Abstract
Diallyl trisulfide (DATS), a metabolic byproduct of garlic, is known to inhibit the growth of breast cancer cells in vitro and in vivo. This study demonstrates that DATS targets breast cancer stem cells (bCSC). Exposure of MCF-7 and SUM159 human breast cancer cells to pharmacological concentrations of DATS (2.5 and 5 μm) resulted in dose-dependent inhibition of bCSC, as evidenced by a mammosphere assay and flow cytometric analysis of aldehyde dehydrogenase 1 (ALDH1) activity and the CD44high/CD24low/epithelial specific antigen-positive fraction. DATS-mediated inhibition of bCSC was associated with a decrease in the protein level of FoxQ1. Overexpression of FoxQ1 in MCF-7 and SUM159 cells increased ALDH1 activity and the CD49f+/CD24− fraction. Inhibition of ALDH1 activity and/or mammosphere formation upon DATS treatment was significantly attenuated by overexpression of FoxQ1. In agreement with these results, stable knockdown of FoxQ1 using small hairpin RNA augmented bCSC inhibition by DATS. Expression profiling for cancer stem cell-related genes suggested that FoxQ1 may negatively regulate the expression of Dachshund homolog 1 (DACH1), whose expression is lost in invasive breast cancer. Chromatin immunoprecipitation confirmed recruitment of FoxQ1 at the DACH1 promoter. Moreover, inducible expression of DACH1 augmented DATS-mediated inhibition of bCSC. Expression of FoxQ1 protein was significantly higher in triple-negative breast cancer cases compared with normal mammary tissues. Moreover, an inverse association was observed between FoxQ1 and DACH1 gene expression in breast cancer cell lines and tumors. DATS administration inhibited ALDH1 activity in vivo in SUM159 xenografts. These results indicate that FoxQ1 is a novel target of bCSC inhibition by DATS.
Keywords: breast cancer, cancer stem cells, chemoprevention, FOXO, natural product
Introduction
The medicinal usefulness of garlic (Allium sativum) has been appreciated throughout recorded history and reviewed extensively (1, 2). For example, the Codex Ebers (an Egyptian medical text) recommended garlic for the treatment of different conditions, such as high blood pressure and abnormal tissue growth. The health-promoting effects of garlic are ascribed to sulfur-containing metabolic byproducts, including allicin, water-soluble organosulfur compounds (e.g. S-allylcysteine and S-allylmercaptocysteine), and lipid-soluble organosulfur compounds, including diallyl trisulfide (DATS),3 diallyl disulfide (DADS), diallyl sulfide (DAS), and ajoene (3, 4). These compounds are generated upon cutting or chewing of garlic and certain other Allium vegetables (3).
Accumulating evidence from population-based case-control studies as well as preclinical work strongly argues for the anticancer activity of garlic and its constituents (5–10). For example, a French case-control study suggested an inverse association between garlic and onion intake and breast cancer risk with an odds ratio of 0.3 (p for trend < 10−6) for the highest versus lowest intake groups (8). Similarly, garlic intake was suggested to be protective against breast cancer in a hospital-based study from Switzerland (9). However, such an association was not evident for garlic supplementation in a Dutch cohort study (10). Nevertheless, the preclinical evidence for the anticancer effect of garlic or its bioactive components against breast cancer is fairly persuasive (11–14). For instance, feeding of garlic powder (20 g/kg of diet) as well as its constituents S-allylcysteine and DADS (57 μmol/kg of diet) for 2 weeks prior to N-methyl-N-nitrosourea treatment significantly inhibited mammary tumor incidence and the cumulative number of tumors in female Sprague-Dawley rats (11). Likewise, feeding of rats with diets supplemented with 2% and 4% garlic powder for 2 weeks before and 2 weeks after 7,12-dimethylbenz[a]anthracene treatment resulted in a significant decrease in mammary tumor incidence and multiplicity in rats (12). Intratumor injection of a purified protein fraction from fresh garlic resulted in a significant increase in CD8+ T lymphocytes and tumor growth inhibition in BALB/c mice (14).
Cellular models have been utilized to delineate the mechanisms by which garlic constituents may inhibit breast cancer growth (15–20). For example, DATS treatment inhibited DNA strand breaks induced by an environmental carcinogen (benzo[a]pyrene) in a non-tumorigenic normal human mammary epithelial cell line (MCF-10A) (16). Interestingly, DATS was found to be a much more potent inducer of apoptotic cell death in MCF-7 cells compared with its monosulfide (DAS) or disulfide (DADS) analogues (18). We have shown previously that breast cancer cells are much more sensitive to cell growth inhibition and apoptosis induction by DATS than normal human mammary epithelial cells (19). Cancer cell selectivity is especially desirable for cancer-preventive agents. DATS was also shown to inhibit estrogen receptor-α (ER-α) activity in MCF-7 and T47D cells (20).
Recent progress in our understanding of the biology of breast cancer highlights a critical role for breast cancer stem cells (bCSC) in tumor initiation and progression (21–23). Therefore, it is only logical to speculate that elimination of both bCSC and proliferating cells constituting the bulk of the tumor mass may be necessary for the treatment and/or prevention of breast cancer. This study was undertaken to test the hypothesis that garlic organosulfides may inhibit bCSC using an ER-α positive (MCF-7) and a triple-negative (SUM159) human breast cancer cell line as a model.
Experimental Procedures
Reagents and Cell Lines
DATS, DADS, and DAS (purity >98%) were purchased from LKT Laboratories (St. Paul, MN) and diluted with DMSO before treatment. E-ajoene and E-propyl-ajoene were synthesized as described previously (24). Reagents for cell culture (culture medium, fetal bovine serum, and antibiotics) were purchased from Life Technologies/Thermo Fisher Scientific (Waltham, MA). The antibody against B-lymphoma Moloney murine leukemia virus insertion region 1 (Bmi-1, catalog no. CS6964, dilution 1:5000) was purchased from Cell Signaling Technology (Danvers, MA). An antibody against Forkhead box Q1 (FoxQ1, catalog no. SC166265, dilution 1:200–1:500) was from Santa Cruz Biotechnology (Dallas, TX). An antibody against urokinase-type plasminogen activator receptor (catalog no. GTX100466, dilution 1:1000) was from GeneTex (Irvine, CA), anti-Dachshund homolog 1 (DACH1) antibody (catalog no. ab176718, dilution 1:1000) was from Abcam (Cambridge, MA), and anti-FLAG and anti-actin antibodies were from Sigma-Aldrich (St. Louis, MO). The MCF-7 cell line was acquired from the American Type Culture Collection and authenticated in 2015. Authenticated stocks of MCF-7 cells were used in this study. SUM159 cells were purchased from Asterand and authenticated by the supplier. MDA-MB-231 cells stably transfected with an inducible DACH1 plasmid were a gift from Dr. Richard G. Pestell (Thomas Jefferson University, Philadelphia, PA) (25). MCF-7 and SUM159 cells were transfected with empty pCMV6 (FLAG-tagged) vector or the same vector encoding for FoxQ1 using FuGENE 6. The pCMV6-FoxQ1 plasmid was purchased from ORIGENE (Rockville, MD). Stably transfected cells were selected by culture in medium supplemented with G418 over a 6-week period. SUM159 cells were stably transfected with 6 μg of a control shRNA or a FoxQ1-targeted shRNA using transfection medium and reagents from Santa Cruz Biotechnology. The SUM159 clone with stable knockdown of FoxQ1 was selected with 5 μg/ml of puromycin for 5 weeks.
Mammosphere Formation Assay
The mammosphere assay was performed as described by us previously (26). Briefly, 1000 cells in medium supplemented with penicillin/streptomycin, B27, insulin, hydrocortisone, epidermal growth factor, basic fibroblast growth factor, 2-mercaptoethanol, and methylcellulose were plated on ultralow attachment plates. The indicated concentrations of DATS were then added to the medium for the primary mammosphere formation assay. Five days later, the first-generation mammospheres were photographed and counted and then disaggregated. Single-cell suspensions were replated for the second-generation mammosphere assay without further treatment with DMSO (control) or DATS. The second-generation mammospheres were scored 7 days after cell plating.
Determination of Aldehyde Dehydrogenase 1 (ALDH1) Activity and Flow Cytometric Analysis of CD44high/CD24low/Epithelial Specific Antigen-positive (ESA+) Population
ALDH1 activity was determined using the Aldefluor kit (Stemcell Technologies, Vancouver, BC, Canada) as recommended by the supplier. For analysis of the CD44high/CD24low/ESA+ population, cells were incubated with anti-ESA (FITC-conjugated), anti-CD24 (phycoerythrin-conjugated), and anti-CD44 (allophycocyanin-conjugated) antibodies in the dark for 30 min at room temperature. Cells were washed with PBS and analyzed using a BD Accuri TM C6 flow cytometer (BD Biosciences).
Western Blotting
Whole cell lysates from control or DATS-treated cells were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting as described by us previously (27). Cells were lysed on ice with a solution (50 mm Tris-HCl (pH 8.0), 1% Triton X-100, 0.1% sodium dodecyl sulfate, 150 mm NaCl, protease inhibitors, and phosphatase inhibitors) for 20 min and then cleared by centrifugation at 14,000 × g for 20 min. The change in protein level was determined by densitometric scanning. The blots were stripped and reprobed with β-actin antibody for normalization. After transfer, the membranes were stained with Ponceau S for protein loading normalization. A band on the Ponceau S-stained membrane close to the size of the desired protein was used for normalization. Two different molecular weight marker protein positions were identified on each immunoblot.
