Abstract
Active DNA repair pathways are crucial for preserving genomic integrity and are likely among the complex mechanisms involved in the normal development of preimplantation embryos. MicroRNAs (miRNA), short non-coding RNAs, are key regulators of gene expression through the post-transcriptional and post-translational modification of mRNA. The association of miRNA expression with infertility or polycystic ovarian syndrome has been widely investigated; however, there are limited data regarding the importance of miRNA regulation in DNA repair during preimplantation embryo development. In this article, we review normal miRNA biogenesis and consequences of aberrant miRNA expression in the regulation of DNA repair in gametes and preimplantation embryos.
Keywords: DNA repair, Embryo development, miRNA, miRNA biogenesis, mRNA expression
Mammalian preimplantation embryo development follows a series of critical events. These events begin at gametogenesis and continue until parturition. Male and female gametes are derived from primordial germ cells during spermatogenesis and oogenesis, respectively. Following fertilization, oocyte and sperm nuclei fuse, resulting in syngamy. In preimplantation embryos, because the cell cycle is short, the risk of genetic errors during replication or segregation is increased [1]. Once DNA damage is detected, cells may undergo apoptosis or activate different DNA repair mechanisms, such as base excision repair (BER), nucleotide excision repair (NER), double strand break repair (DSBR) and mismatch repair (MMR) [2,3,4, 5]. Poor activation of DNA repair may affect embryo implantation, because apoptosis of even a single cell at the cleavage stage is likely to delay embryo development into the blastocyst.
In the early stages of preimplantation embryo development, maternal mRNAs direct DNA repair. In mammals, during the cleavage stage divisions programming of maternal and paternal chromosomes occurs to create the embryonic genome (embryonic genome activation, EGA) and to begin preimplantation embryo development. Upon EGA, remarkable reprogramming of expression occurs. In mammals, these reprogramming events are controlled by transcription, translation, and microRNA (miRNA) regulation [6]. miRNAs form a large family of short non-coding RNAs between 17–25 nucleotides (nts) in length that have been shown to be expressed in preimplantation embryos [7,8,9].
The biological significance of miRNAs and the roles of these small non-coding RNAs in gametes and preimplantation embryos are poorly understood. This review briefly summarizes the biogenesis of miRNAs and their expression in gametes and preimplantation embryos, as well as their role in regulating DNA repair.
MicroRNA Biogenesis
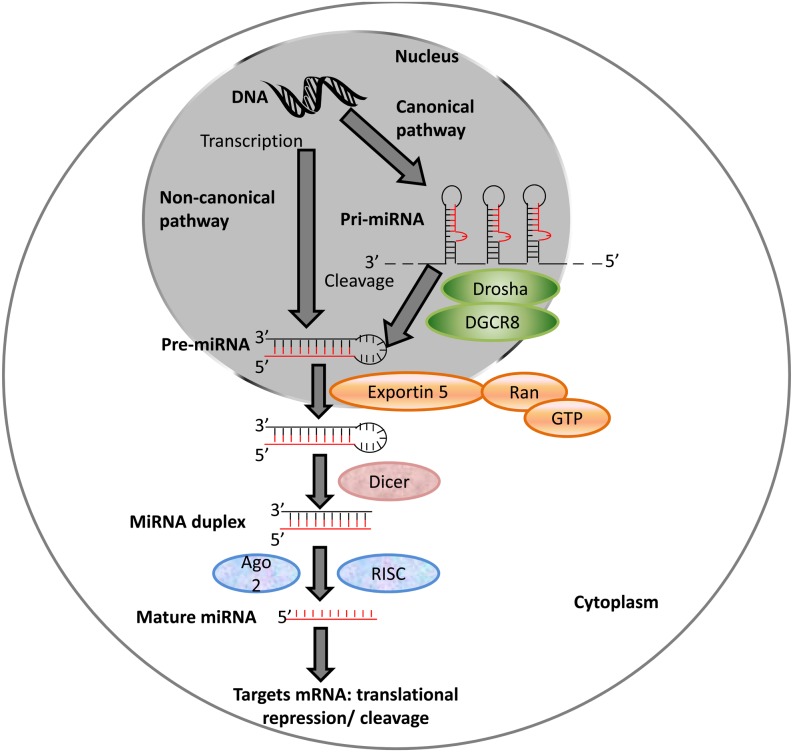
The biogenesis of a small group of miRNAs is induced in an ATM-dependent manner. These miRNAs are associated with KH-type splicing regulatory protein, an AU-rich binding protein involved in Drosha and Dicer processing and in mRNA decay [10,11,12]. Generally, miRNAs are transcribed into long RNAs with a stem-loop structure by RNA polymerase II [13] (Fig. 1). Intergenic miRNAs, which contain their own promoters and regulatory units, are transcribed into pri-miRNA by RNA polymerase II, whereas intronic miRNAs are co-transcribed using host genes from a common promoter [14, 15]. The intergenic pri-miRNAs are processed by Drosha, a 130–160-kDa protein with one dsRNA-binding and two catalytic domains [16]. In the presence of Pasha/DiGeorge syndrome critical region gene 8 (DGCR8), both strands of the hairpin are cut, generating a pre-miRNA product of approximately 70 nt in size [17]. Although both Drosha and DGCR8 were found to be essential for the formation of mature miRNA in Drosophila melanogaster and Caenorhabditis elegans, some pri-miRNAs do not involve these processes by Drosha and DGCR8 and they either use other endonucleases or are directly transcribed into short hairpin structures [18,19,20]. Pre-miRNAs are then transported from the nucleus into the cytoplasm by Exportin-5 (Exp5), which is a nucleocytoplasmic transporter in the karyopherin family and has binding sites for pre-miRNAs in the presence of Ras-related nuclear protein and guanosine triphosphate [21, 22]. Pre-miRNAs are further cleaved by the cytoplasmic RNase endonuclease Dicer, forming 21–22-nt double-stranded structures. The mRNA levels of Dicer in oocytes are higher compared to those in other cell types, suggesting that Dicer plays an important role in the female germline [23]. The crucial role of Dicer in the female germline was also supported in a recent study showing that loss of a splice variant of Dicer, DicerO, led to sterility in female rodents [24]. Reduced and disorganized spindles and incorrect chromosome alignment were observed in Dicer mutant mice and C. elegans oocytes, respectively [7, 25]. Both strands of the pre-miRNA can be associated with Argonaut (Ago)-protein-containing complex and mediated by RNA-induced silencing complex/mi-ribonucleoprotein (RISC/miRNP); however, one strand is mostly degraded [16, 26]. There is no discrimination of which miRNA strand is degraded in mammals. However, the strand that regulates and is involved in the loading of the other strand, miR*, on RISC is typically degraded in mammals [27]. Selection of the strand for degradation may also depend on the stability of the 5′ end and the sequence characteristics such as the bias for A and U [28, 29].
