Abstract
The long noncoding RNA SChLAP1 is overexpressed in a subset of prostate cancers (PCa), and high SChLAP1 expression according to in situ hybridization (ISH) independently predicts biochemical recurrence after radical prostatectomy. Importantly, although biochemical recurrence is a significant clinical outcome, it is not a validated surrogate for PCa-related mortality. Thus, we evaluated the association between SChLAP1 expression and development of lethal PCa in a large cohort of American men with PCa and long-term follow-up. SChLAP1 ISH was performed on tissue microarrays containing representative formalin-fixed, paraffin-embedded PCa tissue from all patients and scored using a semiquantitative method (ISH score range 0–400). Hazard ratios (HRs) for the association between SChLAP1 expression and time to development of lethal PCa were estimated using multivariable Cox regression analysis. Of the 937 patients evaluated, 89 (9.5%) had high SChLAP1 expression (ISH score ≥100), which in patients treated with radical prostatectomy was strongly associated with development of lethal PCa independent of age, Gleason score, pathologic stage, and PTEN status (HR 2.2, 95% confidence interval 1.1–4.1). These results suggest that SChLAP1 may be a useful tissue-based biomarker for identifying PCa patients at higher risk of lethal progression.
Keywords: SChLAP1; Metastatic prostate cancer, long noncoding RNA; In situ hybridization; Lethal prostate cancer
Numerous long noncoding RNAs (lncRNAs) have recently emerged as targets for clinical translation [1,2]. We previously identified 121 novel, differentially expressed prostate cancer (PCa) lncRNAs, including SChLAP1, which is overexpressed in a subset of tumors and associated with more aggressive disease [3–5]. We subsequently validated a novel RNA in situ hybridization (ISH) assay for detection of SChLAP1 in formalin-fixed, paraffin-embedded (FFPE) tissue and used this assay to demonstrate that high SChLAP1 expression independently predicts biochemical PCa recurrence (ie, prostate-specific antigen [PSA] relapse) after radical prostatectomy in patients with clinically localized PCa [6]. Although biochemical recurrence almost universally precedes clinical disease recurrence, many PCa patients with biochemical recurrence do not develop lethal disease [7]. Given the predictive value of SChLAP1 ISH for biochemical recurrence after radical prostatectomy, we sought to investigate whether high SChLAP1 expression on ISH is also associated with development of lethal PCa (PCa-specific mortality or distant metastases).
We evaluated whether tumor SChLAP1 expression is associated with lethal PCa among PCa cases from the Physicians’ Health Study (PHS) and the Health Professionals Follow-up Study (HPFS). ISH was performed [6] on tumor tissue available from a biorepository of archival prostatectomy tissue. Cox proportional hazard models were used to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between SChLAP1 expression and time to development of lethal PCa (defined as cancer-related death or metastases to bone and/or other organs). Further details of the statistical methods are provided in Supplementary Methods. SChLAP1 ISH data were available for 937 PCa patients (607 from the HPFS and 330 from the PHS). Clinicopathologic characteristics for the study population are shown in Supplementary Table 1. Concordant with our previous findings [6], SChLAP1 expression varied among patient samples (Fig. 1). We previously validated a SChLAP1 ISH score threshold of 100 (range 0–400) as a statistically significant predictor of PSA relapse for clinically localized PCa [6]. Using this threshold we categorized cases as high (ISH score ≥100) or low (ISH score <100) SChLAP1 expression and examined the association between SChLAP1 expression and selected clinicopathologic parameters (Table 1). High SChLAP1 expression was associated with higher Gleason score (GS), pathologic tumor stage, and clinical tumor stage (p < 0.005), and with loss of the tumor suppressor PTEN (as assessed by immunohistochemistry; p < 0.0001).
Illustration.