Real-time Quantitative PCR
Total RNA from cells was isolated using the RNeasy kit (Qiagen, Germantown, MD). First-strand cDNA was synthesized using Superscript reverse transcriptase (Life Technologies) with oligo(dT)20 primer. Primers were as follows: Bmi-1, 5′-AAATGCTGGAGAACTGGAAAG-3′ (forward) and 5′-CTGTGGATGAGGAGACTGC-3′ (reverse); FoxQ1, 5′-CGCGGACTTTGCACTTTGAA-3′ (forward) and 5′-AGCTTTAAGGCACGTTTGATGGAG-3′ (reverse); DACH1, 5′-CCTTGACAAACTCTCTCTAACTGG-3′ (forward) and 5′-CTTGAGCTCTGGCATTATCTATGG-3′ (reverse); MYC, 5′-GCCACGTCTCCACACATCAG-3′ (forward) and 5′-TGGTGCATTTTCGGTTGTTG-3′ (reverse); ZEB1, 5′-GCCAATAAGCAAACGATTCTG-3′ (forward) and 5′-TTTGGCTGGATCACTTTCAAG-3′ (reverse); TWIST1, 5′-AGTCCGCAGTCTTACGAGGA-3′ (forward) and 5′-GCCAGCTTGAGGGTCTGAAT-3′ (reverse); and TWIST2, 5′-CAAGCTGAGCAAGATCCAGAC-3′ (forward) and 5′-GGTCATCTTATTGTCCATCTCG-3′ (reverse). The quantitative PCR was done using 2× SYBR Green Master Mix (Applied Biosystems/Life Technologies) with 95 °C (15 s), 60 °C annealing (60 s), and 72 °C (30 s) for 40 cycles. Relative gene expression was calculated using the method described by Livak and Schmittgen (28).
Confocal Microscopy to Determine FoxQ1 Protein Expression in SUM159 Cells
SUM159 cells (6 × 104 cells/well) were plated on glass coverslips in 12-well plates (in triplicate) and allowed to attach by overnight incubation. After 24-h treatment with DMSO or 5 μm DATS, cells were fixed with 2% paraformaldehyde for 1 h, permeabilized with 0.5% Triton X-100 for 10 min, and then blocked with buffer containing 0.5% bovine serum albumin and 0.15% glycine in PBS for 1 h at room temperature. This was followed by incubation with anti-FoxQ1 antibody (1:200 dilution) overnight at 4 °C. The cells were incubated with Alexa Fluor 568-conjugated secondary antibody for 1 h at room temperature. After washing with PBS, the cells were mounted and observed using an Olympus FluoView FV1000 confocal microscope at ×60 objective magnification.
Colony Formation Assay
250 cells were plated in 6-well pates and incubated for 10 days. The cells were fixed in methanol and stained with crystal violet for 30 min. The colonies were counted.
RT2 Profiler PCR Array
Total RNA was extracted using the RNeasy kit (Qiagen), followed by reverse transcription with 1 μg of total RNA and the RT2 First-Strand kit (Qiagen) as recommended by the supplier. To evaluate the effect of FoxQ1 on the expression of genes involved in cancer “stemness” the human cancer stem cell RT2 ProfilerTM PCR array (Qiagen) was used. The change in gene expression was quantified using web-based software provided by the manufacturer. Gene expression with a Ct value of above 35 was considered undetectable. The cutoff was 1.5 -fold change in expression and p ≤ 0.05 (by two-sided Student's t test).
ChIP Assay
The ChIP assay was performed using a kit from Upstate Biotechnology (Lake Placid, NY) according to the protocol of the manufacturer. Briefly, DNA and associated proteins in cells were cross-linked with formaldehyde (final concentration of 1%) for 20 min. The cross-link was quenched by addition of 0.125 m glycine for 5 min. The DNA-protein complexes were sheared by sonication to generate 200- to 600-bp DNA fragments. Sonicated lysates were incubated with anti-FLAG antibody (for FoxQ1 pulldown) or anti-IgG antibody (control) overnight and reverse cross-linked for DNA isolation. The putative FoxQ1 binding sites at the DACH1 promoter were amplified with the following primers: site 1, 5′-CACCAGAATGGCAAAAATTTTATTA-3′ (forward) and 5′-GCTCATGTCTCCAAGTATCTTCAAT-3′ (reverse); site 2, 5′-CCCTTTTTCCACTTTCTAAATCTTC-3′ (forward) and 5′-TCCTAATAGGTTGTGGAAAGAAATG-3′ (reverse).
Immunohistochemistry for Analysis of FoxQ1 Expression in Tissue Microarrays of Normal Breast and Triple-negative Breast Cancer (TNBC)
The differential expression of FoxQ1 between normal and breast tumor was analyzed using breast tissue microarrays from US Biomax (Rockville, MD; catalog no. for normal BRN801a (n = 80) and catalog no. for TNBC-BR487a (n = 48) and BR10011 (n = 57)). Immunohistochemistry was performed as described by us previously (29). At least 10 randomly selected and non-overlapping fields on each tissue section of the tissue microarrays were examined for FoxQ1 expression by the nuclear algorithm of the Aperio ImageScope software (Leica Biosystems, Buffalo Grove, IL). The H score is based on intensity (0, 1+, 2+, and 3+) and percent positivity (0–100%) and calculated using the following formula: H score = (percent of negative cells × 0) + (percent of 1 + cells × 1) + (percent of 2 + cells × 2) + (percent of 3 + cells × 3). Some specimens for normal (n = 23) and TNBC (n = 8) were not available for analysis because of poor staining and/or less than optimal sectioning on the tissue microarray (cracked or folded). Thus, the final number was 57 for normal and 97 for TNBC.
TCGA and Cancer Cell Line Encyclopedia Data
Breast cancer TCGA and Cancer Cell Line Encyclopedia RNA sequencing data were obtained from CGHub through the Pittsburgh Genome Resource Repository (30). Additional breast cancer cell line data were obtained from GEO (GSE48213) (31). Log2 transformed transcript per million values were generated from FASTQ files using lightweight alignment - salmon v0.5.1 (21-kmer size, quasi-mapping, Ensembl v82) (32). PAM50 subtypes were called on TCGA breast cancer expression data using the genefu Bioconductor package (pam50.robust model) (33). PAM50 expression plots were generated with beeswarm from comprehensive R archive network (CRAN).
SUM159 Xenograft Study
Female SCID mice (6 weeks old) were purchased from The Jackson Laboratory (Bar Harbor, ME). Following 48 h of acclimation, the mice were subcutaneously injected on the right flank with exponentially growing SUM159 cells (1 × 106 cells, 0.1-ml suspension in 50% PBS and 50% Matrigel (Corning, Bedford, MA)). The mice were divided into two groups of 10 mice/group. The mice were treated by oral gavage with either vehicle or 2 mg DATS in 100 μl of PBS beginning on the day of tumor cell implantation. Treatment in both groups was performed three times each week (Mondays, Wednesdays, and Fridays) until the conclusion of the study. During the study, animal weights were recorded weekly, and tumor presence/volume measurements were recorded two to three times each week. One mouse from the control group and two mice from the DATS treatment group died within 2 weeks of the study for unknown reasons. The mice were sacrificed after 55 days of treatment. Two mice from the control group were excluded from ALDH1 activity determination because of early sacrifice because of morbidity and/or unusually high activity. The ALDH1 activity was also unusually very high in one sample of the DATS treatment group, and this was excluded from analysis. However, these mice were included for tumor incidence and tumor volume calculations. For analysis of ALDH1 activity and mammosphere formation, primary tumors were processed as described by us previously (26). Briefly, tissues were digested in Dulbecco's modified Eagle's medium supplemented with 300 units/ml collagenase and 100 units/ml hyaluronidase for 3–4 h at 37 °C. Disaggregated cells were suspended in Hanks' balanced salt solution supplemented with 2% fetal bovine serum and ammonium chloride and treated with a solution containing 0.25% trypsin-EDTA, 5 mg/ml dispase, and 0.1 mg/ml DNase1 in Hanks' balanced salt solution. The samples were passed through a 40-μm strainer. The cells were used for determination of ALDH1 activity. For mammosphere formation, cells were plated in ultralow attachment plates in Dulbecco's modified Eagle's medium/F12 supplemented with 2% fetal bovine serum, 10 mmol/liter HEPES, 5% bovine serum albumin, 1% penicillin/streptomycin, 10 μg/ml insulin, 20 ng/ml epidermal growth factor, 20 ng/ml basic fibroblast growth factor, B27, 10 μg/ml heparin, and 2-mercaptoethanol for 7 days. The mammospheres were photographed and scored for each group.
Statistical Analysis
Each experiment was performed at least twice, and quantitative experiments were performed in triplicate. Statistical significance of difference was determined using GraphPad Prism. One-way analysis of variance (ANOVA) with Dunnett's adjustment was used for dose-response comparisons. One-way ANOVA followed by Bonferroni's test was used for multiple group comparisons. Statistical significance of the difference in FoxQ1 level between normal breast and TNBC (immunohistochemistry) was determined by two-sided Student's t test. Statistically significant differences in expression between FoxQ1 and DACH1 in basal and luminal breast cancers were assessed by Mann-Whitney U test. Pearson's R correlation and plots were conducted in R using Log2 transformed transcript per million expression values. The difference was considered significant at p < 0.05.