Fig. 1.
Schematic diagram of miRNA biogenesis. Pri-miRNAs are cleaved by the Drosha/DGCR8 complex producing pre-miRNA. Exp5 transports pre-miRNAs into the cytoplasm that are then processed by Dicer and Ago2/RISC complex to form mature miRNA. Adapted from [13]. Copyright adapted with permission from “Development”, via DOI:- http://dev.biologists.org/content/138/9/1653.long/.
Gametogenesis and miRNA Expression
miRNA expression has been observed as early as oogenesis and spermatogenesis in mouse, bovine, and human (Supplementary Table 1: online only). Two percent of the miRNAs analyzed (15/722) in human oocytes showed different expression levels between GV and MII oocytes, such that four miRNAs, including hsa-miR-602, hsa-miR-193a-5p, hsa-miR-297, and hsa-miR-625, were up-regulated in oocytes matured in vitro compared to immature GV oocytes, whereas 11 miRNAs, including hsa-miR-888, hsa-miR-212, hsa-miR-662, hsa-miR-299-5p, hsa-miR- hsa-miR-339-5pm hsa-miR-20a, hsa-miR-486-5p, hsa-miR-141, hsa-miR-768-5p, hsa-miR-376a, and hsa-miR-15a, were down-regulated in mature oocytes [30]. Ago2-deficient oocytes were matured with abnormal spindles, the chromosomes could not unite properly, and showed reduced expression levels of miRNAs [31].
Similar to the oocyte, sperm carries a range of miRNAs. Approximately 20% of these miRNAs are located in the nuclear or perinuclear region of the sperm, indicating that these miRNAs are transferred to the zygote at the time of fertilization. However, the role of these sperm-borne miRNAs is not clear since they are already present in the mature oocyte (meiosis II) [32].
Aberrant expression of miRNA biogenesis genes causes defects in both oogenesis and spermatogenesis. Complete loss of Dicer in somatic cells in the mouse reproductive tract not only showed reduced expression of miRNAs, but also caused the female mice to become infertile with compromised oocytes and embryo integrity [7, 9, 33,34,35]. Loss of Dicer in the germ-line of male mice (homozygote Dicer) led to decreased fertility resulting from abnormal spermatogenesis. In these mice, the number of germ cells was reduced with abnormal spermatids, abnormal phenotype of spermatocytes with condensed nucleus, abnormal sperm motility, and mutant testes with Sertoli tubules [36]. However, it has been suggested that maternal cytoplasmic Dicer transfer disguised the early abnormal phenotypes [25, 37]. Deletion of Dicer led to the loss of sperm, which may be related to the reduced levels of miRNA production [7, 38,39,40].
Preimplantation Embryos and miRNA Expression
Similar miRNA expression profiles in mature mouse oocytes and early developing embryos indicate that the zygote contains maternally inherited miRNAs [7]. As mouse and bovine embryos undergo cleavage divisions, variation in the expression levels of some miRNAs were observed (Supplementary Table 1). In murine embryos, miRNA expression is reduced by as much as 60% between the 1- and 2-cell stages. However, at the end of the 4-cell stage, the expression of miRNAs were doubled in mouse embryos compared to the 2-cell stage embryo, supporting that EGA begins between the 1-cell and 4-cell stages (Supplementary Table 1) [7]. Although the synthesis and degradation of miRNAs co-occur during mouse preimplantation embryo development, overall miRNA expression increases towards the blastocyst stage [41]. Despite numerous studies in mice, only two studies have analyzed the expression of miRNAs in human blastocysts [9, 42].
Abnormalities in Preimplantation Embryos Resulting from Aberrant miRNA Biogenesis
The correct biogenesis and expression of miRNAs is important in preimplantation embryos, as any changes in the expression of DNA repair genes may cause defective DNA repair in the embryos. The 5′ and 3′ nucleotides of the mature miRNAs are determined by Drosha cleavage, and defects in Drosha cleavage may lead to changes in the seed sequence of miRNAs. Exp5 is fundamental for miRNA expression in human and Drosophila, as the knock-down of Exp5 reduces miRNA and tRNA levels in human and Drosophila, respectively [43]. Exp5 was also suggested to stabilize pre-miRNAs. In the absence of Exp5, no pre-miRNA accumulation was detected in the nucleus [22], while when Exp5 was over-expressed, endogenous and exogenous miRNA levels were increased [44]. Knockout of Ago2 in mouse embryonic fibroblasts and hematopoietic cells decreased the levels of mature miRNAs [38, 45, 46]. Any changes to the miRNA sequence or miRNA expression level will likely alter the regulation of mRNAs, which may cause changes in crucial pathways in preimplantation embryos [47] (Table 1).
Table 1. Functions of miRNA processing genes and defects caused by abnormal functioning of these genes.
| Genes | Functions | Defects caused by abnormal functioning of genes |
| Drosha | • Vital for the formation of mature miRNA [47] | • 5′ and 3′ nucleotides of the mature miRNA are determined by Drosha cleavage and any defects in Drosha cleavage may lead to changes in the seed sequence of miRNAs [47] |
| • miRNA processing by cutting both strands of miRNA forming pre-miRNA product in the presence of DGCR8 [16, 127] | ||
| Exp5 | • Stabilization of pre-miRNAs [44] | • Knockdown of Exp5 was shown to reduce miRNA expression [43] |
| • Transport of pre-miRNAs from the nucleus into the cytoplasm in the presence of Ran and GTP [44] | ||
| Dicer | • Maturation of miRNA [16, 26] | • Cell proliferation defects [36] |
| • Lack of Dicer in Drosophila germ line stem cells postponed the G1/S phase transition | ||
| • Dicer deletion in hippocampal, mouse and zebrafish initiated problems in nervous system and led to inability of forming mature miRNAs that resulted in variations of brain morphogenesis and differentiation of neurons [128, 129] | ||
| • Complete loss of Dicer1 caused reduced expression of miRNAs and infertile female mice [7, 9] | ||
| • Homozygote Dicer1 germ-line mutant male mice caused decreased male fertility [36] | ||
| • Dicer deficiency led to embryo death in mouse around embryonic day 7.5 [7, 25, 56] and in zebrafish [58] | ||
| Ago2 | • Component of miRISC [16, 26] | • In the absence of Ago2, oocytes developed to the mature oocytes but with abnormal spindles, and chromosomes were not able to unite properly with reduced miRNA expression levels [57] |
| • miRNAs associated with Ago2/RISC complex target mRNAs [13] | ||
The early differentiation of the maternal to zygotic transition was normal in Dicer mutant zebrafish and mouse embryos; however, defects were triggered in somatogenesis, morphogenesis that affected gastrulation, and heart development, as well as led to apoptosis in the limb mesoderm [25, 48,49,50,51]. Injection of miR-430, which is expressed at the time of EGA and increases the rate of deadenylation and degradation of maternal mRNAs [49], partially repaired the gastrulation, retinal development, and somatogenesis in zebrafish and C. elegans [25].
