Abstract
5-Hydroxymethylcytosine (5hmC) is an epigenetic modification that is generated by ten-eleven translocation (TET) protein-mediated oxidation of 5-methylcytosine (5mC). 5hmC is associated with transcription regulation and is decreased in many cancers including melanoma. Accumulating evidence has suggested that 5hmC is functionally distinct from 5mC. Ubiquitin-like with PHD and ring finger domains 2 (UHRF2) is the first known specific 5hmC reader that has higher affinity to 5hmC than 5mC, suggesting that UHRF2 might mediate 5hmC's function. Structural analysis has revealed the molecular mechanism of UHRF2–5hmC binding in vitro, but it is not clear how UHRF2 recognizes 5hmC in vivo. In this study, we have identified zinc figure protein 618 (ZNF618) as a novel binding partner of UHRF2. ZNF618 specifically interacts with UHRF2 but not its paralog UHRF1. Importantly, ZNF618 co-localizes with UHRF2 at genomic loci that are enriched for 5hmC. The ZNF618 chromatin localization is independent of its interaction with UHRF2 and is through its first two zinc fingers. Instead, ZNF618 regulates UHRF2 chromatin localization. Collectively, our study suggests that ZNF618 is a key protein that regulates UHRF2 function as a specific 5hmC reader in vivo.
Keywords: 5-hydroxymethylcytosine (5-hmC), chromatin, DNA demethylation, molecular biology, molecular cell biology
Introduction
DNA methylation is an epigenetic modification that regulates multiple cellular processes including gene transcription, genomic imprinting, and X chromosome inactivation. DNA methylation mainly exists as 5-methylcytosine (5mC)3 in CpG context, which is established and maintained by three major DNA methyltransferases in cells (1). 5mC can be oxidized by ten-eleven translocation (TET) proteins to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (2–4). These 5mC derivatives are believed to be intermediates of DNA demethylation (5).
5hmC is much more abundant than others among three 5mC derivatives (6). It is not a favorable substrate for further oxidation and is therefore most stable (7, 8). It is widely present in many cell types and tissues and is particularly enriched in neurons and stem cells (9, 10). 5hmC and 5mC have different genome-wide distributions and are associated with different transcriptional outcome (11–16). Loss of 5hmC, but not 5mC, is a hallmark of many cancers including melanoma (17). These studies suggest that 5hmC is more than just an intermediate of DNA demethylation, but is an important epigenetic modification that is distinct from 5mC.
The function of 5hmC is still not clear, largely due to the lack of knowledge about 5hmC readers. Previous studies have shown that some 5mC readers, such as UHRF1 and MeCP2, can also bind 5hmC (18, 19). These proteins use the same 5mC-binding domain to interact with 5hmC, but the 5hmC binding affinity was similar to or weaker than their 5mC binding affinities (18, 19). Recent proteomic approaches have revealed several 5hmC-specific binding proteins that have higher affinity to 5hmC than 5mC (20). UHRF2 is the best characterized one and its SRA domain has 3-fold higher binding affinity to 5hmC than 5mC (21). Therefore, UHRF2 is a specific 5hmC reader.
UHRF2 is a paralog of UHRF1, which is essential for the maintenance of DNA methylation. UHRF1 loads DNMT1 to hemi-methylated DNA regions after DNA replication (22, 23) to methylate the nascent strand. In addition, UHRF1 interacts with Topoisomerase II, which resolves the catenated DNA after DNA replication to allow efficient DNA methylation (24). Studies have also revealed that UHRF1-mediated histone H3 ubiquitination is important for this process (25, 26). UHRF1 and UHRF2 have the same domain structure and highly similar primary sequences, but their functions are different. Uhrf1 knock-out mice is embryonic lethal and Uhrf1 knock-out ES cells have dramatic decrease in global DNA methylation (27, 28), suggesting that UHRF1 and UHRF2 are not functionally redundant in the maintenance of DNA methylation. UHRF1 is highly expressed in undifferentiated cells while the expression of UHRF2 is low in undifferentiated cells and increases when cells are differentiated (29). Although both UHRF1 and UHRF2 interact with DNMT1, UHRF2 cannot target DNMT1 to pericentric heterochromatin (PCH) in S phase when overexpressed in cells (30).
The most well characterized structural difference between UHRF1 and UHRF2 is at their SRA domains. The SRA domain of UHRF1 has higher affinity to 5mC and that of UHRF2 prefers 5hmC (18, 20, 21). Structural analysis has revealed that subtle differences in UHRF2's SRA domain create a larger pocket that is suitable for 5hmC binding and allow binding of 5hmC on both DNA strands (21). The unique in vitro 5hmC binding ability of UHRF2 SRA domain suggests that UHRF2 could co-localize with 5hmC and mediate 5hmC function, but this possibility has not been tested in vivo. In addition, extra factors might also regulate UHRF2's 5hmC binding in vivo. Here we have identified ZNF618 as a novel binding partner of UHRF2 but not its paralog UHRF1. ZNF618 co-localizes with UHRF2 at genomic loci that are enriched for 5hmC. In addition, ZNF618 facilitates UHRF2 chromatin localization. Therefore, ZNF618 might be a key protein that regulates UHRF2 function as a specific 5hmC reader in vivo.
Experimental Procedures
Cell Culture and RNA Interference
All cells were cultured in DMEM supplemented with 10% FBS. For siRNA-mediated gene knockdown, two identical siRNA transfections separated by 24 h were performed. Cells were harvested 48 h after second transfection for analysis. All siRNAs were purchased from Dharmacon. siRNA sequence for UHRF2 was GGAAGAAGCCUAAAGGACAdTdT and that for ZNF618 was CAGUACUGGUCGUGCGUUAdTdT. siRNA-resistant ZNF618 was generated by introducing silent mutations to the siRNA-targeting sequence.