Results
DATS Treatment Inhibited Self-renewal of bCSC in MCF-7 and SUM159 Cells
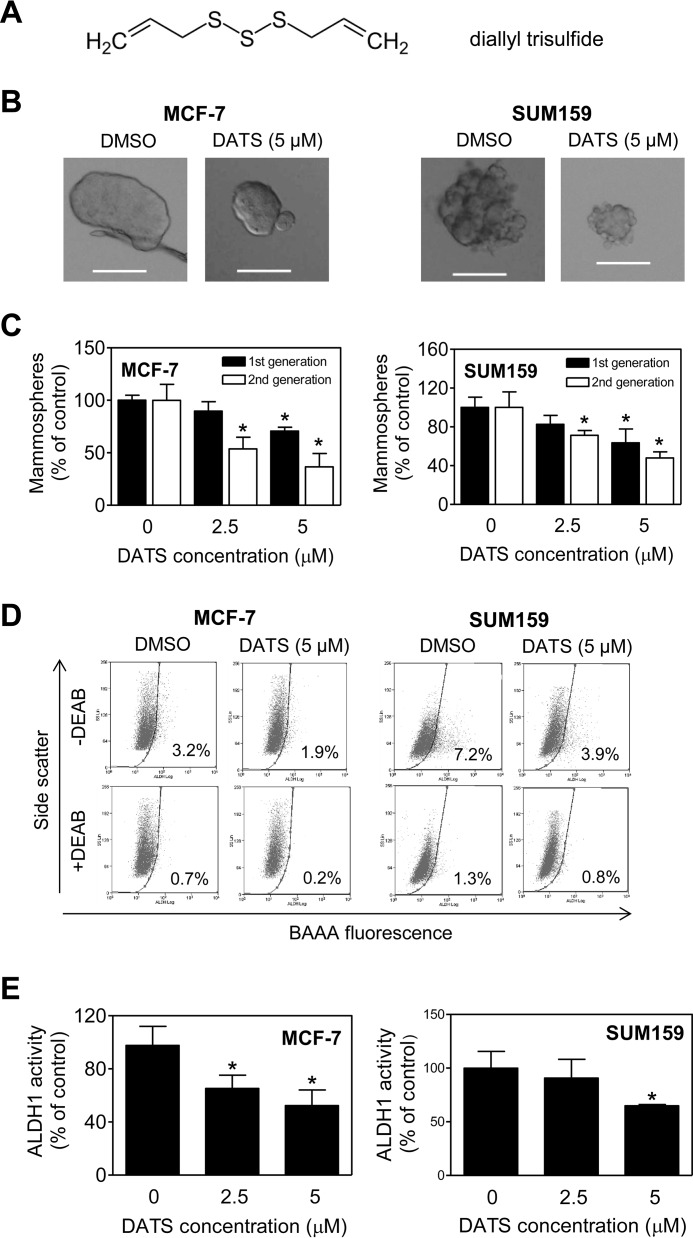
Because DATS is the most abundant water-insoluble sulfur compound in fresh garlic (34), initially we focused on this phytochemical (the structure of DATS is shown in Fig. 1A) to determine its effect on self-renewal of bCSC. Fig. 1B displays first-generation mammospheres resulting from MCF-7 and SUM159 cells after 5 days of cell seeding in the absence (DMSO control) or presence of 5 μm DATS. The first-generation mammosphere frequency was dose-dependently decreased upon DATS treatment in both cell lines compared with the DMSO-treated control (Fig. 1C). The second-generation mammosphere number was also significantly lower in the DATS treatment group compared with the control in both cell lines (Fig. 1C). High ALDH1 activity is another characteristic feature of bCSC. Flow histograms for ALDH1 activity in MCF-7 and SUM159 cells after 72-h treatment with DATS (5 μm) or DMSO (control) are shown in Fig. 1D. The ALDH1 activity was significantly lower in DATS-treated MCF-7 and SUM159 cells compared with the control, especially at the 5 μm dose (Fig. 1E).
FIGURE 1.
DATS treatment inhibits mammosphere formation and ALDH1 activity in MCF-7 (luminal subtype) and SUM159 cells (basal subtype). A, the chemical structure of DATS. B, representative images of first-generation mammospheres resulting after 5 days of cell seeding and treatment of MCF-7 and SUM159 cells with DMSO or 5 μm of DATS (magnification ×100, scale bars = 100 μm). C, quantitation of first- and second-generation mammospheres in DATS-treated MCF-7 and SUM159 cells relative to the DMSO-treated control (mean ± S.D., n = 3). D, representative flow histograms for ALDH1 activity in MCF-7 and SUM159 cells after 72-h treatment with DMSO or 5 μm DATS. The ALDH inhibitor DEAB was used as a control. E, quantitation of ALDH1 activity in MCF-7 and SUM159 cells after DATS treatment. The results shown are relative to the DMSO-treated control (mean ± S.D., n = 3). *, p < 0.05 compared with the DMSO-treated control by one-way ANOVA followed by Dunnett's adjustment. Comparable results were observed in two independent experiments. DEAB, diethylaminobenzaldehyde; BAAA, BODIPYTM-aminoacetaldehyde.
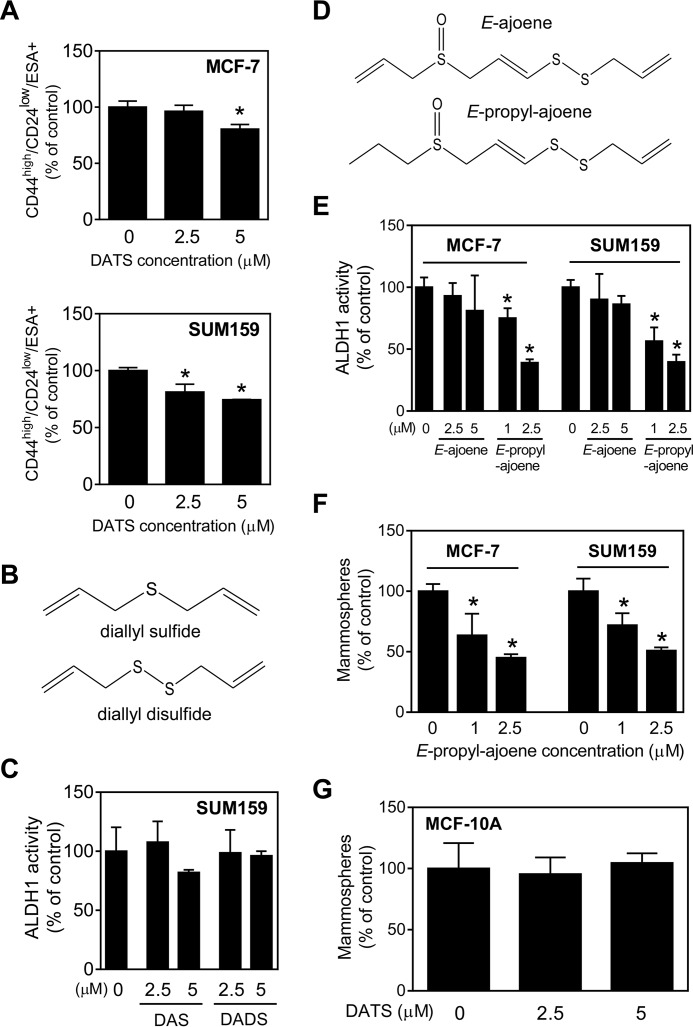
DATS Treatment Decreased the CD44high/CD24low/ESA+ Fraction in MCF-7 and SUM159 Cells
DATS-treated MCF-7 and SUM159 cells showed a significant decrease in CD44high/CD24low/ESA+ fraction in comparison with the control at the 5 μm concentration in both cell lines (Fig. 2A). In contrast, DAS and DADS (the structure is shown in Fig. 2B) were practically inactive for inhibition of ALDH1 activity (Fig. 2C). Ajoene (Fig. 2D), which is derived from a chemical rearrangement of allicin, is undetectable in freshly crushed garlic but present in aged and heated garlic preparations, with abundance of the E-isomer over Z-ajoene (35, 36). In both MCF-7 and SUM159 cells, naturally occurring E-ajoene was unable to inhibit ALDH1 activity (Fig. 2E). On the other hand, inhibition of ALDH1 activity was observed to a level mirroring DATS after substitution of one of the terminal allyl groups with a propyl group in E-ajoene (Fig. 2E). Consistent with these results, mammosphere formation in both MCF-7 and SUM159 cells was significantly and dose-dependently inhibited by E-propyl-ajoene (Fig. 2F). The primary conclusions from these structure-activity analyses are that oligosulfide chain length is a critical structural determinant of bCSC inhibition by DATS and that modulation of bCSC inhibition is feasible by chemical modification of the terminal allyl group in E-ajoene.
FIGURE 2.
DATS treatment decreases CD44high/CD24low/ESA+ fraction in MCF-7 and SUM159 cells. A, quantitation of the CD44high/CD24low/ESA+ fraction in MCF-7 and SUM159 cells relative to the DMSO-treated control. B, the chemical structures of DAS and DADS. C, ALDH1 activity in DAS- or DADS-treated SUM159 cells (72-h treatment) relative to the DMSO-treated control. D, the chemical structures of E-ajoene and E-propyl-ajoene. E, quantitation of ALDH1 activity in MCF-7 and SUM159 cells after 72-h treatment with E-ajoene or E-propyl-ajoene relative to the DMSO-treated control. F, frequency of mammospheres in E-propyl-ajoene-treated MCF-7 and SUM159 cells relative to the DMSO-treated control. G, frequency of mammospheres in DATS-treated MCF-10A cells relative to the DMSO-treated control. All data shown are mean ± S.D. (n = 3). *, p < 0.05 compared with the DMSO-treated control by one-way ANOVA with Dunnett's adjustment. Experiments were repeated at least twice, and representative data from one such experiment are shown.
We have shown previously that a normal human mammary epithelial cell line (MCF-10A) is resistant to cell viability inhibition as well as apoptosis induction by DATS even at the 40 μm concentration (19). In this study, we determined the effect of DATS treatment on the stem cell-like phenotype using this cell line. The mammosphere frequency in MCF-10A cells was much lower compared with breast cancer cells (data not shown). However, DATS exposure failed to inhibit mammosphere formation in MCF-10A cells (Fig. 2G) at concentrations that were highly effective in breast tumor cells, eliciting nearly 50% inhibition (Fig. 1C). These results further underscore the cancer cell selectivity of DATS.
DATS Treatment Decreased the FoxQ1 Protein Level in Breast Cancer Cells
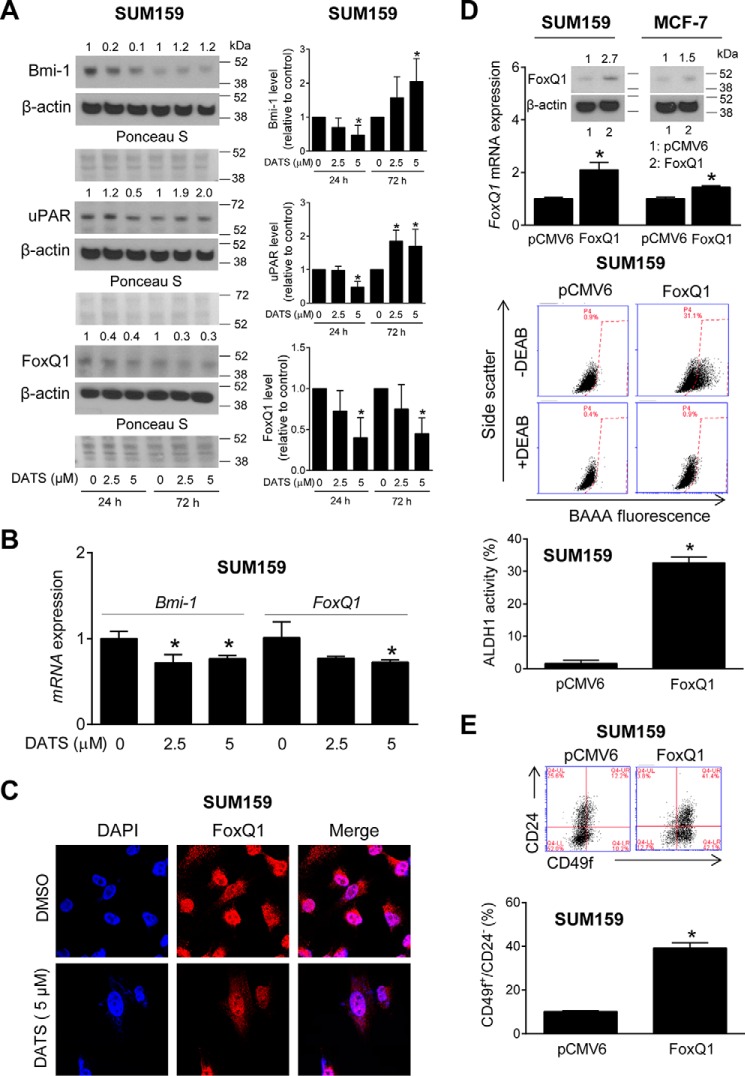
Fig. 3A shows a Western blotting analysis for the effect of DATS treatment on the levels of a panel of proteins implicated in the maintenance of the bCSC phenotype, including Bmi-1 (37), urokinase-type plasminogen activator receptor (38), and FoxQ1 (39). The inhibitory effect of DATS on the protein levels of Bmi-1 and urokinase-type plasminogen activator receptor was transient, followed by an increase in their expression after 72 h of treatment (Fig. 3A). On the other hand, the DATS-mediated down-regulation of FoxQ1 protein was sustained for the duration of the experiment, at least at the 5 μm dose (Fig. 3A). Similar to SUM159 cells, the protein levels of many of the above proteins were increased in MCF-7 cells after 72 h of DATS treatment (data not shown). FoxQ1 protein expression was very low in MCF-7 cells (data not shown). Nevertheless, we focused on FoxQ1 for functional studies of its role in the regulation of bCSC inhibition by DATS.
