Abstract
Transcription is a highly regulated process in all domains of life. In human mitochondria, transcription of the circular genome involves only two promoters, called light strand promoter (LSP) and heavy strand promoter (HSP), located in the opposite DNA strands. Initiation of transcription occurs upon sequential assembly of an initiation complex that includes mitochondrial RNA polymerase (mtRNAP) and the initiation factors mitochondrial transcription factor A (TFAM) and TFB2M. It has been recently suggested that the transcription initiation factor TFAM binds to HSP and LSP in opposite directions, implying that the mechanisms of transcription initiation are drastically dissimilar at these promoters. In contrast, we found that binding of TFAM to HSP and the subsequent recruitment of mtRNAP results in a pre-initiation complex that is remarkably similar in topology and properties to that formed at the LSP promoter. Our data suggest that assembly of the pre-initiation complexes on LSP and HSP brings these transcription units in close proximity, providing an opportunity for regulatory proteins to simultaneously control transcription initiation in both mtDNA strands.
Keywords: mitochondrial DNA (mtDNA), promoter, protein cross-linking, RNA polymerase, transcription, transcription factor, LSP, TFAM, TFB2M, mtRNAP
Introduction
Both strands of human mtDNA encode subunits of protein complexes involved in energy production by oxidative phosphorylation. Transcription of human mitochondrial genome is thus bi-directional and is driven by two closely spaced promoters (1). In the first stage of transcription initiation, mtRNAP2 assembles with TFAM and promoter DNA into a transcription-incompetent pre-IC in which the elements of mtRNAP that are responsible for specific binding and melting of the promoter are not in position to interact with DNA (2). In the second stage, the pre-IC binds transcription factor TFB2M, which causes melting of the promoter and conversion of the pre-IC into a transcription-competent IC (3). Because both strands of human mtDNA encode essential components of the electron transport chain, it is thought that their activity must be coordinated to generate appropriate levels of polycistronic transcripts (4–8).
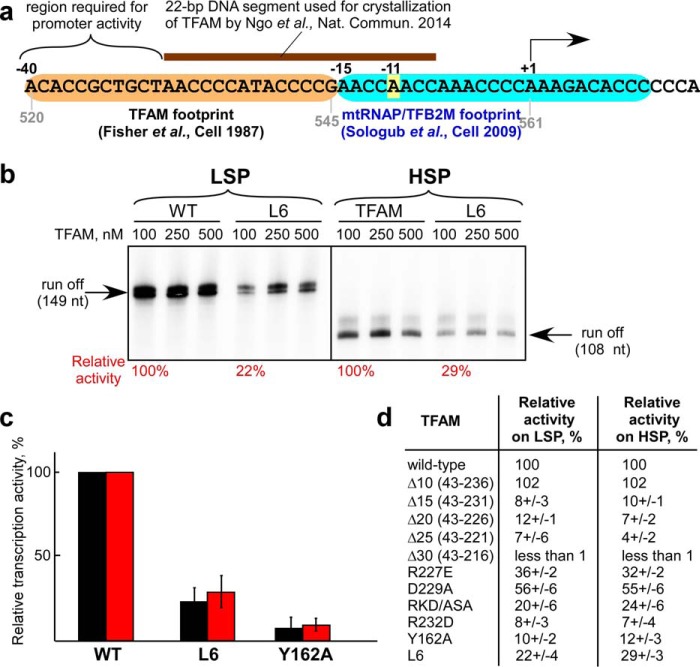
A recent study by Ngo et al. (9) suggested that the IC assembled at HSP has a different structure and opposite orientation as compared with the IC assembled on LSP. However, the 22-bp DNA fragment used by Ngo et al. (9) to represent the HSP contains only a part of the TFAM-binding site that is required for promoter function (see Fig. 1a). The DNase I footprint of TFAM on HSP has been mapped to the region from −15 to −40 relative to the promoter start site (10, 11). Moreover, the exonuclease III (Exo III) footprint of mtRNAP-TFB2M assembled on a pre-melted HSP indicated protection of a 15-bp region upstream of the promoter start site (12) (see Fig. 1a). Consistent with this observation, the −11 base in HSP makes contact with the mtRNAP (13). Importantly, substitution of the region −30 to −40 (which is absent from the oligonucleotides used by Ngo et al. (9) (see Fig. 1a)) with random DNA results in promoter inactivation (14). We conclude that the DNA fragment used by Ngo et al. (9) in their structural studies is incomplete, and that the reported structure likely represents a nonspecific DNA-TFAM complex (similar to that observed by Ngo et al. on an unrelated DNA template). In this work, we compared initiation complexes assembled on LSP and HSP using protein-DNA and protein-protein cross-linking and found that these complexes are very similar in molecular architecture.
