Linguraru MG, Summers RM. Computational anatomy in the abdomen: automated multi-organ and tumor analysis from computed tomography. In: Suzuki K, editor. Computational intelligence in biomedical imaging. New York, NY: Springer; 2014. pp. 107–139. [Google Scholar]
- 7.Liu D, Zhou KS, Bernhardt D, Comaniciu D. 2010 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) San Francisco, CA: IEEE; 2010. Search strategies for multiple landmark detection by sub-modular maximization; pp. 2831–2838. [Google Scholar]
- 8.Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D. Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans Med Imaging. 2013;32:1723–1730. doi: 10.1109/TMI.2013.2265805. [DOI] [PubMed] [Google Scholar]
- 9.Tong T, Wolz R, Wang Z, et al. Discriminative dictionary learning for abdominal multi-organ segmentation. Med Image Anal. 2015;23:92–104. doi: 10.1016/j.media.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Heimann T, van Ginneken B, Styner MA, et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging. 2009;28:1251–1265. doi: 10.1109/TMI.2009.2013851. [DOI] [PubMed] [Google Scholar]
- 11.Jiménez del Toro OA, Goksel O, Menze B, et al. VISCERAL: VISual Concept Extraction Challenge in RAdioLogy—ISBI 2014 Challenge Organization. In: Goksel O, editor. Proceedings of the VISCERAL Challenge at ISBI. Beijing, China: International Symposium on Biomedical Imaging; 2014. pp. 6–15. [Google Scholar]
- 12.Zikic D, Glocker B, Criminisi A. Encoding atlases by randomized classification forests for efficient multi-atlas label propagation. Med Image Anal. 2014;18:1262–1273. doi: 10.1016/j.media.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Okada T, Linguraru MG, Hori M, Summers RM, Tomiyama N, Sato Y. Abdominal multi-organ segmentation from CT images using conditional shape-location and unsupervised intensity priors. Med Image Anal. 2015;26:1–18. doi: 10.1016/j.media.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerrolaza JJ, Reyes M, Summers RM, Gonzalez-Ballester MA, Linguraru MG. Automatic multi-resolution shape modeling of multi-organ structures. Med Image Anal. 2015;25:11–21. doi: 10.1016/j.media.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu C, Oda M, Kitasaka T, et al. Multi-organ segmentation based on spatially-divided probabilistic atlas from 3D abdominal CT images. Med Image Comput Comput Assist Interv. 2013;16:165–172. doi: 10.1007/978-3-642-40763-5_21. [DOI] [PubMed] [Google Scholar]
- 16.Udupa JK, Odhner D, Zhao L, et al. Body-wide hierarchical fuzzy modeling, recognition, and delineation of anatomy in medical images. Med Image Anal. 2014;18:752–771. doi: 10.1016/j.media.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki M, Linguraru MG, Summers RM, Okada K. Analyses of missing organs in abdominal multi-organ segmentation. In: Yoshida H, Sakas G, Linguraru MG, editors. Lecture notes in computer science, vol. 7029. Abdominal imaging: computational and clinical applications. Heidelberg, Germany: Springer; 2012. pp. 256–263. [Google Scholar]
- 18.Linguraru MG, Sandberg JK, Li Z, Shah F, Summers RM. Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation. Med Phys. 2010;37:771–783. doi: 10.1118/1.3284530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linguraru MG, Richbourg WJ, Liu J, et al. Tumor burden analysis on computed tomography by automated liver and tumor segmentation. IEEE Trans Med Imaging. 2012;31:1965–1976. doi: 10.1109/TMI.2012.2211887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linguraru MG, Sandberg JK, Jones EC, Petrick N, Summers RM. Assessing hepatomegaly: automated volumetric analysis of the liver. Acad Radiol. 2012;19:588–598. doi: 10.1016/j.acra.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linguraru MG, Sandberg JK, Jones EC, Summers RM. Assessing splenomegaly: automated volumetric analysis of the spleen. Acad Radiol. 2013;20:675–684. doi: 10.1016/j.acra.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomoshige S, Oost E, Shimizu A, Watanabe H, Nawano S. A conditional statistical shape model with integrated error estimation of the conditions: application to liver segmentation in noncontrast CT images. Med Image Anal. 2014;18:130–143. doi: 10.1016/j.media.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Umetsu S, Shimizu A, Watanabe H, Kobatake H, Nawano S. An automated segmentation algorithm for CT volumes of livers with atypical shapes and large pathological lesions. IEICE Trans Inf Syst. 2014;E97D:951–963. [Google Scholar]
- 24.Zheng YF, Georgescu B, Ling HB, Zhou SK, Scheuering M, Comaniciu D. IEEE conference on computer vision and pattern recognition, 2009: CVPR 2009. Miami, FL: IEEE; 2009. Constrained marginal space learning for efficient 3D anatomical structure detection in medical images; pp. 194–201. [Google Scholar]
- 25.Koga K, Hayashi Y, Hirose T, et al. Development of automated extraction method of biliary tract from abdominal CT volumes based on local intensity structure analysis. In: Ourselin S, Styner MA, editors. Medical imaging 2014: image processing—proceedings of SPIE. Vol. 9034. Bellingham, WA: SPIE; 2014. p. 903448. [Google Scholar]
- 26.Matsuzaki T, Oda M, Kitasaka T, Hayashi Y, Misawa K, Mori K. Automated anatomical labeling of abdominal arteries and hepatic portal system extracted from abdominal CT volumes. Med Image Anal. 2015;20:152–161. doi: 10.1016/j.media.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Selle D, Preim B, Schenk A, Peitgen HO. Analysis of vasculature for liver surgical planning. IEEE Trans Med Imaging. 2002;21:1344–1357. doi: 10.1109/TMI.2002.801166. [DOI] [PubMed] [Google Scholar]
- 28.Pamulapati V, Venkatesan A, Wood BJ, Linguraru MG. Liver segmental anatomy and analysis from vessel and tumor segmentation via optimized graph cuts. In: Yoshida H, Sakas G, Linguraru MG, editors. Lecture notes in computer science, vol. 7029. Abdominal imaging: computational and clinical applications. Heidelberg, Germany: Springer; 2012. pp. 189–197. [Google Scholar]
- 29.Audigier C, Mansi T, Delingette H, et al. Parameter estimation for personalization of liver tumor radiofrequency ablation. In: Yoshida H, Näppi JJ, Saini S, editors. Lecture notes in computer science, vol. 8676. Abdominal imaging: computational and clinical applications. Heidelberg, Germany: Springer; 2014. pp. 3–12. [Google Scholar]