FIGURE 3.
FoxQ1 regulates bCSC. A, Western blotting for Bmi-1, urokinase-type plasminogen activator receptor (uPAR), and FoxQ1 proteins using whole cell lysates from SUM159 cells after treatment with DMSO or DATS. Each immunoblot shows two molecular weight markers. The quantitation of each protein is shown in the right panel (mean ± S.D., n = 4). The numbers above the bands represent changes in each protein level relative to the corresponding DMSO-treated control. B, quantitation of Bmi-1 and FoxQ1 mRNA expression by quantitative PCR in SUM159 cells after 24-h treatment with DMSO or DATS (mean ± S.D., n = 3). *, p < 0.05 compared with the DMSO-treated control by one-way ANOVA with Dunnett's adjustment (A and B). C, confocal images depicting nuclear levels of FoxQ1 protein in SUM159 cells after 24-h treatment with DMSO or DATS at ×60 objective magnification. D, FoxQ1 protein and FoxQ1 mRNA expression in transfected cells. Bottom panels, representative flow histograms for ALDH1 activity in empty vector-transfected or FoxQ1-overexpressing SUM159 cells. The bar graph shows the quantitation of ALDH1 activity in empty vector-transfected or FoxQ1-overexpressing SUM159 cells (mean ± S.D., n = 3). E, representative flow histograms for the CD49f+/CD24− population in empty vector-transfected or FoxQ1-overexpressing SUM159 cells. Bottom panel, quantitation of the CD49f+/CD24− population in empty vector-transfected or FoxQ1-overexpressing SUM159 cells (mean ± S.D., n = 3). *, p < 0.05 compared with pCMV6-transfected cells by two-sided Student's t test (D and E). Comparable results were observed in replicate experiments. DEAB, diethylaminobenzaldehyde; BAAA, BODIPYTM-aminoacetaldehyde.
Down-regulation of FoxQ1 protein upon DATS treatment was accompanied by its transcriptional suppression (Fig. 3B). The FoxQ1 protein was predominantly localized in the nucleus of SUM159 cells, as revealed by confocal microscopy (Fig. 3C). DATS-treated SUM159 cells exhibited a decrease in nuclear FoxQ1 protein level compared with vehicle (DMSO)-treated control cells (Fig. 3C). Accordingly, we focused on FoxQ1 for functional studies to determine its role in bCSC inhibition by DATS.
Overexpression of FoxQ1 Increased the bCSC Fraction in SUM159 Cells
Fig. 3D shows overexpression of FoxQ1 in stably transfected SUM159 and MCF-7 cells at the protein and mRNA levels. The effect of FoxQ1 overexpression on bCSC was studied using SUM159 cells. As can be seen in Fig. 3D, the ALDH1 activity was 20-fold higher in FoxQ1-overexpressing SUM159 cells compared with empty pCMV6 vector-transfected control cells. Likewise, a 4-fold increase in the CD49f+/CD24− fraction was discernible in SUM159 cells transfected with the FoxQ1 plasmid in comparison with empty vector-transfected control cells (Fig. 3E).
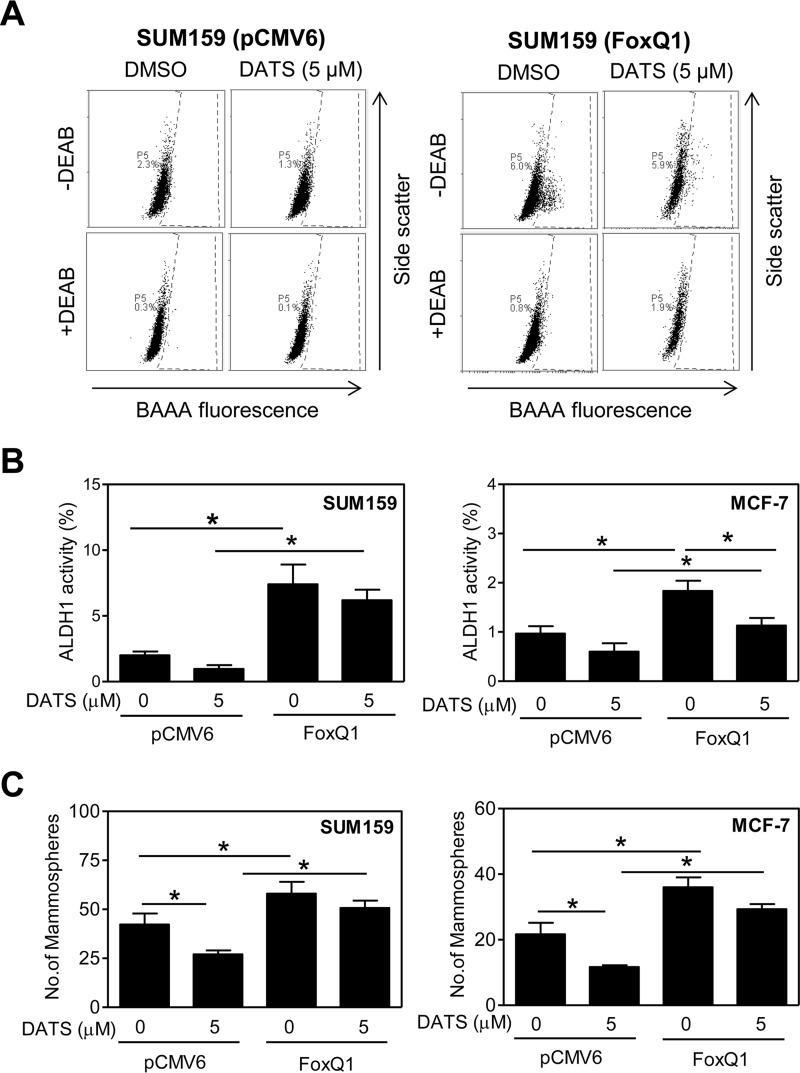
The Effect of FoxQ1 Overexpression on bCSC Inhibition by DATS
Fig. 4A shows flow histograms for the effect of DATS treatment on ALDH1 activity in empty vector-transfected control cells and FoxQ1-overexpressing SUM159 cells. Similar to untransfected cells, ALDH1 activity was decreased upon 72-h treatment with 5 μm DATS in both cell lines (Fig. 4B). In addition, FoxQ1 overexpression conferred partial but statistically significant protection against ALDH1 activity inhibition by DATS in both cell lines (Fig. 4B). Consistent with ALDH1 activity data, DATS-mediated inhibition of mammosphere formation was partially but statistically significantly attenuated by FoxQ1 overexpression in both cells (Fig. 4C).
FIGURE 4.
FoxQ1 overexpression is protective against bCSC inhibition by DATS in MCF-7 and SUM159 cells. A, representative flow histograms for ALDH1 activity in empty vector-transfected or FoxQ1-overexpressing SUM159 cells after 72-h treatment with DMSO or DATS. B, quantitation of ALDH1 activity in empty vector-transfected or FoxQ1-overexpressing MCF-7 and SUM159 cells after 72-h treatment with DMSO or 5 μm DATS (mean ± S.D., n = 3). C, number of mammospheres in empty vector-transfected or FoxQ1-overexpressing MCF-7 and SUM159 cells after 5 (MCF-7) or 7 (SUM159) days of treatment with DMSO or 5 μm DATS (mean ± S.D., n = 3). *, p < 0.05 between the indicated groups by one-way ANOVA followed by Bonferroni's multiple comparisons test. Each experiment was repeated at least twice, and data from one such experiment are shown. DEAB, diethylaminobenzaldehyde; BAAA, BODIPYTM-aminoacetaldehyde.