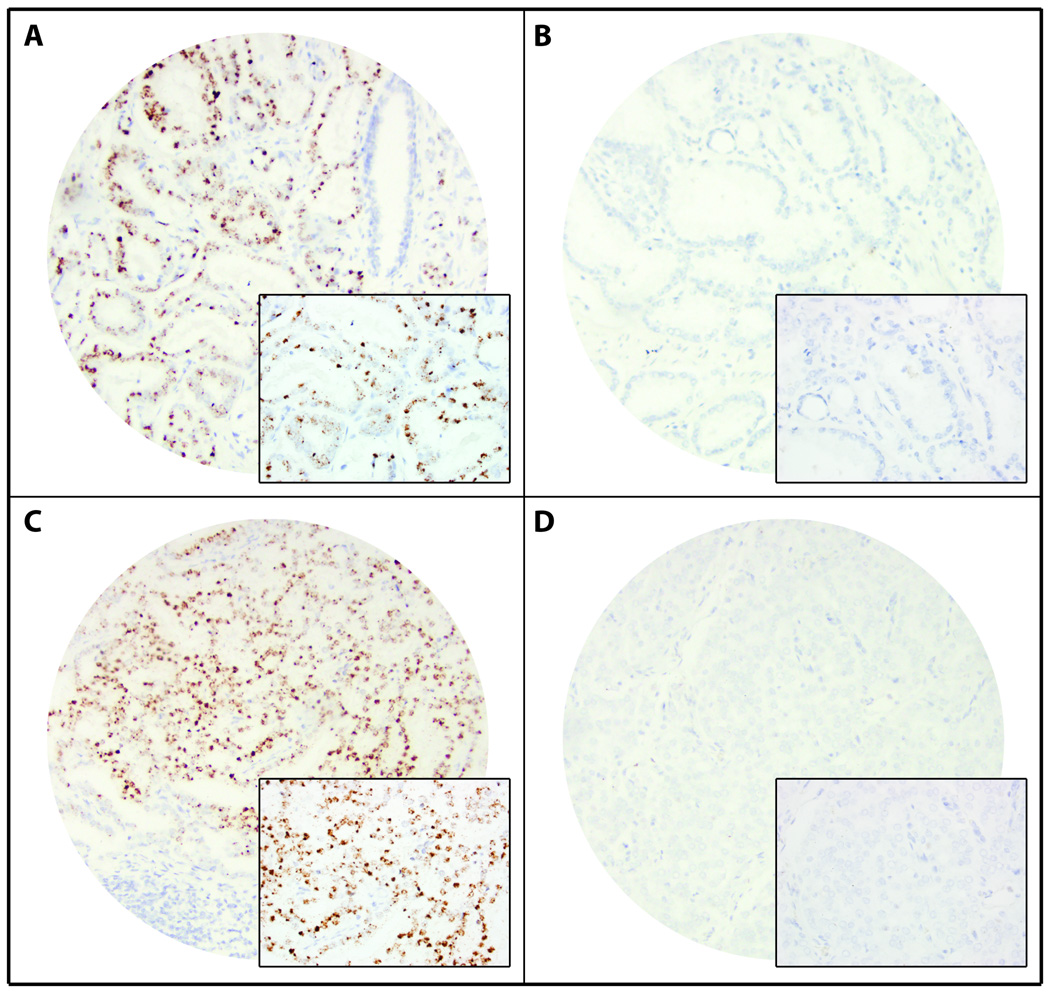
SChLAP1 expression in prostate cancer by in situ hybridization (ISH). Representative images of SChLAP1 ISH demonstrating (A,C) high or (B,D) low SChLAP1 expression in (A,B) low-grade and (C,D) high-grade prostate cancer. Magnification 100× (inset 400×).
Table 1.
Association between SChLAP1 expression by ISH and selected clinicopathologic characteristics among men diagnosed with prostate cancer between 1983 and 2011 in a combined PHS (n = 330) and HPFS (n = 607) cohort
| Characteristic | Overall | SChLAP1 ISH score |
p value a | |
|---|---|---|---|---|
| Low (<100) | High (≥100) | |||
| Participants, n (%) | 937 (100) | 848 (90.5) | 89 (9.5) | |
| Age at diagnosis, yr (mean ± SD) | 66.1 ± 5.9 | 66.0 ± 5.9 | 67.2 ± 5.5 | 0.07 |
| BMI at diagnosis, kg/m2 (mean ± SD) | 25.8 ± 3.5 | 25.9 ± 3.5 | 25.3 ± 3.0 | 0.18 |
| Median PSA at diagnosis, ng/ml (IQR) | 6.5 (4.7–10.0) | 6.5 (4.7–10.0) | 6.0 (4.3–9.7) | 0.35 |
| Gleason score | ||||
| ≤6 | 165 (17.6) | 161 (19.0) | 4 (4.5) | <0.0001 |
| 3 + 4 | 334 (35.7) | 323 (38.1) | 11 (12.4) | |
| 4 + 3 | 227 (24.2) | 193 (22.8) | 34 (38.2) | |
| 8–10 | 211 (22.5) | 171 (20.2) | 40 (44.9) | |
| Pathologic stage, n (%) | ||||
| T2N0/NxM0/Mx | 607 (71.0) | 567 (73.1) | 40 (50.6) | <0.0001 |
| T3N0/NxM0/Mx | 221 (26.0) | 187 (24.2) | 34 (43.0) | |
| T4N1M1 | 25 (3.0) | 20 (2.6) | 5 (6.3) | |
| Clinical stage, n (%) | ||||
| T1/T2N0/NxM0/Mx | 868 (94.3) | 792 (95.2) | 76 (85.4) | 0.002 |
| T3N0/NxM0/Mx | 32 (3.5) | 23 (2.8) | 9 (10.1) | |
| T4/N1/M1 | 21 (2.3) | 17 (2.0) | 4 (4.5) | |
| Specimen type, n (%) | ||||
| Radical prostatectomy | 882 (94.1) | 800 (94.3) | 82 (92.1) | 0.40 |
| TURP | 55 (5.9) | 48 (5.7) | 7 (7.9) | |
| PTEN IHC status, n (%) | ||||
| Intact | 640 (83.0) | 595 (86.1) | 45 (56.3) | <0.0001 |
| Loss | 131 (17.0) | 96 (13.9) | 35 (43.8) | |
PHS = Physicians’ Health Study; HPFS = Health Professionals Follow-up Study; ISH = in situ hybridization; BMI = body mass index; SD = standard deviation; PSA = prostate-specific antigen; TURP = transurethral resection of the prostate; IHC = immunohistochemistry.
Age and BMI at diagnosis between SChLAP1 groups were compared using Student’s t test; PSA level at diagnosis was compared using the Wilcoxon rank-sum test; specimen type and PTEN IHC status were compared using the χ2 test; and Gleason score, pathologic stage, and clinical stage were compared using the Cochran-Armitage test for trends.
We then evaluated the association between SChLAP1 expression and time to development of lethal PCa. Overall, there were 88 lethal PCa events (9.4% of patients; mean follow-up time 12.8 yr). High SChLAP1 expression was associated with a higher risk of lethal PCa in the age-adjusted model (HR 3.3, 95% CI 2.0–5.5; Supplementary Table 2) and remained suggestively associated with lethal PCa after adjusting for GS and clinical stage (HR 1.5, 95% CI 0.9–2.5; Supplementary