Abstract
Chitoporin from the chitinolytic marine Vibrio has been characterized as a trimeric OmpC-like channel responsible for effective chitin uptake. In this study we describe the identification and characterization of a novel OprD-like chitoporin (so-called EcChiP) from Escherichia coli. The gene was identified, cloned, and functionally expressed in the Omp-deficient E. coli BL21 (Omp8) Rosetta strain. On size exclusion chromatography, EcChiP had an apparent native molecular mass of 50 kDa, as predicted by amino acid sequencing and mass analysis, confirming that the protein is a monomer. Black lipid membrane reconstitution demonstrated that EcChiP could readily form stable, monomeric channels in artificial phospholipid membranes, with an average single channel conductance of 0.55 ± 0.01 nanosiemens and a slight preference for cations. Single EcChiP channels showed strong specificity, interacting with long chain chitooligosaccharides but not with maltooligosaccharides. Liposome swelling assays indicated the bulk permeation of neutral monosaccharides and showed the size exclusion limit of EcChiP to be ∼200–300 Da for small permeants that pass through by general diffusion while allowing long chain chitooligosaccharides to pass through by a facilitated diffusion process. Taking E. coli as a model, we offer the first evidence that non-chitinolytic bacteria can activate a quiescent ChiP gene to express a functional chitoporin, enabling them to take up chitooligosaccharides for metabolism as an immediate source of energy.
Keywords: carbohydrate metabolism, chitin, Escherichia coli (E. coli), ion channel, sugar transport, chitin uptake channel, non-chitinolytic bacteria, single channel recordings
Introduction
Escherichia coli is a Gram-negative, heterotrophic bacterium that lives in open environments, such as soil, manure, and water, but the persistence of E. coli populations depends upon the availability of carbon substrates in each natural environment. E. coli usually grows on glucose-enriched nutrients such as starch, cellulose, and hemicellulose (1) but not on chitin polysaccharides as it intrinsically lacks competent chitin-utilization machinery (2, 3). The chitin degradation pathway is known to be highly active in marine Vibrio, the growth of which depends on the utilization of the chitin biomass as their sole source of cellular energy. The chitin degradation pathway of Vibrio incorporates a large number of chitin-degrading enzymes and transporters for chitooligosaccharides and N-acetyl glucosamine (4–6). Roseman and co-workers (6) first reported the identification and molecular cloning of the gene encoding chitoporin (VfChiP)3 from Vibrio furnissii. VfChiP was expressed on induction with (GlcNAc)n, n = 2–6, but was not induced by GlcNAc or by other sugars. In contrast to the parental strain, a mutant strain lacking VfChiP did not grow on GlcNAc3, implying that VfChiP was selective for chitooligosaccharides (6). We recently identified and characterized the chitin uptake channel (so-called VhChiP) from the bioluminescent marine bacterium Vibrio harveyi (7, 8). VhChiP is a trimeric OmpC-like porin located in the outer membrane and responsible for the molecular uptake of chitin breakdown products that are generated by the action of secreted chitinases (4, 9–11). Single-channel recordings and liposome swelling assays confirmed that VhChiP is a sugar-specific channel that is particularly selective for chitooligosaccharides, chitohexaose having the greatest rate of translocation.
Unlike Vibrio species, the chitin catabolic cascade of non-chitinolytic bacteria, such as Candida albicans (12), Xanthomonas campestris (13), Shewanella oneidensis (14), and E. coli (15–17) was innately inactive although presumed to be preserved. Yang et al. (14) proposed the three-step biochemical conversion of GlcNAc (the monomer of chitin) to fructose 6-phosphate in E. coli through sequential phosphorylation, deacetylation, and isomerization-deamination reactions. The gene ChiP (formerly ybfM) encoding chitoporin was previously identified in E. coli and Salmonella sp. as a silent gene controlled by a non-coding small RNA (16). However, this ChiP gene was sporadically expressed as an adaptive strategy for the bacterium to thrive in glucose-deficient environments (16, 18–21). To date, E. coli chitoporin (so-called EcChiP) has not been functionally characterized, and our study used electrophysiological and biochemical approaches to uncover the physiological roles of EcChiP.
Experimental Procedures
Bacterial Strains and Vectors
E. coli strain DH5α, used for routine cloning and plasmid preparations, was obtained from Invitrogen. E. coli BL21(DE3) Omp8 Rosetta (ΔlamBompF::Tn5 ΔompAΔompC) mutant strain was a gift from Professor Dr. Roland Benz, Jacobs University, Bremen, Germany. The pET23d(+) expression vector was a product of Novagen (Merck). pUC57 vector carrying the E. coli ChiP gene was obtained from GenScript USA Inc. Piscataway, NJ.
Chitosugars
Chitooligosaccharides, including chitobiose, chitotriose, chitotetraose, chitopentaose, and chitohexaose were purchased from Dextra Laboratories (Science and Technology Centre, Reading, United Kingdom).
Structural Prediction and Sequence Analysis
Amino acid sequences of four bacterial ChiPs from E. coli (P75733), Salmonella (Q7CQY4), Serratia marcescens (L7ZIP1), and V. harveyi (L0RVU0) were aligned and displayed using the program CLC Main Workbench v6.0. The secondary structure of the E. coli ChiP was constructed by ESPript 3.0 (22) according to the three-dimensional structure of Pseudomonas aeruginosa OprD (pdb 2odj).
Cloning and Sequencing
The nucleotide sequence encoding chitoporin was identified in the E. coli strain K-12 substrain MG1655 chromosome in the NCBI database (gi 49175990), and the ChiP gene was commercially synthesized using the GenScript Gene Synthesis Service. The ChiP DNA fragment ligated in the pUC57 cloning vector was excised and then transferred into the pET23d(+) expression vector using the NcoI and XhoI cloning sites so that the ChiP gene could be expressed under the control of the T7 promoter. The oligonucleotides used for colony detection of the ChiP PCR product were 5′-ATACCATGGCCATGCGTACGTTTAGT-3′ for the forward primer and 5′-AACCTCGAGTCAGAAGATGGTGAA-3′ for the reverse primer (sequences underlined indicate the restriction sites). Nucleotide sequences of sense and antisense strands of the PCR fragment were determined by automated sequencing (First BASE Laboratories SdnBhd, Selangor DarulEhsan, Malaysia).
Protein Expression and Purification
Expression and purification of the recombinant EcChiP were carried out as previously described (8). Briefly, the expression vector pET23d(+), harboring the full-length ChiP gene, was transformed into E. coli BL21(DE3) Omp8 Rosetta strain, which lacks major endogenous Omps, including OmpF, OmpC, OmpA, and LamB. The transformed cells were grown at 37 °C in Luria-Bertani (LB) broth supplemented with 100 μg·ml−1 ampicillin and 25 μg·ml−1 kanamycin. During the exponential growth phase (A600 ∼ 0.6–0.8), EcChiP expression was induced with 0.5 mm final concentration of isopropyl thio-β-d-galactoside (IPTG). After 6 h of additional incubation at 37 °C, the cell pellet was harvested by centrifugation at 2,948 × g for 20 min at 4 °C.
For protein purification, the cell pellet was resuspended in lysis buffer (20 mm Tris-HCl, pH 8.0, 2.5 mm MgCl2, 0.1 mm CaCl2) containing 10 μg·ml−1 RNase A and 10 μg·ml−1 DNase I. Cells were disrupted with a high speed ultrasonic processor (EmulsiFlex-C3, Avestin Europe, Mannheim, Germany) for 10 min. After this, 20% (w/v) SDS solution was added to obtain a final concentration of 2%, and the suspension was further incubated at 50 °C for 60 min with 300 rpm shaking to ensure complete lysis. Cell wall components were removed by centrifugation at 100,000 × g at 4 °C for 1 h. The pellet, containing recombinant EcChiP, was extracted twice with 2.5% (v/v) n-octylpolyoxyethylene (Alexis Biochemicals, Lausanne, Switzerland) in 20 mm phosphate buffer, pH 7.4, and centrifuged again. The supernatant was then dialyzed thoroughly against 20 mm phosphate buffer, pH 7.4, containing 0.2% (v/v) lauryldimethylamine oxide (Sigma).
To obtain protein of high purity, the solubilized EcChiP was subjected to ion-exchange chromatography using a Hitrap Q HP prepacked column (5 × 1 ml) connected to an ÄKTA Prime plus FPLC system (GE Healthcare). Bound proteins were eluted with a linear gradient of 0–1 m KCl in 20 mm phosphate buffer, pH 7.4, containing 0.2% (v/v) lauryldimethylamine oxide. The purity of the eluted fractions was confirmed by SDS-PAGE. Fractions containing EcChiP were pooled and subjected to size exclusion chromatography using a HiPrep 16/60 Sephacryl S-200 high resolution column. The purity of the EcChiP fractions obtained after the size exclusion step was verified by SDS-PAGE before they were pooled, and the protein concentration of the freshly prepared EcChiP was estimated using the Novagen BCA protein assay kit (EMD Chemicals Inc., San Diego, CA).