Antibodies
The following antibodies were generated: anti-UHRF1 (a.a. 14–159), anti-UHRF2 (a.a. 14–159), anti-ZNF618-N (a.a. 1–320, for Western blotting), and anti-ZNF618-C (a.a. 501–861, for immunoprecipitation). The following antibody was purchased: anti-UHRF1 (Santa Cruz, sc-373750), anti-UHRF2 (Santa Cruz Biotechnology, sc-398953),anti-α-tubulin (Genscript, A01410), anti-FLAG (Sigma, F1804), and anti-HA (Covance, MMS-101P).
Tandem Affinity Purification
293T cells stably expressing S-FLAG-streptavidin binding protein (SFB) triple-tagged UHRF2 were generated. Cells from 50 10-cm2 culture dishes were collected and lysed with NTN300 (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, and 0.5% Nonidet P-40). The lysate was collected, diluted with the same volume of ddH2O and incubated with streptavidin beads (Thermo) at 4 °C for 2 h. The beads were then washed three times with NTN (50 mm Tris-HCl pH8.0, 100 mm NaCl, and 0.5% Nonidet P-40), and bound proteins were eluted twice with saturating biotin solutions in NTN. The eluents were combined and incubated with S-protein beads (Millipore) at 4 °C for 2 h. The beads were then washed three times, and the bound proteins were eluted by addition of SDS sample loading buffer. The samples were briefly electrophoresed into 7.5% PAGE, and the entire lane (less than 1 cm) was excised. Mass spectrometry was performed by Taplin mass spectrometry facility in Harvard University.
Immunoprecipitation and Western Blotting
Cells were harvested and lysed with NTN300. The lysate was collected and diluted with the same volume of ddH2O. 1 μg of antibody and 40 μl of protein A beads were added. The mixture was incubated with shaking at 4 °C for 2 h. The beads were then washed three times with NTN, incubated with SDS sample loading buffer, and subjected to polyacrylamide gel electrophoresis (PAGE). For extraction of chromatin-bound proteins, cells were lysed with NTN. The insoluble material was pelleted and resuspended in 0.2 m HCl. The dissolved chromatin-bound proteins were neutralized with 1 m Tris-HCl, pH 8.0 and subjected to PAGE. Western blotting was performed according to standard procedures. All Western blotting experiments were performed for at least three times and representative figures are shown.
Recombinant Protein Expression
GST-tagged SRA domains of UHRF1 (a.a. 409–619) and UHRF2 (a.a. 425–634) were expressed in bacteria BL21(DE3) using standard protocols and purified using glutathione-Sepharose 4B (Thermo). N-terminal SFB-tagged full-length UHRF2 and N-terminal GST-tagged full-length ZNF618 were expressed in insect cells Sf9 using the Bac-to-Bac Baculovirus expression system (Life Technologies) and purified using streptavidin beads (Thermo) and glutathione-Sepharose 4B, respectively.
Immunofluorescence Staining
All procedures were performed at room temperature. Cells were fixed with 3% paraformaldehyde for 10 min and incubated with 0.5% Triton X-100 for 5 min. After PBS wash, the cells were incubated with primary antibody for 1 h, washed with PBS, and incubated with secondary antibody for 30 min. All antibodies were diluted in 5% goat serum in PBS. After PBS wash, cells were briefly incubated with 1 μg/ml Hoechst 33342 and mounted on slides.
Chromatin Immunoprecipitation Sequencing (ChIP-Seq)
ChIP was performed using ChIP assay kit (Millipore) following manufacturer's instructions, except that S-protein beads were used to replace protein A beads. 2 × 107 293T cells stably expressing SFB-tagged UHRF2 and ZNF618 were used for each IP. The DNA after ChIP was de-crosslinked, end processed using Quick Blunting Kit and dA-Tailing Module, and ligated to Illumina-compatible adaptors. The DNA was then amplified by PCR using barcode-containing primers. The products were pooled and sequenced using HiSeq 2500 (Illumina) by Harvard biopolymers facility.
ChIP-Seq Data Analysis
The resulting 50-bp single-ended short reads were aligned to the human reference genome hg19 using Bowtie 2 (version 2.1.0) using default parameters (31). To determine peaks, Model-Based Analysis of ChIP-Seq (MACS, version 1.4.0) was used with default parameters and p value cutoff of 1e−8 (32). HOMER (Hypergeometric Optimization of Motif EnRichmert) Suite (v4.7) was applied to annotate the significant peak regions (33). 5hmC data were obtained from GEO (GSE 67621) (34). 5hmC sites with low reads (<5 reads) were filtered out, and all the other sites were combined as the global 5hmC pattern. To find the distribution of 5hmC in UHRF2- and ZNF618-enriched regions, the BEDTools suite (v2.25.0) was used (35).
