Abstract
Inflammatory breast cancer (IBC) is an extremely lethal cancer that rapidly metastasizes. Although the molecular attributes of IBC have been described, little is known about the underlying metabolic features of the disease. Using a variety of metabolic assays, including 13C tracer experiments, we found that SUM149 cells, the primary in vitro model of IBC, exhibit metabolic abnormalities that distinguish them from other breast cancer cells, including elevated levels of N-acetylaspartate, a metabolite primarily associated with neuronal disorders and gliomas. Here we provide the first evidence of N-acetylaspartate in breast cancer. We also report that the oncogene RhoC, a driver of metastatic potential, modulates glutamine and N-acetylaspartate metabolism in IBC cells in vitro, revealing a novel role for RhoC as a regulator of tumor cell metabolism that extends beyond its well known role in cytoskeletal rearrangement.
Keywords: breast cancer, glutamine, hypoxia-inducible factor (HIF), Rho (Rho GTPase), tumor metabolism, N-acetylaspartate
Introduction
Inflammatory breast cancer (IBC)3 is a highly aggressive cancer that affords the lowest 5-year survival rate of any breast cancer diagnosis, with a large proportion of patients exhibiting nodal metastases at diagnosis (1). IBC disproportionally affects both younger women and African American women and is typically characterized by rapid and specific changes in skin appearance, such as peau d'orange, erythema, skin thickening, and rapid enlargement of the affected breast (2). Instead of forming solid masses, IBC cells tend to form abundant tumor emboli that have the capacity to rapidly invade the lymphatic system and spread to distant sites. Although the molecular attributes of IBC are well described and are suggestive of constitutive adaptation to hypoxia (3), little is known about the underlying metabolic features of the disease.
Metabolic reprogramming is increasingly recognized as a fundamental hallmark of cancer, and efforts to identify clinically viable drugs that target cancer metabolism are underway (4). It is well established that many tumor cells consume glucose and produce lactate at significantly higher rates than the surrounding tissue, even when sufficient oxygen is present, via a phenomenon known as aerobic glycolysis or the Warburg effect (5). It was originally hypothesized that cancer cells utilize aerobic glycolysis because of mitochondrial respiratory dysfunction. However, even though a number of mitochondrial defects in tumor cells have been reported, evidence suggests that most tumor cells have functional tricarboxylic acid (TCA) cycles and electron transport chains (6, 7).
In addition to increased glucose uptake, many cancer cells also consume glutamine at higher rates than normal cells, both in culture and in vivo (8, 9). The Rho GTPase family of proteins has recently been linked to the regulation of glutaminase activity (10). The oncogene RhoC, a GTPase involved in cytoskeletal rearrangement and cell motility, is a driver of the metastatic phenotype exhibited by IBC (11, 12). RhoC expression increases with tumor stage, is a robust predictor of breast cancer aggressiveness and survival (13), and has been demonstrated to partially recapitulate the IBC phenotype (12). In addition to a role in breast cancer metastasis, RhoC increases the metastatic potential in lung, melanoma, pancreatic, and bladder cancers (14–17).To better understand the metabolic alterations associated with IBC, we probed the metabolic phenotypes of a set of breast cancer cell lines, including IBC-derived SUM149 cells, which is the primary in vitro model for the study of IBC. Our results indicate that SUM149 cells exhibit a number of metabolic abnormalities that distinguish them from other breast cancer cells, including a near-complete conversion of glucose to lactate, low mitochondrial respiratory capacity, and a large reductive carboxylation flux from glutamine-derived α-ketoglutarate (AKG) to citrate under normal culture conditions. We also discovered that RhoC is a potent regulator of both glutamine and N-acetylaspartate (NAA) metabolism in SUM149 cells. Although NAA has long been considered a brain-specific metabolite (18), it was recently linked to ovarian cancer (19–21). Here we provide the first evidence of elevated NAA in breast cancer cells and demonstrate that RhoC modulates NAA levels in IBC cells in vitro. Taken together, these findings suggest a number of potential metabolic and molecular vulnerabilities in IBC cells that warrant additional exploration as therapeutic targets.