Peptide Mass Analysis by MALDI-TOF MS
The purified EcChiP was electrophoresed on a 12% polyacrylamide gel, and the EcChiP bands were excised and sent to BGI Tech Solutions (HongKong) Co. Ltd. for MALDI-TOF MS analysis. Briefly, protein in the gel was digested with trypsin and eluted to obtain a peptide mixture, then MALDI-TOF mass spectrographic analysis was performed, and the obtained peptide masses were identified using Mascot software v2.3.02.
Molecular Weight Determination of EcChiP
Standard proteins and dyes of known molecular weight were resolved on a HiPrep 26/60 Sephacryl S-300 HR column. Dextran-2000 was used to obtain the void volume (V0), whereas DNP-lysine was used to calculate the volume of the stationary phase (Vi) and the elution volume of each protein sample, denoted as Ve. The elution of a protein sample was characterized by the distribution coefficient (Kd) derived as in Equation 1,
 |
A calibration curve was created by plotting Kd versus logarithmic values of the corresponding molecular weights of the standard proteins and was used to estimate the molecular weight of EcChiP. The standard proteins used in this experiment were ferritin (440 kDa), catalase (250 kDa), aldolase (158 kDa), bovine serum albumin (66 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and ribonuclease A (13.7 kDa).
Black Lipid Bilayer Measurements of Pore Conductance and Chitin Oligosaccharide Translocation
Black lipid membrane (BLM) reconstitution was carried out in electrolyte containing 1 m KCl and 20 mm HEPES, pH 7.5, at room temperature (25 °C). Solvent-free bilayer (Montal-Mueller type) formation was performed using 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine; Avanti Polar Lipids, Alabaster, AL). First, the aperture was prepainted with a few microliters of 1% (v/v) hexadecane in hexane, then a planar bilayer was formed across the aperture by lowering and raising the liquid level (23). Ionic currents were detected using Ag+/AgCl electrodes with a patch-clamp amplifier connected to a two-electrode bilayer head-stage (PC-ONE plus PC-ONE-50; Dagan Corp., Minneapolis, MN). The BLM setup was operated within a Faraday cage on a vibration-dampening table with an A/D converter (LIH 1600, HEKA Elektronik, Lambrecht, Germany) and was operated using the software PULSE program (HEKA Elektronik, Lambrecht, Germany). One of the electrodes, immersed in 1 m KCl electrolyte on the cis side of the cuvette, was connected to ground, whereas the electrode on the trans side was connected to the amplifier head-stage. EcChiP was always added to the cis side of the cuvette. Conductance values were extracted from the current steps observed at different voltages after the addition of the protein. The ion selectivity of EcChiP was determined using different salt solutions, such as 1 m lithium chloride (LiCl), 1 m cesium chloride (CsCl), and 1 m potassium acetate (KAc).
To investigate sugar translocation, single EcChiP channels were reconstituted in the artificial lipid membrane as described earlier. To prevent multiple insertions during data acquisition, the protein solution in the chamber was gently diluted after the first insertion by sequential additions of the working electrolyte. Then the fully open EcChiP channel was titrated with distinct concentrations of different chitooligosaccharides: chitobiose (GlcNAc2), chitotriose (GlcNAc3), chitotetraose (GlcNAc4), chitopentaose (GlcNAc5) and chitohexaose (GlcNAc6). Each sugar was added to the cis side of the chamber. Fluctuations of ion flow were observed as a result of sugar diffusion through the reconstituted channel and were usually recorded for 2 min at different transmembrane potentials. To test the substrate specificity of the channel toward chitooligosaccharides, maltodextrin sugars were used as controls.
Liposome Swelling Assay
The EcChiP- and VhChiP-reconstituted proteoliposomes were prepared as described elsewhere (24, 25). Soybean l-α-phosphatidylcholine (20 mg/ml, freshly prepared in chloroform) (Sigma) was used to form multilamellar liposomes. For the preparation of proteoliposomes, 200 ng of EcChiP was reconstituted into 200 μl of the liposome suspension by sonication, and then 17% (w/v) dextran (40kDa) was entrapped in the proteoliposomes. d-Raffinose solutions were prepared in phosphate buffer to obtain concentrations of 40, 50, 60, and 70 mm for determination of the isotonic solute concentration. This value was then used for the adjustment of the isotonic concentration for other solutes. To carry out a liposome-swelling assay, 25 μl of the proteoliposome suspension was added to 600 μl of sugar solution, and changes in absorbance at 500 nm were monitored immediately. The apparent absorbance change over the first 60 s was used to estimate the swelling rate (s−1) following the equation ϕ = (1/Ai)dA/dt in which ϕ is the swelling rate, Ai is the initial absorbance, and dA/dt is the rate of absorbance change during the first 60 s. The swelling rate for each sugar was normalized by setting the rate of l-arabinose (150 Da), the smallest sugar, to 100%. The values presented are averages from three to five independent determinations. Protein-free liposomes and proteoliposomes without sugars were used as negative controls. The sugars tested were d-glucose (180 Da), d-mannose (180 Da), d-galactose (180 Da), N-acetylglucosamine (GlcNAc) (221 Da), d-sucrose (342 Da), d-melezitose (522 Da), GlcNAc2 (424 Da), GlcNAc3 (628 Da), GlcNAc4 (830 Da), GlcNAc5 (1034 Da), GlcNAc6 (1237 Da), and maltodextrins.
Results
Cloning, Sequence Analysis, and Structure Prediction
The availability of the nucleotide sequence in the genome of E. coli strain K-12 sub-strain MG1655, (complete genome NCBI reference sequence; NC_000913) allowed the putative amino acid sequence of E. coli ChiP (so-called EcChiP) to be identified. The full-length ChiP gene corresponding to EcChiP was synthesized commercially, for which the target gene was ligated into the NcoI and XhoI cloning sites of the pUC57 cloning vector (GenScript). The nucleotide sequence of the ChiP gene, comprising 1407 bps, was translated to a putative polypeptide of 468 amino acids, including the 26-amino acid signal sequence. The theoretical mass of the full-length EcChiP was 52,780 Da, with a predicted isoelectric point of 4.70.
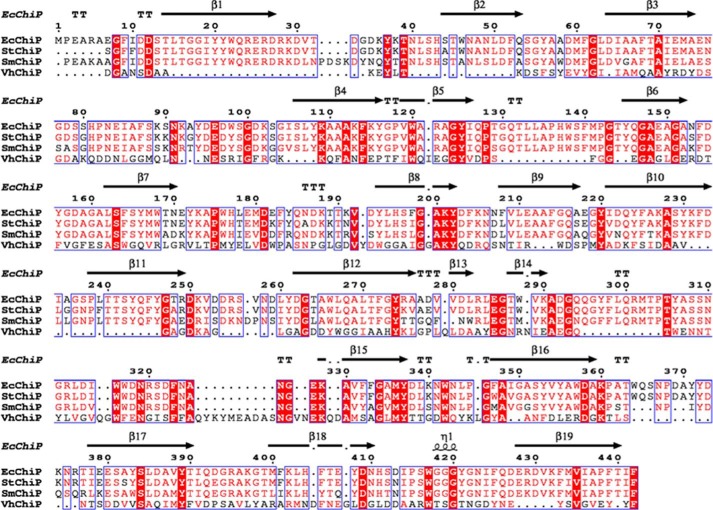
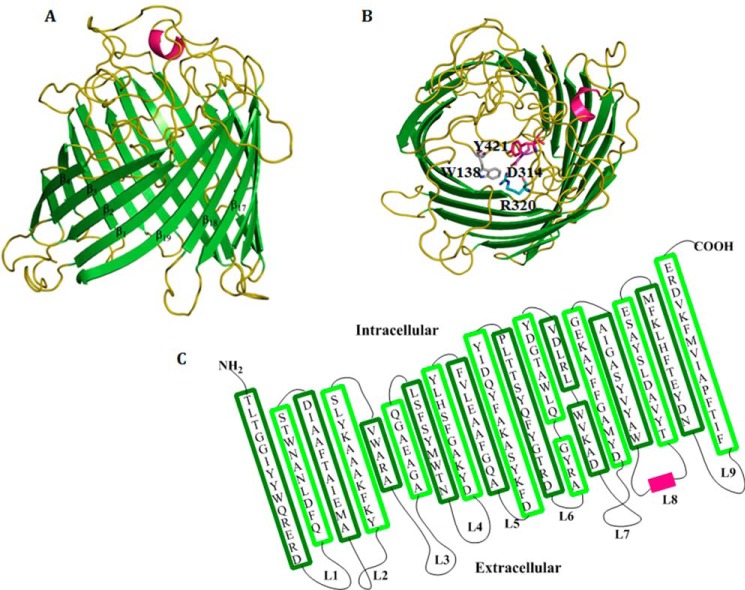
Amino acid sequence comparison of EcChiP with other bacterial ChiPs in the SwissProt/UniProtKB database is presented in Fig. 1. The putative sequence of EcChiP showed highest sequence identity to Salmonella typhimurium ChiP (Q7CQY4) (90%) (18) followed by S. marcescens ChiP (L7ZIP1) (70%) (26). Our sequence analysis indicated that EcChiP had exceptionally low identity with the ChiP sequences from marine Vibrio species, such as Vibrio cholerae ChiP (Q9KTD0) (27), V. furnissii ChiP (Q9KK91) (6), and V. harveyi ChiP (L0RVU0) (8) with 13, 14, and 12% identity, respectively. Unlike marine Vibrio species, E. coli and S. typhimurium are non-chitinolytic bacteria that possess ChiP homologs belonging to the OprD family of porins. The EcChiP amino acid sequence was submitted to the Swiss-model database for homology structure prediction, and the crystal structure of P. aeruginosa OprD (pdb 2odj) (28) was computationally selected as a structure template. Fig. 2A shows a side view of the predicted β-barrel structure of EcChiP, atypically consisting of 19-strands connected by 9 external loops and 9 periplasmic turns. Previous reports of the crystal structures of the maltoporin (LamB) (29) and sucrose-specific porin ScrY (30) suggested that aromatic residues are important for sugar transport. Amino acid residues located within the pore interior, such as Trp-138, Asp-314, Arg-320, and Tyr-421, are predicted to be crucial for sugar transport (residues marked as sticks in Fig. 2B, top view). The predicted transmembrane topology for EcChiP is shown in Fig. 2C. The longest loop (L3, Gly-124 to Tyr-145) found inside the channel lumen presumably acts as the pore-confining loop that controls ion flow.