Deletion of DGCR8 caused cell proliferation defects; however, injection of miR-19, miR-20a, miR-20b, miR-294, and miR-295 prevented these aberrations in mouse embryonic stem cells [52]. A lack of Dicer in Drosophila germ line stem cells postponed the G1/S phase transition [53], suggesting that miRNAs may be vital for stem cells to bypass this checkpoint. Additional studies showed that deletion of Dicer in the developing animals caused aberrations [54, 55]. Dicer deficiency and loss of Ago2 function led to embryo death in mice around embryonic day 7.5 [7, 25, 56] and 9.5 [57], respectively, and in zebrafish [58].
Regulation of mRNAs by miRNAs
miRNAs are either transcribed as separate genes or as miRNA clusters from precursor transcripts, whereas intronic miRNAs are encoded by introns of protein-coding genes, and are formed following pre-mRNA splicing. Approximately 30% of genes in the human genome are estimated to be targeted by miRNAs [59]. Until recently, all studies reported that miRNAs down-regulated their target mRNAs; however, in the past few years, miRNAs were suggested to stabilize their targets [60,61,62,63,64,65,66,67]. This stabilization of mRNAs is not well understood; however, several possible mechanisms have been proposed. miRNA regulation through binding to AU-rich elements of specific proteins [68, 69], miRNA regulation between repression and activation of genes with repression more active in proliferating cells and activation in G1/G0 arrest [70], a computational hypothesis of differential mRNA regulation by miRNAs [61, 62], and competition of pseudogenes with their legitimate genes for the same miRNAs, reducing the down-regulatory effect of miRNAs on their target genes [67], are the main proposed stabilization pathways. Therefore, pseudogenes, non-coding genes, and circular RNAs can function as endogenous decoys for miRNAs [64,65,66,67, 71,72,73]. From a similar perspective, it was also suggested that if an miRNA has multiple mRNA targets, the down-regulatory effect of the miRNAs is reduced [63]. Based on these observations, the competitive endogenous RNA (ceRNA) hypothesis was proposed, in which coding and non-coding RNAs can regulate the mRNA/miRNA association by competing for miRNA binding sites (miRNA response elements) [60]. Similarly to the ceRNA hypothesis, it is also possible that miRNA/miRNA interactions positively regulate mRNAs, such that one miRNA down-regulates another miRNA, leading to increased expression of its target gene [69].
Regulation of Genes Involved in the Cell Cycle by miRNAs Expressed in Preimplantation Embryos
Several miRNAs targeting mRNAs function at cell cycle checkpoints and in DNA repair mechanisms. Proper coordination of the cell cycle is crucial in the response to DNA damage. Cell cycle arrest occurs during replication and at G1/S (first gap phase/DNA synthesis phase) or G2/M (second gap phase/mitosis) checkpoints to activate the correct repair pathway [74]. If the repair mechanisms are unable to repair the damage, which can be caused by inactive DNA repair mechanisms, apoptosis of an embryonic cell can be detrimental to the early developing embryo. Therefore, the correct activation of genes and proteins is critical for fully functioning DNA repair pathways.
Many miRNAs expressed in preimplantation embryos regulate or are regulated by genes functioning at cell cycle checkpoints [75,76,77]. As described above, DNA replication and cell proliferation are shorter in preimplantation embryos. Therefore, the correct regulation of mRNA function during the cell cycle is crucial for avoiding genetic errors, cell cycle arrest, or apoptosis [78].
The tumor suppressor p53 is a DNA damage-induced transcription factor. Recent studies showed that the miR-125b [79], miRNA-380-5p, miR-34, and miR-200 families inhibit the expression of p53 and its related family member p63 in cancer cells [80]. miRNAs expressed in preimplantation embryos, such as miR-16-1, miR-143, and miR-145, were up-regulated by p53- and p68/p72-dependent pathways upon DNA damage in human colon cancer cell lines [81]. Additionally, p53 transcriptionally regulates the miR-192, miR-194, miR-215, and miR-17-92 clusters, and the miR-34 family that targets genes involved in G1/S and G2/M checkpoints [82,83,84,85,86]. Ectopic expression of these miRNAs results in cell cycle arrest in cancer cells and cell lines [79, 82, 83, 87,88,89]. Over-expression of miR-34c, a member of the miR-34 family, suppressed c-Myc expression, which regulates the G1/S cell cycle transition and prevents the replication of damaged DNA in cell lines, whereas inhibition of miR-34c prevents DNA damage-induced S-phase arrest [89]. RB1, another important gene functioning at the G1/S cell cycle transition, has recently been shown to have a direct expression association, indicating a possible stabilization effect of this miRNA on its target mRNAs. miR-21, which is involved in the epithelial-mesenchymal transition and represses tumor suppressors [90], negatively regulates the G1/S phase in laryngeal carcinoma tissues [91]. Members of miR-17-92 in embryonic stem cells [92] and miR-371 and miR-302 clusters in humans [93] were also suggested to control the G1/S transition, and this cluster along with the signaling processes by Oct4, Sox2, and c-Myc may be vital for embryonic cell pluripotency and self-renewal [92]. Additionally, ectopic expression of miR-421 influences the efficiency of the S-phase cell-cycle checkpoint by down-regulating ataxia-telangiectasia mutated (ATM) kinase and increases the sensitivity to ionizing radiation in cell lines [94]. This effect was reversed by blocking the miR-421 and ATM 3′UTR interaction [95]. Similarly, over-expression of miR-18a in colorectal cancer cells suppressed ATM expression [96]. Additionally, over-expression of N-Myc, frequently amplified in neuroblastoma, is capable of inducing miR-421 expression, leading to ATM down-regulation. This suggests that N-Myc-mediated oncogenesis may be associated with the miRNAs in DNA damage response and repair. Up-regulation of miR-101 in the plasmid constructs reduces ATM and DNA-dependent protein kinase protein levels [97]. M059J glioblastoma DNA-PK-deficient cells expressed high levels of miR-100, leading to low expression levels of ATM [98]. Over-expression of miR-143 protects cells from DNA damage-induced death, leading to G2 checkpoint arrest by targeting fragile histidine triad (FHIT) [99]. Figure 2 summarizes the association between miRNAs expressed in mouse, bovine, or human preimplantation embryos and the cell cycle genes and proteins.