Results
UHRF2 Interacts with ZNF618
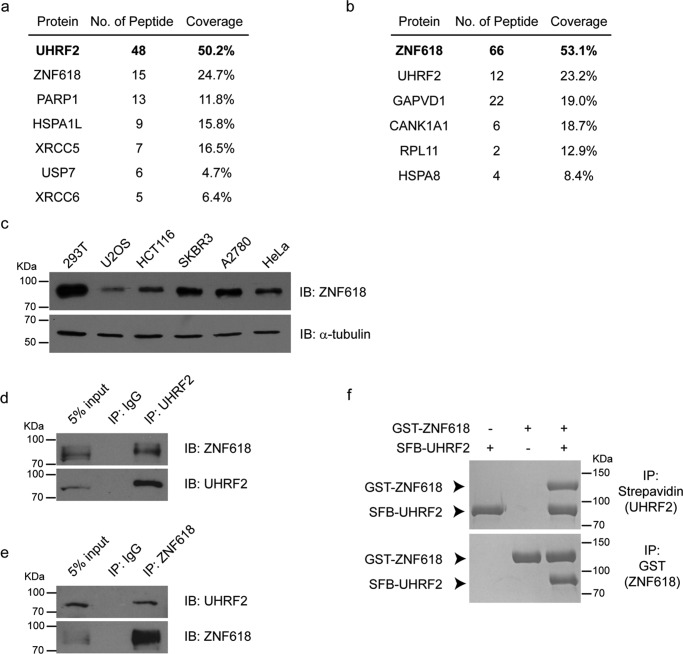
To search for proteins that might regulate its UHRF2 5hmC binding ability in vivo, we stably expressed S-FLAG-streptavidin-binding protein (SFB) triple-tagged UHRF2 in 293T cells and purified its interacting proteins. Through mass spectrometry analyses, zinc finger protein 618 (ZNF618) was identified as the most abundant protein except UHRF2 itself (Fig. 1a). To exclude possible contaminations during protein affinity purification, we performed a reverse protein affinity purification using ZNF618 as the bait and found that UHRF2 was also a major binding partner of ZNF618 (Fig. 1b). ZNF618 is ubiquitously expressed in a number of human cell lines from different tissue origin (Fig. 1c). Reciprocal co-immunoprecipitation (co-IP) experiments confirmed the interaction between endogenous UHRF2 and ZNF618 (Fig. 1, d and e) in 293T cells. Importantly, direct interaction with a stoichiometric ratio of 1:1 between UHRF2 and ZNF618 was readily detected using recombinant proteins expressed from insect cells (Fig. 1f), suggesting that UHRF2 and ZNF618 form a heterodimer.
FIGURE 1.
ZNF618 interacts with UHRF2. a, UHRF2 (shown in bold)-interacting proteins were purified in 293T cells stably expressing SFB-tagged UHRF2. A list of proteins identified by mass spectrometry is shown. b, ZNF618 (shown in bold)-interacting proteins were purified in 293T cells stably expressing SFB-tagged ZNF618. A list of proteins identified by mass spectrometry is shown. c, expression of ZNF618 was detected in a number of human cell lines with different tissue origins: 293T (kidney), U2OS (bone), HCT116 (colon), SKBR3 (breast), A2780 (ovary), and HeLa (cervix). d and e, co-IP was performed using antibodies against endogenous proteins as indicated. e, recombinant SFB-tagged UHRF2 and GST-tagged ZNF618 were expressed in Sf9 cells, purified, and incubated together as indicated. Pull-down was performed using beads as indicated.
UHRF2′ SRA Domain Binds the ZNF618 Zinc Finger Domain
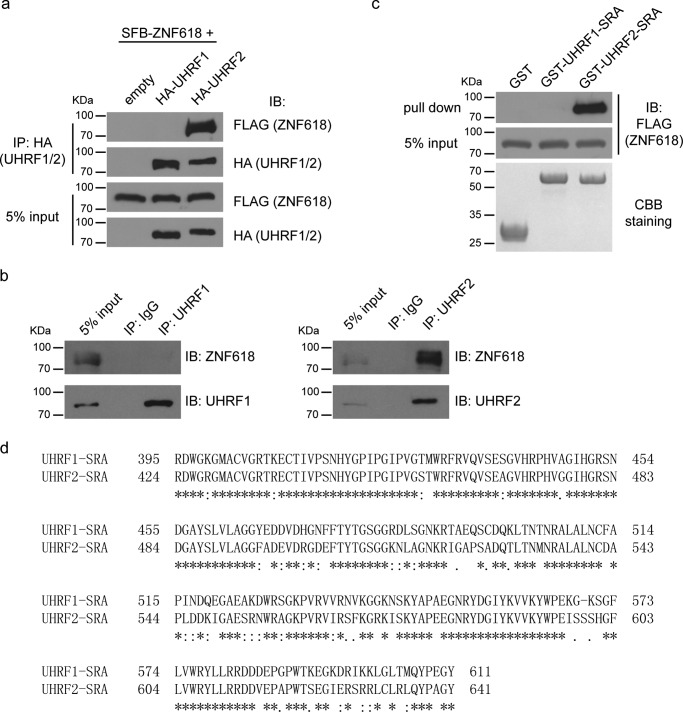
To further validate our results, we mapped the interacting regions between UHRF2 and ZNF618. Similar to UHRF1, UHRF2 has five major domains: UBL, Tudor, PHD, SRA, and RING domains (Fig. 2a). We generated deletion mutants of each domain of UHRF2 and tested their interaction with ZNF618. While deletion of other domains retained UHRF2 ability to interact with ZNF618, deletion of the SRA domains completely abolished the interaction between UHRF2 and ZNF618 (Fig. 2b). We further analyzed the regions on ZNF618 responsible for this interaction. The only known domains on ZNF618 are the four zinc fingers on its N terminus (Fig. 3a). We generated five deletion mutants that covered the entire region of ZNF618, and the first two mutants (D1 and D2) deleted two zinc fingers each. Co-IP experiments revealed the interaction with UHRF2 was abolished when a region containing zinc finger 3 and 4 was deleted from ZNF618 (Fig. 3b). Therefore, the SRA domain of UHRF2 interacts with a region containing zinc finger 3 and 4 of ZNF618.
FIGURE 2.
