Abstract
In Escherichia coli, cytoplasmic copper ions are toxic to cells even at the lowest concentrations. As a defense strategy, the cuprous oxidase CueO is secreted into the periplasm to oxidize the more membrane-permeable and toxic Cu(I) before it can enter the cytoplasm. CueO itself is a multicopper oxidase that requires copper for activity. Because it is transported by the twin-arginine translocation (Tat) pathway, which transports folded proteins, a requirement for cofactor assembly before translocation has been discussed. Here we show that CueO is transported as an apo-protein. Periplasmic CueO was readily activated by the addition of copper ions in vitro or under copper stress conditions in vivo. Cytoplasmic CueO did not contain copper, even under copper stress conditions. In vitro Tat transport proved that the cofactor assembly was not required for functional Tat transport of CueO. Due to the post-translocational activation of CueO, this enzyme contributes to copper resistance not only by its cuprous oxidase activity but also by chelation of copper ions before they can enter the cytoplasm. Apo-CueO was indistinguishable from holo-CueO in terms of secondary structural elements. Importantly, the binding of copper to apo-CueO greatly stabilized the protein, indicating a transformation from an open or flexible domain arrangement with accessible copper sites to a closed structure with deeply buried copper ions. CueO is thus the first example for a natural Tat substrate of such incomplete folding state. The Tat system may need to transport flexibly folded proteins in any case when cofactor assembly or quaternary structure formation occurs after transport.
Keywords: copper, metalloprotein, protein folding, protein translocation, stress response, Tat system
Introduction
In Escherichia coli, several systems are involved in copper homeostasis (1, 2), which serve to supply some copper to the periplasm and the cytoplasmic membrane, whereas copper is removed from the cytoplasm. The only established cytoplasmic copper protein is the regulator CueR, which has been demonstrated to sense copper with zeptomolar (10−21) affinity (3). In the copper-bound state, CueR activates transcription of the copper resistance genes encoding the copper efflux ATPase CopA (4) and the cuprous oxidase CueO (5, 6). CueO itself is a multicopper oxidase family enzyme that contains four copper ions involved in catalysis (6, 7). These copper ions are arranged in mononuclear type 1 and type 2 copper centers, and one binuclear type 3 copper center. One additional copper ion was found in crystals and represents copper bound in the active site (7, 8). The periplasmic oxidation of Cu(I) to Cu(II), as catalyzed by CueO, contributes to copper resistance of E. coli (9). The copper-induced damage to iron-sulfur cluster-containing hydrolases is the reason for the virtual exclusion of copper ions from the cytoplasm and for the absence of cytoplasmic copper-containing enzymes in E. coli (10). Besides CueO, E. coli contains three known copper enzymes in the periplasm: the superoxide dismutase SodC (11), the tyramine oxidase TynA (12), and the Cu(I)-binding protein CusF (13). Proteomic approaches indicated that CueO is the predominant soluble copper protein (14). In addition, two membrane integral copper enzymes are known, the Cyt bo3 ubiquinol oxidase (15) and the NADH-dehydrogenase 2 (16).
Although the periplasmic localization of CueO argues for a periplasmic copper assembly, its mode of translocation into the periplasm does not immediately fit to this idea: CueO is transported by the Tat system, which serves to translocate folded proteins across the cytoplasmic membrane, and it is generally assumed that such Tat substrates tightly fold and assemble their cofactors before translocation (17). However, it is unclear whether Tat substrates with simple copper centers generally need to assemble copper before Tat transport or not. Arguments for both scenarios have been reported (2, 18). As CueO specifically serves to combat copper stress, it was suggested that sufficient copper reaches the cytoplasm under such conditions (2). However, CueO is transported strictly Tat-dependently also without copper stress (19), and a copper content-reducing mutation in either the type 1 or the type 2 copper binding site did not affect transport (20), which prompted us to analyze whether wild type CueO might actually be translocated as an apo-protein.
We examined the transport of CueO in the absence of copper stress as well as during copper stress, purified CueO from cytoplasmic and periplasmic fractions, and analyzed copper content, activity, and the copper assembly process. In addition, we analyzed CueO transport in vitro and examined the effect of copper assembly on CueO folding, stability, and protease sensitivity. The data clearly demonstrate that CueO is translocated without any bound copper, and copper can be readily assembled after transport. Apo-CueO contains already the secondary structural elements folded in a way that suffices for Tat compatibility. However, CueO is markedly stabilized by the copper assembly, most likely reflecting a switch from a flexible domain assembly to a tightly closed state. A structural explanation for these observations is discussed.
Experimental Procedures
Strains and Growth Conditions
E. coli MC4100 (21) was used for physiological studies, and E. coli XL1-Blue Mrf′ Kan (Stratagene) was used for cloning. The bacteria were grown aerobically at 37 °C in LB medium (1% Tryptone, 1% NaCl, 0.5% yeast extract) in the presence of the appropriate antibiotics (100 μg/ml ampicillin, 20 μg/ml chloramphenicol). CueO production was induced with 0.1% rhamnose at an A600 nm of 0.8, and cells were harvested after 3 h post induction.
Genetic Methods and Plasmids
Mature CueO was produced using the rhamnose-inducible vector pBW-matCueO-strep. The full-length precursor form of CueO was produced using the vector pBW-CueO-strep. For both vectors, the respective CueO-encoding regions were amplified by PCR and cloned into the NdeI and BamHI restriction sites of the plasmid pBW22 (22). For signal peptide exchanges by Sec3 signal peptides, a NheI site was generated at the beginning of the mature CueO-encoding domain, the signal peptide-encoding regions of DsbA and MalE were amplified from chromosomal DNA with NdeI and NheI restriction site-generating primers, and the CueO signal peptide-encoding region was substituted. All constructs were confirmed by restriction analyses and sequencing.
Biochemical Methods
SDS-PAGE and protein estimations were carried out by standard methods (23, 24). Subcellular fractionations, Western blotting, and blot developments were carried out as described previously (25). Blots were developed with StrepTactin-HRP conjugate according to the manufacturer's instructions (IBA, Göttingen, Germany). Limited proteolysis was carried out on ice with 1.5 μg/ml trypsin and 0.3 mg/ml CueO for the indicated time intervals and stopped by the addition of 1 volume 2× SDS-PAGE sample buffers and immediate incubation at 95 °C for 5 min.
UV-visible spectra of holo-/apo-CueO were recorded on a JASCO V-650 UV-visible spectra spectrometer in 20 mm Tris/HCl, pH 8.0. The sample was degassed before measurement. For sequential reconstitution of apo-CueO, 120% of the amount needed for full occupancy was added sequentially in 6 steps at 20%, and spectra were recorded. We determined an ϵ280 nm of 61.39 mm−1cm−1, which is ∼16% lower than reported in literature (8), most likely due to protein estimation reasons.