FIGURE 1.
Sequence alignment of EcChiP, showing the secondary structural elements of EcChiP. The amino acid sequences of S. typhimurium ChiP (StChiP), S. marcescens ChiP (SmChiP), and V. harveyi ChiP (VhChiP), without signal peptides, were aligned using CLC Main Workbench 6. The secondary structure of E. coli was constructed by ESPript 3.0 according to the structure of P. aeruginosa OprD (pdb 2odj). β-Strands are marked with black arrows, and an α-helix is marked with black curl. Absolutely conserved residues are highlighted in red.
FIGURE 2.
The Swiss-model structure of E. coli chitoporin. A, schematic of EcChiP viewed from the side. B, top view of the EcChiP modeled structure. Important residues in the pore that may be involved in sugar transport; Trp-138, Asp-314, Arg-320, and Tyr-421 are presented in gray, purple, teal, and pink, respectively, as stick structures. The x-ray structure of OprD from P. aeruginosa (pdb 2odj) was selected as the structure template for E. coli chitoporins. Green, β-strands; olive, loops and turns; hot pink, α-helices. C, the predicted transmembrane topology of EcChiP.
Recombinant Expression, Purification, and Mass Identification
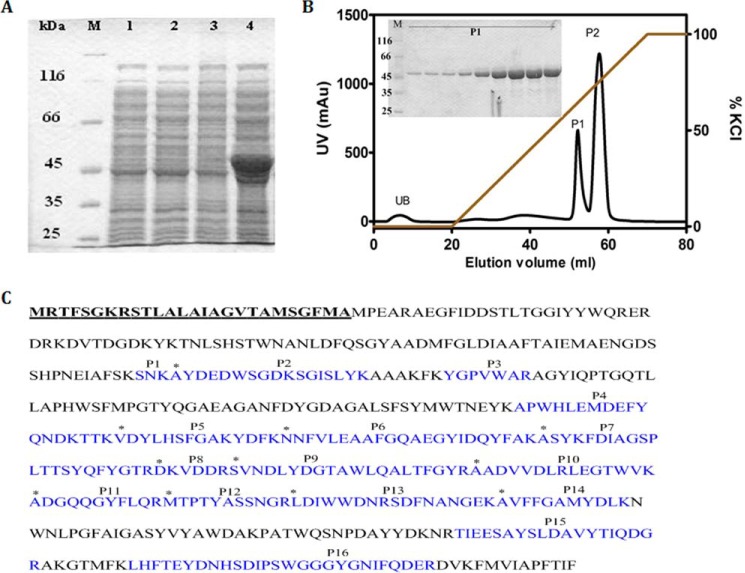
The plasmid pET23d(+) harboring the ChiP gene fragment was designed to express recombinant EcChiP, with the 26-amino acid N-terminal signal sequence aiding channel insertion into the cell wall of the E. coli BL21(DE3) Omp8 Rosetta host. When the transformed cells were grown to exponential phase, expression of the recombinant EcChiP was induced with 0.5 mm IPTG for a period of 6h at 37 °C. Fig. 3A shows SDS-PAGE analysis of whole cell lysates of the Omp-deficient E. coli expressing EcChiP. When compared with cells transformed with the empty vector (lane 1, no induction; and lane 2, IPTG induction), uninduced cells transformed with the pET23d(+)/ChiP vector did not produce the heterologous protein (lane 3), whereas a prominent band of the expected size (50 kDa) appeared on induction with IPTG (lane 4). These results confirmed successful production of EcChiP in the selected host cells.
FIGURE 3.
Recombinant expression, purification, and mass identification. A, SDS-PAGE analysis of whole-cell lysate with and without IPTG induction for E. coli carrying pET23d(+) and pET23d(+)/EcChiP. Lane M, marker proteins; lane 1, E. coli Omp8 Rosetta carrying pET23d(+) without IPTG induction; lane 2, E. coli Omp8 Rosetta carrying pET23d(+) with IPTG induction; lane 3, E. coli Omp8 Rosetta carrying pET23d(+)/EcChiP without IPTG induction; lane 4, E. coli Omp8 Rosetta carrying pET23d(+)/EcChiP with IPTG induction. B, chromatographic profile of EcChiP purification with a Hitrap Q HP prepacked column (5 × 1 ml) connected to an ÄKTA Prime plus FPLC system. Bound proteins were eluted with a linear gradient of 0–1 m KCl in 20 mm phosphate buffer, pH 7.4, containing 0.2% (v/v) lauryldimethylamine oxide. SDS-PAGE analysis of bound fraction P1 is shown in an inset. mAu, milliabsorbance units. C, identification of tryptic digests of the expressed proteins by MALDI-TOF MS. The 16 identified peptides (P1-P16) that gave a complete match with putative peptides of EcChiP are shown in cyan. The N-terminal ends of peptides P2, P5, P6, P7, P8, P9, P10, P11, P12, P13, and P14 are indicated by asterisks. The 26-amino acid signal peptide is in bold and underlined.
For purification of EcChiP, the induced cells were subjected to a two-step detergent extraction. In the first step using 2% (w/v) SDS, most of EcChiP remained in the insoluble fraction, and in the second step EcChiP was solubilized with 2.5% (v/v) n-octylpolyoxyethylene. The protein purity observed after these steps was >90%. EcChiP was further subjected to ion-exchange chromatography using a Hitrap Q HP pre-packed column. Fig. 3B shows the chromatographic profile, indicating that EcChiP fractions were eluted in two peaks (P1 and P2) by an applied gradient of 0–1 m KCl. SDS-PAGE analysis shows that the EcChiP fractions in the first peak (P1) were highly purified (Fig. 3B, inset), whereas the fractions in P2 included some contaminants (not shown); P2 may, therefore, contain EcChiP bound to other proteins. Peaks P1 and P2 were, therefore, applied separately to a HiPrep 16/60 Sephacryl S-200 high resolution exclusion chromatography column for final purification. The highly purified EcChiP obtained after gel filtration chromatography was subjected to in-gel digestion for MALDI-TOF MS analysis. A MASCOT database search identified 16 peptides (designated P1-P16) that belonged to the internal sequences of the putative chitoporin from E. coli (gi 251784171 ref YP_002998475.1) (Fig. 3C, sequences in cyan). The sequence coverage for the identified peptides was 50%. These results confirmed that the 50-kDa protein expressed in E. coli BL21(DE3) Omp8 Rosetta host was EcChiP.