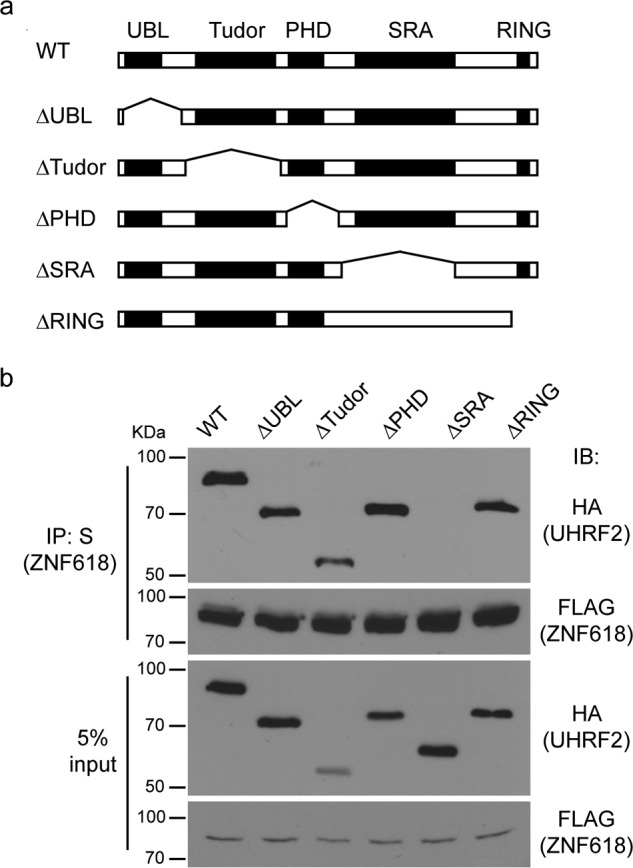
The UHRF2 SRA domain interacts with ZNF618. a, schematic illustration of full-length UHRF2 and its deletion mutants is shown. b, 293T cells stably expressing SFB-tagged ZNF618 were transfected with HA-tagged UHRF2 and its deletion mutants, and co-IP was performed.
FIGURE 3.
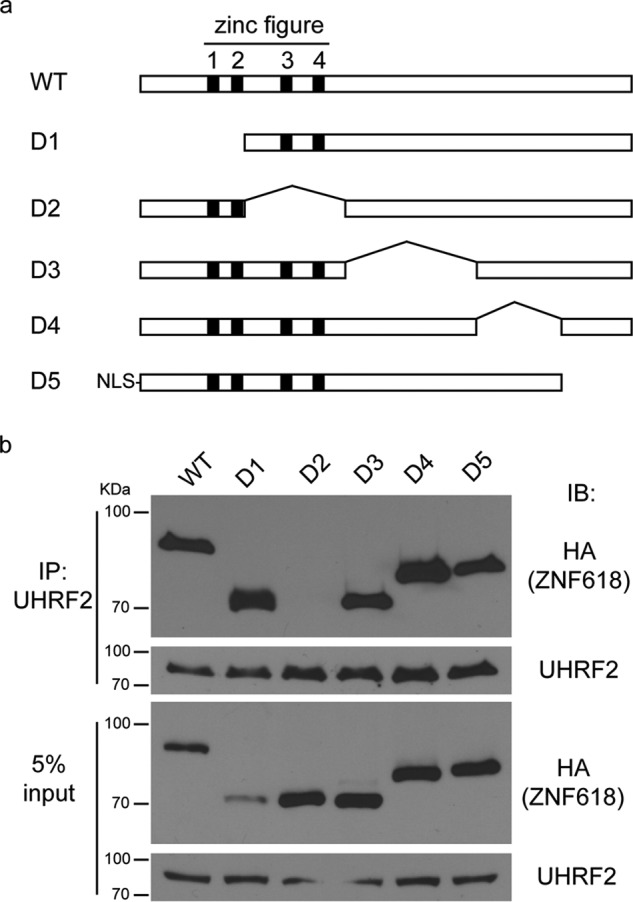
A region containing zinc finger 3 and 4 of ZNF618 interacts with UHRF2. a, schematic illustration of full-length ZNF618 and its deletion mutants is shown. b, 293T cells were transfected with HA-tagged ZNF618 and its deletion mutants, and co-IP was performed.
ZNF618 Specifically Interacts with UHRF2 but Not UHRF1
UHRF2 is a paralog of UHRF1. These two proteins have over 80% sequence similarities and share similar domain structures. Since we did not identify ZNF618 as a UHRF1 binding partner during our previous studies (24), we wonder if ZNF618 is a specific interacting partner of UHRF2. To test this possibility, we examined the interaction between exogenously expressed ZNF618 and UHRF1 and UHRF2. Indeed, SFB-tagged ZNF618 could co-immunoprecipitate with HA-tagged UHRF2 but not HA-tagged UHRF1 (Fig. 4a). Examination of endogenous proteins also revealed that ZNF618 interacted with UHRF2 but not UHRF1 (Fig. 4b). To further confirm our findings, we generated GST-tagged recombinant SRA domains of UHRF1 and UHRF2 and tested their interaction with ZNF618. The SRA domain of UHRF2, but not that of UHRF1, could robustly pull down SFB-tagged ZNF618 (Fig. 4c). These results suggest that ZNF618 is indeed a specific binding partner of UHRF2 but not UHRF1. The SRA domains of UHRF1 and UHRF2 are largely similar (Fig. 4d). Future structural analysis will reveal the molecular mechanism how ZNF618 distinguishes these two proteins.
FIGURE 4.
ZNF618 interacts with UHRF2 but not UHRF1. a, 293T cells stably expressing SFB-tagged ZNF618 were transfected with HA-tagged UHRF1 or UHRF2, and co-IP was performed. b, Co-IP was performed using antibodies against endogenous proteins as indicated. c, GST-tagged SRA domain of UHRF1 or UHRF2 were used to pull-down SFB-tagged ZNF618 stably expressed in 293T cells. d, alignment of the SRA domains of UHRF1 and UHRF2 was performed using Clustal Omiga.