Activity assays were performed in 20 mm Tris/HCl, pH 8.0, at 20 °C using 1 mm concentrations of the artificial chromogenic substrate dimethoxyphenol (DMP) and the indicated concentrations of CuSO4. The reaction was either started by the addition of DMP or by the addition of CuSO4 (in case of activations of apo-CueO). Oxidation of DMP was monitored at 470 nm. Enzyme activities were corrected for copper-only catalyzed oxidation rates and calculated using a millimolar extinction coefficient of 14.8 mm−1 cm−1. Quantitative copper and zinc determinations were carried out by inductively coupled plasma mass spectrometry.
For in vitro Tat transport assays, inverted membrane vesicles (IMVs) were prepared in accordance with published protocols (26, 27) with indicated modifications. Briefly, E. coli strain DADE/pABS-tatABC (28) was grown in phosphate-buffered medium (27) supplemented with 25 μg/ml chloramphenicol. Overnight cultures were inoculated into fresh medium (1:100) and grown at 37 °C for 4–5 h until the A600 reached ∼1.5. Cells were collected by centrifugation at 5000 × g for 10 min at 4 °C, and the cell pellet was resuspended in ice-cold buffer A (50 mm triethanolamine acetate, pH 7.5, 250 mm sucrose, 1 mm EDTA-KOH, pH 7.0, and 1× protease inhibitor mixture (PIC, P8465, Sigma); 12 ml of buffer A for 1 liter culture) supplied with freshly added 1 mm PMSF, 10 μg/ml DNase I, 0.1 mg/ml lysozyme, and 1 mm DTT. The cell suspension was incubated on ice for 20 min before cell disruption by two French press passages at 8000 p.s.i. to produce IMVs. After removal of cell debris by sedimentation at 8,000 × g for 10 min at 4 °C, the IMV suspension was layered over a 3-step sucrose gradient (0.77 m, 1.44 m, and 2.2 m sucrose, buffered with 50 mm triethanolamine acetate, pH 7.5, 1 mm EDTA-KOH, pH 7.0, 1 mm DTT, and 1× protease-inhibitor mix HP (Serva, Germany) and centrifuged for 16 h at 82,000 × g in a swing-out rotor. The band that formed between the 0.77 m and the 1.44 m sucrose layers was collected and diluted 1:4 with buffer B (50 mm triethanolamine acetate, 250 mm sucrose, 1 mm DTT and 1× protease inhibitor cocktail) and centrifuged for 1 h at 108,000 × g to collect the IMVs in the pellet. The pellet was resuspended in an appropriate volume of buffer B to have an A280 ≈ 50–60, and aliquots were frozen immediately with liquid nitrogen.
A standard in vitro transport reaction was set as per 200 μl of reaction volume containing 200 nm concentrations of apo-preCueO-strep or holo-preCueO-strep and IMVs at a final concentration of 10 units of A280 in transport buffer (20 mm HEPES, 40 mm KAc, 5 mm MgAc2, 250 mm sucrose) supplied with 50 μg/ml BSA, 1× protease-inhibitor mix HP with/without EDTA, 2.5 mm ATP, 25 mm creatine phosphate, 10 μg/ml creatine phosphokinase. In some experiments, 50 μm carbonyl cyanide m-chlorophenylhydrazone or in vitro refolded precursor of the high potential iron-sulfur protein (preHiPIP (25)) of the indicated concentrations was included in the reactions. The reactions were incubated at 30 °C for 45 min with gentle agitation and then quenched in an ice bath for 2 min. The IMVs were then purified through a 0.5 m sucrose cushion (20 mm HEPES, 0.5 m sucrose, 40 mm KAc, 5 mm MgAc2) by centrifugation at 130,000 × g for 45 min. The pellets were resuspended in 200 μl of transport buffer, and half of this IMV suspension was digested with 100 μg/ml thermolysin for 30 min on ice. After termination of the protease digestion by the addition of 10 mm EDTA, the IMV suspension as well as the other half of the mock-treated IMV suspension was mixed with 100 μl of 2× SDS sample buffer. Samples were boiled before separation on SDS-PAGE gels, and the CueO bands were visualized in Western blots with StrepTactin-HRP conjugates followed by ECL detection.
Differential scanning calorimetry (DSC) analyses were carried out using a Nano-DSC (TA Instruments) according to the manufacturer's instructions in a total volume of 600 μl containing 5.8 μm CueO in 10 mm Tris/HCl, pH 8.0. The equilibration time was 600 s, and heating was at 1 K/min at a pressure of 3 bar. Circular dichroism (CD) spectra were recorded on a JASCO J-815 spectropolarimeter (sample temperature 20 °C, Peltier thermostat, data interval 1 nm, scanning speed 50 nm/min, 1 nm bandwidth, 20 scans averaging, cell path length 0.1 cm) with a sample concentration of 5.8 μm.
Results
CueO Folds without Detectable Copper Recruitment in the Cytoplasm of E. coli
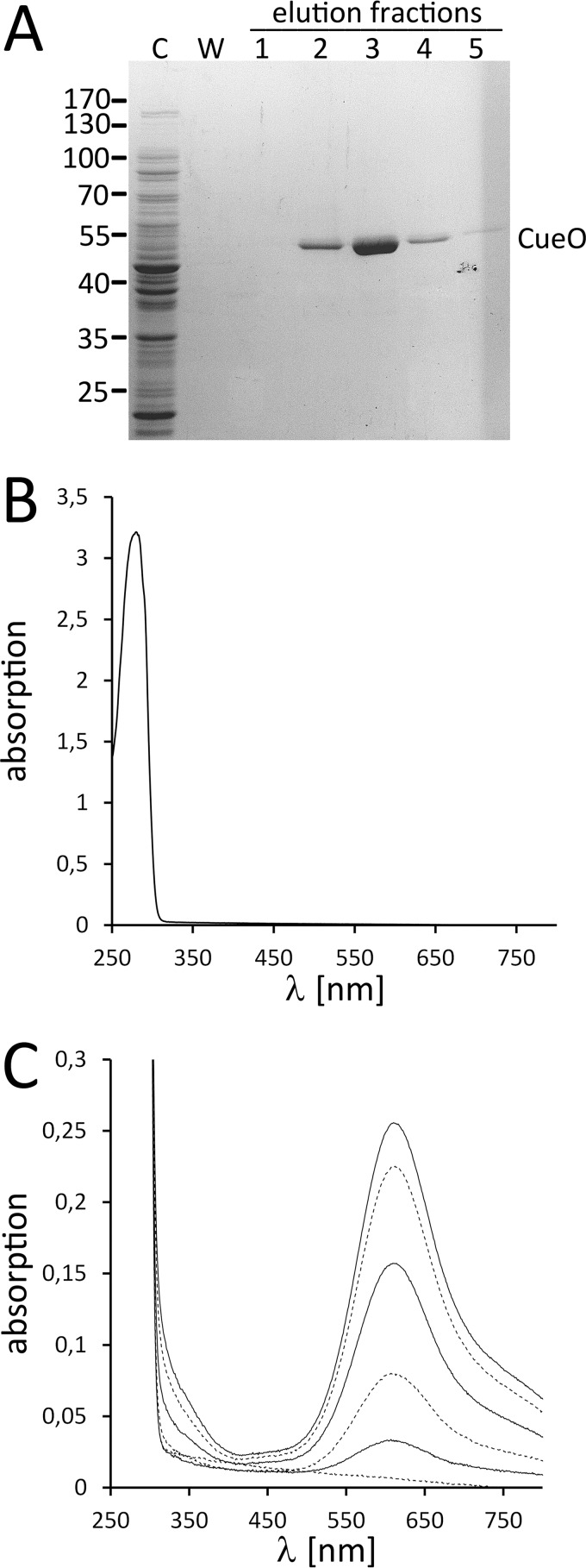