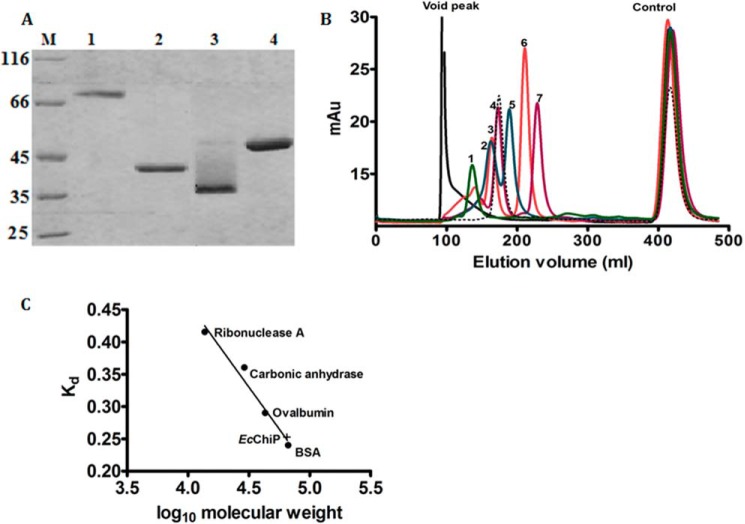
Determination of the Native State of the EcChiP Channel
All chitoporins identified in marine Vibrio species are trimeric channels (8). In the next series of experiments we investigated the native state of EcChiP. Unlike VhChiP (8), EcChiP did not migrate on SDS-PAGE gel to the position corresponding to a trimer under non-denaturing conditions. Fig. 4A is an SDS-PAGE analysis showing the migrations of VhChiP (lane 1, unheated; lane 2, heated) and EcChiP (lane 3, unheated; lane 4, heated). Intact VhChiP migrated with an apparent molecular mass close to 100 kDa, indicating a trimer (lane 1), but the dissociated subunits migrated close to 40 kDa (lane 2). Different results were observed with the E. coli sample; native EcChiP migrated with an apparent molecular mass of ∼35 kDa (lane 3), indicating a monomeric, folded structure according to literature review (31, 32). After boiling, the apparent molecular mass increased to nearly 50 kDa, presumably due to unfolding of the protein (lane 4). Gel filtration chromatography was used to confirm the monomeric structure of native EcChiP. Fig. 4B shows a chromatographic profile of the protein standards eluted from a HiPrep 26/60 Sephacryl S-300 pre-packed column. EcChiP was eluted at a position between ovalbumin (43 kDa) and bovine serum albumin (66 kDa) (Fig. 4B, black dotted line), and its apparent molecular mass estimated from its distribution coefficient (Kd) was ca. 60 kDa (Fig. 4C), consistent with a monomeric molecule. The slightly greater molecular mass than the expected size of EcChiP (50 kDa) may be added by the molecular mass the detergent lauryldimethylamine oxide (micelle molecular mass 17 kDa) (33), which was included to maintain the protein solubility.
FIGURE 4.
SDS-PAGE analysis of EcChiP and molecular weight determination. A, SDS-PAGE analysis of purified EcChiP, with VhChiP for comparison. Lane M, marker proteins; Lane 1,VhChiP (unheated); lane 2, VhChiP (heated); lane 3, EcChiP (unheated); lane 4, EcChiP (heated). B, size exclusion chromatogram of standard proteins with EcChiP. Standards were run separately, together with DNP-lysine (control). Protein standards: lane 1, ferritin (440 kDa); lane 2, catalase (250 kDa); lane 3, aldolase (158 kDa); lane 4, bovine serum albumin (66 kDa); lane 5, ovalbumin (43 kDa); lane 6, carbonic anhydrase (29 kDa); lane 7, ribonuclease A (13.7 kDa). Void peak, elution peak of blue dextran 2000. Control, elution peak for DNP-lysine. EcChiP was eluted (black dotted line) as a monomer within the 43–66 kDa range. C, calibration curve to determine the Kd value of EcChiP.
Channel-forming Properties of EcChiP
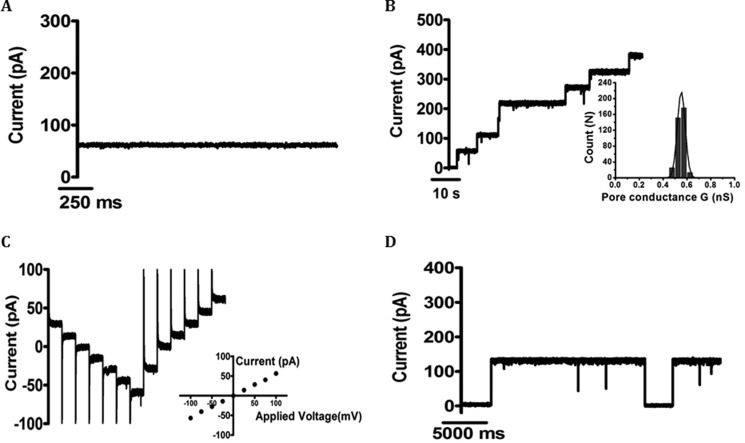
To examine the pore-forming properties of the isolated channel, EcChiP was reconstituted into artificial planar phospholipid membranes. An abrupt increase in ion current in response to an externally applied potential was observed soon after the addition of the protein, and the induced current remained steady throughout the subsequent period of data acquisition (usually 2 min). The BLM results clearly demonstrated that EcChiP was a channel-forming protein. Fig. 5A is a representative ion current recording of ∼50 pA at +100 mV, signifying a characteristic single EcChiP insertion under a given condition (<2 ng·ml−1 EcChiP added on the cis side of the chamber filled with 1 m KCl, pH 7.5). This channel insertion behavior was observed consistently throughout our study. In Fig. 5B, we show typical ion current traces obtained from multiple insertions in the presence of a high added concentration of EcChiP (>10 ng·ml−1) in the same electrolyte solution. The Fig. 5B inset shows the Gaussian distribution of the pore conductance, derived from 365 channel insertions. The value was fitted with a mean conductance of 0.54 ± 0.04 nS, which corresponded well with the value obtained from the slope of the I-V curve shown in Fig. 5C, inset. For individual EcChiP channels, currents were recorded at potentials from −100 to +100 mV, increased in 25-mV steps, as shown in Fig. 5C. The plot of current as a function of transmembrane potential was constructed from 17 independent single channel insertions. The conductance of the pore (slope of the curve) was constant over the entire voltage range scanned, yielding the conductance value of 0.55 ± 0.01 nS. EcChiP was shown to be a relatively stable channel at both negative and positive potentials with a threshold for channel gating observed at approximately −200 mV and +200 mV. An example of channel gating at +200 mV is shown in Fig. 5D.
FIGURE 5.
Single-channel recordings of EcChiP in artificial lipid membranes. Lipid bilayers were formed across a 70 μm aperture by the lowering and raising technique, using 5 mg·ml−1 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine in n-pentane and 1 m KCl in 20 mm HEPES, pH 7.5, on both sides of the chamber. The protein was added to the cis side. A, fully open EcChiP current trace at +100 mV. B, multiple channel insertions; Inset, histogram of the conductance steps observed with 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine artificial bilayer for 365 independent channel insertions. The black line represents a single Gaussian fit. C, stepwise ramping of the potential for single insertion. Inset, I-V plot for a single EcChiP single channel. The average current values were obtained by varying the potential from −100 mV to +100 mV in 25 mV steps for 17 independent channel insertions. D, gating behavior of EcChiP at high potential (+200 mV).
Single-channel experiments were also performed with salts other than KCl to obtain information on the ion selectivity of EcChiP; the results are summarized in Table 1. Replacement of Cl− by CH3COO−, a less mobile anion, slightly reduced the single channel conductance from 0.5 to 0.4 nS. However, replacement of K+ by the less mobile cation Li+ resulted in a much larger decrease, from 0.5 nS to 0.25 nS, indicating a preference of EcChiP for cations (Table 1). Although Li+ and CH3COO− and K+ and Cl− have similar aqueous mobilities (34, 35), the conductance of EcChiP channel was lower in LiCl than in KAc. This result supports the conclusion that the EcChiP channel was cation-selective.
TABLE 1.
Average single channel conductance (G) of EcChiP in different salt solutions
The pH of the aqueous salt solutions was around 7.5. G was calculated from the single channel recording by averaging single events as indicated within the parentheses. The applied membrane potential was +100 mV.
| Aqueous salt solution | Single channel conductance (G) |
|---|---|
| nS | |
| 1 m KCl | 0.54 ± 0.04 (n = 365)a |
| 1 m KAc | 0.40 ± 0.03 (n = 71) |
| 1 m CsCl | 0.60 ± 0.04 (n = 87) |
| 1 m LiCl | 0.25 ± 0.02 (n = 65) |
a n represents the number of BLM measurements in which the data were acquired in this experiment.