ZNF618 Co-localizes with UHRF2 on Chromatin
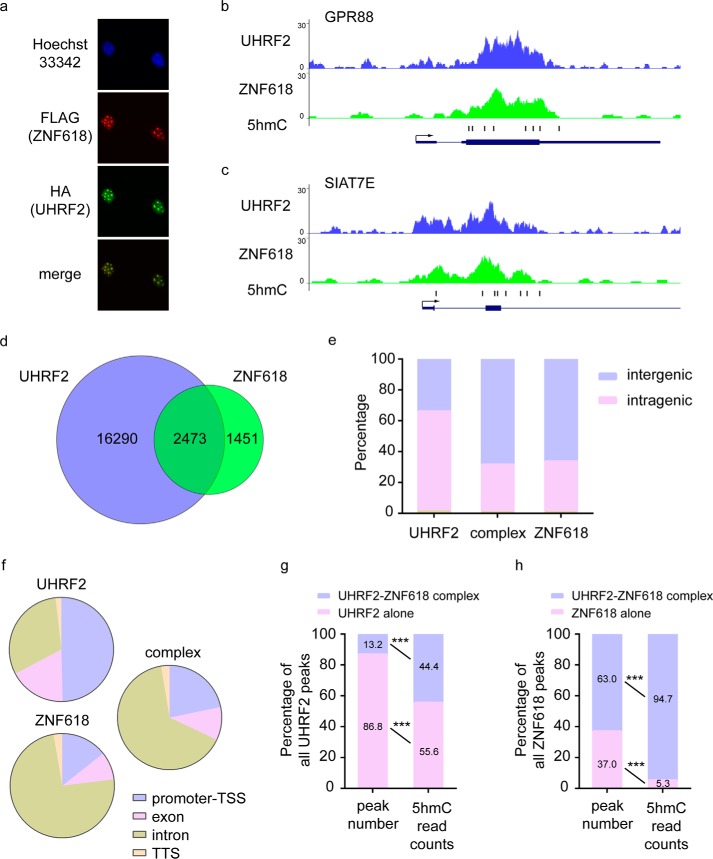
The interaction between UHRF2 and ZNF618 suggests that these two proteins co-localize in cells and function together. It has been shown that UHRF2 localizes to chromatin through concerted actions of its Tudor, PHD, and SRA domains (29, 30). Consistent with previous reports, UHRF2 could be readily observed at pericentric heterochromatin (PCH) when transfected into mouse embryonic fibroblasts (MEFs) (Fig. 5a). In agreement with its interaction with UHRF2, ZNF618 also localized at PCH when transfected into MEFs and co-localized with UHRF2 (Fig. 5a). To analyze the genome-wide chromatin distribution of these two proteins, we performed chromatin immunoprecipitation-sequencing (ChIP-Seq) experiments. S protein beads were used to immunoprecipitate SFB-tagged UHRF2 and ZNF618 from 293T cells stably expressing them, and the bound chromatin DNA was subjected to high-throughput sequencing. In agreement with the interaction of these two proteins, significant overlap of the peaks was observed (Fig. 5, b–d), which demonstrated that these two proteins indeed formed a complex. 63.0% of ZNF618 peaks overlapped with UHRF2, indicating that a large portion of ZNF618 forms a functional complex with UHRF2. Much more peaks were obtained for UHRF2 and 13.2% overlapped with ZNF618 (Fig. 5d), suggesting that UHRF2 has broader distribution than ZNF618 on chromatin and has ZNF618-independent functions. To characterize the chromatin binding pattern of these two proteins, we analyzed the features of the genome-wide distribution of these two proteins. While UHRF2 was enriched at intragenic regions, ZNF618 was enriched at intergenic regions (Fig. 5e). Interestingly, the overlapping peaks between UHRF2 of ZNF618 (UHRF2-ZNF618 complex) distributed preferentially at intergenic regions, which were similar to those of ZNF618 (Fig. 5e). The overlapping intragenic peaks (UHRF2-ZNF618 complex) also had similar features over genes as those of ZNF618 (Fig. 5f). These results suggested that ZNF618 contributes significantly to the localization of UHRF2-ZNF618 complex at these genomic loci.
FIGURE 5.
ZNF618 is localized at pericentric heterochromatin (PCH). a, SFB-tagged ZNF618 and HA-tagged were co-transfected into MEF. Immunofluorescence staining was performed using indicated antibodies. b and c, typical UCSC genome browser views of UHRF2 and ZNF618 peaks at GPR88 and SIAT7E genes are shown. 5hmC profiles are also shown. Arrows denote promoter orientations. Boxes denote exons, and lines denote introns. d, overlap between UHRF2 and ZNF618 peaks is shown. The overlapping peaks are defined as peaks for UHRF2-ZNF618 complex. e, genome-wide peak distribution of UHRF2, ZNF618, and UHRF2-ZNF618 complex is shown. f, intragenic peak distribution of UHRF2, ZNF618, and UHRF2-ZNF618 complex is shown. g, UHRF2 peaks were separated into two groups: those overlapped with ZNF618 (UHRF2-ZNF618 complex) and those did not (UHRF2 alone). The peak number and the 5hmC read counts within peaks in each group were calculated, and the percentages are summarized. h, ZNF618 peaks were separated into two groups: those overlapped with UHRF2 (UHRF2-ZNF618 complex) and those did not (ZNF618 alone). The peak number and the 5hmC read counts within peaks in each group were calculated, and the percentages are summarized.
Because UHRF2 SRA domain is a specific 5hmC reader (20, 21), the direct binding between the UHRF2 SRA domain and ZNF618 suggests that the UHRF2 SRA domain is occupied by ZNF618 in this complex and is no longer available for 5hmC binding. To test if UHRF2-ZNF618 complex is depleted from 5hmC-containing genomic loci, we obtained 5hmC information in 293T cells (34) and analyzed their positions and levels in UHRF2 peaks. We separated UHRF2 peaks into two groups: those overlapped with ZNF618 (UHRF2-ZNF618 complex) and those did not (UHRF2 alone). Surprisingly, peaks of UHRF2-ZNF618 complex contained almost half (44.4%) of all 5hmC read counts within all UHRF2 peaks, despite that they were only 13.2% of all UHRF2 peaks (Fig. 5g). On the contrary, although 86.8% of all UHRF2 peaks had UHRF2 alone, they contained only 55.6% of all 5hmC read counts (Fig. 5g). This observation revealed that UHRF2-ZNF618 complex was not depleted from, but instead enriched at, 5hmC-containing genomic loci, suggesting that ZNF618 positively regulates UHRF2 5hmC binding. To test if ZNF618 itself could bind 5hmC, we analyzed the positions and levels of 5hmC in ZNF618 peaks. Interestingly, the 5hmC read counts were almost exclusively present (94.7%) in peaks of UHRF2-ZNF618 complex (Fig. 5h). Although more than one-third (37.0%) of all ZNF618 peaks had ZNF618 alone, they barely contained any 5hmC read counts (5.35%) (Fig. 5g). Therefore, it is unlikely that ZNF618 directly binds 5hmC. It is possible that ZNF618 binds DNA adjacent to 5hmC and facilitates UHRF2 5hmC binding.
