Abstract
We report the main characteristics of Haloferax massiliensis strain Arc-HrT (= CSUR P974) isolated from stool specimen of a 22-year-old Amazonian obese female patient.
Keywords: Culturomics, genomics, Haloferax massiliensis, taxonogenomics, taxonomy
In December 2013, we successfully isolated the strain Arc-Hr from a stool specimen of a 22-year-old Amazonian obese female patient. Strain Arc-Hr was isolated through the culturomics study, which aims to cultivate and isolate all prokaryotes colonizing the human gut [1]. The patient provided a signed informed consent, and the agreements of the National Ethics Committee of the IFR48 (Marseille, France) were obtained under number 09-022. Strain Arc-Hr was not correctly identified using our matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) on a Microflex spectrometer (Bruker Daltonics, Leipzig, Germany) [2].
The isolate of the strain Arc-Hr was obtained by aerobic culture of the stool specimen into 100 mL of a homemade liquid medium [3] in a flask incubated at 37°C with stirring at low speed. The pure culture of this halophilic archaea grew aerobically after 7-day incubation at 37°C. Strain Arc-Hr exhibits positive catalase and oxidase activities. The growing colonies on the halophilic medium were red, opaque and 0.5 to 1 mm in diameter. Cells were Gram-negative cocci, nonmotile and non–spore forming with a diameter of 0.9 μm. The 16S rRNA gene was sequenced using the primers as previously described [4] using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France). Strain Arc-Hr exhibited 99.3% sequence similarity with Haloferax volcanii JCM 8879 (GenBank accession no. NR113448) [5], the phylogenetically closest validated species with standing in nomenclature (Fig. 1) [6]. Because 16S rRNA gene sequence comparison has been proven to poorly discriminate Haloferax species, we sequenced the complete genome of strain Arc-Hr. Genome comparison demonstrated that the draft genome of the strain Arc-Hr is larger than those of Haloferax volcanii (NR113448) Haloferax gibbonsii (NR113441), Haloferax lucentense (EU308258) and Haloferax alexandrinus (NR113438) (4.35, 3.9, 3.62, 2.95 and 2.85 Mb respectively). The G+C content of strain Arc-Hr is smaller than those of H. alexandrinus, H. lucentense, H. volcanii and H. gibbonsii (65.36, 66, 66.4, 66.6 and 67.1% respectively). The gene content of the strain Arc-Hr is larger than those of H. alexandrinus, H. lucentense, H. gibbonsii and H. volcanii (3911, 3770, 3593, 2997 and 2917 coding genes, respectively). On the basis of this genome comparison information, we propose to classify the strain Arc-Hr as a new member of the genus Haloferax within the family Haloferacaceae in the Euryarchaeota phylum [7], [8], and consequently, we formally propose the creation of Haloferax massiliensis sp. nov., with strain Arc-Hr as the type strain of Haloferax massiliensis sp. nov.
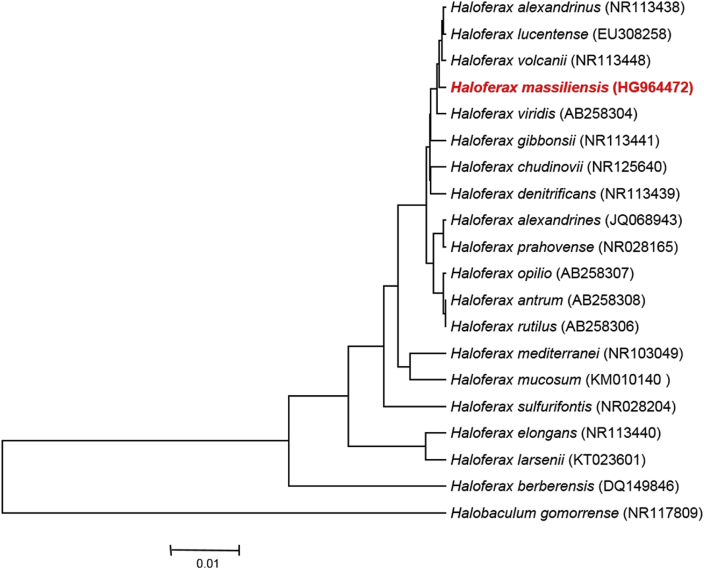
Fig. 1.
Phylogenetic tree showing position of Haloferax massiliensis strain Arc-HrT (= CSUR P974) relative to other phylogenetically close members of family Haloferacaceae. GenBank accession numbers are indicated in parentheses. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values (≥95%) obtained by repeating analysis 500 times to generate majority consensus tree. Scale bar indicates 1% nucleotide sequence divergence.
MALDI-TOF MS spectrum
The MALDI-TOF MS spectrum of Haloferax massiliensis strain Arc-HrT is available at http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database.
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in GenBank under accession number HG964472.
Deposit in a culture collection
Strain Arc-HrT was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under number CSUR P974.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khelaifia S., Lagier J.C., Bibi F., Azhar E.I., Croce O., Padmanabhan R. Microbial culturomics to map halophilic bacterium in human gut: genome sequence and description of Oceanobacillus jeddahense sp. nov. OMICS. 2016;20:248–258. doi: 10.1089/omi.2016.0004. [DOI] [PubMed] [Google Scholar]
- 4.Asker D., Ohta Y. Haloferax alexandrinus sp. nov., an extremely halophilic canthaxanthin-producing archaeon from a solar saltern in Alexandria (Egypt) Int J Syst Evol Microbiol. 2002;52:729–738. doi: 10.1099/00207713-52-3-729. [DOI] [PubMed] [Google Scholar]
- 5.Hartman A.L., Norais C., Badger J.H., Delmas S., Haldenby S., Madupu R. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010;19 doi: 10.1371/journal.pone.0009605. 5:e9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Euzeby J. Validation list n° 143. Int J Syst Evol Microbiol. 2012;62:1–4. [Google Scholar]
- 7.Gupta R.S., Naushad S., Baker S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int J Syst Evol Microbiol. 2015;65:1050–1069. doi: 10.1099/ijs.0.070136-0. [DOI] [PubMed] [Google Scholar]
- 8.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]