Investigation of Chitooligosaccharide Interactions with EcChiP
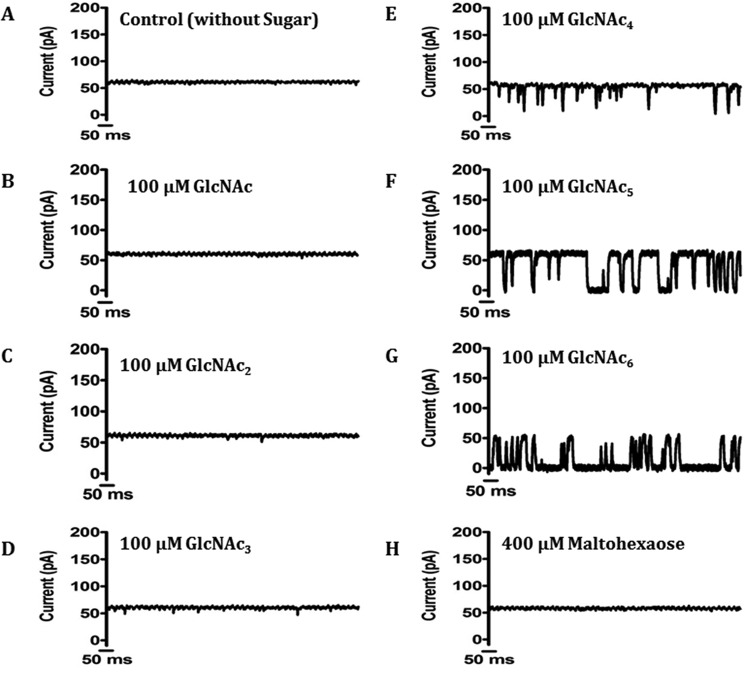
In this set of experiments we performed single-channel measurements in the presence of different chitooligosaccharides to address the substrate specificity of the newly isolated channel. Fig. 6A is a control trace, showing a stably opening channel of conductance ∼55 pA at +100 mV in the absence of ligand. The addition of the chitooligosaccharides chitotetraose, pentaose, and hexaose resulted in frequent current blockages in EcChiP, reflecting strong sugar-channel interactions (Fig. 6, E–G). We observed no fluctuation of ion current on the addition of N-acetylglucosamine, chitobiose and chitotriose (Fig. 6, B, C and D), and addition of structurally related maltohexaose (Fig. 6H) showed no fluctuation of ion current even at a concentration 200-fold greater than that of the chitosugars, indicating that the EcChiP channel was highly specific for chitooligosaccharides.
FIGURE 6.
Current recordings of single EcChiP channels in solutions of different chitooligosaccharides of various chain lengths. Ion current fluctuations were monitored for 120 s at applied potentials of ±100mV. Here, only current traces for 1 s at +100 mV are presented. A, a fully open state of EcChiP before sugar addition. Then GlcNAc (N-acetylglucosamine) (B), chitobiose (GlcNAc2) (C), chitotriose (GlcNAc3) (D), chitotetraose (GlcNAc4) (E), chitopentaose (GlcNAc5) (F), or chitohexaose (GlcNAc6) (G) were added on the cis side of the chamber to a final concentration of 100 μm. H, control recording with maltohexaose at a concentration of 400 μm.
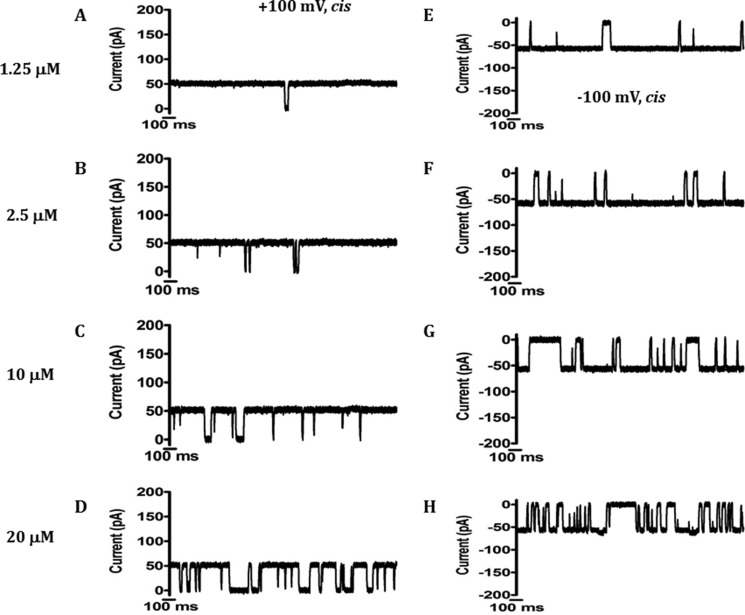
LamB has been the subject of intensive studies on sugar binding (36–38). Similarly, our BLM data showed that EcChiP interacted with chitosugars to various extents depending on the sizes and the types of the sugars, as shown in Fig. 6. Next, we selected chitohexaose as a substrate to study ion current fluctuation at different sugar concentrations. Fig. 7 shows current recordings obtained from a single EcChiP channel in the presence of several discrete concentrations of chitohexaose. These traces, recorded at +100 mV, indicated increasing numbers of blocking events as concentrations of chitohexaose were increased from 1.25 to 20 μm, leading to a gradual reduction in the average conductance of the channel (Fig. 7, A–D). Similar results were obtained at −100 mV, with the channel more susceptible to sugar occlusion at negative voltages (Fig. 7, E–H). At the highest sugar concentration (20 μm), we observed that the sugar molecules fully occupied the channel, leading to more frequent decreases in ion current to zero (Fig. 7, D and H). We did not detect three-stage transient blockages with EcChiP measurements on sugar addition, which are usually observed for trimeric channels (8, 36–39). These results provide further evidence that EcChiP acts as a monomeric channel.
FIGURE 7.
Conductance of the same single EcChiP channel with increasing chitohexaose concentration at positive and negative potentials. Ion current fluctuations were monitored for 120 s at applied potentials of ±100 mV with sugar addition on the cis side. Here only current traces for 2 s are presented with four different concentrations at +100mV (A–D) and at −100mV (E–H).
Determination of Channel Specificity Using a Liposome-swelling Assay
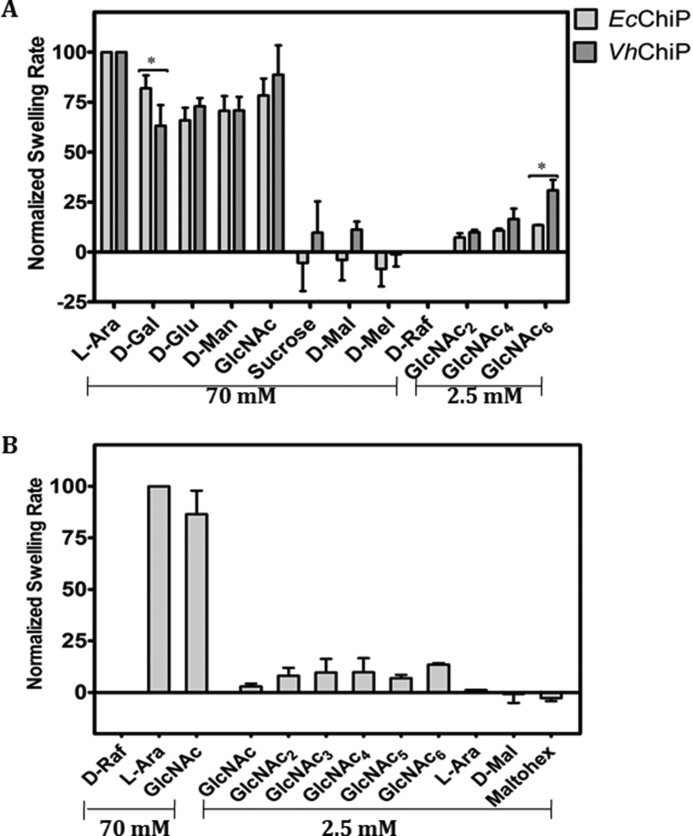
Proteoliposome swelling assays were performed to evaluate the bulk permeation of neutral solutes through the EcChiP channel. EcChiP-containing proteoliposomes were prepared according to the protocol described elsewhere (24, 25). Swelling of the proteoliposomes caused by diffusion of solute molecules through the protein channel resulted in a decrease in apparent absorbance at 500 nm, whereas under isotonic conditions constant absorbance was maintained. In this assay we used d-raffinose (504 Da), a branched sugar that is unable to diffuse through the porin, to establish the isotonic concentration and enable the comparison of diffusion rates. l-Arabinose (150 Da), the smallest sugar tested in this experiment, had the highest diffusion rate through EcChiP, and the swelling rates in the other sugars tested were normalized relative to that in l-arabinose, which was set to 100%.