In a first approach, we aimed to find out whether CueO can fold to a functional copper-containing protein inside the cytoplasm without copper stress. For this purpose we produced signal peptide-deficient CueO in the cytoplasm of E. coli, purified the C-terminally Strep-tagged enzyme by affinity chromatography, and analyzed the copper content (Fig. 1). Early studies on the sequence of copper site assembly in CueO already demonstrated that type 1 and type 3 copper assemble first followed by type 2 copper (29, 30). Type 1 copper is blue due to a thiolate ligand charge transfer band near 600 nm, type 3 binuclear copper shows a weak absorbance near 330 nm, and type 2 copper hardly absorbs any light (8). As the light-absorbing copper centers are assembled first, UV-visible absorption spectra should readily indicate whether copper is bound to CueO or not. Cytoplasmic CueO did not show any copper absorption, indicating the absence of copper (Fig. 1B). This spectroscopic result was confirmed by quantitative inductively coupled plasma mass spectrometry analyses that indicated a copper content below the detection limit, which was at 0.016 copper atoms per CueO molecule. As Zn2+ can occupy copper sites without contributing to the visible spectrum due to its d10 electron configuration, we also analyzed the zinc content, which was also below the detection limit of 0.026 zinc atoms per CueO molecule. Together, these data showed that cytoplasmic CueO contained neither copper nor zinc.
FIGURE 1.
Cytoplasmic CueO does not contain detectable copper. A, affinity purification of Strep-tagged CueO from the cytoplasm of strain MC4100/pBW-matCueO-strep. Coomassie-stained SDS-PAGE of purification fractions are shown. C, cytoplasm; W, last wash fraction. The CueO band and the positions of marker proteins are indicated. B, UV-visible absorption spectrum of CueO as purified from the cytoplasm, showing the absence of copper cofactor absorption. C, titration of CuSO4 to apo-CueO (as shown in B) and gradual increase of the type 3 and type 1 copper absorption signals.
We then titrated substoichiometric amounts of CuSO4 to CueO and determined the gradual occupancy of the type 1 and type 3 copper sites. The assembly of the copper centers was readily detectable in the spectra (Fig. 1C). As expected for sites with similar affinity and accessibility, the assembly of type 1 and type 3 copper sites proceeded in parallel. At the end of the titration, the absorbance ratio A280 nm/A610 nm = 12.15 of the reconstituted enzyme indicated >96% occupancy of the type 1 copper site (8).
As previously structurally characterized, CueO with reduced copper content was only deficient in type 2 copper and still contained most type 1 and type 3 copper (30), it was possible that copper was essential for folding. We thus addressed the folded state of cytoplasmic apo-CueO that did not contain any copper. Apo-CueO was stable in solution, not showing any tendency to aggregate, which already implies a folded structure with hidden hydrophobic core regions. In agreement with this expectation, CD spectroscopy indicated a dominant contribution of β-sheet structures, as recognized by the minimum near 213 nm, and some α-helical structure most likely contributes to the crossing of the zero line near 200 nm (Fig. 2) (31). The spectrum was unaltered by copper assembly, indicating that occupancy of the copper sites does not significantly affect secondary structural elements, which are already formed in apo-CueO (Fig. 2). Together, the CD analyses show that copper-free apo-CueO folds to a large extent in the cytoplasm of E. coli.
FIGURE 2.
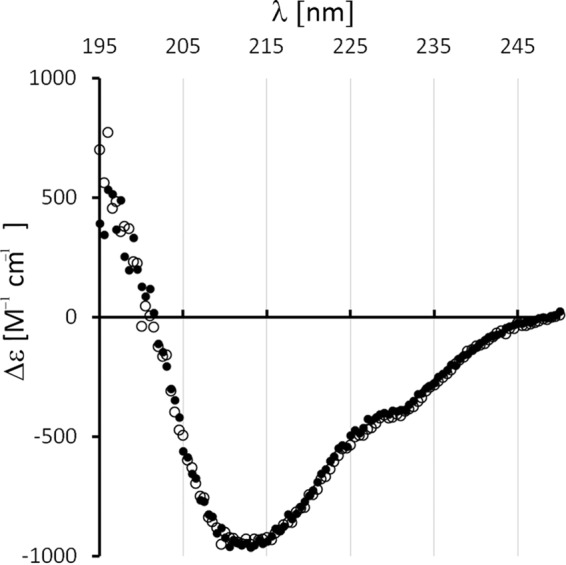
CueO folds already in the complete absence of copper cofactors. Far UV circular dichroism spectra of cytoplasmic apo-CueO (open circles) and copper-reconstituted holo-CueO (filled circles).
Folded Purified Cytoplasmic apo-CueO Can Be Readily Activated by Externally Added Copper Ions
Correct and complete copper assembly should result in active enzyme. To our knowledge, cytoplasmic copper-free apo-CueO has never been isolated, and we thus tested whether it could be converted to active holo-CueO by the addition of copper (Fig. 3). CuSO4 activated the enzyme immediately, indicating that copper can be rapidly assembled into the correct sites of CueO without the aid of specific assembly factors. The specific activity of copper-activated CueO was ∼12.5 units/mg with the chromogenic substrate DMP in the presence of 1 mm CuSO4. This is well in the range of CueO activities with similar assay setups described in literature (30, 32), and as already reported in these studies, reconstituted CueO could be further activated by higher copper concentrations (21.2 units/mg in the presence of 2.5 mm CuSO4), possibly due to the occupancy of additional copper sites (32). Cu(I) is the natural substrate of CueO, and the occupancy of the active site by Cu(II) at high copper concentrations exposes redox-active copper more near the surface and thereby most likely enhances the activity with organic chromogenic substrates. Due to its activity-enhancing effect, the occupied active site of CueO was initially regarded as a regulatory copper site (8, 32).
FIGURE 3.