FIGURE 1.
Characterization of the initiation complex assembled on human HSP promoter. a, schematic illustration of the footprinting data on HSP. Black numbers indicate positions of the bases in promoter DNA relative to the start site (+1), and gray numbers indicate the positions according to the reference (Cambridge) mtDNA. The −11 base in the template DNA stand that interacts with mtRNAP is highlighted in yellow. b, Y162A TFAM mutant is defective in run-off transcription assays on both mitochondrial promoters. nt, nucleotides. c, TFAM variants Y162A and L6 are deficient in transcription on both LSP (black bars) and HSP (red bars). Error bars indicate means ± S.E. d, TFAM variants deficient in transcription on LSP are also defective on HSP.
Results and Discussion
In support of their model, Ngo et al. (7, 9) presented variants of TFAM (Y162A and L6 (K136A, H137A, K139A, R140A, K146A, K147A)) that could not support transcription on LSP but were as active on the HSP promoter as the wild-type TFAM. We repeated these experiments and found that, contrary to the study by Ngo et al. (9), these mutants were similarly defective in transcription initiation on both LSP and HSP (Fig. 1, b and c). In addition, the C-terminal deletion mutants of TFAM that were defective in transcription initiation on LSP were defective in experiments involving HSP (Fig. 2d), further supporting a similar mode of TFAM binding to both mitochondrial promoters.
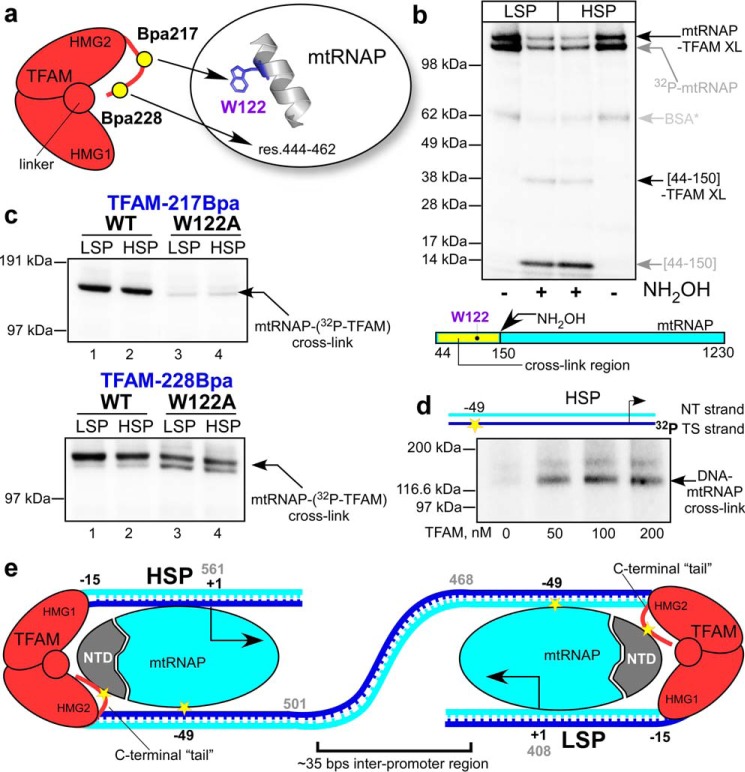
FIGURE 2.
Topology of the pre-initiation complexes assembled on LSP and HSP. a, schematics of TFAM interactions with mtRNAP on LSP. Two major interactions involve TFAM region next to residue 217 and Trp-122 in the N-terminal extension region of mtRNAP (residues (res.) 120–134, predicted α-helix) and TFAM region next to residue 228 and mtRNAP region 444–463. b, the C-terminal region of TFAM interacts with the N-terminal extension region of mtRNAP in the pre-ICs assembled on LSP or HSP promoter. The pre-ICs were UV-irradiated, treated with hydroxylamine (NH2OH), and resolved using SDS-PAGE. The radioactive species (∼37 kDa) that appear upon the treatment of the pre-IC with hydroxylamine represent TFAM cross-link to the region 44–150 of mtRNAP. XL, cross-link. c, MtRNAP residue Trp-122 is a primary interaction target for TFAM in complexes assembled on both promoters. Upper panel, cross-linking performed using 32P-labeled Bpa217 TFAM and WT (lanes 1 and 2) or Trp-122 mtRNAP (lanes 3 and 4). Lower panel, cross-linking performed as above but using 32P-labeled Bpa228 TFAM. d, MtRNAP interacts with the −49 base of the HSP promoter. TS, DNA template strand; NT, DNA non-template strand. e, transcription initiation complexes assemble in close proximity on human mtDNA. Interactions probed in this study are indicated by the yellow stars.