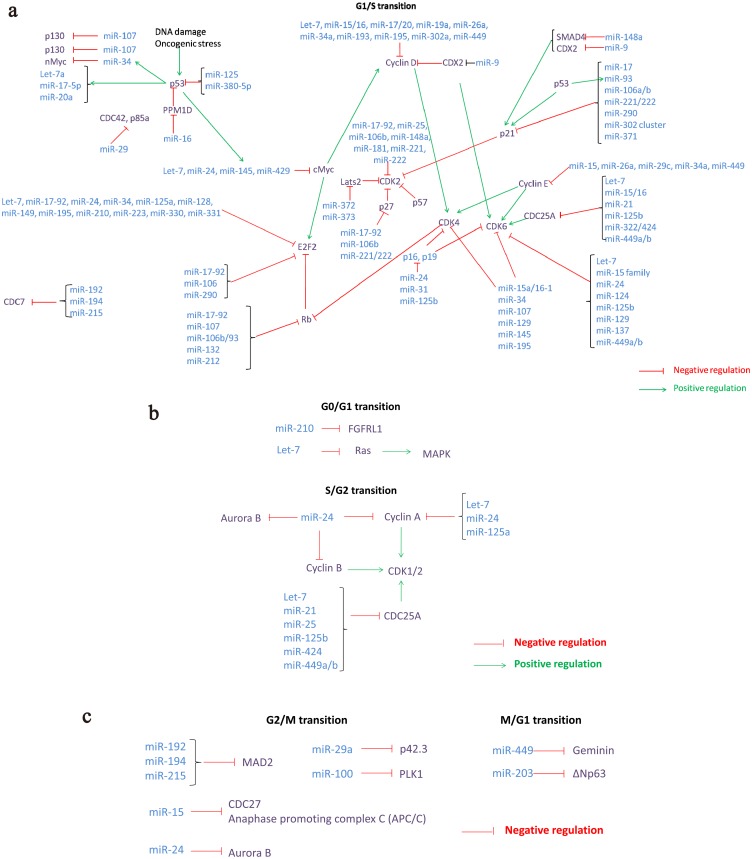
Fig. 2.
miRNAs involved in the regulation of cell cycle checkpoint genes at G0, S, G2, and M phases. a) miRNAs involved in the regulation of cell cycle checkpoint genes at the G1/S transition. b) miRNAs involved in the regulation of cell cycle checkpoint genes at G0/G1 and S/G2 transitions. c) miRNAs involved in the regulation of cell cycle checkpoint genes at G2/M and M/G1 transitions. Main cycles of checkpoint, including G1 (first gap phase), S (synthesis phase), G2 (second gap phase), M (mitosis), and G0 are shown. Cell cycle regulators and effectors, which regulate and/or are regulated by miRNAs expressed in preimplantation embryo development, are grouped according to the cell cycle phase. Positive and negative relationship between miRNAs and mRNAs are shown according to the cell cycle: a) miRNAs involved in the regulation of cell cycle checkpoint genes at G1/S transition. b) miRNAs involved in the regulation of cell cycle checkpoint genes at the G0/G1 and S/G2 transitions c) miRNAs involved in the regulation of cell cycle checkpoint genes at the G2/M and M/G1 transitions [75, 80, 93, 95, 97, 118,119,120,121,122,123,124,125,126].
Cell cycle checkpoint genes play an important role in activating the appropriate DNA repair pathways. It is possible that DNA repair pathways are not fully functional in preimplantation embryos. However, several genes associated with these pathways, including nucleotide excision repair (Fig. 3), base excision repair (Fig. 4), mismatch repair (Fig. 5), and double-strand break repair (Fig. 6), are regulated by miRNAs and these genes expressed during preimplantation embryo development.
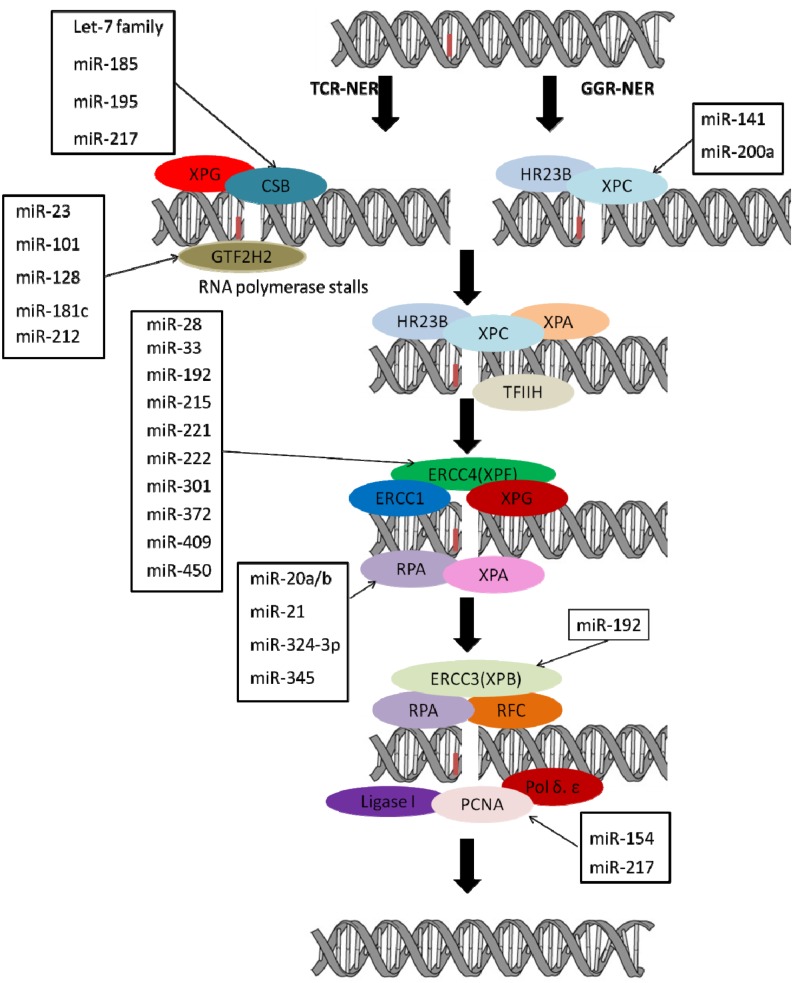
Fig. 3.
Schematic diagram of nucleotide excision repair pathway with genes and miRNAs regulating these genes. Genes involved in nucleotide excision repair and miRNAs predicted to regulate these genes are shown (http://www.microrna.org/microrna/home.do, http://www.targetscan.org/, http://mirdb.org/miRDB/). Several miRNAs targeting or suggested to target the initial sensor genes and genes functioning at later stages of nucleotide excision repair pathway.