ZNF618 Regulates UHRF2 Chromatin Localization
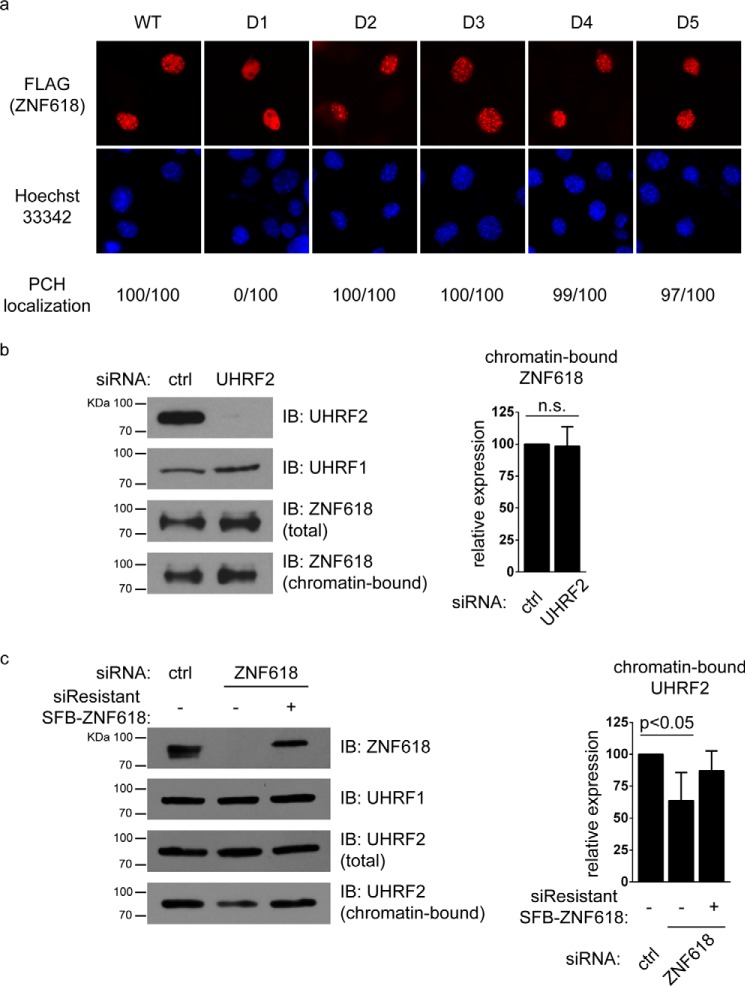
To gain more insight into how ZNF618 functions with UHRF2 in 5hmC binding, we examined how ZNF618 is recruited to chromatin. We utilized 5 deletion mutants of ZNF618 (Fig. 3a) and examined the region of ZNF618 responsible for its PCH localization. Surprisingly, deletion of the region containing zinc finger 3 and 4 (in D2), the UHRF2-interacting region, did not affect ZNF618 localization at PCH (Fig. 6a). Instead, ZNF618 localization at PCH was completely abolished when its N-terminal region containing zinc finger 1 and 2 was deleted (in D1) (Fig. 6a). It is likely that ZNF618 is recruited to chromatin through its N-terminal two zinc fingers. In agreement with these observations in MEFs, UHRF2 depletion did not affect protein levels of chromatin-bound ZNF618 (Fig. 6b), suggesting that UHRF2 is not required for ZNF618 chromatin localization. Interestingly, the chromatin-bound fractions of UHRF2 was decreased when ZNF618 was depleted, which could be rescued by siRNA-resistant ZNF618 (Fig. 6c), suggesting that ZNF618 regulates UHRF2 chromatin localization. Together with previous observations, this result suggests that ZNF618 might contribute to UHRF2 5hmC binding and chromatin retention.
FIGURE 6.
ZNF618 regulates UHRF2 chromatin binding. a, MEF cells were transfected with SFB-tagged ZNF618 and its deletion mutants. Immunofluorescence staining was performed using indicated antibodies. PCH localization was scored in 100 successfully transfected cells from 2 different experiments. b and c, 293T cells were transfected with siRNA and siRNA-resistant construct as indicated. Western blotting was performed using antibodies as indicated.
Discussion
In this study, we have identified ZNF618 as a specific binding partner of UHRF2 but not UHRF1. ZNF618 localizes to chromatin using its N-terminal region containing two zinc fingers and interacts with the UHRF2 SRA domain through a region containing zinc finger 3 and 4. ZNF618 co-localizes with UHRF2 at genomic loci that are enriched for 5hmC and regulates UHRF2's chromatin localization. These observations suggest that ZNF618 is a functional partner of UHRF2.