To address the differences between the EcChiP channel and the chitooligosaccharide-specific porins from the OmpC family, we compared our data with those obtained with VhChiP-incorporating proteoliposomes. The two chitoporins showed similar diffusion rates for small sugars such as d-glucose, d-mannose, and d-galactose (180 Da) and N-acetylglucosamine (GlcNAc, 221 Da) (Fig. 8A). However, d-sucrose (342 Da), maltose (360 Da), and d-melezitose (522 Da) showed no diffusion through EcChiP. In contrast, d-sucrose and maltose permeated VhChiP, albeit with very low diffusion rates, whereas d-melezitose was impermeant. When EcChiP was tested with long chain chitooligosacharides it was found that all neutral chitooligosaccharides were permeant (Fig. 8B), whereas maltosugars (maltose and maltahexaose) did not show permeation. The results obtained from the proteoliposome swelling assays additionally confirmed the high selectivity of EcChiP for chitooligosaccharides.
FIGURE 8.
Proteoliposome swelling assays. In each preparation multilamellar liposomes were reconstituted with 200 ng of EcChiP or VhChiP. d-Raffinose was used to determine the isotonic concentrations that produced no change in absorbance at 500 nm of the proteoliposome suspension over 60s. The swelling rate in l-arabinose was set to 100% to obtain normalized swelling rates. The permeability of channels was assumed to be directly proportional to the swelling rate. A, permeation of different types of sugar through EcChiP- and VhChiP-containing proteoliposomes. Differences between the two data sets were evaluated using a t test. Statistically significant differences (p < 0.05) are marked with an asterisk (*). Values are the means ± S.D., obtained from three-five independent sets of experiments. B, permeation of chitooligosaccharides through EcChiP. Maltodextrins were used as controls. l-Ara, l-arabinose; d-Gal, d-galactose; d-Glu, d-glucose; d-Man, d-mannose; GlcNAc, N-acetylglucosamine; d-Mal, d-maltose; d-Mel, d-melezitose; d-Raf, d-raffinose; GlcNAc2, chitobiose; GlcNAc3, chitotriose; GlcNAc4, chitotetraose; GlcNAc5, chitopentaose; GlcNAc6, chitochexaose; Maltohex, maltohexaose.
Discussion
Chitin is one of the most abundant naturally occurring polysaccharides, and chitin turnover by marine Vibrio species is essential for the recycling of carbon and nitrogen in marine ecosystems (40). Vibrio species possess competent chitin degradation and uptake systems that allow the bacteria to metabolize chitinous materials, generating catabolic intermediates that can be used as their sole source of energy (5, 41–47). In marked contrast, E. coli is a non-chitinolytic bacterium living primarily in the gastrointestinal tract of animals, and its generation of cellular energy relies on glucose-enriched nutrients. Although the ChiP gene, encoding a chitoporin that is responsible for the uptake of chitin-derived chitooligosaccharides, is evolutionarily conserved, it is usually quiescent in non-chitinolytic bacteria. A previous report on Salmonella and E. coli (16, 18) showed that in the absence of any inducer, the ChiP gene (formerly ybfM) was constantly suppressed by forming a DNA-RNA duplex with a conserved small RNA, namely ChiX. However, silencing was relieved in the presence of chitooligosaccharides, as these sugars produced accumulation of anti-ChiX small RNA that paired with ChiX, allowing the ChiP gene to be expressed. Another study reported co-localization of the genes for ChiP and Hex (encoding β-N-acetylglucosaminidase) in the chromosomes of Yersinia and Serratia species (14). This suggested a sequential action of ChiP and β-N-acetylglucosaminidase in chitin uptake and chitin degradation, respectively, and E. coli and Salmonella ChiPs have been proposed to be involved in the uptake of chitobiose, an end product of chitin breakdown that is readily transported through the inner membrane by the phosphoenolpyruvate transferase system.
In the present study we identified the ChiP gene, encoding a hypothetical outer-membrane chitoporin (EcChiP) from the genome of the E. coli strain K-12, substrain MG1655. Amino acid sequence analysis showed that EcChiP had exceptionally low sequence identity (<14%) to all ChiPs from the OmpC family, such as VhChiP from V. harveyi and VfChiP from V. furnissii (6, 8). This suggested that the ChiP genes from E. coli and from Vibrio sp. did not share common ancestors, and further sequence analysis showed that EcChiP was similar to SmChiP from S. marcescens (75% identity), both of which are members of the OprD family.
The recombinant EcChiP displayed quite different channel behavior from other sugar-specific porins. Its most distinctive feature was that it formed a monomeric channel rather than the trimeric channel observed with other known ChiPs and that the channel was stably open over a wide range of external membrane potentials, with only occasional gating at high voltages (±200 mV). At 0.55 ± 0.01 nS, the single channel conductance of EcChiP was approximately ⅓ that of the well studied VhChiP (1.8 ± 0.3 nS) (8), consistent with our observation that EcChiP formed a monomeric channel, whereas VhChiP worked as a trimer. Comparison with the monomeric OprD from P. aeruginosa, a basic amino acid uptake channel (28), revealed that P. aeruginosa OprD had a narrow central constriction zone and displayed a much smaller conductance (28 pS) than that of EcChiP under the same electrolyte conditions (1 m KCl and pH 7.5). This suggests differences in the amino acids that line the channel interior and regulate the net ion flow in EcChiP as compared with those in OprD.
Measuring changes in ion flow upon varying the cationic/anionic species could provide some information regarding ion selectivity. For examples, Benz and co-workers (34, 35) used LiCl and KAc to test the channel selectivity of the maltodextrin-specific channel LamB and the glucose-inducible channel OprB. Both channels showed a preference for cations over anions. Following their method, our channel exhibited similar preference. We also measured the K+/Cl− selectivity by observing changes in reverse membrane potential at zero current under a 0.1–3.0 m gradient of KCl, yielding a Pc/Pa ratio of 2.8, which was slightly lower than the value obtained for the trimeric VhChiP (Pc/Pa = 3.2) (48). Nonetheless, the ion selectivity study obtained from both techniques confirmed that EcChiP was a cationic-selective channel.
We further examined sugar-channel interactions with various chitooligosaccharides. Our BLM data showed that EcChiP interacted strongly with long-chain chitooligosaccharides but not with maltooligosaccharides, implying that the channel was specific for chitooligosaccharide uptake. Strong interaction with the higher molecular weight substrates is also a characteristic of other sugar-specific channels, such as LamB (37, 38, 49), VhChiP (8, 48), and CymA (50). Consistent with this is an earlier in vivo study that showed no growth of S. marcescens expressing the null ChiP mutant in the presence of chitooligosaccharides larger than chitotriose (26). Both results confirmed the physiological roles of the OprD-related ChiP in chitooligosaccharide uptake.
EcChiP was tested for its ability to transport neutral sugars of various sizes by use of a liposome swelling assay. All the monosaccharides tested could permeate into EcChiP-reconstituted liposomes. Similar results were obtained with VhChiP. Neither channel allowed the passage of neutral sugars of >221 Da, such as maltose, sucrose, melezitose, and raffinose, reflecting the size exclusion limit for small molecules that traverse the channel by general diffusion. In our BLM measurements, we did not observe the occlusion of EcChiP by GlcNAc, presumably because the short-lived blocking events (<100 μs) produced a residence time too short to be resolved by the currently available BLM setup. This is also the case when molecules with a molecular weight below the size exclusion limit pass through the channel without interacting with it. However, the behavior of the EcChiP channel was not equivalent to that of other known nonspecific porins, such as BpsOmp38 from Burkholderia pseudomallei (25, 51, 52) and OmpF from E. coli (53), which typically have a size exclusion limit of ∼650 Da. In liposome swelling experiments, in agreement with the electrophysiological data, EcChiP showed sugar-selective behavior, allowing the bulk permeation of chitooligosaccharides at rates that depended on the sizes of the sugar chains, longer chain chitooligosaccharides (chitotetraose, pentaose, and hexaose) tending to show greater permeation rates than short chain sugars such as chitobiose and chitotriose. Additionally, the channel operated even at the low sugar concentration of 2.5 mm, a characteristic of solute-specific channels that has been reported for other well characterized porins, including E. coli LamB (54, 55) and V. harveyi ChiP (8). As shown in Fig. 8A, the rate of permeation of chitohexaose through VhChiP was much greater than those for other sugars, whereas the permeation rates of chitotetraose, pentaose, and hexaose through EcChiP were comparable. Both the liposome swelling assays and the BLM data generally showed the lower affinity of EcChiP than of VhChiP for the same sugars, and suggested high substrate specificity of the Vibrio channel and broad substrate specificity of the E. coli channel. This is not surprising, as VhChiP uses chitin as its sole source of energy, so the channel has evolved to provide every efficient chitooligosaccharide uptake, enabling the bacterium to thrive even in rough seas. On the other hand, E. coli uses mainly glucose as a nutrient, its ChiP functioning only under certain environmental conditions, such as a scarcity of glucose in the growth medium.