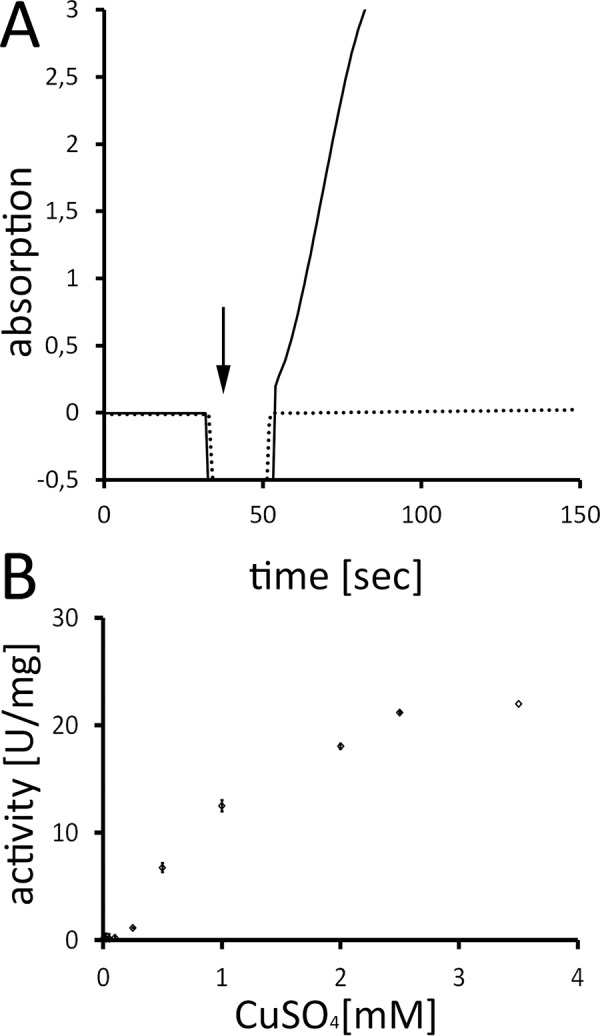
CueO is readily activated by externally added copper. A, exemplary spectroscopic assay of DMP-oxidase activity with apo-CueO from cytoplasm. Note that the enzyme was inactive until CuSO4 was added (indicated by an arrow), which activates the enzyme without requirement of additional factors (solid line). In the negative control, CuSO4 was added to an assay lacking enzyme (dotted line). Assay specifications: CueO, 0.6 μm; CuSO4, 250 μm; DMP, 1 mm; activity, 1.5 units/mg. B, CueO DMP oxidase activity depends on external copper concentrations. The diagram shows the specific activity of DMP oxidation at a range of copper concentrations. All data points are corrected for non-enzymatic copper effects and reflect the result of five replicates. The standard deviation is indicated.
CueO Assembles Copper in the Periplasm under Copper Stress Conditions
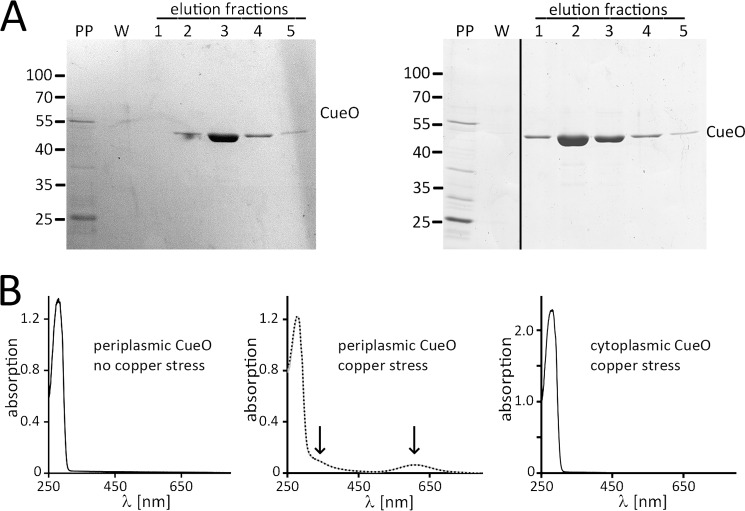
We then purified Strep-tagged CueO from the periplasmic fraction of cells grown in medium without or with added copper, analyzed its copper content (Fig. 4, A and B), and determined its activity. Purified periplasmic CueO migrated in a single mature protein band in SDS-PAGE analyses, indicating complete signal peptide cleavage (Fig. 4A). UV-visible spectra showed that CueO from the periplasmic fraction did not contain copper when cells were grown without copper stress, whereas periplasmic CueO contained copper when cells were grown in the presence of 1.5 mm CuSO4 (Fig. 4B). The spectrum showed typical absorption characteristics of the CueO holo-enzyme and was not altered by the addition of further exogenous copper, indicating that the type 1 and type 3 copper sites were fully occupied. There was some increased absorbance at ∼330 nm that might relate to occupancy of additional copper sites in the periplasm in vivo. Despite an estimated type 1 copper content of ∼83%, purified periplasmic holo-CueO had low activity (0.08 unit/mg), most likely due to loss of type 2 copper during purification, as type 2 copper is known to have lower affinity (30), and the loss of type 2 copper during purification is a known phenomenon also for other copper-enzymes (33). We also purified CueO from the cytoplasmic fraction of copper-stressed cells and found no assembled copper (Fig. 4B), indicating that (i) our washing steps before subcellular fractionations were sufficient to avoid copper contamination and that (ii) cytoplasmic CueO had no detectable access to copper even under copper stress conditions.
FIGURE 4.
Periplasmic CueO assembles copper under copper stress conditions. A, purification of CueO from the periplasmic fraction of cells grown without (left) or with (right) copper stress (LB + 1.5 mm CuSO4). Note that periplasmic CueO is homogeneously matured. Coomassie-stained SDS-PAGE of purification fractions is shown. PP, periplasm; W, last wash fraction. B, UV-visible absorption spectra of the periplasmic CueO preparations shown in A, from MC4100/pBW-CueO-strep grown without (left spectrum) or with (middle spectrum) copper stress, the latter showing the absorption of copper cofactors (indicated by arrows) in the protein as purified. Further copper addition (100 μm) did not alter the spectrum (dotted line). As a control, the spectrum of CueO as purified from the cytoplasm of copper-stressed MC4100/pBW-matCueO-strep is also shown, which shows the absence of copper absorption (right spectrum).
CueO Can Be Translocated in Vitro without Assembled Copper Centers
We examined CueO transport also in vitro, using inverted membrane vesicles in a cell-free system. As shown in Fig. 5A, the holo-form of CueO was translocated with similar efficiency as the apo form, indicating that the copper cofactor assembly is not sensed by the Tat system. Note that the assay monitors vesicle-associated and imported proteins, as soluble external proteins are separated by centrifugation through a sucrose cushion. Apo-CueO had a stronger affinity to vesicle surfaces, which resulted in significantly stronger protease-sensitive (not translocated) precursor signals. Nevertheless, the translocation efficiency was very similar for holo- and apo-CueO. Apparently apo-CueO is sufficiently folded for Tat transport, which agrees with the above-described CD data that indicate no obvious structural differences between the two forms (Fig. 2). Indicative for Tat transport, abolishing the proton motive force by the addition of the uncoupler carbonyl cyanide m-chlorophenylhydrazone completely blocked transport of both forms of CueO. To further address Tat dependence of the observed transport, we carried out competition experiments, using the model Tat substrate HiPIP (25). HiPIP precursor was fully folded in vitro and added to the transport reactions in increasing concentrations (Fig. 5B). Already, the addition of 1 μm HiPIP precursor resulted in strongly diminished transport of holo- as well as apo-CueO. Increase of HiPIP concentrations to up to 4 μm completely inhibited CueO transport. These competition experiments demonstrate unambiguously the Tat dependence of holo- and apo-CueO transport in vitro. Together, these data indicate that Tat systems cannot sense the lack of copper in CueO, as CueO can fold in the absence of copper to a translocation-compatible conformation.