Although UHRF1 and UHRF2 have the same domain structure and highly similar primary sequences, they have non-redundant functions (30). Both of their Tudor and PHD domains bind H3K9me3 (29, 30), which is unlikely to contribute to their functional difference. Subtle structural difference in their SRA domains results in their different affinities to 5hmC (18, 20, 21), but the 3-fold affinity difference in 5hmC binding alone is unlikely to determine the functional difference between these two proteins. Since ZNF618 specifically interacts with UHRF2 and co-localizes with UHRF2 at genomic loci that are enriched for 5hmC, the UHRF2-ZNF618 complex might have much higher affinity to 5hmC than UHRF1 in vivo. Therefore, ZNF618 could be the key protein that determines the functional difference between UHRF1 and UHRF2. Evolutionary analysis also supports the idea that ZNF618 promotes the functional divergence between these two proteins. UHRF1 and UHRF2 genes are evolved from the same ancestor, which co-exists with DNMT1 gene in lower organisms such as Volvox carteri (green alga), Physcomitrella patens (moss), Crassostrea gigas (oyster), and Apis mellifera (honey bee) (Fig. 7). In vertebrates, the UHRF gene is duplicated into UHRF1 and UHRF2 genes. Coincidently, ZNF618 gene only exists in vertebrates (Fig. 7), suggesting that the role of ZNF618 is associated with the duplication of UHRF gene during evolution.
FIGURE 7.
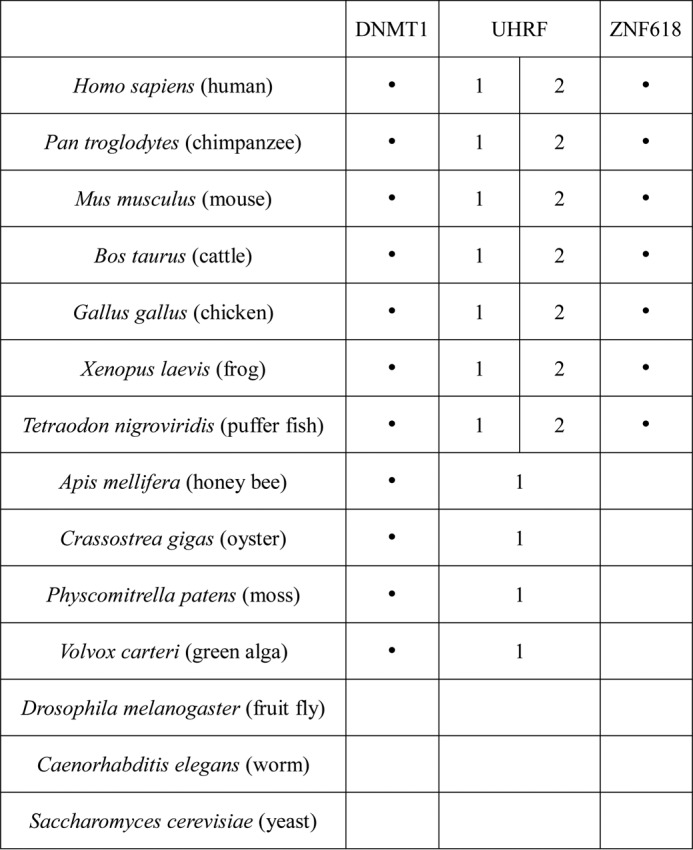
ZNF618 evolves together with UHRF2. The presence or absence of DNMT1, UHRF1/2, and ZNF618 in various species is summarized.
It is interesting that UHRF2 uses the same 5hmC-binding SRA domain to bind ZNF618 (20, 21). To our surprise, ZNF618 binding does not abolish, but instead facilitates UHRF2's binding to 5hmC. From these observations, we speculate that the SRA domain of UHRF2 uses two pockets to bind 5hmC and ZNF618 at the same time. In fact, studies have shown that the SRA domain of UHRF1 could bind 5mC and DNMT1 using two different pockets (22, 23). In this scenario, dual binding of 5hmC and ZNF618 will not only strengthen the chromatin binding of UHRF2, but might also provide additional regulation where UHRF2 is localized through the ZNF618 N-terminal two zinc figures recognizing specific genomic sequences. Future structural analysis will help clarify how UHRF2 coordinates 5hmC and ZNF618 binding.
It is noteworthy that ZNF618 is not absolutely required for UHRF2's chromatin localization. This is consistent with previous reports that the SRA domain is not absolutely required for UHRF2 chromatin localization (30). Indeed, we have found that a large portion of UHRF2 does not co-localize with ZNF618. Therefore, there are additional factors that regulate UHRF2's chromatin localization and function independently of ZNF618. Similar as UHRF1, UHRF2 has Tudor and PHD domains that cooperatively recognize H3K9 di/tri-methylation (29, 30), and mutation of either of these two domains dramatically affects UHRF2 localization at PCH (30). It is likely that these two domains coordinate with the SRA domain to regulate UHRF2's chromatin localization. In addition, we have identified USP7 as an interacting partner of UHRF2 (Fig. 1b), which is known to interact with UHRF1 and regulate its chromatin binding (36). It is possible that USP7 also regulates UHRF2's chromatin localization similarly.
Taken together, we have identified ZNF618 as a novel interacting protein of UHRF2 that regulates UHRF2 function as a specific 5hmC reader in vivo. ZNF618 could be the key protein that determines the functional difference between UHRF1 and UHRF2. Therefore, our study is critical for understanding the molecular function of UHRF2 in vivo.
Author Contributions
L. L. and X. Y. conceived the experiment. Y. L., H. K., G. K., and L. L. performed the experiment. Y. L., L. L., and X. Y. analyzed the data. Y. L., L. L., and X. Y. wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
This work was funded by Zhejiang Provincial Natural Science Foundation of China (LR15H040001, to L. Y.), National Natural Science Foundation of China (81471494, to L. Y.), and National Institutes of Health (CA130899, CA132755, CA187209, and GM108647, to X. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The ChIP-Seq raw datasets for this article have been submitted to GEO under accession number GSE80037.
- 5mC
- 5-methylcytosine
- UHRF
- ubiquitin-like with PHD and ring finger domain
- TET
- ten-eleven translocation
- 5hmC
- 5-hydroxymethylcytosine
- MEF
- mouse embryonic fibroblasts
- ZNF
- zinc finger
- PCH
- pericentric heterochromatin.