Taking all of our data together, we reconstructed the chitooligosaccharide utilization pathway of E. coli based on the GlcNAc utilization pathway suggested previously (14, 15). As shown in Fig. 9, E. coli chitoporin facilitates the uptake of extracellular chitooligosaccharides into the periplasm. The breakdown of high molecular weight chitosugars (chitotriose, chitotetraose, chitopentaose, and chitohexaose) within the periplasm may be initiated by an uncharacterized β-N-acetylglucosaminidase (Hex), yielding GlcNAc and GlcNAc2. In the subsequent step GlcNAc is transported through the inner membrane by a GlcNAc-specific phosphoenolpyruvate transferase system transporter, forming GlcNAc-6-phosphate, whereas GlcNAc2 is transported and phosphorylated by the (GlcNAc2)-specific enzyme II permease of a different phosphoenolpyruvate transferase system. Utilization of chitobiose is further mediated by the Chb-BCARFG gene products of the Chb operon (56–58). The deacetylase ChbG removes one acetyl group from chitobiose-6-phosphate, generating monoacetyl chitobiose-6-phosphate, which is then the substrate for a β-glucosidase, ChbF. Its product, GlcNAc 6-phosphate (15), is deacetylated to GlcN 6-phosphate by NagA and then deaminated by NagB to fructose 6-phosphate. This final product of the pathway is metabolized as a carbon source for the bacterial cells. In conclusion, our study is the first elucidation of the physiological function of the OprD-like ChiP and provides an insight into how non-chitinolytic bacteria can utilize chitin as an alternative source of cellular energy during the scarcity of glucose-rich nutrients.
FIGURE 9.
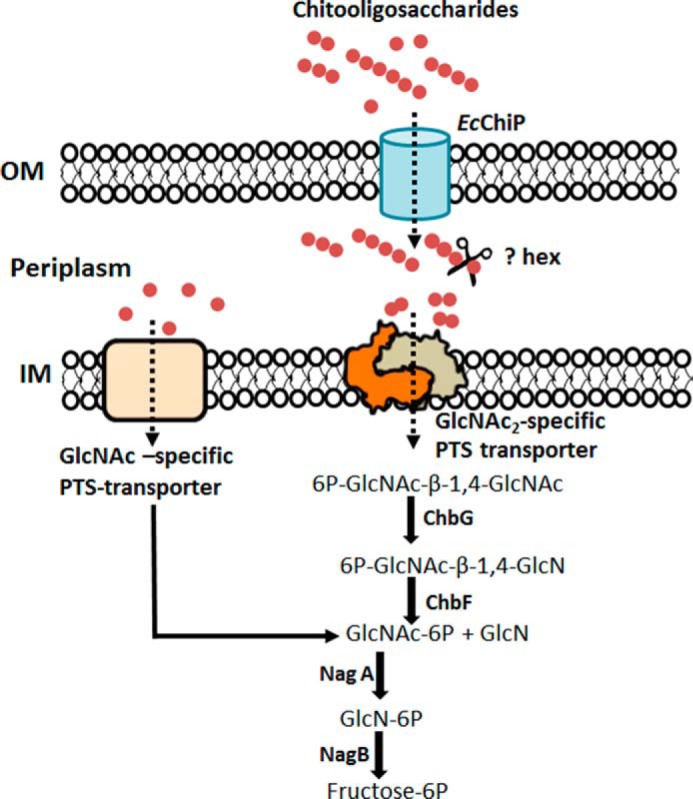
The chitooligosaccharide utilization pathway in E. coli. The scheme is based on the GlcNAc-utilization pathway proposed by Yang et al. (14) and Verma and Mahadevan (15). Solid arrows denote enzymic reactions, and dotted arrows denote the direction of sugar transport. PTS, phosphoenolpyruvate transferase system; OM, outer membrane; IM, inner membrane; ?Hex, uncharacterized β-N-acetylglucosaminidase (EC 3.2.1.96); ChbG, chitooligosaccharide monodeacetylase (EC 3.5.1.105); ChbF, monoacetyl-chitobiose 6-phosphate hydrolase (EC 3.2.1.86); NagA, N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25); NagB, glucosamine-6-phosphate deaminase (EC 3.5.1.10).
Author Contributions
H. S. M. S. designed, performed, and analyzed all the experiments and co-wrote the paper. WS conceived, designed, and coordinated the study and co-wrote and revised the paper.
Acknowledgments
We acknowledge the Biochemistry Laboratory of the Center for Scientific and Technological Equipment and Suranaree University of Technology for providing all research facilities. We greatly appreciate critical proofreading of this manuscript by Dr. David Apps, Centre for Integrative Physiology, School of Biomedical Sciences, University of Edinburgh, United Kingdom.
This work was supported by Suranaree University of Technology and the Office of the Higher Education Commission under the National Research University project of Thailand. The authors declare that they have no conflicts of interest with the contents of this article.
- VfChiP
- V. furnissii chitoporin
- EcChiP
- E. coli chitoporin
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- BLM
- black lipid membrane
- GlcNAc2
- chitobiose
- GlcNAc3
- chitotriose
- GlcNAc4
- chitotetraose
- GlcNAc5
- chitopentaose
- GlcNAc6
- chitohexaose
- nS
- nanosiemen(s).
References
- 1. Kim B. H., and Gadd G. M. (2008) Bacterial Physiology and Metabolism, Cambridge University Press, Cambridge, New York [Google Scholar]
- 2. Francetic O., Belin D., Badaut C., and Pugsley A. P. (2000) Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19, 6697–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Francetic O., Badaut C., Rimsky S., and Pugsley A. P. (2000) The ChiA (YheB) protein of Escherichia coli K-12 is an endochitinase whose gene is negatively controlled by the nucleoid-structuring protein H-NS. Mol. Microbiol. 35, 1506–1517 [DOI] [PubMed] [Google Scholar]
- 4. Suginta W., Chuenark D., Mizuhara M., and Fukamizo T. (2010) Novel β-N-acetylglucosaminidases from Vibrio harveyi 650: cloning, expression, enzymatic properties, and subsite identification. BMC Biochem. 11, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li X., and Roseman S. (2004) The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc. Natl. Acad. Sci. U.S.A. 101, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keyhani N. O., Li X. B., and Roseman S. (2000) Chitin catabolism in the marine bacterium Vibrio furnissii: identification and molecular cloning of a chitoporin. J. Biol. Chem. 275, 33068–33076 [DOI] [PubMed] [Google Scholar]
- 7. Suginta W., Chumjan W., Mahendran K. R., Janning P., Schulte A., and Winterhalter M. (2013) Molecular uptake of chitooligosaccharides through chitoporin from the marine bacterium Vibrio harveyi. PLoS ONE 8, e55126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suginta W., Chumjan W., Mahendran K. R., Schulte A., and Winterhalter M. (2013) Chitoporin from Vibrio harveyi, a channel with exceptional sugar specificity. J. Biol. Chem. 288, 11038–11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meekrathok P., and Suginta W. (2016) Probing the catalytic mechanism of Vibrio harveyi GH20 β-N-acetylglucosaminidase by chemical rescue. PLoS ONE 11, e0149228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suginta W., and Sritho N. (2012) Multiple roles of Asp-313 in the refined catalytic cycle of chitin degradation by Vibrio harveyi chitinase A. Biosci. Biotechnol. Biochem. 76, 2275–2281 [DOI] [PubMed] [Google Scholar]
- 11. Sritho N., and Suginta W. (2012) Role of Tyr-435 of Vibrio harveyi chitinase A in chitin utilization. Appl. Biochem. Biotechnol. 166, 1192–1202 [DOI] [PubMed] [Google Scholar]
- 12. Biswas S., Van Dijck P., and Datta A. (2007) Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71, 348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boulanger A., Déjean G., Lautier M., Glories M., Zischek C., Arlat M., and Lauber E. (2010) Identification and regulation of the N-acetylglucosamine utilization pathway of the plant pathogenic bacterium Xanthomonas campestris pv. campestris. J. Bacteriol. 192, 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang C., Rodionov D. A., Li X., Laikova O. N., Gelfand M. S., Zagnitko O. P., Romine M. F., Obraztsova A. Y., Nealson K. H., and Osterman A. L. (2006) Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 281, 29872–29885 [DOI] [PubMed] [Google Scholar]
- 15. Verma S. C., and Mahadevan S. (2012) The chbG gene of the chitobiose (chb) operon of Escherichia coli encodes a chitooligosaccharide deacetylase. J. Bacteriol. 194, 4959–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rasmussen A. A., Johansen J., Nielsen J. S., Overgaard M., Kallipolitis B., and Valentin-Hansen P. (2009) A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol. Microbiol. 72, 566–577 [DOI] [PubMed] [Google Scholar]
- 17. Peri K. G., Goldie H., and Waygood E. B. (1990) Cloning and characterization of the N-acetylglucosamine operon of Escherichia coli. Biochem. Cell Biol. 68, 123–137 [DOI] [PubMed] [Google Scholar]
- 18. Figueroa-Bossi N., Valentini M., Malleret L., Fiorini F., and Bossi L. (2009) Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 23, 2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valentin-Hansen P., Johansen J., and Rasmussen A. A. (2007) Small RNAs controlling outer membrane porins. Curr. Opin. Microbiol. 10, 152–155 [DOI] [PubMed] [Google Scholar]
- 20. Vogel J., and Papenfort K. (2006) Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9, 605–611 [DOI] [PubMed] [Google Scholar]
- 21. Plumbridge J., Bossi L., Oberto J., Wade J. T., and Figueroa-Bossi N. (2014) Interplay of transcriptional and small RNA-dependent control mechanisms regulates chitosugar uptake in Escherichia coli and Salmonella. Mol. Microbiol. 92, 648–658 [DOI] [PubMed] [Google Scholar]