Experimental Procedures
Cell Culture and Reagents
MCF10A cells were maintained in 50:50 DMEM:F12 medium supplemented with 5% horse serum, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 0.02 μg/ml epidermal growth factor, and 0.1 μg/ml cholera toxin. MCF7 cells were maintained in DMEM, and MDA231 cells were maintained in RPMI1640, both supplemented with 10% FBS. SUM149 and SUM149 shRNA cells were maintained in F12 medium supplemented with 5% FBS, 5 μg/ml insulin, and 1 μg/ml hydrocortisone. Cells were grown in 5% CO2 except the SUM149 WT and SUM149 shRNA cell lines, which were grown in 10% CO2. For the 13C isotope, extracellular flux, and proliferation assay experiments, all cells were grown in base DMEM (Sigma-Aldrich, D5030) lacking glutamine, glucose, sodium bicarbonate, and pyruvate, which was supplemented with the cell line-specific supplements described above and 44 mm sodium bicarbonate as well as glutamine, glucose, and sodium pyruvate as indicated. RhoC (3430), Cdc42 (2466), and citrate synthase (14309) antibodies were purchased from Cell Signaling Technology. RhoA (sc-418), pyruvate carboxylase (sc-67021), GLS1 (sc-100533), GLS2 (sc-168424), and Actin (sc-1616) antibodies were purchased from Santa Cruz Biotechnology, and HIF-1α (610958) antibody was purchased from BD Biosciences. The GS (Invitrogen, PA5–29737) antibody was a gift from Robert W. O'Rourke (University of Michigan). Cobalt(II) chloride hexahydrate (255599) was purchased from Sigma-Aldrich.
Extracellular Flux Measurements
Glucose, lactate, and glutamine concentrations were measured in media using a YSI 2950D biochemistry analyzer. The difference in metabolite concentrations in conditioned cell culture media from concentrations in incubated control medium not exposed to cells (but otherwise treated the same) were normalized to total protein. Oxygen consumption rates (OCRs) were measured using a Seahorse XF24 flux analyzer and XF Cell Mito stress test kits (Seahorse Bioscience) according to the protocol of the manufacturer. Cells were seeded on 24-well assay plates at a density of 50,000 cells/well, except for MDA231 cells, which were seeded at 80,000 cells/well, and allowed to adhere overnight. Growth medium was replaced with unbuffered, serum-free DMEM supplemented with 11 mm glucose, 2.5 mm glutamine, and 10 mm sodium pyruvate.
shRNA Stable Cell Lines
Stable shRNA cell lines were generated via lentiviral transduction using pGIPZ shRNA clones (GE Healthcare). Following infection, cells were sorted for GFP expression and grown as a population of transduced cells. SUM149 shRhoC pSM2c has been described previously (22). Both SUM149 shRhoC pGIPZ and pSM2c contain the same shRNA sequence. SUM149 shRhoC pSM2c was used for isotope labeling experiments and extracellular flux experiments. SUM149 shRhoC pGIPZ was used for extracellular flux measurements and RT-qPCR.