- 30.Frericks BB, Caldarone FC, Nashan B, et al. 3D CT modeling of hepatic vessel architecture and volume calculation in living donated liver transplantation. Eur Radiol. 2004;14:326–333. doi: 10.1007/s00330-003-2161-8. [DOI] [PubMed] [Google Scholar]
- 31.Shim H, Chang S, Tao C, Wang JH, Kaya D, Bae KT. Semiautomated segmentation of kidney from high-resolution multidetector computed tomography images using a graph-cuts technique. J Comput Assist Tomogr. 2009;33:893–901. doi: 10.1097/RCT.0b013e3181a5cc16. [DOI] [PubMed] [Google Scholar]
- 32.Okada T, Linguraru MG, Hori M, Summers RM, Tomiyama N, Sato Y. Abdominal multi-organ CT segmentation using organ correlation graph and prediction-based shape and location priors. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Lecture notes in computer science, vol. 8151. Medical image computing and computer-assisted intervention: MICCAI 2013—part III. Berlin, Germany: Springer-Verlag; 2013. pp. 275–282. [DOI] [PubMed] [Google Scholar]
- 33.Zhou XR, Watanabe A, Zhou XN, et al. Automatic organ segmentation on torso CT images by using content-based image retrieval. In: Haynor DR, Ourselin S, editors. Medical imaging 2012: image processing—proceedings of SPIE. Vol. 8314. Bellingham, WA: SPIE; 2012. p. 831405. [Google Scholar]
- 34.Chen X, Summers RM, Cho M, Bagci U, Yao J. An automatic method for renal cortex segmentation on CT images: evaluation on kidney donors. Acad Radiol. 2012;19:562–570. doi: 10.1016/j.acra.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth HR, Farag A, Lu L, Turkbey EB, Summers RM. Deep convolutional networks for pancreas segmentation in CT imaging. In: Ourselin S, Styner MA, editors. Medical imaging 2015: image processing—proceedings of SPIE. Vol. 9413. Bellingham, WA: SPIE; 2015. p. 94131G. [Google Scholar]
- 36.Roth HR, Lu L, Farag A, et al. DeepOrgan: multi-level deep convolutional networks for automated pancreas segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. Lecture notes in computer science, vol. 9349. Medical image computing and computer-assisted intervention: MICCAI 2015—part I. Geneva, Switzerland: Springer International Publishing; 2015. pp. 556–564. [Google Scholar]
- 37.Shimizu A, Kimoto T, Kobatake H, Nawano S, Shinozaki K. Automated pancreas segmentation from three-dimensional contrast-enhanced computed tomography. Int J Comput Assist Radiol Surg. 2010;5:85–98. doi: 10.1007/s11548-009-0384-0. [DOI] [PubMed] [Google Scholar]
- 38.Farag A, Liu J, Summers RM. Automatic segmentation of abdominal vessels for improved pancreas localization. In: Mello-Thomas CR, Kupinski MA, editors. Medical imaging 2014: image perception, observer performance, and technology assessment—proceedings of SPIE. Vol. 9037. Bellingham, WA: SPIE; 2014. p. 90371M. [Google Scholar]
- 39.Farag A, Lu L, Turkbey E, Liu J, Summers RM. A bottom-up approach for automatic pancreas segmentation in abdominal CT scans. In: Yoshida H, Näppi JJ, Saini S, editors. Lecture notes in computer science, vol. 8676. Abdominal imaging: computational and clinical applications. Geneva, Switzerland: Springer International Publishing; 2014. pp. 103–113. [Google Scholar]
- 40.Karasawa K, Oda M, Hayashi Y, et al. Pancreas segmentation from 3D abdominal CT images using patient-specific weighted-subspatial probabilistic atlases. In: Ourselin S, Styner MA, editors. Medical imaging 2015:image processing—proceedings of SPIE. Vol. 9413. Bellingham, WA: SPIE; 2015. p. 94131A. [Google Scholar]
- 41.Karasawa K, Kitasaka T, Oda M, et al. Structure specific atlas generation and its application to pancreas segmentation from contrasted abdominal CT volumes. In: Menze B, Langs G, Montillo A, et al., editors. MICCAI 2015 workshop on medical computer vision: algorithms for big data. Cambridge, MA: Springer; 2015. [Google Scholar]
- 42.Cha K, Hadjiiski L, Chan HP, Cohan RH, Caoili EM, Zhou C. Detection of urinary bladder mass in CT urography with SPAN. Med Phys. 2015;42:4271–4284. doi: 10.1118/1.4922503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cha K, Hadjiiski L, Chan HP, Caoili EM, Cohan RH, Zhou C. CT urography: segmentation of urinary bladder using CLASS with local contour refinement. Phys Med Biol. 2014;59:2767–2785. doi: 10.1088/0031-9155/59/11/2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadjiiski L, Chan HP, Cohan RH, et al. Urinary bladder segmentation in CT urography (CTU) using CLASS. Med Phys. 2013;40:111906. doi: 10.1118/1.4823792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hadjiiski L, Weizer AZ, Alva A, et al. Treatment response assessment for bladder cancer on CT based on computerized volume analysis, World Health Organization criteria, and RECIST. AJR. 2015;205:348–352. doi: 10.2214/AJR.14.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C, Zheng Y, Birkbeck N, et al. Precise segmentation of multiple organs in CT volumes using learning-based approach and information theory. Med Image Comput Comput Assist Interv. 2012;15:462–469. doi: 10.1007/978-3-642-33418-4_57. [DOI] [PubMed] [Google Scholar]
- 47.Feng Q, Foskey M, Chen W, Shen D. Segmenting CT prostate images using population and patient-specific statistics for radiotherapy. Med Phys. 2010;37:4121–4132. doi: 10.1118/1.3464799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S, Lovelock DM, Radke RJ. Segmenting the prostate and rectum in CT imagery using anatomical constraints. Med Image Anal. 2011;15:1–11. doi: 10.1016/j.media.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Lu C, Chelikani S, Papademetris X, et al. An integrated approach to segmentation and nonrigid registration for application in image-guided pelvic radiotherapy. Med Image Anal. 2011;15:772–785. doi: 10.1016/j.media.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas B, Coradi T, Scholz M, et al. Automatic segmentation of thoracic and pelvic CT images for radiotherapy planning using implicit anatomic knowledge and organ-specific segmentation strategies. Phys Med Biol. 2008;53:1751–1771. doi: 10.1088/0031-9155/53/6/017. [DOI] [PubMed] [Google Scholar]