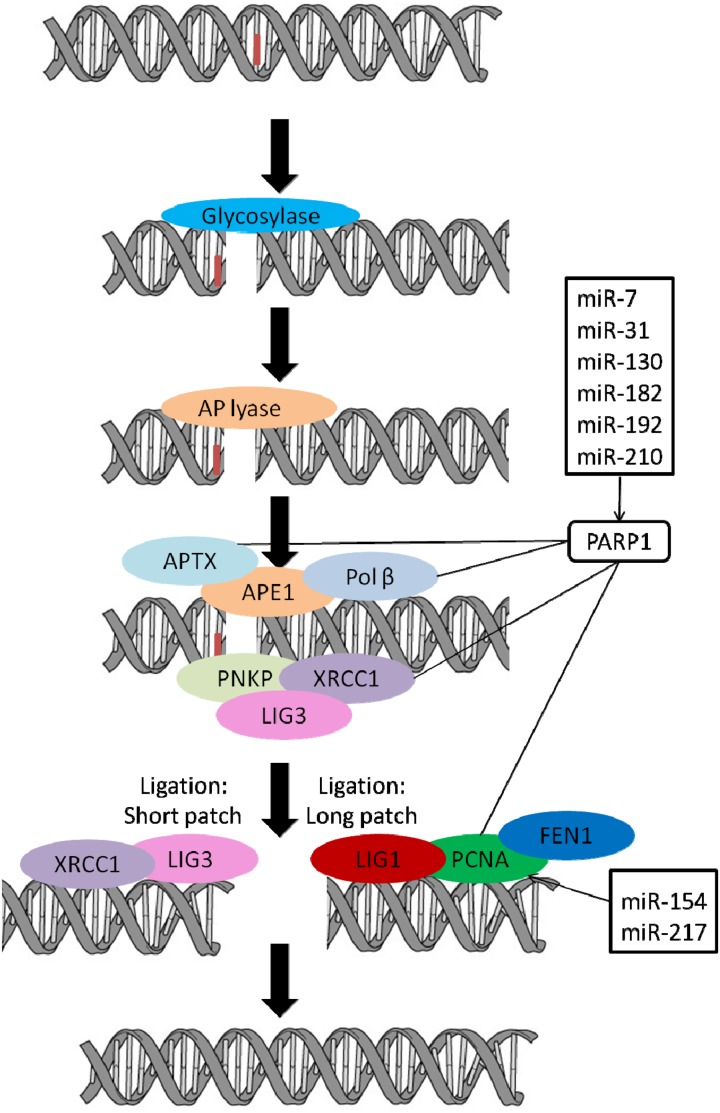
Fig. 4.
Schematic diagram of base excision repair pathway with genes and miRNAs regulating these genes. Genes involved in base excision repair and miRNAs predicted to regulate these genes are shown (http://www.microrna.org/microrna/home.do, http://www.targetscan.org/, http://mirdb.org/miRDB/). Several miRNAs were shown to target PARP1, which interacts with several genes involved in base excision repair and genes functioning at later stages of the base excision repair pathway. Bioinformatics studies also showed that PCNA is regulated by two miRNAs. Although a direct relationship among miRNAs and base excision repair genes, proteins, and polymerases has not been established, base excision repair components may be indirectly regulated by miRNA-regulated PARP1.
Fig. 5.
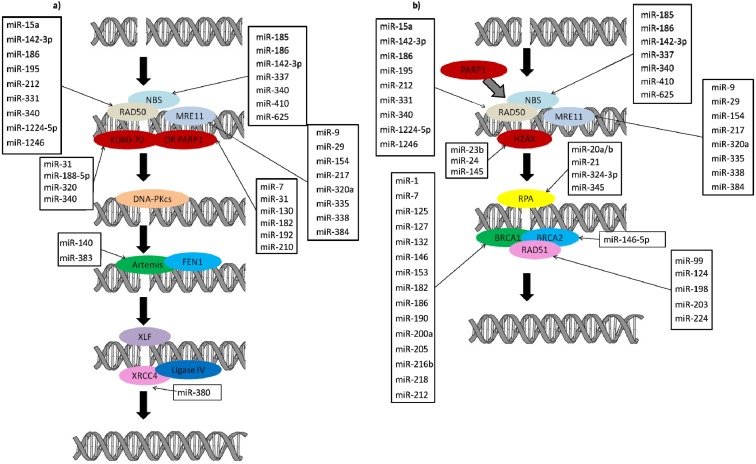
Schematic diagram of the double-strand break repair pathways with genes and miRNAs regulating these genes. Genes involved in a) non-homologous end joining and b) homologous recombination pathways, and miRNAs predicted to regulate these genes are shown (http://www.microrna.org/microrna/home.do, http://www.targetscan.org/, http://mirdb.org/miRDB/). Several miRNAs target the initial sensor genes of non-homologous end joining and homologous recombination repair pathways. Several more miRNAs regulate the expression of genes functioning at later stages of both non-homologous end joining and homologous recombination repair pathways.
Fig. 6.
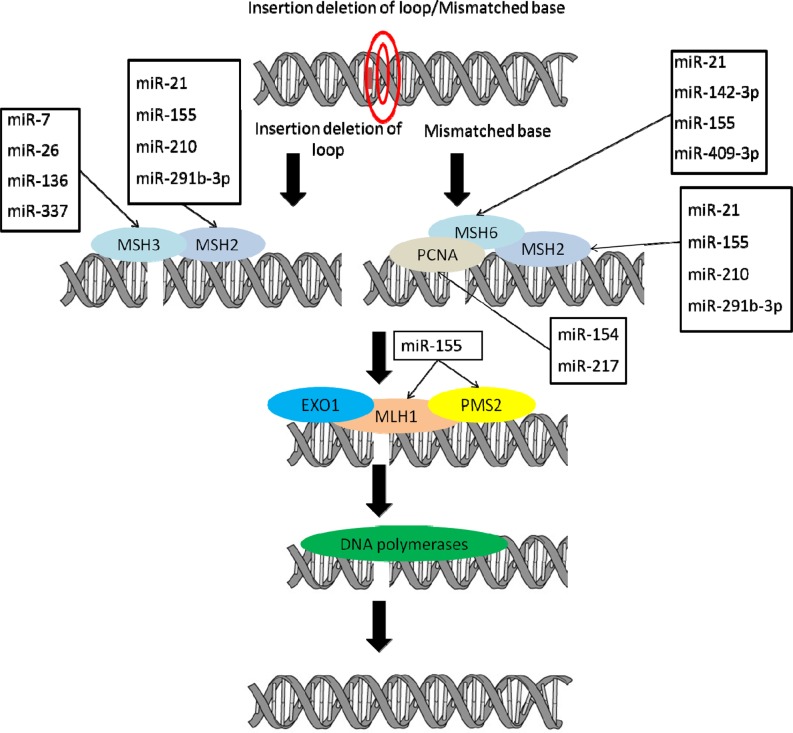
Schematic diagram of mismatch repair pathway with genes and miRNAs regulating these genes. Genes involved in mismatch repair and miRNAs predicted to regulate these genes are shown (http://www.microrna.org/microrna/home.do, http://www.targetscan.org/, http://mirdb.org/miRDB/). Multiple miRNAs have been suggested to regulate the expression of mismatch repair sensor genes (MSH2, MSH3, MSH6, PCNA) and genes functioning at later stages of mismatch repair (MLH1 and PMS2).