- 22. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Montal M., and Mueller P. (1972) Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. U.S.A. 69, 3561–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshimura F., and Nikaido H. (1985) Diffusion of beta-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob. Agents Chemother. 27, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aunkham A., Schulte A., Winterhalter M., and Suginta W. (2014) Porin involvement in cephalosporin and carbapenem resistance of Burkholderia pseudomallei. PLoS ONE 9, e95918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takanao S., Honma S., Miura T., Ogawa C., Sugimoto H., Suzuki K., and Watanabe T. (2014) Construction and basic characterization of deletion mutants of the genes involved in chitin utilization by Serratia marcescens 2170. Biosci. Biotechnol. Biochem. 78, 524–532 [DOI] [PubMed] [Google Scholar]
- 27. Meibom K. L., Li X. B., Nielsen A. T., Wu C. Y., Roseman S., and Schoolnik G. K. (2004) The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U.S.A. 101, 2524–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Biswas S., Mohammad M. M., Patel D. R., Movileanu L., and van den Berg B. (2007) Structural insight into OprD substrate specificity. Nat. Struct. Mol. Biol. 14, 1108–1109 [DOI] [PubMed] [Google Scholar]
- 29. Wang Y. F., Dutzler R., Rizkallah P. J., Rosenbusch J. P., and Schirmer T. (1997) Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin. J. Mol. Biol. 272, 56–63 [DOI] [PubMed] [Google Scholar]
- 30. Forst D., Welte W., Wacker T., and Diederichs K. (1998) Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose. Nat. Struct. Biol. 5, 37–46 [DOI] [PubMed] [Google Scholar]
- 31. Conlan S., Zhang Y., Cheley S., and Bayley H. (2000) Biochemical and biophysical characterization of OmpG: A monomeric porin. Biochemistry 39, 11845–11854 [DOI] [PubMed] [Google Scholar]
- 32. van den Berg B. (2012) Structural basis for outer membrane sugar uptake in pseudomonads. J. Biol. Chem. 287, 41044–41052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strop P., and Brunger A. T. (2005) Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 14, 2207–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saravolac E. G., Taylor N. F., Benz R., and Hancock R. E. (1991) Purification of glucose-inducible outer membrane protein OprB of Pseudomonas putida and reconstitution of glucose-specific pores. J. Bacteriol. 173, 4970–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Benz R., Schmid A., and Vos-Scheperkeuter G. H. (1987) Mechanism of sugar transport through the sugar-specific LamB channel of Escherichia coli outer membrane. J. Membr. Biol. 100, 21–29 [DOI] [PubMed] [Google Scholar]
- 36. Ranquin A., and Van Gelder P. (2004) Maltoporin: sugar for physics and biology. Res. Microbiol. 155, 611–616 [DOI] [PubMed] [Google Scholar]
- 37. Kullman L., Winterhalter M., and Bezrukov S. M. (2002) Transport of maltodextrins through maltoporin: a single-channel study. Biophys. J. 82, 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bezrukov S. M., Kullman L., and Winterhalter M. (2000) Probing sugar translocation through maltoporin at the single channel level. FEBS Lett. 476, 224–228 [DOI] [PubMed] [Google Scholar]
- 39. Schwarz G., Danelon C., and Winterhalter M. (2003) On translocation through a membrane channel via an internal binding site: kinetics and voltage dependence. Biophys. J. 84, 2990–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zobell C. E., and Rittenberg S. C. (1938) The occurrence and characteristics of chitinoclastic bacteria in the sea. J. Bacteriol. 35, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bassler B. L., Yu C., Lee Y. C., and Roseman S. (1991) Chitin utilization by marine bacteria: degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266, 24276–24286 [PubMed] [Google Scholar]
- 42. Bassler B. L., Gibbons P. J., Yu C., and Roseman S. (1991) Chitin utilization by marine bacteria: chemotaxis to chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266, 24268–24275 [PubMed] [Google Scholar]
- 43. Yu C., Lee A. M., Bassler B. L., and Roseman S. (1991) Chitin utilization by marine bacteria: a physiological function for bacterial adhesion to immobilized carbohydrates. J. Biol. Chem. 266, 24260–24267 [PubMed] [Google Scholar]
- 44. Keyhani N. O., and Roseman S. (1999) Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473, 108–122 [DOI] [PubMed] [Google Scholar]
- 45. Park J. K., Keyhani N. O., and Roseman S. (2000) Chitin catabolism in the marine bacterium Vibrio furnissii. Identification, molecular cloning, and characterization of a N,N′-diacetylchitobiose phosphorylase. J. Biol. Chem. 275, 33077–33083 [DOI] [PubMed] [Google Scholar]
- 46. Hunt D. E., Gevers D., Vahora N. M., and Polz M. F. (2008) Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 74, 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pruzzo C., Vezzulli L., and Colwell R. R. (2008) Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 10, 1400–1410 [DOI] [PubMed] [Google Scholar]
- 48. Chumjan W., Winterhalter M., Schulte A., Benz R., and Suginta W. (2015) Chitoporin from the marine bacterium Vibrio harveyi: probing the essential roles of trp136 at the surface of the constriction zone. J. Biol. Chem. 290, 19184–19196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Danelon C., Brando T., and Winterhalter M. (2003) Probing the orientation of reconstituted maltoporin channels at the single-protein level. J. Biol. Chem. 278, 35542–35551 [DOI] [PubMed] [Google Scholar]
- 50. Bhamidimarri S. P., Prajapati J. D., van den Berg B., Winterhalter M., and Kleinekathöfer U. (2016) Role of electroosmosis in the permeation of neutral molecules: CymA and cyclodextrin as an example. Biophys. J. 110, 600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Siritapetawee J., Prinz H., Krittanai C., and Suginta W. (2004) Expression and refolding of Omp38 from Burkholderia pseudomallei and Burkholderia thailandensis and its function as a diffusion porin. Biochem. J. 384, 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Siritapetawee J., Prinz H., Samosornsuk W., Ashley R. H., and Suginta W. (2004) Functional reconstitution, gene isolation, and topology modelling of porins from Burkholderia pseudomallei and Burkholderia thailandensis. Biochem. J. 377, 579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saint N., Lou K. L., Widmer C., Luckey M., Schirmer T., and Rosenbusch J. P. (1996) Structural and functional characterization of OmpF porin mutants selected for larger pore size. II: functional characterization. J. Biol. Chem. 271, 20676–20680 [PubMed] [Google Scholar]
- 54. Dumas F., Koebnik R., Winterhalter M., and Van Gelder P. (2000) Sugar transport through maltoporin of Escherichia coli: role of polar tracks. J. Biol. Chem. 275, 19747–19751 [DOI] [PubMed] [Google Scholar]
- 55. Van Gelder P., Dumas F., Rosenbusch J. P., and Winterhalter M. (2000) Oriented channels reveal asymmetric energy barriers for sugar translocation through maltoporin of Escherichia coli. Eur. J. Biochem. 267, 79–84 [DOI] [PubMed] [Google Scholar]
- 56. Keyhani N. O., Bacia K., and Roseman S. (2000) The transport/phosphorylation of N,N′-diacetylchitobiose in Escherichia coli: characterization of phospho-IIB(Chb) and of a potential transition state analogue in the phosphotransfer reaction between the proteins IIA(Chb) AND IIB(Chb). J. Biol. Chem. 275, 33102–33109 [DOI] [PubMed] [Google Scholar]
- 57. Keyhani N. O., Boudker O., and Roseman S. (2000) Isolation and characterization of IIAChb, a soluble protein of the enzyme II complex required for the transport/phosphorylation of N,N′-diacetylchitobiose in Escherichia coli. J. Biol. Chem. 275, 33091–33101 [DOI] [PubMed] [Google Scholar]
- 58. Keyhani N. O., Wang L. X., Lee Y. C., and Roseman S. (2000) The chitin disaccharide, N,N′-diacetylchitobiose, is catabolized by Escherichia coli and is transported/phosphorylated by the phosphoenolpyruvate:glycose phosphotransferase system. J. Biol. Chem. 275, 33084–33090 [DOI] [PubMed] [Google Scholar]