Table 2). For patients who underwent radical prostatectomy, high SChLAP1 expression was significantly associated with a higher risk of lethal PCa independent of age at diagnosis, GS, and pathologic stage (HR 2.1, 95% CI 1.1–3.8; Supplementary Table 2). High SChLAP1 expression was similarly independently associated with PCa-specific death (Supplementary Table 3). Adjusting for PTEN status did not meaningfully alter the association between SChLAP1 expression and lethal PCa (HR 2.2, 95% CI 1.1–4.1; Supplementary Table 4). In all multivariable models, GS and tumor stage were significantly associated with the outcome. High SChLAP1 expression was associated with lethal PCa among patients with non-advanced clinical tumor stage (T1/T2N0/NxM0/Mx; HR 2.1, 95% CI 1.1–3.7) but not among patients with advanced clinical tumor stage (T3/4N1M1). Similarly, high SChLAP1 expression was associated with lethal PCa among patients with relatively low-grade tumors (GS ≤7; HR 4.0, 95% CI 1.7–9.3) but not among patients with high-grade tumors (GS >7; Supplementary Table 5).
In the current study, we found high SChLAP1 expression on ISH was associated with lethal PCa independent of GS and pathologic stage. We also found that SChLAP1 expression was associated with lethal PCa independent of tumor PTEN status, a known poor prognostic factor for PCa patients [8,9]. Furthermore, the association between high SChLAP1 expression and lethal PCa was most apparent in patients with non-advanced clinical stage and relatively low-grade tumors; owing to our limited sample size in these stratified analyses, however, these findings need to be interpreted with caution. Therefore, our results indicate that further clinical investigation of SChLAP1 is warranted in the drive to improve patient risk stratification and identify patients who might benefit from adjuvant therapy after radical prostatectomy. Further investigation of SChLAP1 could also improve identification of patients with non-advanced clinical stage and relatively low-grade tumors who might be candidates for active surveillance. A recent study reported higher SChLAP1 expression in PCa lymph node metastases, but no prognostic association between SChLAP1 ISH and biochemical recurrence or cancer-specific mortality was detected [10]. The lack of predictive value in that study may be due to differences in cohort characteristics and composition and/or the relatively larger number of patients in our current cohort. Furthermore, Bottcher et al [10] used a simple binary definition for SChLAP1 expression by ISH (positive or negative), whereas we used a semiquantitative scoring method (ISH score range 0–400) and empirically defined a threshold for high SChLAP1 expression (ISH score ≥100) [10]. Additional studies are needed to establish the reproducibility of our semiquantitative/threshold approach to SChLAP1 ISH evaluation before incorporation into routine clinical practice.