- 51.Bondar ML, Hoogeman M, Schillemans W, Heijmen B. Intra-patient semi-automated segmentation of the cervix-uterus in CT-images for adaptive radiotherapy of cervical cancer. Phys Med Biol. 2013;58:5317–5332. doi: 10.1088/0031-9155/58/15/5317. [DOI] [PubMed] [Google Scholar]
- 52.Eisenhauer EA, Therasse P, Bodgaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1. 1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 53.Liu J, Feng CH, Hua J, Yao JH, White JM, Summers RM. 9th IEEE International Symposium on Biomedical Imaging (ISBI) Barcelona, Spain: IEEE; 2012. Automatic detection and segmentation of abdominopelvic lymph nodes on computed tomography scans; pp. 1455–1458. [Google Scholar]
- 54.Liu JM, Hua J, Yao JH, White JM, Summers RM. Computer-aided abdominal lymph node detection using contrast-enhanced CT images. In: Summers RM, van Ginneken B, editors. Medical imaging 2011: computer-aided diagnosis—proceedings of SPIE. Vol. 7963. Bellingham, WA: SPIE; 2011. p. 796313. [Google Scholar]
- 55.Liu JM, White JM, Summers RM. Computer-aided lymph node detection in abdominal CT images. In: Karssemeijer N, Summers RM, editors. Medical imaging 2010: computer-aided diagnosis—proceedings of SPIE. Vol. 7624. Bellingham, WA: SPIE; 2010. p. 76240U. [Google Scholar]
- 56.Barbu A, Suehling M, Xu X, Liu D, Zhou SK, Comaniciu D. Automatic detection and segmentation of lymph nodes from CT data. IEEE Trans Med Imaging. 2012;31:240–250. doi: 10.1109/TMI.2011.2168234. [DOI] [PubMed] [Google Scholar]
- 57.Seff A, Lu L, Cherry KM, et al. 2D view aggregation for lymph node detection using a shallow hierarchy of linear classifiers. In: Golland P, Hata N, Barillot C, Hornegger J, Howe R, editors. Lecture notes in computer science, vol. 8673. Medical image computing and computer-assisted intervention: MICCAI 2014—part I. Heidelberg, Germany: Springer; 2014. pp. 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roth HR, Lu L, Seff A, et al. A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations. In: Golland P, Hata N, Barillot C, Hornegger J, Howe R, editors. Lecture notes in computer science, vol. 8673. Medical image computing and computer-assisted intervention: MICCAI 2014—part I. Heidelberg, Germany: Springer; 2014. pp. 520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roth HR, Lu L, Liu J, et al. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging. 2015 May 12; doi: 10.1109/TMI.2015.2482920. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beichel RR, Wang Y. Computer-aided lymph node segmentation in volumetric CT data. Med Phys. 2012;39:5419–5428. doi: 10.1118/1.4742845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seff A, Lu L, Barbu A, Roth H, Shin HC, Summers RM. Leveraging mid-level semantic boundary cues for automated lymph node detection. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. Lecture notes in computer science, vol. 9350. Medical image computing and computer-assisted intervention: MICCAI 2015. Heidelberg, Germany: Springer; 2015. pp. 53–61. [Google Scholar]
- 62.Conze PH, Rousseau F, Noblet V, Heitz F, Memeo R, Pessaux P. Semi-automatic liver tumor segmentation in dynamic contrast-enhanced CT scans using random forests and supervoxels. In: Zhou L, Wang L, Wang Q, Shi Y, editors. Lecture notes in computer science, vol. 9352. Machine learning in medical imaging. Heidelberg, Germany: Springer; 2015. pp. 212–219. [Google Scholar]
- 63.Miccai 2008 Segmentation Workshop. [Accessed April 10, 2016];NA-MIC website. No authors listed. www.na-mic.org/Wiki/index.php/Miccai_2008_Segmentation_Workshop.
- 64.Moltz JH, Bornemann L, Kuhnigk JM, et al. Advanced segmentation techniques for lung nodules, liver metastases, and enlarged lymph nodes in CT scans. IEEE J Selected Top Signal Processing. 2009;3:122–134. [Google Scholar]
- 65.Zhou JY, Wong DK, Ding F, et al. Liver tumour segmentation using contrast-enhanced multi-detector CT data: performance benchmarking of three semiautomated methods. Eur Radiol. 2010;20:1738–1748. doi: 10.1007/s00330-010-1712-z. [DOI] [PubMed] [Google Scholar]
- 66.Summers RM, Agcaoili CML, McAuliffe MJ, et al. Proceedings of 2001 international conference on image processing. Vol. 2. Thessaloniki, Greece: IEEE; 2001. Helical CT of von Hippel–Lindau: semi-automated segmentation of renal lesions; pp. 293–296. [Google Scholar]
- 67.Liu JF, Wang SJ, Linguraru MG, Yao JH, Summers RM. Computer-aided detection of exophytic renal lesions on non-contrast CT images. Med Image Anal. 2015;19:15–29. doi: 10.1016/j.media.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu JF, Wang SJ, Turkbey EB, Linguraru MG, Yao JH, Summers RM. Computer-aided detection of renal calculi from noncontrast CT images using TV-flow and MSER features. Med Phys. 2015;42:144–153. doi: 10.1118/1.4903056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen X, Summers RM, Yao J. Kidney tumor growth prediction by coupling reaction-diffusion and biomechanical model. IEEE Trans Biomed Eng. 2013;60:169–173. doi: 10.1109/TBME.2012.2222027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Linguraru MG, Wang SJ, Shah F, et al. Automated noninvasive classification of renal cancer on multiphase CT. Med Phys. 2011;38:5738–5746. doi: 10.1118/1.3633898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong KCL, Summers RM, Kebebew E, Yao JH. Tumor growth prediction with reaction-diffusion and hyperelastic biomechanical model by physiological data fusion. Med Image Anal. 2015;25:72–85. doi: 10.1016/j.media.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong KC, Summers RM, Kebebew E, Yao J. Pancreatic tumor growth prediction with multiplicative growth and image-derived motion. Inf Process Med Imaging. 2015;24:501–513. doi: 10.1007/978-3-319-19992-4_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, Wang S, Linguraru MG, Yao J, Summers RM. Tumor sensitive matching flow: a variational method to detecting and segmenting perihepatic and perisplenic ovarian cancer metastases on contrast-enhanced abdominal CT. Med Image Anal. 2014;18:725–739. doi: 10.1016/j.media.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dachman AH, Obuchowski NA, Hoffmeister JW, et al. Effect of computer-aided detection for CT colonography in a multireader, multicase trial. Radiology. 2010;256:827–835. doi: 10.1148/radiol.10091890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Summers RM, Yao JH, Pickhardt PJ, et al. Computed tomographic virtual colonoscopy computer-aided polyp detection in a screening population. Gastroenterology. 2005;129:1832–1844. doi: 10.1053/j.gastro.2005.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Halligan S, Taylor SA, Dehmeshki J, et al. Computer-assisted detection for CT colonography: external validation. Clin Radiol. 2006;61:758–763. doi: 10.1016/j.crad.2006.02.015. discussion, 764–755. [DOI] [PubMed] [Google Scholar]