FIGURE 5.
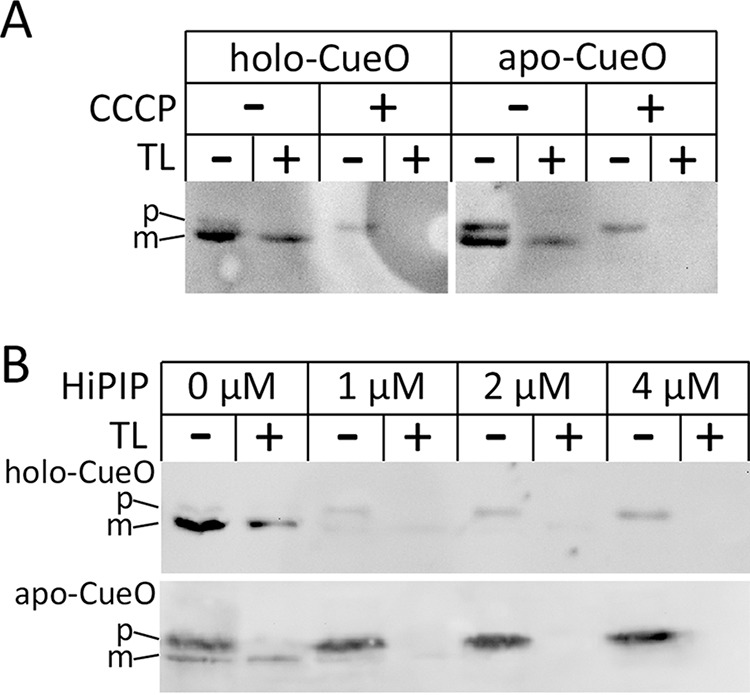
In vitro translocation of holo- and apo-CueO into inverted membrane vesicles. A, transport of holo- and apo-CueO and its dependence of the ΔpH. Vesicle-associated CueO was detected by Western blotting. p and m indicate precursor and mature forms of CueO, respectively. The CueO precursor band is accessible by externally added thermolysin (TL), whereas translocated mature CueO is protected by the vesicle membrane. Note that the addition of 50 μm carbonyl cyanide m-chlorophenylhydrazone (CCCP) completely abolished transport. B, competition of CueO and HiPIP transport indicates Tat dependence. HiPIP precursor was in vitro folded from inclusion bodies, purified to >99% homogeneity, and added with indicated concentrations to the transport assays.
Sec Transport of E. coli CueO Results in Aggregation
We exchanged the Tat signal peptide of CueO by a co-translational (DsbASP-CueO) and a post-translational Sec signal peptide (MalESP-CueO) and analyzed the effects of these exchanges. While all DsbASP-CueO and MalESP-CueO ended up in inclusion bodies, wild type CueO with its natural Tat signal peptide was fully soluble under the same conditions (Fig. 6). We conclude that Sec signal peptides do not permit proper folding of CueO. The inclusion bodies showed no indication for the presence of precursor, which suggests that they represent aggregations in the periplasm.
FIGURE 6.
Sec signal peptides do not support folding of CueO. The signal peptide of CueO was substituted by the Sec signal peptides of DsbA (co-translational transport) and MalE (post-translational transport; some additional residues were included to ensure proper cleavage), and the solubility of the protein products was monitored by Western blotting using cell debris/inclusion body fractions (IB) and soluble fractions (S). The signal peptides of all constructs are shown at the left of the respective blots, with their h-regions underlined and the cleavage site indicated by an arrow. DnaK was detected in the soluble fraction as control. Note that the soluble wt-CueO is rendered insoluble when its authentic Tat signal peptide is exchanged by a Sec signal peptide. The lower blot compares the inclusion body signal of MalE(SP)-CueO and DsbA(SP)-CueO (IB) with a mixture of precursor and mature CueO. The processing of the two Sec signal peptides is suggestive for aggregation after transport.
The Assembly of Copper Ions Drastically Stabilizes CueO
Having folded mature apo- and holo-forms of CueO in our hands, we analyzed their thermal stability by DSC. To our surprise, apo- and holo-CueO showed considerable differences (Fig. 7A). Although apo-CueO denatured at ∼58 °C, holo-CueO denatured at >81 °C. This shift by >23 K reflects a dramatic stabilization of CueO by cofactor assembly, implying a higher degree of flexibility in folded apo-CueO due to the lack of the interdomain copper ions. This conclusion is further supported by limited proteolysis experiments that clearly demonstrate the existence of multiple additional trypsin-accessible cleavage sites in apo-CueO (Fig. 7B).
FIGURE 7.
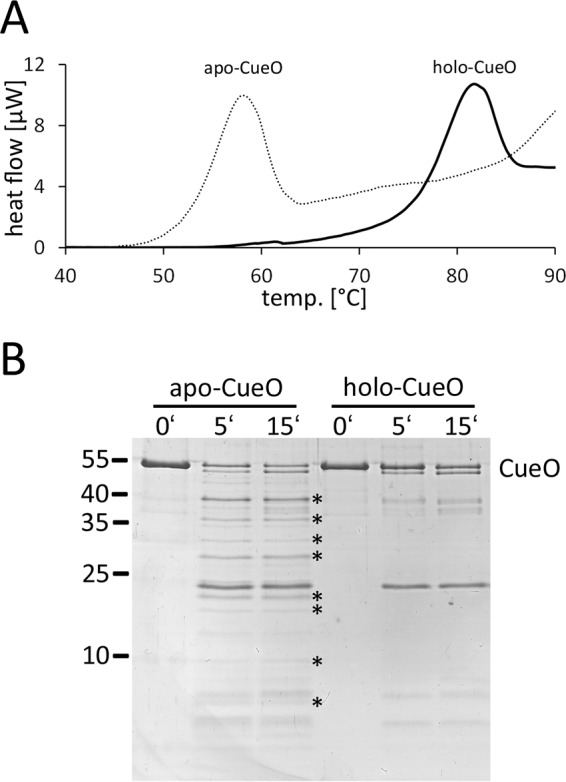
Copper assembly stabilizes CueO and results in decreased protease sensitivity. A, DSC analysis of apo-CueO (dotted line) and holo-CueO (solid line), indicating a shift of denaturation temperature upon cofactor assembly. B, limited proteolysis of apo-CueO and holo-CueO, showing that a number of trypsin-generated proteolysis products occurs only in the apo-CueO (indicated by asterisks).