13C Labeling Experiments
Cells were grown to ∼60–70% confluence in 6-cm dishes in unlabeled growth media. Prior to the start of the experiment, cells were incubated for 1 h in fresh unlabeled DMEM (except where indicated). Cells were changed to fresh DMEM containing 13C-labeled substrate(s). An equivalent number of additional plates of each cell line were grown in parallel in chemically identical media for total protein analysis. After 5 h (unless otherwise indicated), cells were rapidly washed with double-distilled H2O, quenched with liquid nitrogen, and stored at −80 °C for subsequent LC-MS and GC-MS analysis. Samples were extracted using an organic solvent/water mixture and analyzed by LC-MS as described previously (23). A second portion of the sample extracts was also used for GC-MS analysis as described previously (24). Reported metabolites were measured by LC-MS, except pyruvate, glutamine, glutamate, and AKG, which were measured by GC-MS. Reported citrate data represent the sum of citrate and isocitrate pools. M0, M1, M2, M3, M4, M5, and M6 reported from LC-MS and GC-MS analysis indicate the presence of 0–6 13C-enriched atoms. For total metabolite pool sizes, the average of the total peak intensities was normalized by total protein. All data were corrected for natural abundance of stable isotopes (25, 26). For steady-state labeling experiments, 5 h was selected as the treatment time after verification that an isotopic steady state was achieved between 4 and 6 h in the cell lines used in this study.
RT-qPCR
RT-qPCR was carried out essentially as described previously (27). For preparation of cDNA, total RNA was isolated from the indicated cells using the RNeasy mini kit (Qiagen) following the instructions of the manufacturer. Reverse transcriptase reactions were carried out using the reverse transcription system (Promega) following the protocol of the manufacturer. Quantitative PCR analysis employed the following primers (5′ → 3′): ASPA, CCAAGATATCAGCTCTCAGAACC (forward) and TCCCTCAAATATGCGACCAC (reverse); RIMKLA, CTGATGGACGGATGCAGAG (forward) and AGGAGATCAATGCCACAGAAG (reverse); RIMKLB, GAGGCTGAAGTTCTGGAGTTC and TCATGGCGAATAAGATGGCT (reverse); NAT8L, CGAGCAGTACTACATGAAGCC (forward) and AACGTGAGTCCACAGACATC (reverse); RPL22, TGCTCTTGCTCCTTTCGATG (forward) and GAAGATGGAATCATGGATGCTG (reverse); and RPL30, CACCAGTTTTAGCCAACATAGC (forward) and GATCAGACAAGGCAAAGCGA (reverse). qPCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen) and a Step-One Plus real-time PCR system (Applied Biosystems). All qPCR reactions were performed in triplicate. Ct values were normalized to RPL22 and RPL30. Analysis of three biological replicate experiments was performed using REST 2009 software (Qiagen).
Proliferation Assay
Assays that measured the incorporation of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma) were carried out as described previously (12). Prior to the start of the assay, SUM149 WT and shRhoC cells were plated at 6000 and 5000 cells/well, respectively, and grown in normal growth medium for 48 h. At day 0, cells were changed to DMEM containing either 20 mm glucose, 4 mm glutamine, or both. Medium was replaced at day 3 to replenish glucose and/or glutamine. Three independent experiments consisting of five technical replicates at each time point were performed.
Imaging
Cells were plated on coverslips and mounted in a flow chamber (Warner Instruments) on an Olympus IX71 inverted microscope equipped with a ×60/1.2 numerical aperture water immersion objective. The chamber was perfused at 0.3 ml/min, and the temperature was maintained at 33 °C using inline solution and chamber heaters (Warner Instruments). Excitation was provided by a TILL Polychrome V monochromator set to 10% output. Pyruvate kinase M2 activity reporter (PKAR) mCerulean-mCitrine FRET biosensors (28) were excited by light filtered with an ET430/24× bandpass filter and reflected via an ET-CFP/YFP/mCherry dichroic (Chroma). Fluorescence emission was split with a 495LP dichroic mounted in a DV2 beamsplitter (Photometrics) and reported as the ratio of emission through ET535/30m and ET470/24m bandpass filters (R535/470). A 458LP dichroic (Semrock Inc.) was used to measure NADH and NADPH, collectively referred to as NAD(P)H, excited by 365 nm. Emission through an ET470/24m filter (Chroma) was collected with a Photometrics QuantEM:512SC camera at 0.2 Hz.