DNA Repair Genes and miRNA Expression
DNA repair is a multi-protein repair system and miRNAs regulate the expression of genes involved in different repair mechanisms. Several studies have reported the down-regulatory effects of miRNAs on their target mRNAs, resulting in defective repair. However, many of these studies were performed in tumors, cancer cells, and cell lines. Since there is only one published study describing the possible regulatory roles of miRNAs on repair genes in gametes or embryos [8], these studies in tumors, cells, or cell lines may provide a foundation for understanding the possible relationship between miRNAs and mRNAs during preimplantation development. In this part of the review, we will summarize the studies demonstrating the effects of miRNA expression on DNA repair mechanisms.
In a recently published study reporting the possible regulatory roles of miRNAs on their target repair genes, it was shown that the expression of hsa-miR-23b was inversely associated with the expression of its target nucleotide excision repair gene GTF2H2, indicating a possible down-regulatory role of this miRNA on repair gene [8]. Furthermore, forced expression of another miRNA, miR-192, impaired nucleotide excision repair because increased expression of this miRNA was shown to down-regulate ERCC3 (XPB) and ERCC4 (XPF) in hepatoma HepG2.2.15 cells [100]. Similarly, the down-regulatory effects of miRNAs on genes involved in double-strand break repair, including inverse expression association of hsa-miR-128 with its target genes DCLRE1A and RAD50 in human blastocysts [8], were reported. Over-expression of miR-24 and miR-138 reduced the expression of H2AX, leading to an increased sensitivity to ionizing radiation and reduced repair capacity [88, 101, 102]; over-expression of miR-138 inhibited homologous recombination, leading to an increased sensitivity to DNA damaging agents [102]. Moreover, miR-23a, miR-23b [103], miR-24 [88], and miR-145 [104], target the initial sensor gene H2AX, which is involved in double-strand break repair. In breast cancer cell lines and tumors, miR-18a was over-expressed and the ectopic expression of miR-18a down-regulated ATM by interacting with the 3′UTR of the gene. Over-expression of miR-18a reduced homologous recombination and DNA repair in breast cancer cells and, as expected, inhibition of miR-18a improved the homologous recombination and DNA repair efficiency [105]. miR-182 targets BRCA1; over-expression of this miRNA may be involved in the up-regulation of BRCA1 in sporadic breast tumors and increase the sensitivity to PARP1 inhibition in cultured cells or in xenograft models [106]. miR-99 [107] and siSNF2H, which facilitates homologous recombination and the non-homologous end joining repair pathways [108], affect the localization of BRCA1 and Rad51 to DNA damage sites and therefore influence DNA repair efficiency [109]. Forced expression of miR-210 suppressed RAD52 expression, while over-expression of miR-373 decreased RAD23B and RAD52 [110]. Both of these miRNAs are induced in a hypoxia-inducible factor-1α-dependent manner, indicating an association between the miRNA and DNA repair pathways [110, 111]. Another study showed that the tumor suppressor miR-146, which is expressed in cleavage stage embryos, reduced the expression of BRCA1 in tumor tissues [112]. In silico studies showed that the conserved binding sites for miR-205 within BRCA1 and this miRNA and miR-146b were down-regulated in BRCA1 tumors [107].
Studies suggest that miR-155, which is expressed at different cleavage stages in mouse preimplantation embryos, down-modulates the mismatch repair heterodimer proteins MSH2-MSH6, MLH1, and PMS2 [113]. Mismatch repair recognition protein complex, involving hMSH2 and hMSH6, is down-regulated by increased miR-21 expression in colorectal tumors [114].
Studies suggest that defects in mismatch repair post-replication cause defects in microsatellites, which are short tandem repetitive DNA sequences [115]. Differential expression of miRNAs, including miR-31, miR-625, miR-196b, miR-181c [116], and several members of the miR-17-92 family [117], between tumors with microsatellite instability and/or the absence of protein expression of hMLH1 and tumors with no microsatellite instability and normal protein expression of hMLH1 were reported. Additionally, 39 miRNAs showed differential expression levels between normal colon tissue and tumor specimens. Up-regulation of miRNAs are thought to directly or indirectly lower the tumor suppressor protein expression, leading to improper cell division and tumor formation [116].
Although the expression of several genes involved in repair mechanisms was suggested to be regulated by miRNAs, the possible association between these miRNAs and mRNAs must be further analyzed in gametes and embryos.
Conclusion
Short non-coding RNAs, miRNAs, have gained attention for their regulatory roles of mRNAs and their involvement in many diseases, such as cancers and infertility. Studies of miRNA expression in preimplantation embryos have recently increased to understand their regulatory roles on gene expression. A limited number of articles have been published regarding miRNA expression in gametes and preimplantation embryos. Most of these expressed miRNAs have been associated with DNA repair and aberrant expression of these miRNAs. However, further studies are required to understand the complete contribution of miRNAs in preimplantation embryos. Therefore, the analysis of miRNA expression may identify a regulatory role for these small non-coding RNAs in the expression of DNA repair genes affecting the overall repair activity of the embryo.