- 77.Dehmeshki J, Halligan S, Taylor SA, et al. Computer assisted detection software for CT colonography: effect of sphericity filter on performance characteristics for patients with and without fecal tagging. Eur Radiol. 2007;17:662–668. doi: 10.1007/s00330-006-0430-z. [DOI] [PubMed] [Google Scholar]
- 78.Summers RM, Johnson CD, Pusanik LM, Malley JD, Youssef AM, Reed JE. Automated polyp detection at CT colonography: feasibility assessment in a human population. Radiology. 2001;219:51–59. doi: 10.1148/radiology.219.1.r01ap0751. [DOI] [PubMed] [Google Scholar]
- 79.Yoshida H, Nappi J, MacEneaney P, Rubin DT, Dachman AH. Computer-aided diagnosis scheme for the detection of polyps in CT colonography. Radio Graphics. 2002;22:963–979. doi: 10.1148/radiographics.22.4.g02jl16963. [DOI] [PubMed] [Google Scholar]
- 80.Wang ZA, Liang ZR, Li LH, et al. Reduction of false positives by internal features for polyp detection in CT-based virtual colonoscopy. Med Phys. 2005;32:3602–3616. doi: 10.1118/1.2122447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Göktürk SB, Tomasi C, Acar B, et al. A statistical 3-D pattern processing method for computer-aided detection of polyps in CT colonography. IEEE Trans Med Imaging. 2001;20:1251–1260. doi: 10.1109/42.974920. [DOI] [PubMed] [Google Scholar]
- 82.Acar B, Beaulieu CF, Göktürk SB, et al. Edge displacement field-based classification for improved detection of polyps in CT colonography. IEEE Trans Med Imaging. 2002;21:1461–1467. doi: 10.1109/TMI.2002.806405. [DOI] [PubMed] [Google Scholar]
- 83.Sundaram P, Zomorodian A, Beaulieu C, Napel S. Colon polyp detection using smoothed shape operators: preliminary results. Med Image Anal. 2008;12:99–119. doi: 10.1016/j.media.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 84.Mang T, Peloschek P, Plank C, et al. Effect of computer-aided detection as a second reader in multidetector-row CT colonography. Eur Radiol. 2007;17:2598–2607. doi: 10.1007/s00330-007-0608-z. [DOI] [PubMed] [Google Scholar]
- 85.Suzuki K. Machine learning in computer-aided diagnosis of the thorax and colon in CT: a survey. IEICE Trans Inf Syst. 2013;E96D:772–783. doi: 10.1587/transinf.e96.d.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang S, Summers RM. Machine learning and radiology. Med Image Anal. 2012;16:933–951. doi: 10.1016/j.media.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Summers RM. Improving the accuracy of CTC interpretation: computer-aided detection. Gastrointest Endosc Clin N Am. 2010;20:245–257. doi: 10.1016/j.giec.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boone D, Mallett S, McQuillan J, Taylor SA, Altman DG, Halligan S. Assessment of the incremental benefit of computer-aided detection (CAD) for interpretation of CT colonography by experienced and inexperienced readers. PLoS One. 2015;10:e0136624. doi: 10.1371/journal.pone.0136624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu HB, Fan Y, Lu HB, Liang ZR. Improved curvature estimation for computer-aided detection of colonic polyps in CT colonography. Acad Radiol. 2011;18:1024–1034. doi: 10.1016/j.acra.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aman JM, Yao JH, Summers RM. Prediction of polyp histology on CT colonography using content-based image retrieval. In: Karssemeijer N, Summers RM, editors. Medical imaging 2010: computer-aided diagnosis—proceedings of SPIE. Vol. 7624. Bellingham, WA: SPIE; 2010. p. 76240D. [Google Scholar]
- 91.Song B, Zhang G, Lu H, et al. Volumetric texture features from higher-order images for diagnosis of colon lesions via CT colonography. Int J Comput Assist Radiol Surg. 2014;9:1021–1031. doi: 10.1007/s11548-014-0991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu JM, Kabadi S, Van Uitert R, Petrick N, Deriche R, Summers RM. Improved computer-aided detection of small polyps in CT colonography using interpolation for curvature estimation. Med Phys. 2011;38:4276–4284. doi: 10.1118/1.3596529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Näppi J, Yoshida H. Adaptive correction of the pseudo-enhancement of CT attenuation for fecal-tagging CT colonography. Med Image Anal. 2008;12:413–426. doi: 10.1016/j.media.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsagaan B, Näppi J, Yoshida H. Nonlinear regression-based method for pseudoenhancement correction in CT colonography. Med Phys. 2009;36:3596–3606. doi: 10.1118/1.3147201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, Yao J, Summers RM. Scale-based scatter correction for computer-aided polyp detection in CT colonography. Med Phys. 2008;35:5664–5671. doi: 10.1118/1.3013552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Summers RM, Handwerker LR, Pickhardt PJ, et al. Performance of a previously validated CT colonography computer-aided detection system in a new patient population. AJR. 2008;191:168–174. doi: 10.2214/AJR.07.3354. [DOI] [PubMed] [Google Scholar]
- 97.Halligan S, Mallett S, Altman DG, et al. Incremental benefit of computer-aided detection when used as a second and concurrent reader of CT colonographic data: multiobserver study. Radiology. 2011;258:469–476. doi: 10.1148/radiol.10100354. [DOI] [PubMed] [Google Scholar]
- 98.Taylor SA, Charman SC, Lefere P, et al. CT colonography: investigation of the optimum reader paradigm by using computer-aided detection software. Radiology. 2008;246:463–471. doi: 10.1148/radiol.2461070190. [DOI] [PubMed] [Google Scholar]