TFAM makes two distinct contacts with mtRNAP in the presence of LSP, as revealed by site-specific cross-linking (2, 3): residue 217 cross-links to the residue Trp-122 in the N-terminal extension region of the mtRNAP, and residue 228 cross-links to the promoter-binding domain of mtRNAP, region 444–462 (Fig. 2a). To clarify whether the topology of the HSP pre-IC is distinct from that assembled on LSP, we looked at major interactions found in the LSP pre-IC: one involving the C terminus of TFAM and the N-terminal region of mtRNAP and the other involving the upstream promoter DNA and mtRNAP (2). We mapped interactions of TFAM having a photo-reactive amino acid, Bpa, incorporated in the C terminus of TFAM at position 217 with a 32P-labeled variant of mtRNAP (Fig. 2b). The cross-link between mtRNAP and TFAM can only be detected when DNA (either promoter or nonspecific) is present (2). Treatment of the mtRNAP-TFAM cross-link with hydroxylamine (which cleaves the peptide bond between Asn and Gly residues) resulted in generation of labeled species containing the N-terminal fragment of mtRNAP (residues 44–150) and TFAM (Fig. 2b, lanes 2 and 3), regardless of which promoter DNA was used for the assembly of the complex. We previously demonstrated that the absolutely conserved residue Trp-122 of mtRNAP likely serves as the primary target for Bpa cross-link when the pre-IC is assembled on LSP (2). Indeed, the efficiency of cross-link between TFAM and the W122A variant of mtRNAP was about 20 times lower as compared with WT mtRNAP (Fig. 2c, upper panel, lanes 1 and 3). We repeated this experiment using the pre-ICs assembled on the HSP and found a similar effect for W122A substitution on cross-link efficiency (Fig. 2c, upper panel, lanes 2 and 4). We therefore conclude that the Trp-122 residue constitutes the primary point of interaction between mtRNAP and TFAM at the HSP promoter as well, suggesting that the topology of the mtRNAP-TFAM complex is similar on both promoters. As expected, another point of contact, between TFAM residue 228 and mtRNAP region 444–463, was unaffected by W122A substitution as the similar efficiency of cross-linking was observed in the case of both LSP and HSP (Fig. 2c, lower panel).
An important characteristic of the pre-IC is the interaction between the far upstream region of promoter DNA and mtRNAP. This interaction is revealed by means of photo cross-linking of 32P-labeled DNA having 4-thio UMP at base −49 with mtRNAP in the presence of the nonspecific DNA competitor (2). We found that, similar to LSP (2, 6, 7), TFAM bends the HSP promoter to bring the upstream region in close proximity to mtRNAP, as evident from DNA-mtRNAP cross-linking (Fig. 2d).
Our data reveal identical point contacts at a single-amino acid and a single-DNA base resolution in complexes assembled on both mitochondrial promoters. We therefore conclude that the pre-ICs assembled on HSP and LSP have similar overall topology and suggest that regulation of transcription initiation at these promoters occurs in a similar manner and serves to ensure coordinated expression of mitochondrial genes from both mtDNA strands.
In human mtDNA, LSP and HSP are located just 150 bp apart from each other. Our model predicts that two transcription units assemble in a close proximity to each other (Fig. 2e), and therefore their activity may be regulated simultaneously by yet unidentified transcription factors(s).
Experimental Procedures
Transcription Assays
Run-off transcription assays were performed using synthetic DNA oligonucleotides as described previously (2). Transcription reactions were carried out using previously described (2) PCR templates (50 nm), mtRNAP (50 nm), TFAM (50 nm), and TFB2M (50 nm) in a transcription buffer containing 40 mm Tris (pH = 7.9), 10 mm MgCl2, and 10 mm DTT in the presence of ATP (0.3 mm), GTP (0.3 mm), CTP (0.3 mm), UTP (0.01 mm), and 0.3 μCi of [α-32P]UTP (800 Ci/mmol) to produce 149-nucleotide (LSP) or 108-nucleotide (HSP) run-off RNA products. Reactions were carried out at 35 °C for 30 min and stopped by the addition of an equal volume of 95% formamide/0.05 m EDTA. The products were resolved by 20% PAGE containing 6 m urea and visualized by PhosphorImagerTM (GE Health).
Proteins
Purification of TFAM, TFB2M, and mtRNAP was described previously (2). TFAM mutants Y162A and L6 (7) were generated using the QuikChange mutagenesis kit (Agilent). For mapping experiments, a variant of mtRNAP having an Asn-Gly pair (hydroxylamine cleavage site) at position 150 and an engineered PKA site (2) was used.