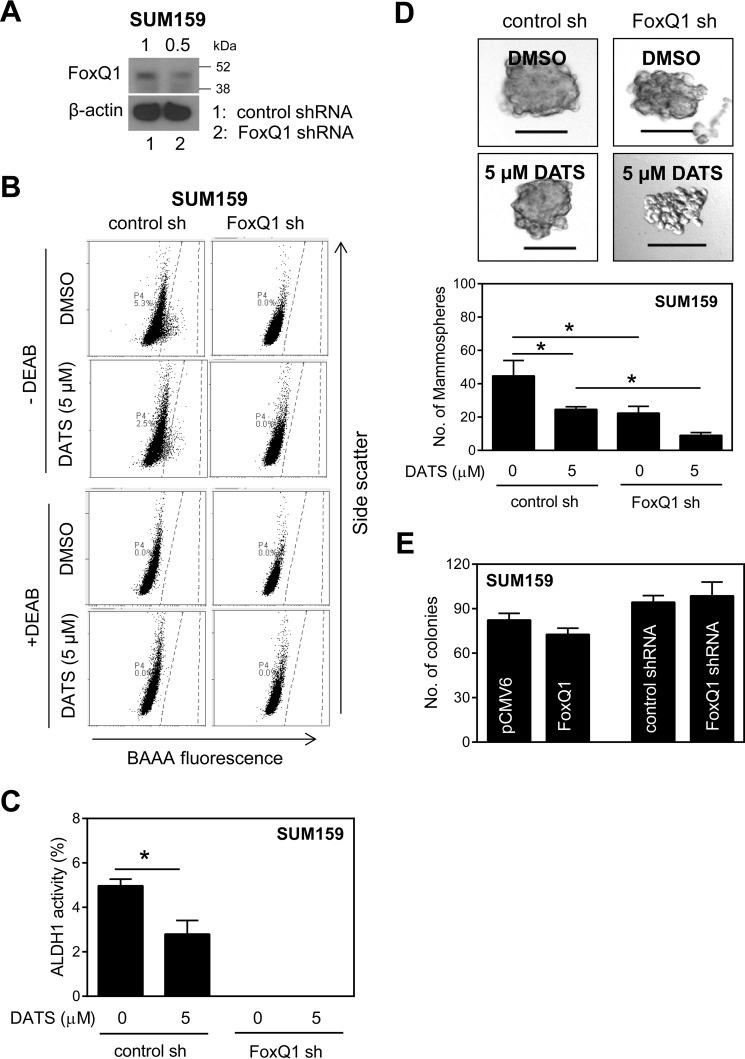
FoxQ1 Knockdown Augmented DATS-mediated Inhibition of bCSC in SUM159 Cells
The contribution of FoxQ1 in bCSC inhibition by DATS was further tested by its knockdown using SUM159 cells. The level of FoxQ1 protein was decreased by 50% in cells stably transfected with a FoxQ1-targeted shRNA (Fig. 5A). FoxQ1 knockdown alone resulted in complete inhibition of ALDH1 activity (Fig. 5, B and C) and significant inhibition of mammosphere formation (Fig. 5D). DATS-mediated inhibition of mammosphere formation was significantly augmented by FoxQ1 knockdown (Fig. 5D). These results provided additional experimental evidence for the involvement of FoxQ1 in bCSC inhibition by DATS.
FIGURE 5.
DATS-mediated inhibition of bCSC is augmented by knockdown of FoxQ1 in SUM159 cells. A, Western blotting analysis showing FoxQ1 knockdown. B, representative flow histograms for ALDH1 activity in SUM159 cells stably transfected with a control shRNA or a FoxQ1-targeted shRNA and treated for 72 h with DMSO or 5 μm DATS. C, quantitation of ALDH1 activity in SUM159 cells stably transfected with a control shRNA or a FoxQ1-targeted shRNA and treated for 72 h with DMSO or 5 μm DATS (mean ± S.D., n = 3). D, representative mammosphere images after 7 days of treatment with DMSO or 5 μm DATS in SUM159 cells stably transfected with a control shRNA or a FoxQ1-targeted shRNA (×100 magnification, scale bars = 100 μm). Bottom panel, quantitation of mammosphere number in SUM159 cells stably transfected with a control shRNA or a FoxQ1-targeted shRNA (mean ± S.D., n = 3). *, p < 0.05 between the indicated groups by one-way ANOVA followed by Bonferroni's multiple comparisons test (C and D). E, number of colonies in FoxQ1-overexpressing or FoxQ1 knockdown SUM159 cells (mean ± S.D., n = 3). DEAB, diethylaminobenzaldehyde; BAAA, BODIPYTM-aminoacetaldehyde.
SUM159 Cell Proliferation Was Not Affected by FoxQ1 Expression
Previous work has demonstrated inhibition of breast cancer cell proliferation in vitro upon DATS treatment (19). Therefore, it was of interest to determine whether these effects were altered by FoxQ1 expression status. Because FoxQ1 overexpression or its knockdown failed to alter colony formation (Fig. 5E), the effect of DATS treatment on colony formation was not investigated.
FoxQ1 Overexpression Altered the Expression of Genes Involved in Cancer Stem Cell Maintenance
To gain insights into the mechanism(s) downstream of FoxQ1 in bCSC inhibition by DATS, we determined the effect of FoxQ1 overexpression on genes (84-gene array) involved in cancer stem cell maintenance. FoxQ1 overexpression resulted in a significant (p ≤ 0.05) up-regulation of 11 genes (ABCG2, ALDH1A1, CD24, DDR1, DLL1, IL8, ITGA2, ITGA6, MYC, PLAT, and TWIST2) and down-regulation of 16 genes (ALCAM, DACH1, DKK1, EPCAM, FOXA2, FZD7, JAK2, KITLG, KLF17, MUC1, POU5F1, PROM1, PTPRC, STAT3, TWIST1, and ZEB1) (Fig. 6, A and B). Consistent with the results of ALDH1 activity determination, FoxQ1-overexpressing cells exhibited a significant increase in the expression of ALDH1A1 and ITGA6 (CD49f) (Fig 6B). Quantitative RT-PCR for a panel of genes was performed to confirm the results of the RT2 profiler PCR array. As can be seen in Fig. 6C, FoxQ1 overexpression resulted in significant down-regulation of DACH1 and ZEB1 but up-regulation of MYC and TWIST2 (Fig. 6C). DATS treatment in empty vector-transfected SUM159 cells resulted in a statistically significant increase in DACH1 mRNA level that was abolished in the FoxQ1-overexpressing variant (Fig. 6D). A modest but significant decrease in MYC mRNA level was observed upon DATS treatment in empty vector-transfected control SUM159 cells, but this effect was restored in FoxQ1-overexpressing cells (Fig. 6D). In accordance with Fig. 6C, we observed suppression of DACH1 protein in FoxQ1-overexpressing SUM159 cells and an increase in DACH1 protein expression in SUM159 cells transfected with a FoxQ1-targeted shRNA (Fig. 6E). These results suggested that DACH1 might be the regulator of bCSC inhibition downstream of FoxQ1 in DATS-treated cells.
FIGURE 6.
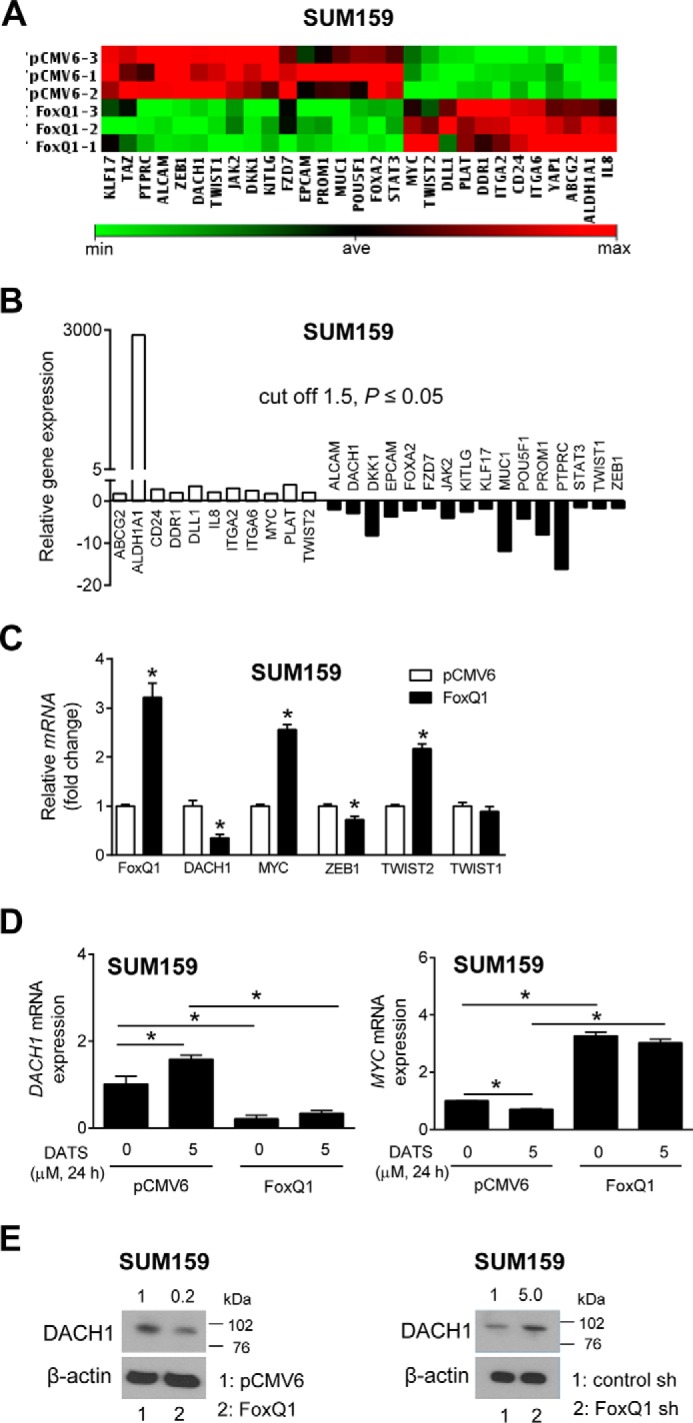
DACH1 is a novel target of FoxQ1. A, cluster analysis showing the difference in gene expression between empty vector-transfected control cells and FoxQ1-overexpressing SUM159 cells. min, minimum; ave, average; max, maximum. B, change in expression of genes related to the cancer stem cell-like phenotype in FoxQ1-overexpressing SUM159 cells relative to empty vector-transfected control cells. Three independently prepared samples from each group were used for gene expression profiling. The cutoff was 1.5-fold change and p ≤ 0.05 by two-sided Student's t test. C, quantitation of FoxQ1, DACH1, MYC, ZEB1, TWIST2, and TWIST1 mRNA expression by quantitative PCR in empty vector-transfected cells or FoxQ1-overexpressing SUM159 cells (mean ± S.D., n = 3). *, p < 0.05 by two-sided Student's t test. D, quantitation of DACH1 and MYC mRNA expression in DATS-treated (24 h) empty vector-transfected cells or FoxQ1-overexpressing SUM159 cells (mean ± S.D., n = 3). *, p < 0.05 between the indicated groups by one-way ANOVA followed by Bonferroni's multiple comparisons test. E, Western blotting for DACH1 expression using lysates from FoxQ1-overexpressing and FoxQ1 knockdown SUM159 cells.