- 99.Yoshida H, Nappi J. CAD in CT colonography without and with oral contrast agents: progress and challenges. Comput Med Imaging Graph. 2007;31:267–284. doi: 10.1016/j.compmedimag.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 100.Zalis ME, Blake MA, Cai W, et al. Diagnostic accuracy of laxative-free computed tomographic colonography for detection of adenomatous polyps in asymptomatic adults: a prospective evaluation. Ann Intern Med. 2012;156:692–702. doi: 10.7326/0003-4819-156-10-201205150-00005. [DOI] [PubMed] [Google Scholar]
- 101.Cai W, Lee JG, Zalis ME, Yoshida H. Mosaic decomposition: an electronic cleansing method for inhomogeneously tagged regions in noncathartic CT colonography. IEEE Trans Med Imaging. 2011;30:559–574. doi: 10.1109/TMI.2010.2087389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lu L, Jian B, Wu DJ, Wolf M. A new algorithm of electronic cleansing for weak faecal-tagging CT colonography. In: Wu G, Zhang D, Shen D, Yan P, Suzuki K, Wang F, editors. Lecture notes in computer science, vol. 8184. Machine learning in medical imaging. Geneva, Switzerland: Springer International Publishing; 2013. pp. 57–65. [Google Scholar]
- 103.Cai W, Kim SH, Lee JG, Yoshida H. Informatics in radiology: dual-energy electronic cleansing for fecal-tagging CT colonography. RadioGraphics. 2013;33:891–912. doi: 10.1148/rg.333125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cai W, Lee JG, Zhang D, Kim SH, Zalis M, Yoshida H. Electronic cleansing in fecal-tagging dual-energy CT colonography based on material decomposition and virtual colon tagging. IEEE Trans Biomed Eng. 2015;62:754–765. doi: 10.1109/TBME.2014.2364837. [DOI] [PubMed] [Google Scholar]
- 105.Tachibana R, Nappi JJ, Yoshida H. Application of pseudo-enhancement correction to virtual monochromatic CT colonography. Abdom Imaging. 2014;8676:169–178. doi: 10.1007/978-3-319-13692-9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nguyen TB, Wang SJ, Anugu V, et al. Distributed human intelligence for colonic polyp classification in computer-aided detection for CT colonography. Radiology. 2012;262:824–833. doi: 10.1148/radiol.11110938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McKenna MT, Wang SJ, Nguyen TB, Burns JE, Petrick N, Summers RM. Strategies for improved interpretation of computer-aided detections for CT colonography utilizing distributed human intelligence. Med Image Anal. 2012;16:1280–1292. doi: 10.1016/j.media.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang A, Roy D, Franaszek M, Summers RM. Proceedings of the IEEE visualization conference. Minneapolis, MN: IEEE; 2005. Teniae coli guided navigation and registration for virtual colonoscopy; pp. 279–285. [Google Scholar]
- 109.Huang A, Roy DA, Summers RM, et al. Teniae coli-based circumferential localization system for CT colonography: feasibility study. Radiology. 2007;243:551–560. doi: 10.1148/radiol.2432060353. [DOI] [PubMed] [Google Scholar]
- 110.Huang A, Summers RM, Roy D. Synchronous navigation for CT colonography. In: Manduca A, Amini AA, editors. Medical imaging 2006: physiology, function, and structure from medical images—proceedings of SPIE. Vol. 6143. Bellingham, WA: SPIE; 2006. p. 614315. [Google Scholar]
- 111.Hampshire T, Roth H, Hu M, et al. Automatic prone to supine haustral fold matching in CT colonography using a Markov random field model. Med Image Comput Comput Assist Interv. 2011;14:508–515. doi: 10.1007/978-3-642-23623-5_64. [DOI] [PubMed] [Google Scholar]
- 112.Roth H, McClelland J, Modat M, et al. Establishing spatial correspondence between the inner colon surfaces from prone and supine CT colonography. Med Image Comput Comput Assist Interv. 2010;13:497–504. doi: 10.1007/978-3-642-15711-0_62. [DOI] [PubMed] [Google Scholar]
- 113.Roth HR, McClelland JR, Boone DJ, et al. Registration of the endoluminal surfaces of the colon derived from prone and supine CT colonography. Med Phys. 2011;38:3077–3089. doi: 10.1118/1.3577603. [DOI] [PubMed] [Google Scholar]
- 114.Boone DJ, Halligan S, Roth HR, et al. CT colonography: external clinical validation of an algorithm for computer-assisted prone and supine registration. Radiology. 2013;268:752–760. doi: 10.1148/radiol.13122083. [DOI] [PubMed] [Google Scholar]
- 115.Helbren E, Roth HR, Hampshire TE, et al. CT colonography: clinical evaluation of a method for automatic coregistration of polyps at follow-up surveillance studies. Radiology. 2014;273:417–424. doi: 10.1148/radiol.14140473. [DOI] [PubMed] [Google Scholar]
- 116.Cherry KM, Peplinski B, Kim L, et al. Sequential Monte Carlo tracking of the marginal artery by multiple cue fusion and random forest regression. Med Image Anal. 2015;19:164–175. doi: 10.1016/j.media.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wei ZS, Yao JH, Wang SJ, et al. Feasibility of using the marginal blood vessels as reference landmarks for CT colonography. AJR. 2014;202:W50–W58. doi: 10.2214/AJR.12.10463. web. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dachman AH. Comparison of optical colonoscopy and CT colonography for polyp detection. AJR. 2009;193:1289–1290. doi: 10.2214/AJR.09.3311. [DOI] [PubMed] [Google Scholar]
- 119.Summers RM, Swift JA, Dwyer AJ, Choi JR, Pickhardt PJ. Normalized distance along the colon centerline: a method for correlating polyp location on CT colonography and optical colonoscopy. AJR. 2009;193:1296–1304. doi: 10.2214/AJR.09.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Duncan JE, McNally MP, Sweeney WB, et al. CT colonography predictably overestimates colonic length and distance to polyps compared with optical colonoscopy. AJR. 2009;193:1291–1295. doi: 10.2214/AJR.09.2365. [DOI] [PubMed] [Google Scholar]
- 121.Liu JM, Chang KW, Yao JH, Summers RM. Predicting polyp location on optical colonoscopy from CT colonography by minimal-energy curve modeling of the colonoscope path. IEEE Trans Biomed Eng. 2012;59:3531–3540. doi: 10.1109/TBME.2012.2217960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang W, Liu J, Yao J, et al. Mesenteric vasculature-guided small bowel segmentation on 3-D CT. IEEE Trans Med Imaging. 2013;32:2006–2021. doi: 10.1109/TMI.2013.2271487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nguyen T, Summers RM. Roadmapping the small bowel. [Accessed April 10, 2016];YouTube website. www.youtube.com/watch?v=8rXxMAamhg8. Published 2011.