Statistics
Statistical analysis was performed using GraphPad Prism software (San Diego, CA). When total protein normalization was performed, reported mean ± S.E. values also include propagated uncertainty from total protein measurements.
Results
SUM149 Cells Operate Near Maximal Glycolytic and Mitochondrial Capacity
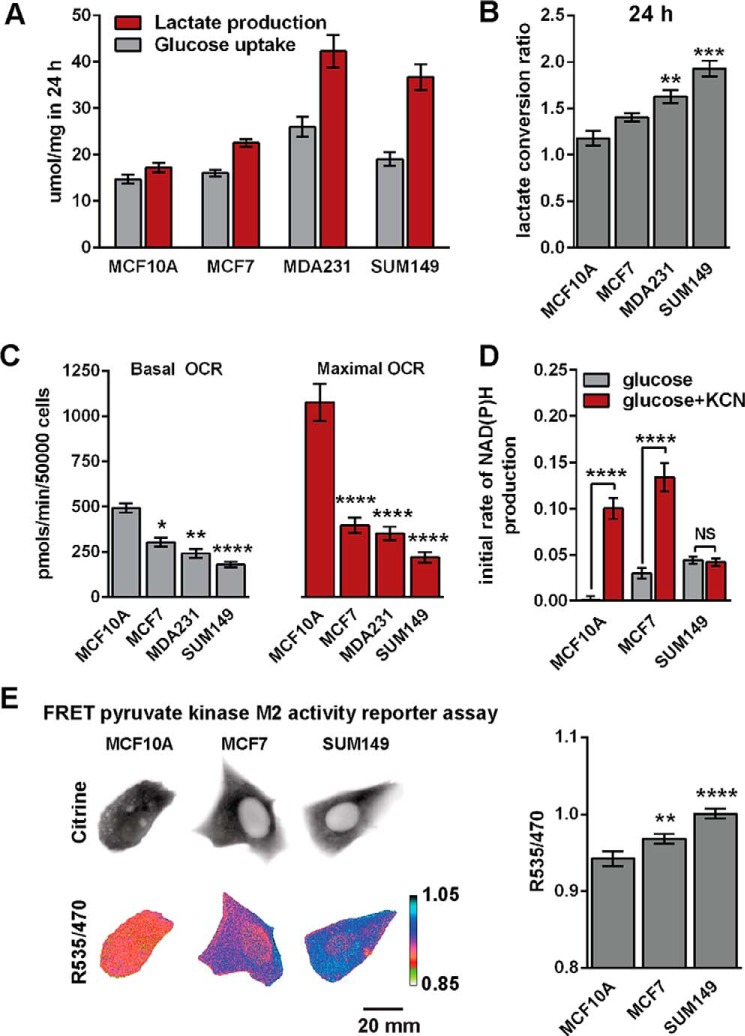
Glucose uptake and lactate production were measured in MCF10A (normal-like mammary epithelial), MCF7 (estrogen and progesterone receptor-positive breast cancer), MDA-MB-231 (MDA231, metastatic triple-negative breast cancer), and SUM149 (metastatic triple-negative IBC) cells by quantifying changes in glucose and lactate concentrations in cell culture media after a 24-h incubation period (Fig. 1A).
FIGURE 1.
SUM149 cells are highly glycolytic. A, extracellular glucose consumption and lactate production rates were measured after 24 h (n = 3). B, the lactate conversion ratio was calculated by dividing lactate production by glucose consumption in A. C, basal and maximal OCRs were measured via mitochondrial stress tests. Data are from three to six independent experiments. D, NAD(P)H fluorescence was measured in response to 20 mm glucose and 20 mm glucose +5 mm KCN, which inhibits cytochrome c oxidase. Cells were deprived of glucose for 30 min prior to recording. Data represent the average of eight to nine fields for each cell line with >100 cells/field. The initial rate of NAD(P)H production is plotted for the first 3 min of glucose and glucose + KCN application. E, cytosolic pyruvate kinase M2 activity was measured with a FRET reporter (PKAR). Representative images of cells transfected with the FRET PKAR are presented for three cell lines. The higher the FRET reading, the more likely PKM2 is in the highly active tetrameric state. Cytosolic PKM2 activity was quantified based on the FRET efficiency (R535/470) of PKAR. Values are the mean ± S.E. Statistical differences within groups were confirmed by ANOVA, and pairwise differences were assessed via Tukey's (B and E) or Sidak's (C and D) multiple comparison tests. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; NS, not significant. Reported pairwise differences are compared with MCF10A, except in D, where differences between glucose and glucose + KCN are reported.