FoxQ1 Regulated DACH1 Transcription
Because expression of DACH1 was decreased in FoxQ1-overexpressing cells, we first determined the effect of DACH1 on the bCSC fraction. MDA-MB-231 breast cancer cells stably transfected with a Ponasterone A-inducible DACH1 expression vector demonstrated inhibition of ALDH1 activity upon addition of Ponasterone A (Fig. 7A). In addition, DATS-mediated inhibition of ALDH1 activity was augmented by DACH1 overexpression (Fig. 7A). A similar trend was observed for mammosphere formation (Fig. 7A). Next, we performed a ChIP assay to determine whether DACH1 is a target of FoxQ1 using SUM159 cells. Bieller et al. (40) have identified the FoxQ1 binding sequence of 5′-A(A/T)TGTTTA(G/T)(A/T)T-3′. Two putative FoxQ1 binding sites were found in the DACH1 promoter (Fig. 7B). The ChIP assay revealed recruitment of FoxQ1 at both sites of the DACH1 promoter (Fig. 7C). Collectively, these results indicated that DACH1 expression was negatively regulated by FoxQ1.
FIGURE 7.
FoxQ1 is a negative regulator of DACH1 expression. A, ALDH1 activity or number of mammospheres in inducible DACH1-overexpressing MDA-MB-231 cells (mean ± S.D., n = 3). Ponasterone A (Pon A, 2 μm) was used to induce DACH1 expression. Inset, Western blotting for DACH1 protein. *, p < 0.05 between the indicated groups by one-way ANOVA followed by Bonferroni's multiple comparisons test. B, putative FoxQ1 binding sites on the DACH1 promoter. C, ChIP assay showing recruitment of FoxQ1 at the DACH1 promoter. The results shown are mean ± S.D. (n = 3). *, p < 0.05 between the indicated groups by two-sided Student's t test. D, representative immunohistochemical image for FoxQ1 expression in a normal breast section and a TNBC section (×200 magnification, scale bars = 100 μm). E, quantitation of FoxQ1 expression in normal breast and TNBC sections. The result shown is the mean H score for FoxQ1 expression (error bar, S.D.). Statistical significance of difference compared with normal breast section was analyzed by two-sided Student's t test.
FoxQ1 Was Overexpressed in TNBC Compared with Normal Breast
To determine the association of FoxQ1 expression with clinical features, we performed immunohistochemical analysis using commercially available tissue microarrays of TNBC and normal breast. Fig. 7D shows representative immunohistochemical image for FoxQ1 expression in a normal breast specimen and a TNBC section. Expression of FoxQ1 was very low in normal breast. On the other hand, FoxQ1 expression was statistically significantly higher in the nucleus of TNBC (n = 97) compared with normal breast tissue (n = 57, p = 0.01 by two-sided Student's t test) (Fig. 7E). These observations provide strong evidence for the importance of FoxQ1 in aggressive types of breast cancer.
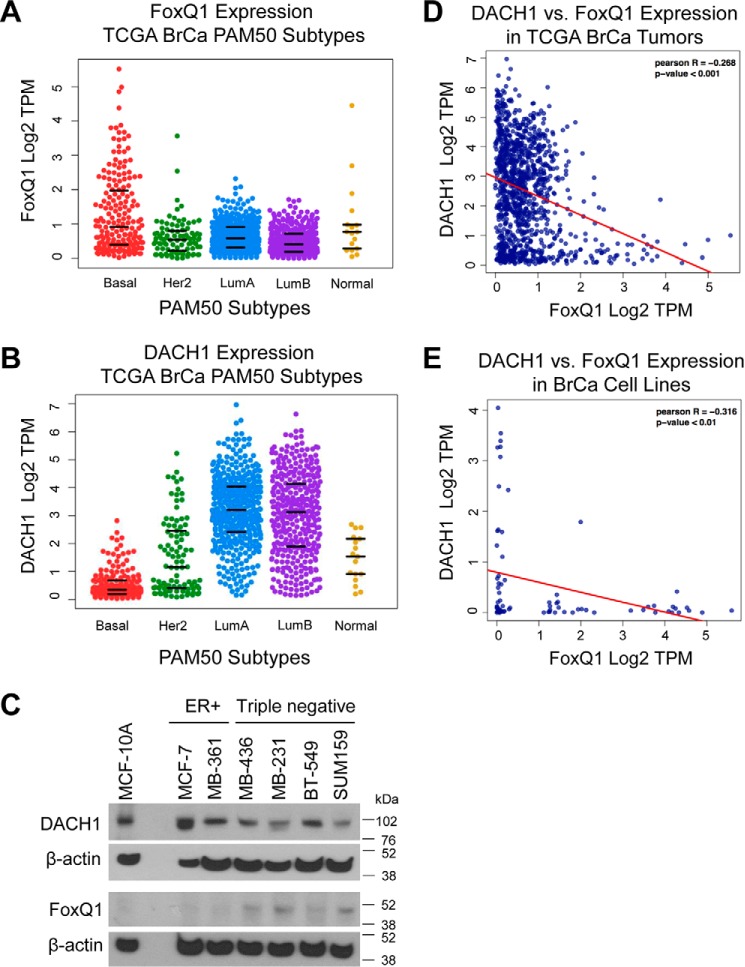
Inverse Association between FoxQ1 and DACH1 Expression in Breast Tumors and Breast Cancer Cell Lines
To further explore the clinical relevance of our findings, we analyzed RNA sequencing expression data in an expanded cohort of human tumors (n = 1078) from The Cancer Genome Atlas (TCGA) and cell lines (n = 77) from the Cancer Cell Line Encyclopedia and Gene Expression Omnibus. We found that FoxQ1 expression was higher in basal-type breast tumors compared with luminal breast cancers (p < 0.001) (Fig. 8A), consistent with our findings of elevated FoxQ1 protein measured by immunohistochemistry in TNBC (Fig. 7E). On the other hand, DACH1 expression was higher in luminal A/B and normal-like breast tumors than in the basal subtype (Fig. 8B). Western blotting confirmed the relatively higher expression of FoxQ1 protein in TNBC cell lines compared with MCF-10A or luminal-type breast cancer cells (MCF-7 and MDA-MB-361) (Fig. 8C). Furthermore, DACH1 and FoxQ1 showed a statistically significant negative correlation in tumors and cell lines (Fig. 8, D and E), supporting our data on FoxQ1 overexpression being associated with lower DACH1 levels.
FIGURE 8.
FoxQ1 and DACH1 exhibit inverse correlation in expression in breast tumors and breast cancer cell lines. A, FoxQ1 expression in molecular subtypes of breast cancer (TCGA, n = 1078). B, DACH1 expression in molecular subtypes of breast cancer (TCGA, n = 1078). The bottom, center, and top lines in panels A and B represent the 25th percentile, median, and 75th percentile, respectively. C, immunoblots for expression of DACH1 and FoxQ1 proteins in normal and breast cancer cell lines. D, DACH1 versus FoxQ1 expression in TCGA breast tumors. E, DACH1 versus FoxQ1 expression in breast cancer cell lines (n = 77). TPM, transcript per million.
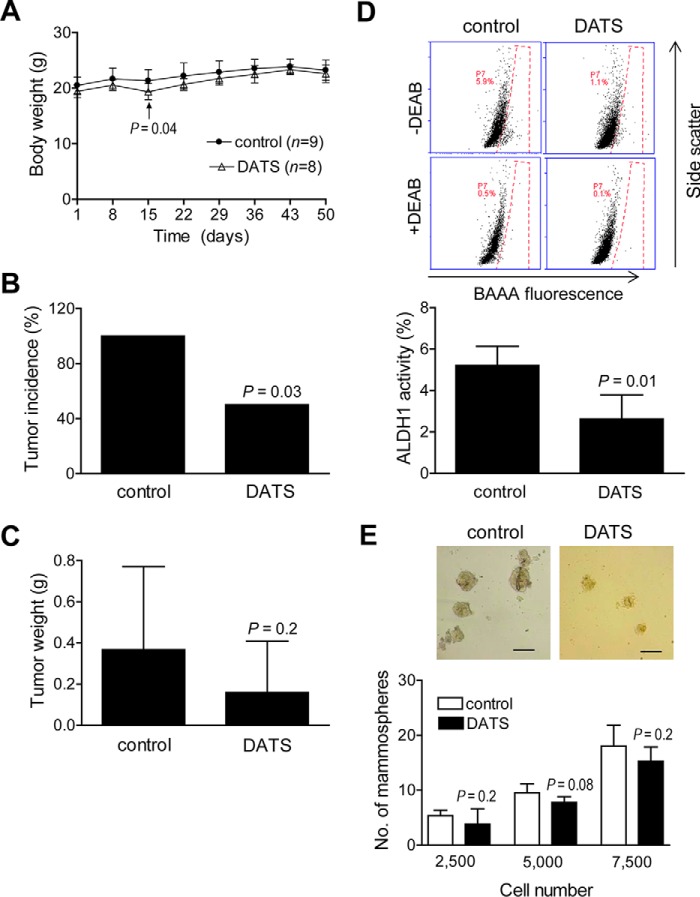
DATS Administration Decreased the bCSC Fraction in Vivo
We next determined the in vivo efficacy of DATS for inhibition of bCSC using a SUM159 xenograft model. The body weights of the mice of both groups did not differ significantly except on day 15 (Fig. 9A). The tumor incidence was 100% in vehicle-treated control mice, but it was reduced by 50% in the DATS treatment group (Fig. 9B, p = 0.03 by Fisher's exact test). The average wet tumor weight was also lower in the DATS treatment group compared with the control group, but the difference did not reach statistical significance because of the large data scatter as well as small sample size in the DATS treatment group (Fig. 9C, p = 0.2 by Student's t test). Fig. 9D shows flow histograms for ALDH1 activity in tumor cells from the control and DATS treatment groups. The ALDH1 activity was about 50% lower in tumor cells from DATS-treated mice compared with the vehicle-treated control (p = 0.01 by Student's t test). Fig. 9E depicts mammospheres resulting from representative tumor of a vehicle-treated control mouse and a DATS-treated mouse. The number of mammospheres in the DATS treatment group was generally lower compared with the control at each seeding density, but the difference was not significant, likely because of the small sample size. Together, these results provide in vivo evidence for bCSC inhibition by DATS.
FIGURE 9.