Protein-Protein Cross-linking Using Artificial Photo-reactive Amino Acid (Bpa)
Bpa was incorporated in TFAM at residue 217 or 228. Expression of Bpa-containing proteins was performed as described previously (2). The pre-ICs were assembled using 32P-labeled Bpa-TFAM (100 nm), mtRNAP (300 nm), and the LSP (300 nm) or the HSP (300 nm) promoters. The cross-linking was activated by UV irradiation at 312 nm for 15 min at room temperature.
Mapping of the Cross-linking Sites in mtRNAP
Hydroxylamine cleavage of pre-IC was performed as described previously (2). The pre-ICs (200 nm) assembled using 32P-labeled NG150-mtRNAP were treated with hydroxylamine in the presence of BSA (1 mg/ml) for 4 h, and the products of the reaction were resolved using a 4–12% Bis-Tris NuPAGE gel (Invitrogen).
Protein-DNA Photo Cross-linking
Promoter-mtRNAP cross-link with 4-thio-UTP at base −49 in the LSP promoter was generated as described previously (2). For HSP-mtRNAP cross-linking, the oligonucleotide containing photo-reactive probe (4-thio UMP) at the base −49 in the 32P-labeled template strand was used (Midland Scientific). Transcription complexes (250 nm) were formed as described above and UV-irradiated (312 nm) for 15 min at room temperature in the presence of nonspecific oligonucleotides (10 μm). Cross-linking products were resolved using a 4–12% Bis-Tris NuPAGE gel (Invitrogen) and visualized by PhosphorImagerTM (GE Health).
Author Contributions
Y. I. M. designed and performed experiments. Y. I .M. and D. T. analyzed the data, prepared the figures, and wrote the manuscript.
Acknowledgments
We thank members of the Temiakov lab for helpful discussions and Drs. W. T. McAllister and M. Anikin for critical reading of the manuscript.
This work was supported in part by National Institutes of Health Grant RO1GM104231 (to D. T.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- mtRNAP
- mitochondrial RNA polymerase
- TFAM
- mitochondrial transcription factor A
- LSP
- light strand promoter
- HSP
- heavy strand promoter
- pre-IC
- pre-initiation complex
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- Bpa
- p-benzoyl-l-phenylalanine.
References
- 1. Falkenberg M., Larsson N. G., and Gustafsson C. M. (2007) DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76, 679–699 [DOI] [PubMed] [Google Scholar]
- 2. Morozov Y. I., Agaronyan K., Cheung A. C., Anikin M., Cramer P., and Temiakov D. (2014) A novel intermediate in transcription initiation by human mitochondrial RNA polymerase. Nucleic Acids Res. 42, 3884–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morozov Y. I., Parshin A. V., Agaronyan K., Cheung A. C., Anikin M., Cramer P., and Temiakov D. (2015) A model for transcription initiation in human mitochondria. Nucleic Acids Res. 43, 3726–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hällberg B. M., and Larsson N. G. (2014) Making proteins in the powerhouse. Cell Metab. 20, 226–240 [DOI] [PubMed] [Google Scholar]
- 5. Posse V., Hoberg E., Dierckx A., Shahzad S., Koolmeister C., Larsson N. G., Wilhelmsson L. M., Hällberg B. M., and Gustafsson C. M. (2014) The amino terminal extension of mammalian mitochondrial RNA polymerase ensures promoter specific transcription initiation. Nucleic Acids Res. 42, 3638–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rubio-Cosials A., Sidow J. F., Jiménez-Menéndez N., Fernández-Millán P., Montoya J., Jacobs H. T., Coll M., Bernadó P., and Solà M. (2011) Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 18, 1281–1289 [DOI] [PubMed] [Google Scholar]
- 7. Ngo H. B., Kaiser J. T., and Chan D. C. (2011) The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 18, 1290–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wallace D. C. (2007) Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76, 781–821 [DOI] [PubMed] [Google Scholar]
- 9. Ngo H. B., Lovely G. A., Phillips R., and Chan D. C. (2014) Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 5, 3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fisher R. P., and Clayton D. A. (1988) Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 8, 3496–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fisher R. P., Topper J. N., and Clayton D. A. (1987) Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell 50, 247–258 [DOI] [PubMed] [Google Scholar]
- 12. Sologub M., Litonin D., Anikin M., Mustaev A., and Temiakov D. (2009) TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell 139, 934–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Litonin D., Sologub M., Shi Y., Savkina M., Anikin M., Falkenberg M., Gustafsson C. M., and Temiakov D. (2010) Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 285, 18129–18133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Topper J. N., and Clayton D. A. (1989) Identification of transcriptional regulatory elements in human mitochondrial DNA by linker substitution analysis. Mol. Cell. Biol. 9, 1200–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]