References
- 1. Li E., Bestor T. H., and Jaenisch R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 [DOI] [PubMed] [Google Scholar]
- 2. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., and Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., He C., and Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. He Y. F., Li B. Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C. X., Zhang K., He C., and Xu G. L. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohli R. M., and Zhang Y. (2013) TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song C. X., Yi C., and He C. (2012) Mapping recently identified nucleotide variants in the genome and transcriptome. Nat. Biotechnol. 30, 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu L., Lu J., Cheng J., Rao Q., Li Z., Hou H., Lou Z., Zhang L., Li W., Gong W., Liu M., Sun C., Yin X., Li J., Tan X., Wang P., Wang Y., Fang D., Cui Q., Yang P., He C., Jiang H., Luo C., and Xu Y. (2015) Structural insight into substrate preference for TET-mediated oxidation. Nature 527, 118–122 [DOI] [PubMed] [Google Scholar]
- 8. Hashimoto H., Pais J. E., Zhang X., Saleh L., Fu Z. Q., Dai N., Corrêa I. R. Jr., Zheng Y., and Cheng X. (2014) Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506, 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Globisch D., Münzel M., Müller M., Michalakis S., Wagner M., Koch S., Brückl T., Biel M., and Carell T. (2010) Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5, e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kriaucionis S., and Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song C. X., Szulwach K. E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C. H., Zhang W., Jian X., Wang J., Zhang L., Looney T. J., Zhang B., Godley L. A., Hicks L. M., Lahn B. T., Jin P., and He C. (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 29, 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu H., D'Alessio A. C., Ito S., Wang Z., Cui K., Zhao K., Sun Y. E., and Zhang Y. (2011) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25, 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ficz G., Branco M. R., Seisenberger S., Santos F., Krueger F., Hore T. A., Marques C. J., Andrews S., and Reik W. (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 [DOI] [PubMed] [Google Scholar]
- 14. Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A. J., Zheng L., Zhang H., Huang S., Min J., Nicholson T., Chen T., Xu G., Shi Y., Zhang K., and Shi Y. G. (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 42, 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pastor W. A., Pape U. J., Huang Y., Henderson H. R., Lister R., Ko M., McLoughlin E. M., Brudno Y., Mahapatra S., Kapranov P., Tahiliani M., Daley G. Q., Liu X. S., Ecker J. R., Milos P. M., Agarwal S., and Rao A. (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stroud H., Feng S., Morey Kinney S., Pradhan S., and Jacobsen S. E. (2011) 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 12, R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lian C. G., Xu Y., Ceol C., Wu F., Larson A., Dresser K., Xu W., Tan L., Hu Y., Zhan Q., Lee C. W., Hu D., Lian B. Q., Kleffel S., Yang Y., Neiswender J., Khorasani A. J., Fang R., Lezcano C., Duncan L. M., Scolyer R. A., Thompson J. F., Kakavand H., Houvras Y., Zon L. I., Mihm M. C. Jr., Kaiser U. B., Schatton T., Woda B. A., Murphy G. F., and Shi Y. G. (2012) Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150, 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frauer C., Hoffmann T., Bultmann S., Casa V., Cardoso M. C., Antes I., and Leonhardt H. (2011) Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS ONE 6, e21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mellén M., Ayata P., Dewell S., Kriaucionis S., and Heintz N. (2012) MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spruijt C. G., Gnerlich F., Smits A. H., Pfaffeneder T., Jansen P. W., Bauer C., Münzel M., Wagner M., Müller M., Khan F., Eberl H. C., Mensinga A., Brinkman A. B., Lephikov K., Müller U., Walter J., Boelens R., van Ingen H., Leonhardt H., Carell T., and Vermeulen M. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 [DOI] [PubMed] [Google Scholar]
- 21. Zhou T., Xiong J., Wang M., Yang N., Wong J., Zhu B., and Xu R. M. (2014) Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2. Mol. Cell 54, 879–886 [DOI] [PubMed] [Google Scholar]
- 22. Bashtrykov P., Jankevicius G., Jurkowska R. Z., Ragozin S., and Jeltsch A. (2014) The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. J. Biol. Chem. 289, 4106–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berkyurek A. C., Suetake I., Arita K., Takeshita K., Nakagawa A., Shirakawa M., and Tajima S. (2014) The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. J. Biol. Chem. 289, 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu L. Y., Kuang H., Korakavi G., and Yu X. (2015) Topoisomerase II regulates the maintenance of DNA methylation. J. Biol. Chem. 290, 851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F., Koseki H., and Nakanishi M. (2013) Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502, 249–253 [DOI] [PubMed] [Google Scholar]
- 26. Qin W., Wolf P., Liu N., Link S., Smets M., La Mastra F., Forné I., Pichler G., Hörl D., Fellinger K., Spada F., Bonapace I. M., Imhof A., Harz H., and Leonhardt H. (2015) DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 25, 911–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bostick M., Kim J. K., Estève P. O., Clark A., Pradhan S., and Jacobsen S. E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 28. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., and Koseki H. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 29. Pichler G., Wolf P., Schmidt C. S., Meilinger D., Schneider K., Frauer C., Fellinger K., Rottach A., and Leonhardt H. (2011) Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J. Cell Biochem. 112, 2585–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J., Gao Q., Li P., Liu X., Jia Y., Wu W., Li J., Dong S., Koseki H., and Wong J. (2011) S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 21, 1723–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grosser C., Wagner N., Grothaus K., and Horsthemke B. (2015) Altering TET dioxygenase levels within physiological range affects DNA methylation dynamics of HEK293 cells. Epigenetics 10, 819–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quinlan A. R., and Hall I. M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z. M., Rothbart S. B., Allison D. F., Cai Q., Harrison J. S., Li L., Wang Y., Strahl B. D., Wang G. G., and Song J. (2015) An allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1. Cell Reports 12, 1400–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]