DATS administration inhibits bCSC in vivo. A, body weights over time for control mice and those treated with DATS. The results shown are mean ± S.D. (n = 9 for the control and 8 for the DATS group except on day 50, where n = 8 for the control). Statistical significance of difference was analyzed by two-sided Student's t test. B, tumor incidence in the control and DATS treatment groups. The p value was calculated by two-sided Fisher's exact test. C, average tumor weight in control and DATS-treated mice. The results shown are mean ± S.D. (n = 9 for the control and 8 for the DATS group). The p value was calculated by two-sided Student's t test. D, representative flow histograms for ALDH1 activity. Bottom panel, percentage of ALDH1 activity from tumors of control and DATS-treated mice (mean ± S.D., n = 7 for the control and 3 for the DATS group). The p value was calculated by two-sided Student's t test. E, representative images of mammospheres from the control and DATS treatment groups (×100 magnification, scale bars = 300 μm). Bottom panel, number of mammospheres from tumors of control and DATS-treated mice. The results shown are mean ± S.D. (n = 7 for the control and 4 for the DATS group). The p value was calculated by two-sided Student's t test. DEAB, diethylaminobenzaldehyde; BAAA, BODIPYTM-aminoacetaldehyde.
Discussion
A non-endocrine preventive strategy effective against different subtypes of breast cancer is still a clinical need because currently available options are ineffective against ER-negative disease (41–43). Rare but serious and often life-threatening side effects are another limitation of the clinically used breast cancer preventive interventions such as tamoxifen (41, 42). DATS appears promising for the prevention of breast cancer based on the following considerations: population-based studies suggest that garlic intake may lower breast cancer risk (8, 9); dietary administration of garlic powder inhibits the incidence of chemically induced mammary cancer in rats (11, 12); DATS treatment inhibits the growth of cultured human breast cancer cells irrespective of the ER-α status (19); oral administration of DATS inhibits in vivo growth of MCF-7 xenografts in mice (18); a clinical intervention with 200 mg of synthetic DATS (Allitridum) every day plus 100 μg of selenium every other day for 1 month showed acceptable tolerance (44); and bCSC fraction is decreased upon treatment with DATS in breast cancer cells representing luminal and basal subtypes, and DATS administration causes a decrease in ALDH1 activity in vivo in SUM159 xenografts (this study). Noticeably, bCSC inhibition by DATS is evident at a non-cytotoxic concentration. For example, ALDH1 activity is inhibited by about 50% in the presence of 5 μm DATS (this study), whereas a much higher concentration is necessary to induce apoptosis in proliferating mammary tumor cells (19). Nevertheless, the DATS concentration required to inhibit bCSC is likely achievable because the maximum blood concentration was shown to be ∼31 μm in rats after a single intravenous injection of 10 mg of DATS (45).
This study reveals that FoxQ1 partly contributes to bCSC inhibition by DATS. We found protection against DATS-mediated inhibition of ALDH1 activity and mammosphere formation by FoxQ1 overexpression, and this effect was not a cell line-specific response. Conversely, FoxQ1 knockdown sensitizes bCSC to DATS. The role of FoxQ1 in mammary cancer development is still not fully understood, but its expression is increased in the aggressive basal-like breast cancer subtype (39). This study also reveals FoxQ1 overexpression in TNBC specimens compared with normal breast. FoxQ1 expression is also strongly associated with poor survival in breast cancer patients (39). FoxQ1 overexpression in epithelial ovarian cancer cells relative to normal cells has also been noted (46). Here we show that DACH1 is a novel target of FoxQ1 and that DATS-mediated inhibition of bCSC is regulated by the FoxQ1-DACH1 signaling axis. DACH1 expression is very high in normal human breast epithelium but lost in TNBC (47). Analyses of the TCGA and breast cancer cell line databases indicate higher DACH1 expression in luminal and normal-like breast cancers than in the basal subtype of breast cancer (Fig. 8). This study also reveals a negative correlation between FoxQ1 and DACH1 expression in breast tumors and breast cancer cell lines. However, it is still unclear whether the negative correlation between FoxQ1 and DACH1 expression is unique for breast cancer.
Induced expression of FoxQ1 in mammary epithelial cells results in epithelial-mesenchymal transition and gain of stem cell-like properties (39, 48). Epithelial-mesenchymal transition induction by Snail or Twist overexpression in mammary epithelial cells is accompanied by enrichment in the CD44high/CD24low population with an increased capacity for self-renewal (49). Prior work from our own laboratory has revealed inhibition of the biochemical features of epithelial-mesenchymal transition upon DATS treatment, at least in the MDA-MB-231 breast cancer cell line (19). The possibility that bCSC inhibition by DATS engages Snail and/or Twist cannot be excluded. The mechanism underlying DATS-mediated down-regulation of FoxQ1 is also unclear and requires further investigation. A recent study showed that FoxQ1 is a direct target of Wnt in colorectal cancer (50). NAC1 protein has been shown to be essential and sufficient for activation of FoxQ1 transcription in ovarian cancer (51). Therefore, it is also possible that DATS treatment suppresses Wnt and/or NAC1 to down-regulate FoxQ1 in breast cancer cells.
Inhibition of bCSC by DATS in MCF-7 cells with low/undetectable FoxQ1 protein expression attests to alternate mechanism(s) in bCSC inhibition by DATS. Expression and activity of ER-α is decreased by DATS treatment (20). Previous studies have shown that ER-α signaling regulates bCSC through miR-140 targeting of SOX2 (52). Estrogen promotes the stem cell-like properties and invasiveness of ER-positive breast cancer cells via Gli1 activation (53). It is possible that down-regulation of ER-α contributes to bCSC inhibition by DATS. However, further work is necessary to experimentally test this possibility.
In conclusion, this study shows, for the first time, that DATS treatment inhibits the bCSC fraction in vitro and in vivo. We show further that bCSC inhibition by DATS is partly regulated by the FoxQ1-DACH1 signaling axis. These results, together with findings published previously (18–20), suggest that DATS may be useful for the prevention of breast cancer.
Author Contributions
S. H. K. and S. V. S. conceived the study. S. H. K., C. H. K., N. P., A. V. L., and S. V. S. coordinated the study and performed the experiments. S. H. K., C. H. K., N. P., A. V. L., and S. V. S. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
This research project used the Flow Cytometry Facility, Cell and Tissue Imaging Facility, and Animal Facility supported in part by the Cancer Center Support Grant P30 CA047904, National Cancer Institute.
This work was supported by NCI, National Institutes of Health Grant RO1 CA142604 (to S. V. S.). This research was supported in part by the University of Pittsburgh Center for Simulation and Modeling and the Pittsburgh Genome Resource Repository (PGRR). Development and use of the PGRR has been approved by the University of Pittsburgh Institutional Review Board (PRO12090374). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DATS
- diallyl trisulfide
- DADS
- diallyl disulfide
- DAS
- diallyl sulfide
- ER-α
- estrogen receptor-α
- bCSC
- breast cancer stem cell(s)
- ESA
- epithelial specific antigen
- TNBC
- triple-negative breast cancer
- TCGA
- The Cancer Genome Atlas
- ALDH
- aldehyde dehydrogenase
- ANOVA
- analysis of variance.
References
- 1. Rivlin R. S. (2001) Historical perspective on the use of garlic. J. Nutr. 131, 951S–954S [DOI] [PubMed] [Google Scholar]
- 2. Rahman K. (2001) Historical perspective on garlic and cardiovascular disease. J. Nutr. 131, 977S–979S [DOI] [PubMed] [Google Scholar]
- 3. Block E. (1985) The chemistry of garlic and onions. Sci. Am. 252, 114–119 [DOI] [PubMed] [Google Scholar]
- 4. Agarwal K. C. (1996) Therapeutic actions of garlic constituents. Med. Res. Rev. 16, 111–124 [DOI] [PubMed] [Google Scholar]
- 5. Milner J. A. (2006) Preclinical perspectives on garlic and cancer. J. Nutr. 136, 827S–831S [DOI] [PubMed] [Google Scholar]
- 6. Herman-Antosiewicz A., Powolny A. A., and Singh S. V. (2007) Molecular targets of cancer chemoprevention by garlic-derived organosulfides. Acta Pharmacol. Sin. 28, 1355–1364 [DOI] [PubMed] [Google Scholar]
- 7. Antony M. L., and Singh S. V. (2011) Molecular mechanisms and targets of cancer chemoprevention by garlic-derived bioactive compound diallyl trisulfide. Indian J. Exp. Biol. 49, 805–816 [PMC free article] [PubMed] [Google Scholar]
- 8. Challier B., Perarnau J. M., and Viel J. F. (1998) Garlic, onion and cereal fibre as protective factors for breast cancer: a French case-control study. Eur. J. Epidemiol. 14, 737–747 [DOI] [PubMed] [Google Scholar]
- 9. Levi F., La Vecchia C., Gulie C., and Negri E. (1993) Dietary factors and breast cancer risk in Vaud, Switzerland. Nutr. Cancer 19, 327–335 [DOI] [PubMed] [Google Scholar]
- 10. Dorant E., van den Brandt P. A., and Goldbohm R. A. (1995) Allium vegetable consumption, garlic supplement intake, and female breast carcinoma incidence. Breast Cancer Res. Treat. 33, 163–170 [DOI] [PubMed] [Google Scholar]
- 11. Schaffer E. M., Liu J. Z., Green J., Dangler C. A., and Milner J. A. (1996) Garlic and associated allyl sulfur components inhibit N-methyl-N-nitrosourea induced rat mammary carcinogenesis. Cancer Lett. 102, 199–204 [DOI] [PubMed] [Google Scholar]
- 12. Liu J., Lin R. I., and Milner J. A. (1992) Inhibition of 7,12-dimethylbenz[a]anthracene-induced mammary tumors and DNA adducts by garlic powder. Carcinogenesis 13, 1847–1851 [DOI] [PubMed] [Google Scholar]
- 13. Gued L. R., Thomas R. D., and Green M. (2003) Diallyl sulfide inhibits diethylstilbestrol-induced lipid peroxidation in breast tissue of female ACI rats: implications in breast cancer prevention. Oncol. Rep. 10, 739–743 [PubMed] [Google Scholar]