- 124.Zhang WD, Liu JM, Yao JH, Summers RM. Reducing false positives of small bowel segmentation on CT scans by localizing colon regions. In: Aylward S, Hadjiiski LM, editors. Medical imaging 2014: computer-aided diagnosis—proceedings of SPIE. Vol. 9035. Bellingham, WA: SPIE; 2014. p. 90350Z. [Google Scholar]
- 125.Sainani NI, Nappi JJ, Sahani DV, Yoshida H. Computer-aided detection of small bowel strictures in CT enterography. In: Summers RM, van Ginneken B, editors. Medical imaging 2011: computer-aided diagnosis—proceedings of SPIE. Vol. 7963. Bellingham, WA: SPIE; 2011. p. 79632J. [Google Scholar]
- 126.Wei Z, Zhang W, Liu J, Wang S, Yao J, Summers RM. IEEE 10th international symposium on biomedical imaging. San Francisco, CA: IEEE; 2013. Computer-aided detection of colitis on computed tomography using a visual codebook; pp. 141–144. [Google Scholar]
- 127.Liu J, Lay N, Lu L, et al. Colitis detection on abdominal CT scans by rich feature hierarchies. In: Tourassi GD, Armato SG, editors. Medical imaging 2016: computer-aided diagnosis—proceedings of SPIE. Vol. 9785. Bellingham, WA: SPIE; 2016. p. 97851N. [Google Scholar]
- 128.Yao J, O’Connor SD, Summers RM. Automated spinal column extraction and partitioning. In: Kovacevic J, Meijering E, editors. 3rd IEEE international symposium on biomedical imaging: nano to macro. Arlington, VA: IEEE; 2006. pp. 390–393. [Google Scholar]
- 129.Klinder T, Ostermann J, Ehm M, Franz A, Kneser R, Lorenz C. Automated model-based vertebra detection, identification, and segmentation in CT images. Med Image Anal. 2009;13:471–482. doi: 10.1016/j.media.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 130.Kelm BM, Wels M, Zhou SK, et al. Spine detection in CT and MR using iterated marginal space learning. Med Image Anal. 2013;17:1283–1292. doi: 10.1016/j.media.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 131.Summers RM, Baecher N, Yao J, et al. Feasibility of simultaneous computed tomographic colonography and fully automated bone mineral densitometry in a single examination. J Comput Assist Tomogr. 2011;35:212–216. doi: 10.1097/RCT.0b013e3182032537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pickhardt PJ, Lee LJ, del Rio AM, et al. Simultaneous screening for osteoporosis at CT colonography: bone mineral density assessment using MDCT attenuation techniques compared with the DXA reference standard. J Bone Miner Res. 2011;26:2194–2203. doi: 10.1002/jbmr.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Carberry GA, Pooler BD, Binkley N, Lauder TB, Bruce RJ, Pickhardt PJ. Unreported vertebral body compression fractures at abdominal multidetector CT. Radiology. 2013;268:120–126. doi: 10.1148/radiol.13121632. [DOI] [PubMed] [Google Scholar]
- 134.Yao JH, Burns JE, Wiese T, Summers RM. Quantitative vertebral compression fracture evaluation using a height compass. In: van Ginneken B, Novak CL, editors. Medical imaging 2012: computer-aided diagnosis—proceedings of SPIE. Vol. 8315. Bellingham, WA: SPIE; 2012. p. 83151X. [Google Scholar]
- 135.Al-Helo S, Alomari RS, Ghosh S, et al. Compression fracture diagnosis in lumbar: a clinical CAD system. Int J Comput Assist Radiol Surg. 2013;8:461–469. doi: 10.1007/s11548-012-0796-0. [DOI] [PubMed] [Google Scholar]
- 136.Ghosh S, Alomari RS, Chaudhary V, Dhillon G. Automatic lumbar vertebra segmentation from clinical CT for wedge compression fracture diagnosis. In: Summers RM, van Ginneken B, editors. Medical imaging 2011: computer-aided diagnosis—proceedings of SPIE. Vol. 7963. Bellingham, WA: SPIE; 2011. p. 796303. [Google Scholar]
- 137.Wang Y, Yao J, Roth H, Burns J, Summers R. Multiatlas segmentation with joint label fusion of osteoporotic vertebral compression fractures on CT. In: Vrtovec T, Yao J, Glocker B, et al., editors. Proceedings of 3rd MICCAI workshop & challenge on computational methods and clinical applications for spine imaging. Munich, Germany: Springer; 2015. pp. 76–86. [Google Scholar]
- 138.Genant HK, Wu CY, van Kuijk C, Nevitt MC. Vertebral fracture assessment using a semiquantitative technique. J Bone Miner Res. 1993;8:1137–1148. doi: 10.1002/jbmr.5650080915. [DOI] [PubMed] [Google Scholar]
- 139.Burns JE, Yao J, Munoz H, Summers RM. Automated detection, localization, and classification of traumatic vertebral body fractures in the thoracic and lumbar spine at CT. Radiology. 2016;278:64–73. doi: 10.1148/radiol.2015142346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Denis F. The three column spine and its significance in the classification of acute thoracolumbar spinal injuries. Spine. 1983;8:817–831. doi: 10.1097/00007632-198311000-00003. [DOI] [PubMed] [Google Scholar]
- 141.Roth HR, Wang Y, Yao J, Lu L, Burns JE, Summers RM. Medical imaging 2016: computer-aided diagnosis—proceedings of SPIE. Vol. 9785. Bellingham, WA: SPIE; 2016. Deep convolutional networks for automated detection of posterior element fractures on spine CT. [Google Scholar]
- 142.Munoz HE, Yao JH, Burns JE, Pham Y, Stieger J, Summers RM. Vertebral degenerative disc disease severity evaluation using random forest classification. In: Aylward S, Hadjiiski LM, editors. Medical imaging 2014: computer-aided diagnosis—proceedings of SPIE. Vol. 9035. Bellingham, WA: SPIE; 2014. p. 90353A. [Google Scholar]
- 143.Munoz HE, Yao JH, Burns JE, Summers RM. Detection of vertebral degenerative disc disease based on cortical shell unwrapping. In: Novak CL, Aylward S, editors. Medical imaging 2013: computer-aided diagnosis—proceedings of SPIE. Vol. 8670. Bellingham, WA: SPIE; 2013. p. 86700C. [Google Scholar]