Supplementary
References
- 1.Iwamori N, Naito K, Sugiura K, Tojo H. Preimplantation-embryo-specific cell cycle regulation is attributed to the low expression level of retinoblastoma protein. FEBS Lett 2002; 526: 119–123. [DOI] [PubMed] [Google Scholar]
- 2.Haber JE. Partners and pathwaysrepairing a double-strand break. Trends Genet 2000; 16: 259–264. [DOI] [PubMed] [Google Scholar]
- 3.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 2000; 19: 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell 2001; 8: 1163–1174. [DOI] [PubMed] [Google Scholar]
- 5.Vinson RK, Hales BF. DNA repair during organogenesis. Mutat Res 2002; 509: 79–91. [DOI] [PubMed] [Google Scholar]
- 6.Bell CE, Calder MD, Watson AJ. Genomic RNA profiling and the programme controlling preimplantation mammalian development. Mol Hum Reprod 2008; 14: 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev 2007; 21: 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tulay P, Naja RP, Cascales-Roman O, Doshi A, Serhal P, SenGupta SB. Investigation of microRNA expression and DNA repair gene transcripts in human oocytes and blastocysts. J Assist Reprod Genet 2015; 32: 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCallie B, Schoolcraft WB, Katz-Jaffe MG. Aberration of blastocyst microRNA expression is associated with human infertility. Fertil Steril 2010; 93: 2374–2382. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Lu X. Non-coding RNAs in DNA damage response. Am J Cancer Res 2012; 2: 658–675. [PMC free article] [PubMed] [Google Scholar]
- 11.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009; 459: 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell 2011; 41: 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development 2011; 138: 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Issabekova A, Berillo O, Regnier M, Anatoly I. Interactions of intergenic microRNAs with mRNAs of genes involved in carcinogenesis. Bioinformation 2012; 8: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014; 15: 509–524. [DOI] [PubMed] [Google Scholar]
- 16.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–114. [DOI] [PubMed] [Google Scholar]
- 17.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010; 11: 537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008; 22: 2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007; 130: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature 2007; 448: 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science 2004; 303: 95–98. [DOI] [PubMed] [Google Scholar]
- 22.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003; 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev 2007; 21: 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 2013; 155: 807–816. [DOI] [PubMed] [Google Scholar]
- 25.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005; 132: 4653–4662. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz DS, Zamore PD. Why do miRNAs live in the miRNP? Genes Dev 2002; 16: 1025–1031. [DOI] [PubMed] [Google Scholar]
- 27.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 2011; 12: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 2010; 16: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 2009; 36: 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu YW, Wang B, Ding CH, Li T, Gu F, Zhou C. Differentially expressed micoRNAs in human oocytes. J Assist Reprod Genet 2011; 28: 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneda M, Tang F, O’Carroll D, Lao K, Surani MA. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin 2009; 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amanai M, Brahmajosyula M, Perry AC. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol Reprod 2006; 75: 877–884. [DOI] [PubMed] [Google Scholar]
- 33.Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology 2008; 149: 6207–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaraja AK, Andreu-Vieyra C, Franco HL, Ma L, Chen R, Han DY, Zhu H, Agno JE, Gunaratne PH, DeMayo FJ, Matzuk MM. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol 2008; 22: 2336–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nothnick WB. The role of micro-RNAs in the female reproductive tract. Reproduction 2012; 143: 559–576. [DOI] [PubMed] [Google Scholar]
- 36.Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biol Reprod 2008; 79: 696–703. [DOI] [PubMed] [Google Scholar]
- 37.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001; 293: 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O’Carroll D, Das PP, Tarakhovsky A, Miska EA, Surani MA. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 2008; 3: e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Z, Jian Z, Shen SH, Purisima E, Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res 2007; 35: 152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu M-F, Lu M-H, Tang Y, Yu HY, Sun SH. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 2009; 28: 2719–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Bai W, Zhang L, Yin G, Wang X, Wang J, Zhao H, Han Y, Yao YQ. Determination of microRNAs in mouse preimplantation embryos by microarray. Dev Dyn 2008; 237: 2315–2327. [DOI] [PubMed] [Google Scholar]
- 42.Rosenbluth EM, Shelton DN, Sparks AE, Devor E, Christenson L, Van Voorhis BJ. MicroRNA expression in the human blastocyst. Fertil Steril 2013; 99: 855–861: e3. [DOI] [PubMed] [Google Scholar]
- 43.Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res 2006; 34: 4711–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 2005; 11: 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007; 131: 1097–1108. [DOI] [PubMed] [Google Scholar]
- 46.O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 2007; 21: 1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004; 432: 235–240. [DOI] [PubMed] [Google Scholar]
- 48.Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev 2005; 15: 410–415. [DOI] [PubMed] [Google Scholar]
- 49.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006; 312: 75–79. [DOI] [PubMed] [Google Scholar]
- 50.Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol 2006; 16: 2135–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005; 436: 214–220. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 2007; 39: 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature 2005; 435: 974–978. [DOI] [PubMed] [Google Scholar]
- 54.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science 2005; 308: 833–838. [DOI] [PubMed] [Google Scholar]
- 55.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 2009; 16: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet 2003; 35: 215–217. [DOI] [PubMed] [Google Scholar]
- 57.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004; 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- 58.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science 2005; 309: 310–311. [DOI] [PubMed] [Google Scholar]
- 59.Wan G, Mathur R, Hu X, Zhang X, Lu X. miRNA response to DNA damage. Trends Biochem Sci 2011; 36: 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seitz H. Redefining microRNA targets. Curr Biol 2009; 19: 870–873. [DOI] [PubMed] [Google Scholar]
- 62.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004; 5: 396–400. [DOI] [PubMed] [Google Scholar]
- 63.Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol 2010; 6: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB. A 3′-untranslated region (3’UTR) induces organ adhesion by regulating miR-199a* functions. PLoS ONE 2009; 4: e4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010; 465: 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 2007; 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- 67.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 2010; 328: 1563–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2001; 2: 237–246. [DOI] [PubMed] [Google Scholar]
- 69.Carroll AP, Tran N, Tooney PA, Cairns MJ. Alternative mRNA fates identified in microRNA-associated transcriptome analysis. BMC Genomics 2012; 13: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007; 318: 1931–1934. [DOI] [PubMed] [Google Scholar]
- 71.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science 2013; 340: 440–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30: 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–338. [DOI] [PubMed] [Google Scholar]
- 74.Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair (Amst) 2004; 3: 997–1007. [DOI] [PubMed] [Google Scholar]