Discussion
Many Tat-dependently translocated redox proteins are translocated together with cofactors (34). Although the Sec system can transport only unfolded proteins (35), the Tat pathway permits the use of cytoplasmic cofactor assembly pathways for extracytoplasmic enzymes (36–38). However, copper centers do not fall into this category.
Enzymes with type 1-, type 2-, or type 3 copper centers apparently do not require specific assembly factors, but still they are often Tat dependently translocated (39, 40). The more complex copper centers of the N2O reductase NosZ require a periplasmic copper assembly machinery, albeit this enzyme is a Tat substrate (41). NosZ contains a binuclear cysteine-bridged CuA center and a tetranuclear CuZ center, which to our knowledge is the only copper-sulfur cluster so far found in nature. The periplasmic assembly of CuA and CuZ sites has been taken as evidence against a cytoplasmic copper assembly into more simple copper enzymes, especially when Tat- and Sec-dependently translocated homologs exist in different organisms (18). However, albeit a periplasmic assembly of these copper sites surely has been regarded as more likely, this issue has not been clarified experimentally, and a cytoplasmic assembly of copper into the Tat-dependently translocated CueO homologs was discussed (2). The potential contribution of cytoplasmic copper assembly to copper detoxification has been regarded as low due to the highly efficient CopA-mediated export of copper from the cytoplasm (2).
We now clarified this issue with CueO, a Tat substrate with all three very common type 1-, type 2-, and type 3 copper sites. Transport of CueO does indeed not require copper assembly. Consequently, CueO does not “wait” for copper in the cytoplasm. A cytoplasmic assembly would have seriously limited its copper-protective catalytic function in the periplasm. CueO picks up copper in the periplasm, the first compartment where copper enters the cell, and this activates the enzyme. CueO thus contributes 2-fold to copper resistance: (i) it decreases the concentration of free copper ions in the periplasm, and (ii) it oxidizes Cu(I) to less membrane-permeable Cu(II). Both strategies serve to prevent copper entry into the cytoplasm. As no copper assembly into CueO could be detected in the cytoplasm even under copper stress conditions, the amount of copper that enters the cytoplasm is below detection limits (Fig. 4B). This is in line with the zeptomolar affinity of the cytoplasmic copper sensor CueR (3), and the view that the ATP-dependent copper removal by CopA must be very effective. CueO homologs that are involved in copper homeostasis characteristically contain a methionine-rich domain (2), and additional copper binding to this domain was already observed in copper-soaked CueO crystals (32). It is possible that this domain serves to increase the copper binding capacity of CueO under copper stress conditions in vivo.
We found that E. coli CueO requires cytoplasmic conditions for folding, as Sec dependently translocated CueO aggregates. Aggregation was observed with signal peptides originating from Sec substrates representing both Sec transport modes, co-translational (DsbA) and post-translational (MalE). The cytoplasm contains a set of chaperones that is often essential for protein folding. An earlier work already indicated an essential role of the cytoplasmic chaperone DnaK, as CueO requires DnaK for being Tat-dependently translocated (19). The role of DnaK relates to the mature domain of CueO and not to the signal peptide (19). Proteome-wide analyses of the roles of the chaperones DnaK, trigger factor (TF), and GroEL identified CueO in aggregates that form in the absence of DnaK/DnaJ and TF (42). CueO also aggregates in the absence of GroEL, a folding chaperone that accepts partially folded proteins from DnaK, especially in the case of multidomain proteins such as CueO, indicating a rather complex folding pathway for CueO in which multiple cytoplasmic chaperones can be involved (42). Because there exist natural CueO homologs with Sec signal peptides in some δ-proteobacteria like Myxococcus or Sorangium species and in many Firmicutes, the requirement for cytoplasmic folding chaperones must depend on specific differences between these proteins.
Also the periplasmic copper nitrite reductase (NirK) occurs in Tat- or Sec-dependently translocated variants, depending on the organism (39, 40, 43). Tat- and Sec-NirK enzymes are highly similar on the sequence level, and it could be that Tat-NirK requires cytoplasmic chaperones for folding, similar to Tat-CueO.
Another Tat-dependently translocated copper enzyme is tyrosinase. In Streptomyces species, this type 3 copper-containing protein is transported by the aid of a Tat-signal peptide-containing chaperone, and there is evidence that copper assembly induces the release of the active enzyme from its chaperone (44). Although not concluded at that time, these data clearly agree with the view that copper is incorporated after transport also in streptomycetes and, therefore, possibly in most or all bacteria.
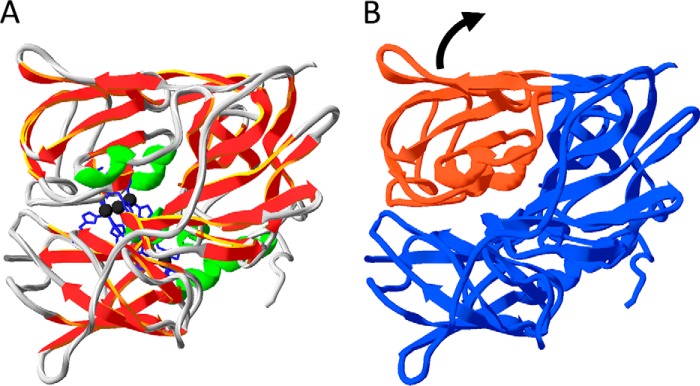
Apo-CueO is sufficiently folded to be translocated by the Tat system. CueO crystal structures with different copper site occupancies (7, 30, 32) suggest that the structure of the apo form is likely highly similar to that of the holo-form, especially when uniform structures are stabilized in crystals. In solution, apo-CueO can be more flexible within or between the folded domains. This fully agrees with our CD data (that show no difference in secondary structure) and our DSC and limited proteolysis data (that show striking differences in stability and protease accessibility). As CueO is clearly stabilized and compacted by the assembly of its copper cofactors (Fig. 7), copper acquisition can be regarded as the last folding step, and the apo form of CueO is actually an incomplete folded state. Based on the known structure of holo-CueO, it is clear that the copper centers in CueO result in a stable contact of two CueO domains that associate to form a deep and narrow cleft (Fig. 8). Three copper ions (type 3 and type 2) are ligated from both sides of this cleft and thereby tightly connect the domains. With bound copper, the surface of CueO is closed, and the copper ions are deeply buried in the enzyme. CueO must open this large cleft to bind type 3 and type 2 copper (Fig. 8). In the absence of copper, it is, therefore, likely that the two domains are more variable in their position. This flexible “open” state of CueO can readily take up copper to switch to the “closed” state with increased stability and less protease sensitivity (Fig. 7). The assembly of type 1 copper likely requires similar flexibility. Thermal unfolding has been studied before with the multicopper oxidase Fet3p from yeast, which behaved differently, showing sequential unfolding of the domains, albeit also in these studies the domain-connecting role of type 3 copper has been recognized (45). Yeast does not possess a Tat system, and Fet3p has a predicted eukaryotic Sec signal peptide and a C-terminal membrane anchor that places this enzyme on the outer surface of the yeast cytoplasmic membrane. It is thus a Sec-type multicopper oxidase, and the observed differences might relate to folding characteristics that are specific for Sec versus Tat-dependently translocated multicopper oxidases.