- 144.Yao J, Muno H, Burns JE, Lu L, Summers RM. Computer aided detection of spinal degenerative osteophytes on sodium fluoride PET/CT. In: Yao J, Klinder T, Li S, editors. Lecture notes in computational vision and biomechanics, vol. 17. Computational methods and clinical applications for spine imaging. Geneva, Switzerland: Springer International Publishing; 2014. pp. 51–60. [Google Scholar]
- 145.O’Connor SD, Yao JH, Summers RM. Lytic metastases in thoracolumbar spine: computer-aided detection at CT—preliminary study. Radiology. 2007;242:811–816. doi: 10.1148/radiol.2423060260. [DOI] [PubMed] [Google Scholar]
- 146.Burns JE, Yao J, Wiese TS, Munoz HE, Jones EC, Summers RM. Automated detection of sclerotic metastases in the thoracolumbar spine at CT. Radiology. 2013;268:69–78. doi: 10.1148/radiol.13121351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hammon M, Dankerl P, Tsymbal A, et al. Automatic detection of lytic and blastic thoracolumbar spine metastases on computed tomography. Eur Radiol. 2013;23:1862–1870. doi: 10.1007/s00330-013-2774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu JM, Pattanaik S, Yao JH, et al. Computer aided detection of epidural masses on computed tomography scans. Comput Med Imaging Graph. 2014;38:606–612. doi: 10.1016/j.compmedimag.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pattanaik S, Liu J, Yao J, et al. Epidural masses detection on computed tomography using spatially-constrained gaussian mixture models. In: Yao J, Klinder T, Li S, editors. Lecture notes in computational vision and biomechanics, vol. 17. Computational methods and clinical applications for spine imaging. Geneva, Switzerland: Springer International Publishing; 2014. pp. 99–108. [Google Scholar]
- 150.Meng K, Lee CH, Saremi F. Metabolic syndrome and ectopic fat deposition: what can CT and MR provide? Acad Radiol. 2010;17:1302–1312. doi: 10.1016/j.acra.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 151.Yoshizumi T, Nakamura T, Yamane M, et al. Abdominal fat: standardized technique for measurement at CT. Radiology. 1999;211:283–286. doi: 10.1148/radiology.211.1.r99ap15283. [DOI] [PubMed] [Google Scholar]
- 152.Yao J, Sussman DL, Summers RM. Medical imaging 2011: biomedical applications in molecular, structural, and functional imaging—proceedings of SPIE. Vol. 7965. Bellingham, WA: SPIE; 2011. Fully automated adipose tissue measurement on abdominal CT; p. 79651Z. [Google Scholar]
- 153.Tong Y, Udupa JK, Torigian DA. Optimization of abdominal fat quantification on CT imaging through use of standardized anatomic space: a novel approach. Med Phys. 2014;41:063501. doi: 10.1118/1.4876275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yamaji T, Iwasaki M, Sasazuki S, et al. Visceral fat volume and the prevalence of colorectal adenoma. Am J Epidemiol. 2009;170:1502–1511. doi: 10.1093/aje/kwp311. [DOI] [PubMed] [Google Scholar]
- 155.Johnson KT, Harmsen WS, Limburg PJ, Carston MJ, Johnson CD. Visceral fat analysis at CT colonography. Acad Radiol. 2006;13:963–968. doi: 10.1016/j.acra.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 156.Ryckman EM, Summers RM, Liu J, Munoz del Rio A, Pickhardt PJ. Visceral fat quantification in asymptomatic adults using abdominal CT: is it predictive of future cardiac events? Abdom Imaging. 2015;40:222–226. doi: 10.1007/s00261-014-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Summers RM, Liu JM, Sussman DL, et al. Association between visceral adiposity and colorectal polyps on CT colonography. AJR. 2012;199:48–57. doi: 10.2214/AJR.11.7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu JM, Pattanaik S, Yao JH, et al. Associations among pericolonic fat, visceral fat, and colorectal polyps on CT colonography. Obesity (Silver Spring, Md) 2015;23:408–414. doi: 10.1002/oby.20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Boutin RD, Yao L, Canter RJ, Lenchik L. Sarcopenia: current concepts and imaging implications. AJR. 2015;205:W255–W266. doi: 10.2214/AJR.15.14635. web. [DOI] [PubMed] [Google Scholar]
- 160.Psutka SP, Boorjian SA, Moynagh MR, et al. Decreased skeletal muscle mass is associated with an increased risk of mortality after radical nephrectomy for localized renal cell cancer. J Urol. 2016;195:270–276. doi: 10.1016/j.juro.2015.08.072. [DOI] [PubMed] [Google Scholar]
- 161.Psutka SP, Boorjian SA, Moynagh MR, et al. Mortality after radical cystectomy: impact of obesity versus adiposity after adjusting for skeletal muscle wasting. J Urol. 2015;193:1507–1513. doi: 10.1016/j.juro.2014.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Psutka SP, Carrasco A, Schmit GD, et al. Sarcopenia in patients with bladder cancer undergoing radical cystectomy: impact on cancer-specific and all-cause mortality. Cancer. 2014;120:2910–2918. doi: 10.1002/cncr.28798. [DOI] [PubMed] [Google Scholar]
- 163.Tamandl D, Paireder M, Asari R, Baltzer PA, Schoppmann SF, Ba-Salamah A. Markers of sarcopenia quantified by computed tomography predict adverse long-term outcome in patients with resected oesophageal or gastro-oesophageal junction cancer. Eur Radiol. 2015 Sep 3; doi: 10.1007/s00330-015-3963-1. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 164.Prado CM, Lieffers JR, McCargar LJ, et al. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: a population-based study. Lancet Oncol. 2008;9:629–635. doi: 10.1016/S1470-2045(08)70153-0. [DOI] [PubMed] [Google Scholar]