- 75.Noto JM, Peek RM. The role of microRNAs in Helicobacter pylori pathogenesis and gastric carcinogenesis. Front Cell Infect Microbiol 2011; 1: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jurado JO, Pasquinelli V, Alvarez IB, Peña D, Rovetta AI, Tateosian NL, Romeo HE, Musella RM, Palmero D, Chuluyán HE, García VE. IL-17 and IFN-γ expression in lymphocytes from patients with active tuberculosis correlates with the severity of the disease. J Leukoc Biol 2012; 91: 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Massirer KB, Perez SG, Mondol V, Pasquinelli AE. The miR-35-41 family of microRNAs regulates RNAi sensitivity in Caenorhabditis elegans. PLoS Genet 2012; 8: e1002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 2008; 9: 297–308. [DOI] [PubMed] [Google Scholar]
- 79.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res 2008; 14: 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eggert A, Schulte JH. A small kiss of death for cancer. Nat Med 2010; 16: 1079–1081. [DOI] [PubMed] [Google Scholar]
- 81.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature 2009; 460: 529–533. [DOI] [PubMed] [Google Scholar]
- 82.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res 2008; 68: 10105–10112. [DOI] [PubMed] [Google Scholar]
- 83.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Ørntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 2008; 68: 10094–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008; 14: 2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pichiorri F, Suh S-S, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM, Jr, Hofmainster C, Alder H, Garofalo M, Di Leva G, Volinia S, Lin HJ, Perrotti D, Kuehl M, Aqeilan RI, Palumbo A, Croce CM. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010; 18: 367–381. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447: 1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg 2008; 135: 255–260, discussion :260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS, Chang KW. miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol 2010; 46: 204–208. [DOI] [PubMed] [Google Scholar]
- 89.Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, Elia A, Kress TR, Dickens M, Clemens MJ, Heery DM, Gaestel M, Eilers M, Willis AE, Bushell M. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci USA 2010; 107: 5375–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 2011; 12: 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, Li X, Tang H. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res 2009; 19: 828–837. [DOI] [PubMed] [Google Scholar]
- 92.Gunaratne PH. Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells? Curr Stem Cell Res Ther 2009; 4: 168–177. [DOI] [PubMed] [Google Scholar]
- 93.Kim VN. Cell cycle micromanagement in embryonic stem cells. Nat Genet 2008; 40: 1391–1392. [DOI] [PubMed] [Google Scholar]
- 94.Mishra PJ, Song B, Mishra PJ, Wang Y, Humeniuk R, Banerjee D, Merlino G, Ju J, Bertino JR. MiR-24 tumor suppressor activity is regulated independent of p53 and through a target site polymorphism. PLoS ONE 2009; 4: e8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc–regulated microRNA-421. Proceedings of the National Academy of Sciences2010. [DOI] [PMC free article] [PubMed]
- 96.Wu CW, Dong YJ, Liang QY, He XQ, Ng SS, Chan FK, Sung JJ, Yu J. MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer. PLoS ONE 2013; 8: e57036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, Mao H, Hao C, Olson JJ, Curran WJ, Wang Y. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS ONE 2010; 5: e11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ng WL, Yan D, Zhang X, Mo YY, Wang Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst) 2010; 9: 1170–1175. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 99.Lin YX, Yu F, Gao N, Sheng JP, Qiu JZ, Hu BC. microRNA-143 protects cells from DNA damage-induced killing by downregulating FHIT expression. Cancer Biother Radiopharm 2011; 26: 365–372. [DOI] [PubMed] [Google Scholar]
- 100.Xie QH, He XX, Chang Y, Sun SZ, Jiang X, Li PY, Lin JS. MiR-192 inhibits nucleotide excision repair by targeting ERCC3 and ERCC4 in HepG2.2.15 cells. Biochem Biophys Res Commun 2011; 410: 440–445. [DOI] [PubMed] [Google Scholar]
- 101.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol 2009; 16: 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res 2011; 9: 1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tsai YS, Lin CS, Chiang SL, Lee CH, Lee KW, Ko YC. Areca nut induces miR-23a and inhibits repair of DNA double-strand breaks by targeting FANCG. Toxicol Sci 2011; 123: 480–490. [DOI] [PubMed] [Google Scholar]
- 104.Revel A, Achache H, Stevens J, Smith Y, Reich R. MicroRNAs are associated with human embryo implantation defects. Hum Reprod 2011; 26: 2830–2840. [DOI] [PubMed] [Google Scholar]
- 105.Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J, Li M, Li J. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS ONE 2011; 6: e25454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854. [DOI] [PubMed] [Google Scholar]
- 107.Tanic M, Zajac M, Gómez-López G, Benítez J, Martínez-Delgado B. Integration of BRCA1-mediated miRNA and mRNA profiles reveals microRNA regulation of TRAF2 and NFκB pathway. Breast Cancer Res Treat 2012; 134: 41–51. [DOI] [PubMed] [Google Scholar]
- 108.Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, Yasui A. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell 2010; 40: 976–987. [DOI] [PubMed] [Google Scholar]
- 109.Mueller AC, Sun D, Dutta A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 2013; 32: 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 2009; 69: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of microRNA expression: the hypoxic component. Cell Cycle 2007; 6: 1426–1431. [PubMed] [Google Scholar]
- 112.Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, Lidereau R, Mikaélian I, Mazoyer S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med 2011; 3: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McPhee F, Friborg J, Levine S, Chen C, Falk P, Yu F, Hernandez D, Lee MS, Chaniewski S, Sheaffer AK, Pasquinelli C. Resistance analysis of the hepatitis C virus NS3 protease inhibitor asunaprevir. Antimicrob Agents Chemother 2012; 56: 3670–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T, Nuovo GJ, Fishel R, Croce CM. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci USA 2010; 107: 21098–21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laghi L, Bianchi P, Malesci A. Differences and evolution of the methods for the assessment of microsatellite instability. Oncogene 2008; 27: 6313–6321. [DOI] [PubMed] [Google Scholar]
- 116.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle 2008; 7: 2485–2492. [DOI] [PubMed] [Google Scholar]
- 117.Lanza G, Ferracin M, Gafà R, Veronese A, Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM, Negrini M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer 2007; 6: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 2012; 12: 613–626. [DOI] [PubMed] [Google Scholar]
- 119.Biggar KK, Storey KB. The emerging roles of microRNAs in the molecular responses of metabolic rate depression. J Mol Cell Biol 2011; 3: 167–175. [DOI] [PubMed] [Google Scholar]
- 120.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta 2011; 1812: 592–601. [DOI] [PubMed] [Google Scholar]
- 121.Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology 2012; 143: 35–47.e2. [DOI] [PubMed] [Google Scholar]
- 122.Zhang H, Li Y, Lai M. The microRNA network and tumor metastasis. Oncogene 2010; 29: 937–948. [DOI] [PubMed] [Google Scholar]
- 123.Ryazansky SS, Gvozdev VA. Small RNAs and cancerogenesis. Biochemistry (Mosc) 2008; 73: 514–527. [DOI] [PubMed] [Google Scholar]
- 124.Liang LH, He XH. Macro-management of microRNAs in cell cycle progression of tumor cells and its implications in anti-cancer therapy. Acta Pharmacol Sin 2011; 32: 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol 2011; 3: 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res 2010; 70: 7176–7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell 2007; 28: 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barbato C, Giorgi C, Catalanotto C, Cogoni C. Thinking about RNA? MicroRNAs in the brain. Mamm Genome 2008; 19: 541–551. [DOI] [PubMed] [Google Scholar]
- 129.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci 2008; 28: 4322–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.