FIGURE 8.
Flexibility of folded apo-CueO domains likely promotes copper assembly. Structure of CueO using the coordinates of PDB file 1KV7 and the program Swiss-PDB viewer (50). A, type 2 and type 3 copper ions (black) connect domains in holo-CueO, resulting in a closed structure. B, the same structure without the copper ions visualizes the deep cleft between CueO domains, which in apo-CueO likely results in a domain flexibility (indicated by an arrow) that facilitates copper assembly.
Our data for the first time demonstrate that the Tat system can naturally accept proteins that are intrinsically much less compact than the final active protein. It is very likely that this ability of the Tat system to translocate intermediately folded proteins is highly important not only for copper proteins but also for proteins that after transport need to form quaternary structures with independently translocated subunits. Examples include a multitude of Tat substrates, ranging from many oxidoreductases in bacteria, such as formate dehydrogenases or dimethyl sulfoxide reductases, to various subunits of photosystems in higher plants. To our knowledge, a lower stability and flexible domain structure of transported folded proteins that are not allowed to reach their final conformation before transport has not been recognized so far as an issue for Tat transport. With artificial Tat substrates, the Tat system has been shown to tolerate unfolded proteins up to a size of ∼25 kDa if there are no larger hydrophobic patches exposed (46). Linker domains are tolerated (47, 48) as well as globular domains in tandem (49). The Tat system is more tolerant than often thought, and our results suggest that this tolerance can be highly important to allow interactions and folding events after transport that are crucial for the physiological functions of the transported proteins. Future studies will hopefully help to understand these aspects in more detail.
Author Contributions
P. S. and B. H. performed the experiments, contributed to the preparation of the figures, and analyzed the data together with T. B. T. B. conceived and coordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Antje Vogel for construction of vectors and Inge Reupke for technical support.
The authors declare that they have no conflicts of interest with the contents of this article.
- Sec system
- general secretory system
- DMP
- dimethoxyphenol
- DSC
- differential scanning calorimetry
- IMV
- inverted cytoplasmic membrane vesicle
- Tat
- twin-arginine translocation.
References
- 1. Yamamoto K., and Ishihama A. (2005) Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56, 215–227 [DOI] [PubMed] [Google Scholar]
- 2. Rensing C., and Grass G. (2003) Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213 [DOI] [PubMed] [Google Scholar]
- 3. Changela A., Chen K., Xue Y., Holschen J., Outten C. E., O'Halloran T. V., and Mondragón A. (2003) Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387 [DOI] [PubMed] [Google Scholar]
- 4. Stoyanov J. V., Hobman J. L., and Brown N. L. (2001) CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39, 502–511 [DOI] [PubMed] [Google Scholar]
- 5. Singh S. K., Grass G., Rensing C., and Montfort W. R. (2004) Cuprous oxidase activity of CueO from Escherichia coli. J. Bacteriol. 186, 7815–7817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Djoko K. Y., Chong L. X., Wedd A. G., and Xiao Z. (2010) Reaction mechanisms of the multicopper oxidase CueO from Escherichia coli support its functional role as a cuprous oxidase. J. Am. Chem. Soc. 132, 2005–2015 [DOI] [PubMed] [Google Scholar]
- 7. Roberts S. A., Weichsel A., Grass G., Thakali K., Hazzard J. T., Tollin G., Rensing C., and Montfort W. R. (2002) Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sakurai T., and Kataoka K. (2007) Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem. Rec. 7, 220–229 [DOI] [PubMed] [Google Scholar]
- 9. Tree J. J., Kidd S. P., Jennings M. P., and McEwan A. G. (2005) Copper sensitivity of cueO mutants of Escherichia coli K-12 and the biochemical suppression of this phenotype. Biochem. Biophys. Res. Commun. 328, 1205–1210 [DOI] [PubMed] [Google Scholar]
- 10. Macomber L., and Imlay J. A. (2009) The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gort A. S., Ferber D. M., and Imlay J. A. (1999) The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol. Microbiol. 32, 179–191 [DOI] [PubMed] [Google Scholar]
- 12. Hanlon S. P., and Cooper R. A. (1995) Cellular location influences copper-dependent topaquinone formation for phenylethylamine oxidase expressed in Escherichia coli K-12. FEMS Microbiol. Lett. 133, 271–275 [DOI] [PubMed] [Google Scholar]
- 13. Kittleson J. T., Loftin I. R., Hausrath A. C., Engelhardt K. P., Rensing C., and McEvoy M. M. (2006) Periplasmic metal-resistance protein CusF exhibits high affinity and specificity for both Cu(I) and AgI. Biochemistry 45, 11096–11102 [DOI] [PubMed] [Google Scholar]
- 14. Sevcenco A. M., Krijger G. C., Pinkse M. W., Verhaert P. D., Hagen W. R., and Hagedoorn P. L. (2009) Development of a generic approach to native metalloproteomics: application to the quantitative identification of soluble copper proteins in Escherichia coli. J. Biol. Inorg. Chem. 14, 631–640 [DOI] [PubMed] [Google Scholar]
- 15. Watmough N. J., Cheesman M. R., Butler C. S., Little R. H., Greenwood C., and Thomson A. J. (1998) The dinuclear center of cytochrome bo3 from Escherichia coli. J. Bioenerg. Biomembr. 30, 55–62 [DOI] [PubMed] [Google Scholar]
- 16. Rapisarda V. A., Chehín R. N., De Las Rivas J., Rodríguez-Montelongo L., Farías R. N., and Massa E. M. (2002) Evidence for Cu(I)-thiolate ligation and prediction of a putative copper-binding site in the Escherichia coli NADH dehydrogenase-2. Arch. Biochem. Biophys. 405, 87–94 [DOI] [PubMed] [Google Scholar]
- 17. Hou B., and Brüser T. (2011) The Tat-dependent protein translocation pathway. Biomol. Concepts 2, 507–523 [DOI] [PubMed] [Google Scholar]
- 18. Berks B. C., Sargent F., and Palmer T. (2000) The Tat protein export pathway. Mol. Microbiol. 35, 260–274 [DOI] [PubMed] [Google Scholar]
- 19. Graubner W., Schierhorn A., and Brüser T. (2007) DnaK plays a pivotal role in Tat targeting of CueO and functions beside SlyD as a general Tat signal binding chaperone. J. Biol. Chem. 282, 7116–7124 [DOI] [PubMed] [Google Scholar]