The strengths of this study include the relatively large cohort size and the long follow-up allowing assessment of PCa-specific mortality, while the limitations include the relatively low overall rate of lethal events and the TMA-based study design.
In summary, high SChLAP1 expression on ISH independently predicts lethal PCa in a large cohort of American men with PCa during long-term follow-up, suggesting that SChLAP1 ISH is a promising biomarker for identifying patients at higher risk of lethal PCa progression.
Supplementary Material
Home Message.
High SChLAP1 expression by ISH independently predicts lethal disease in a large cohort of American men with prostate cancer and long-term follow-up, suggesting that SChLAP1 ISH may be a promising biomarker for identifying patients at higher risk of lethal progression.
Acknowledgments
We are grateful to the participants and staff of the PHS and HPFS for their valuable contributions. In addition, we would like to thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. The Dana-Farber/Harvard Cancer Center Tissue Microarray Core Facility constructed the TMAs used in this project, and we would like to thank Chungdak Li for her expertise.
Financial disclosures: Arul M. Chinnaiyan certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Felix Y. Feng serves on advisory boards for GenomeDx Biosciences and Nanostring Technologies. The University of Michigan has filed a patent on lncRNAs in prostate cancer, including SChLAP1, in which Arul M. Chinnaiyan is named as a co-inventor. GenomeDx Biosciences has a license for detection SChLAP1 for molecular analysis of clinical prostate cancer tissue samples.
Funding/Support and role of the sponsor: This work was supported in part by the Prostate Cancer Foundation (R.M., F.Y.F., L.A.M., and A.M.C.), as well as the National Cancer Institute Prostate SPORE (P50 CA69568 to A.M.C. and P50 CA090381 to P.W.K. and L.A.M.) and Early Detection Research Network (UO1 CA113913 to A.M.C.). Additional support for the PHS and HPFS cohorts is provided by the National Cancer Institute (P01 CA055075, R01 CA133891, R01 CA141298, and UM1 CA167552). The sponsors played a role in the design and conduct of the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: Arul M. Chinnaiyan had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Mehra, Mucci, Chinnaiyan.
Acquisition of data: Mehra, Chinnaiyan, Udager, X Cao.
Analysis and interpretation of data: Mehra, Udager, Ahearn, Petimar, Mucci.
Drafting of the manuscript: Mehra, Chinnaiyan, Udager, Ahearn, Petimar, Mucci.
Critical revision of the manuscript for important intellectual content: Mehra, Chinnaiyan, Udager, Feng, Loda, Kantoff.
Statistical analysis: Ahearn, Petimar, Mucci.
Obtaining funding: Mehra, Chinnaiyan, Mucci.
Administrative, technical, or material support: Mehra, Chinnaiyan, Udager, Cao.
Supervision: Mehra, Chinnaiyan, Mucci.
Other: None.
The remaining authors have nothing to disclose.
References
- 1.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Udager AM, Alva A, Mehra R. Current and proposed molecular diagnostics in a genitourinary service line laboratory at a tertiary clinical institution. Cancer J. 2014;20:29–42. doi: 10.1097/PPO.0000000000000017. [DOI] [PubMed] [Google Scholar]
- 3.Prensner JR, Zhao S, Erho N, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prensner JR, Iyer MK, Sahu A, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lncRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehra R, Shi Y, Udager AM, et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16:1121–1127. doi: 10.1016/j.neo.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 8.Cuzick J, Yang ZH, Fisher G, et al. Prognostic value of PTEN loss in men with conservatively managed localised prostate cancer. Br J Cancer. 2013;108:2582–2589. doi: 10.1038/bjc.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lotan TL, Gurel B, Sutcliffe S, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17:6563–6573. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottcher R, Hoogland AM, Dits N, et al. Novel long non-coding RNAs are specific diagnostic and prognostic markers for prostate cancer. Oncotarget. 2015;6:4036–4050. doi: 10.18632/oncotarget.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.