- 165.Zhang WD, Liu JM, Yao JH, Summers RM. Segmenting the thoracic, abdominal and pelvic musculature on CT scans combining atlas-based model and active contour model. In: Novak CL, Aylward S, editors. Medical imaging 2013: computer-aided diagnosis—proceedings of SPIE. Vol. 8670. Bellingham, WA: SPIE; 2013. p. 867008. [Google Scholar]
- 166.Isgum I, van Ginneken B, Olree M. Automatic detection of calcifications in the aorta from CT scans of the abdomen: 3D computer-aided diagnosis. Acad Radiol. 2004;11:247–257. doi: 10.1016/s1076-6332(03)00673-1. [DOI] [PubMed] [Google Scholar]
- 167.Zhang WD, Liu JM, Yao JH, Summers RM. IEEE 10th international symposium on biomedical imaging. San Francisco, CA: IEEE; 2013. Automatic anatomical labeling of abdominal arteries for small bowel evaluation on 3D CT scans; pp. 210–213. [Google Scholar]
- 168.Matsuzaki T, Oda M, Kitasaka T, Hayashi Y, Misawa K, Mori K. A method for automated anatomical labeling of abdominal veins extracted from 3D CT images. In: Ourselin S, Haynor DR, editors. Medical imaging 2013: image processing—proceedings of SPIE. Vol. 8669. Bellingham, WA: SPIE; 2013. p. 86691Y. [Google Scholar]
- 169.Mori K, Oda M, Egusa T, et al. Automated nomenclature of upper abdominal arteries for displaying anatomical names on virtual laparoscopic images. In: Liao H, Edwards PJ, Pan X, Fan Y, Yang GZ, editors. Lecture notes in computer science, vol. 6326. Medical imaging and augmented reality. Heidelberg, Germany: Springer; 2010. pp. 353–362. [Google Scholar]
- 170.Morin-Roy F, Kauffmann C, Tang A, et al. Impact of contrast injection and stent-graft implantation on reproducibility of volume measurements in semiautomated segmentation of abdominal aortic aneurysm on computed tomography. Eur Radiol. 2014;24:1594–1601. doi: 10.1007/s00330-014-3175-0. [DOI] [PubMed] [Google Scholar]
- 171.Liu JM, Zhao J, Hoffman J, et al. Mediastinal lymph node detection on thoracic CT scans using spatial prior from multi-atlas label fusion. In: Aylward S, Hadjiiski LM, editors. Medical imaging 2014: computer-aided diagnosis—proceedings of SPIE. Vol. 9035. Bellingham, WA: SPIE; 2014. p. 90350M. [Google Scholar]
- 172.Cherry KM, Wang SJ, Turkbey EB, Summers RM. Abdominal lymphadenopathy detection using random forest. In: Aylward S, Hadjiiski LM, editors. Medical imaging 2014: computer-aided diagnosis—proceedings of SPIE. Vol. 9035. Bellingham, WA: SPIE; 2014. p. 90351G. [Google Scholar]
- 173.Thong W, Kadoury S, Piche N, Pal CJ. 1st workshop on deep learning in medical image analysis (DALMIA2015) Munich, Germany: Springer; 2015. Convolutional networks for kidney segmentation in contrast-enhanced CT scans; pp. 9–16. [Google Scholar]
- 174.Shin HC, Lu L, Kim L, Seff A, Yao J, Summers RM. 2015 IEEE conference on computer vision and pattern recognition (CVPR) Boston, MA: IEEE; 2015. Interleaved text/image deep mining on a large-scale radiology database; pp. 1090–1099. [Google Scholar]
- 175.Pathak SD, Kim W, Munasinghe I, Criminisi A, White S, Siddiqui K. Linking DICOM pixel data with radiology reports using automatic semantic annotation. In: Boonn WW, Liu BJ, editors. Medical imaging 2012: advanced pacs-based imaging informatics and therapeutic applications—proceedings of SPIE. Vol. 8319. Bellingham, WA: SPIE; 2012. p. 83190D. [Google Scholar]
- 176.Bagci U, Udupa JK, Mendhiratta N, et al. Joint segmentation of anatomical and functional images: applications in quantification of lesions from PET, PET-CT, MRI-PET, and MRI-PET-CT images. Med Image Anal. 2013;17:929–945. doi: 10.1016/j.media.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.The Cancer Imaging Archive. [Accessed December 14, 2015]; www.cancerimagingarchive.net/
- 178.Visceral. [Accessed December 14, 2015]; www.visceral.eu/
- 179.Roth HR, Summers RM. CT lymph nodes. [Accessed December 14, 2015];Cancer imaging archive website. wiki.cancerimagingarchive.net/display/Public/CT+Lymph+Nodes.
- 180.Xu Z. Multi-atlas labeling beyond the cranial vault: workshop and challenge. [Accessed December 11, 2015];Synapse website. www.synapse.org/#!Synapse:syn3193805/wiki/217789.
- 181.Lake BM, Salakhutdinov R, Tenenbaum JB. Human-level concept learning through probabilistic program induction. Science. 2015;350:1332–1338. doi: 10.1126/science.aab3050. [DOI] [PubMed] [Google Scholar]
- 182.Welling RD, Azene EM, Kalia V, et al. White paper report of the 2010 RAD-AID conference on international radiology for developing countries: identifying sustainable strategies for imaging services in the developing world. J Am Coll Radiol. 2011;8:556–562. doi: 10.1016/j.jacr.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Summers RM. How perceptual factors affect the use and accuracy of CAD for interpretation of CT images. In: Samei E, Krupinski E, editors. The handbook of medical image perception and techniques. chapter 23. Cambridge, UK: Cambridge University Press; 2009. pp. 313–321. [Google Scholar]
- 184.Petrick N, Sahiner B, Armato SG, et al. Evaluation of computer-aided detection and diagnosis systems. Med Phys. 2013;40:087001. doi: 10.1118/1.4816310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huo Z, Summers RM, Paquerault S, et al. Quality assurance and training procedures for computer-aided detection and diagnosis systems in clinical use. Med Phys. 2013;40:077001. doi: 10.1118/1.4807642. [DOI] [PMC free article] [PubMed] [Google Scholar]