- 20. Grass G., and Rensing C. (2001) CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286, 902–908 [DOI] [PubMed] [Google Scholar]
- 21. Casadaban M. J. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104, 541–555 [DOI] [PubMed] [Google Scholar]
- 22. Wilms B., Hauck A., Reuss M., Syldatk C., Mattes R., Siemann M., and Altenbuchner J. (2001) High-cell-density fermentation for production of l-N-carbamoylase using an expression system based on the Escherichia coli rhaBAD promoter. Biotechnol. Bioeng. 73, 95–103 [DOI] [PubMed] [Google Scholar]
- 23. Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 24. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25. Brüser T., Yano T., Brune D. C., and Daldal F. (2003) Membrane targeting of a folded and cofactor-containing protein. Eur. J. Biochem. 270, 1211–1221 [DOI] [PubMed] [Google Scholar]
- 26. Müller M., and Blobel G. (1984) In vitro translocation of bacterial proteins across the plasma membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 81, 7421–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moser M., Panahandeh S., Holzapfel E., and Müller M. (2007) In vitro analysis of the bacterial twin-arginine-dependent protein export. Methods Mol. Biol. 390, 63–79 [DOI] [PubMed] [Google Scholar]
- 28. Berthelmann F., Mehner D., Richter S., Lindenstrauss U., Lünsdorf H., Hause G., and Brüser T. (2008) Recombinant expression of tatABC and tatAC results in the formation of interacting cytoplasmic TatA tubes in Escherichia coli. J. Biol. Chem. 283, 25281–25289 [DOI] [PubMed] [Google Scholar]
- 29. Galli I., Musci G., and Bonaccorsi di Patti M. C. (2004) Sequential reconstitution of copper sites in the multicopper oxidase CueO. J. Biol. Inorg. Chem. 9, 90–95 [DOI] [PubMed] [Google Scholar]
- 30. Li X., Wei Z., Zhang M., Peng X., Yu G., Teng M., and Gong W. (2007) Crystal structures of E. coli laccase CueO at different copper concentrations. Biochem. Biophys. Res. Commun. 354, 21–26 [DOI] [PubMed] [Google Scholar]
- 31. Greenfield N., and Fasman G. D. (1969) Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8, 4108–4116 [DOI] [PubMed] [Google Scholar]
- 32. Roberts S. A., Wildner G. F., Grass G., Weichsel A., Ambrus A., Rensing C., and Montfort W. R. (2003) A labile regulatory copper ion lies near the T1 copper site in the multicopper oxidase CueO. J. Biol. Chem. 278, 31958–31963 [DOI] [PubMed] [Google Scholar]
- 33. Howes B. D., Abraham Z. H., Lowe D. J., Brüser T., Eady R. R., and Smith B. E. (1994) EPR and electron nuclear double resonance (ENDOR) studies show nitrite binding to the type 2 copper centers of the dissimilatory nitrite reductase of Alcaligenes xylosoxidans (NCIMB 11015). Biochemistry 33, 3171–3177 [DOI] [PubMed] [Google Scholar]
- 34. Berks B. C., Palmer T., and Sargent F. (2005) Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8, 174–181 [DOI] [PubMed] [Google Scholar]
- 35. Eser M., and Ehrmann M. (2003) SecA-dependent quality control of intracellular protein localization. Proc. Natl. Acad. Sci. U.S.A. 100, 13231–13234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Genest O., Neumann M., Seduk F., Stöcklein W., Méjean V., Leimkühler S., and Iobbi-Nivol C. (2008) Dedicated metallochaperone connects apoenzyme and molybdenum cofactor biosynthesis components. J. Biol. Chem. 283, 21433–21440 [DOI] [PubMed] [Google Scholar]
- 37. Kaluarachchi H., Chan Chung K. C., and Zamble D. B. (2010) Microbial nickel proteins. Nat. Prod. Rep. 27, 681–694 [DOI] [PubMed] [Google Scholar]
- 38. Py B., and Barras F. (2010) Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 8, 436–446 [DOI] [PubMed] [Google Scholar]
- 39. Nishiyama M., Suzuki J., Kukimoto M., Ohnuki T., Horinouchi S., and Beppu T. (1993) Cloning and characterization of a nitrite reductase gene from Alcaligenes faecalis and its expression in Escherichia coli. J. Gen. Microbiol. 139, 725–733 [DOI] [PubMed] [Google Scholar]
- 40. Chen J. Y., Chang W. C., Chang T., Chang W. C., Liu M. Y., Payne W. J., and LeGall J. (1996) Cloning, characterization, and expression of the nitric oxide-generating nitrite reductase and of the blue copper protein genes of Achromobacter cycloclastes. Biochem. Biophys. Res. Commun. 219, 423–428 [DOI] [PubMed] [Google Scholar]
- 41. Zumft W. G. (2005) Biogenesis of the bacterial respiratory CuA, Cu-S enzyme nitrous oxide reductase. J. Mol. Microbiol. Biotechnol. 10, 154–166 [DOI] [PubMed] [Google Scholar]
- 42. Calloni G., Chen T., Schermann S. M., Chang H. C., Genevaux P., Agostini F., Tartaglia G. G., Hayer-Hartl M., and Hartl F. U. (2012) DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 1, 251–264 [DOI] [PubMed] [Google Scholar]
- 43. Prudêncio M., Eady R. R., and Sawers G. (1999) The blue copper-containing nitrite reductase from Alcaligenes xylosoxidans: cloning of the nirA gene and characterization of the recombinant enzyme. J. Bacteriol. 181, 2323–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsai T. Y., and Lee Y. H. (1998) Roles of copper ligands in the activation and secretion of Streptomyces tyrosinase. J. Biol. Chem. 273, 19243–19250 [DOI] [PubMed] [Google Scholar]
- 45. Sedlák E., Ziegler L., Kosman D. J., and Wittung-Stafshede P. (2008) In vitro unfolding of yeast multicopper oxidase Fet3p variants reveals unique role of each metal site. Proc. Natl. Acad. Sci. U.S.A. 105, 19258–19263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Richter S., Lindenstrauss U., Lücke C., Bayliss R., and Brüser T. (2007) Functional Tat transport of unstructured, small, hydrophilic proteins. J. Biol. Chem. 282, 33257–33264 [DOI] [PubMed] [Google Scholar]
- 47. Cline K., and McCaffery M. (2007) Evidence for a dynamic and transient pathway through the TAT protein transport machinery. EMBO J. 26, 3039–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lindenstrauss U., and Brüser T. (2009) Tat transport of linker-containing proteins in Escherichia coli. FEMS Microbiol. Lett. 295, 135–140 [DOI] [PubMed] [Google Scholar]
- 49. Fan E., Jakob M., and Klösgen R. B. (2010) One signal is enough: stepwise transport of two distinct passenger proteins by the Tat pathway across the thylakoid membrane. Biochem. Biophys. Res. Commun. 398, 438–443 [DOI] [PubMed] [Google Scholar]
- 50. Guex N., and Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]