- 14. Ebrahimi M., Mohammad Hassan Z., Mostafaie A., Zare Mehrjardi N., and Ghazanfari T. (2013) Purified protein fraction of garlic extract modulates cellular immune response against breast transplanted tumors in BALB/c mice model. Cell J. 15, 65–75 [PMC free article] [PubMed] [Google Scholar]
- 15. Modem S., DiCarlo S. E., and Reddy T. R. (2012) Fresh garlic extract induces growth arrest and morphological differentiation of MCF7 breast cancer cells. Genes Cancer 3, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nkrumah-Elie Y. M., Reuben J. S., Hudson A., Taka E., Badisa R., Ardley T., Israel B., Sadrud-Din S. Y., Oriaku E., and Darling-Reed S. F. (2012) Diallyl trisulfide as an inhibitor of benzo(a)pyrene-induced precancerous carcinogenesis in MCF-10A cells. Food Chem. Toxicol. 50, 2524–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bauer D., Mazzio E., Soliman K. F., Taka E., Oriaku E., Womble T., and Darling-Reed S. (2014) Diallyl disulfide inhibits TNFα-induced CCL2 release by MDA-MB-231 cells. Anticancer Res. 34, 2763–2770 [PMC free article] [PubMed] [Google Scholar]
- 18. Na H. K., Kim E. H., Choi M. A., Park J. M., Kim D. H., and Surh Y. J. (2012) Diallyl trisulfide induces apoptosis in human breast cancer cells through ROS-mediated activation of JNK and AP-1. Biochem. Pharmacol. 84, 1241–1250 [DOI] [PubMed] [Google Scholar]
- 19. Chandra-Kuntal K., Lee J., and Singh S. V. (2013) Critical role for reactive oxygen species in apoptosis induction and cell migration inhibition by diallyl trisulfide, a cancer chemopreventive component of garlic. Breast Cancer Res. Treat. 138, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hahm E. R., and Singh S. V. (2014) Diallyl trisulfide inhibits estrogen receptor-α activity in human breast cancer cells. Breast Cancer Res. Treat. 144, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Velasco-Velázquez M. A., Homsi N., De La Fuente M., and Pestell R. G. (2012) Breast cancer stem cells. Int. J. Biochem. Cell Biol. 44, 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Brien C. S., Farnie G., Howell S. J., and Clarke R. B. (2011) Breast cancer stem cells and their role in resistance to endocrine therapy. Horm. Cancer 2, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shah M., and Allegrucci C. (2012) Keeping an open mind: highlights and controversies of the breast cancer stem cell theory. Breast Cancer 4, 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaschula C. H., Hunter R., Stellenboom N., Caira M. R., Winks S., Ogunleye T., Richards P., Cotton J., Zilbeyaz K., Wang Y., Siyo V., Ngarande E., and Parker M. I. (2012) Structure-activity studies on the anti-proliferation activity of ajoene analogues in WHCO1 oesophageal cancer cells. Eur. J. Med. Chem. 50, 236–254 [DOI] [PubMed] [Google Scholar]
- 25. Wu K., Chen K., Wang C., Jiao X., Wang L., Zhou J., Wang J., Li Z., Addya S., Sorensen P. H., Lisanti M. P., Quong A., Ertel A., and Pestell R. G. (2014) Cell fate factor DACH1 represses YB-1-mediated oncogenic transcription and translation. Cancer Res. 74, 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S. H., Sehrawat A., and Singh S. V. (2013) Dietary chemopreventative benzyl isothiocyanate inhibits breast cancer stem cells in vitro and in vivo. Cancer Prev. Res. 6, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao D., Srivastava S. K., Lew K. L., Zeng Y., Hershberger P., Johnson C. S., Trump D. L., and Singh S.V. (2003) Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits proliferation of human prostate cancer cells by causing G2/M arrest and inducing apoptosis. Carcinogenesis 24, 891–897 [DOI] [PubMed] [Google Scholar]
- 28. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 29. Hahm E. R., Lee J., Kim S. H., Sehrawat A., Arlotti J. A., Shiva S. S., Bhargava R., and Singh S. V. (2013) Metabolic alterations in mammary cancer prevention by withaferin A in a clinically relevant mouse model. J. Natl. Cancer Inst. 105, 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilks C., Cline M. S., Weiler E., Diehkans M., Craft B., Martin C., Murphy D., Pierce H., Black J., Nelson D., Litzinger B., Hatton T., Maltbie L., Ainsworth M., Allen, P. et al. (2014) The Cancer Genomics Hub (CGHub): overcoming cancer through the power of torrential data. Database (Oxford) 10.1093/database/bau093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Daemen A., Griffith O. L., Heiser L. M., Wang N. J., Enache O. M., Sanborn Z., Pepin F., Durinck S., Korkola J. E., Griffith M., Hur J. S., Huh N., Chung J., Cope L., Fackler, M. J. et al. (2013) Modeling precision treatment of breast cancer. Genome Biol. 14, R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patro R., Duggal G., and Kingsford C. (2015) Accurate, fast, and model-aware transcript expression quantification with Salmon. bioRxiv 10.1101/021592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gendoo D. M., Ratanasirigulchai N., Schröder M. S., Paré L., Parker J. S., Prat A., and Haibe-Kains B. (2016) Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 32, 1097–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu T. H., Wu C. M., and Liou Y. C. (1989) Volatile compounds from garlic. J. Agric. Food Chem. 37, 725–730 [Google Scholar]
- 35. Block E., Ahmad S., Jain M.K., Crecely R.W., Apitz-Castro R., and Cruz M.R. (1984) The chemistry of alkyl thiosulfate esters: 8: (E, Z)-Ajoene: a potent antithrombotic agent from garlic. J. Am. Chem. Soc. 106, 8295–8296 [Google Scholar]
- 36. Naznin M. T., Akagawa M., Okukawa K., Maeda T., and Morita N. (2008) Characterization of E- and Z-ajoene obtained from different varieties of garlics. Food Chem. 106, 1113–1119 [Google Scholar]
- 37. Liu S., Dontu G., Mantle I. D., Patel S., Ahn N. S., Jackson K. W., Suri P., and Wicha M. S. (2006) Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 66, 6063–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jo M., Eastman B. M., Webb D. L., Stoletov K., Klemke R., and Gonias S. L. (2010) Cell signaling by urokinase-type plasminogen activator receptor induces stem cell-like properties in breast cancer cells. Cancer Res. 70, 8948–8958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qiao Y., Jiang X., Lee S. T., Karuturi R. K., Hooi S. C., and Yu Q. (2011) FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 71, 3076–3086 [DOI] [PubMed] [Google Scholar]
- 40. Bieller A., Pasche B., Frank S., Gläser B., Kunz J., Witt K., and Zoll B. (2001) Isolation and characterization of the human forkhead gene FOXQ1. DNA Cell Biol. 20, 555–561 [DOI] [PubMed] [Google Scholar]
- 41. Fisher B., Costantino J. P., Wickerham D. L., Redmond C. K., Kavanah M., Cronin W. M., Vogel V., Robidoux A., Dimitrov N., Atkins J., Daly M., Wieand S., Tan-Chiu E., Ford L., Wolmark N., et al. (1998) Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 study. J. Natl. Cancer Inst. 90, 1371–1388 [DOI] [PubMed] [Google Scholar]
- 42. Cauley J. A., Norton L., Lippman M. E., Eckert S., Krueger K. A., Purdie D. W., Farrerons J., Karasik A., Mellstrom D., Ng K. W., Stepan J. J., Powles T. J., Morrow M., Costa A., Silfen S. L., et al. (2001) Continued breast cancer risk reduction in postmenopausal women treated with raloxifene: 4-year results from the MORE trial: multiple outcomes of raloxifene evaluation. Breast Cancer Res. Treat. 65, 125–134 [DOI] [PubMed] [Google Scholar]
- 43. Goss P. E., Ingle J. N., Alés-Martínez J. E., Cheung A. M., Chlebowski R. T., Wactawski-Wende J., McTiernan A., Robbins J., Johnson K. C., Martin L. W., Winquist E., Sarto G. E., Garber J. E., Fabian C. J., Pujol P., et al. (2011) Exemestane for breast-cancer prevention in postmenopausal women. N. Eng. J. Med. 364, 2381–2391 [DOI] [PubMed] [Google Scholar]
- 44. Li H., Li H. Q., Wang Y., Xu H. X., Fan W. T., Wang M. L., Sun P. H., and Xie X. Y. (2004) An intervention study to prevent gastric cancer by micro-selenium and large dose of allitridum. Chin. Med. J. 117, 1155–1160 [PubMed] [Google Scholar]
- 45. Sun X., Guo T., He J., Zhao M., Yan M., Cui F., and Deng Y. (2006) Determination of the concentration of diallyl trisulfide in rat whole blood using gas chromatography with electron-capture detection and identification of its major metabolite with gas chromatography mass spectrometry. Yakugaku Zasshi. 126, 521–527 [DOI] [PubMed] [Google Scholar]
- 46. Gao M., Shih I. M., and Wang T. L. (2012) The role of forkhead box q1 transcription factor in ovarian epithelial carcinomas. Int. J. Mol. Sci. 13, 13881–13893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu K., Jiao X., Li Z., Katiyar S., Casimiro M. C., Yang W., Zhang Q., Willmarth N. E., Chepelev I., Crosariol M., Wei Z., Hu J., Zhao K., and Pestell R. G. (2011) Cell fate determination factor Dachshund reprograms breast cancer stem cell function. J. Biol. Chem. 286, 2132–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang H., Meng F., Liu G., Zhang B., Zhu J., Wu F., Ethier S. P., Miller F., and Wu G. (2011) Forkhead transcription factor Foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 71, 1292–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., and Weinberg R. A. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Christensen J., Bentz S., Sengstag T., Shastri V. P., and Anderle P. (2013) FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS ONE 8, e60051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao M., Wu R. C., Herlinger A. L., Yap K., Kim J. W., Wang T. L., and Shin I. M. (2014) Identification of the NAC1-regulated genes in ovarian cancer. Am. J. Pathol. 184, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y., Eades G., Yao Y., Li Q., and Zhou Q. (2012) Estrogen receptor α signaling regulates breast-tumor initiating cells by down-regulating miR-140 which targets the transcription factor SOX2. J. Biol. Chem. 287, 41514–41522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun Y., Wang Y., Fan C., Gao P., Wang X., Wei G., and Wei J. (2014) Estrogen promotes stemness and invasiveness of ER-positive breast cancer cells through Gli1 activation. Mol. Cancer 13, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]