The very aggressive MDA231 and SUM149 cells produced the most lactate over this time period. The 24-h lactate conversion ratio (defined as lactate production rate divided by glucose consumption rate) for SUM149 cells was 1.98 ± 0.08 (n = 3) (Fig. 1B), which is nearly identical to the stoichiometric ratio of 2.0 expected when all glucose taken up by a cell is converted to lactate. These data suggest that very little glucose consumed by SUM149 cells undergoes mitochondrial oxidization. We next probed mitochondrial respiratory capacity using a Seahorse XF24 extracellular flux analyzer. Although SUM149 cells exhibited the lowest basal and lowest maximal oxygen OCRs among the tested cell lines (Fig. 1C), these cells also used most of their mitochondrial respiratory capacity. Basal OCR as a proportion of maximal OCR was 81.9% ± 12.3% in SUM149 cells.
To further probe glycolytic and metabolic capacity, we measured NADH and NADPH, collectively referred to as NAD(P)H, which are spectrally inseparable metabolites that are strongly fluorescent in their reduced forms (29). NAD(P)H fluorescence provides a readout of the reduction of NAD and NADP generated by glycolysis, the oxidative branch of the pentose phosphate pathway, and reactions associated with the TCA cycle in the mitochondria. Potassium cyanide (KCN) treatment rapidly blocks mitochondrial oxygen consumption by strongly inhibiting cytochrome c oxidase/complex IV. Glucose-deprived cells were stimulated with 20 mm glucose followed by addition of 5 mm KCN after 12 min. Compared with MCF10A, the NAD(P)H fluorescence in MCF7 and SUM149 increased at a significantly higher rate immediately following stimulation with 20 mm glucose. However, SUM149 cells were least affected immediately following the inhibition of mitochondrial respiration via KCN (Fig. 1D). These data indicate that SUM149 cells rely heavily on non-mitochondrial NAD(P)H production compared with MCF10A or MCF7 cells.
SUM149 Cells Have High Pyruvate Kinase M2 Activity
The end products of glycolysis are NADH, ATP, and pyruvate. Pyruvate can be further oxidized in mitochondria or converted to lactate in the cytosol. In either case, glucose-derived carbon is first shunted through a rate-limiting glycolytic reaction catalyzed by pyruvate kinase (PK) that converts phosphoenolpyruvate to pyruvate. Studies show that decreased activity of the M2 isoform of PK (PKM2) promotes cellular proliferation during tumorigenesis (30–32). The activity of PKM2 is linked to its quaternary structure. The tetrameric forms of all PK isoforms have high catalytic activity, whereas the monomeric and dimeric forms of the M2 isoform have low catalytic activity (33, 34). PKAR biosensors provide a FRET-based readout of the PKM2 tetramer-to-dimer (or monomer) ratio in living cells (28). PKAR FRET directly correlated with the extent of lactate production observed in the MCF10A, MCF7, and SUM149 cell lines (Fig. 1E). The highest PKAR FRET was observed in SUM149 cells, indicating that these cells exhibited the most PKM2 oligomerization and, therefore, were inferred to have the highest PKM2 activity. This result is consistent with the high lactate conversion ratio observed in SUM149 cells.
Glucose Is Not A Predominant Source of Acetyl-CoA in SUM149 Cells
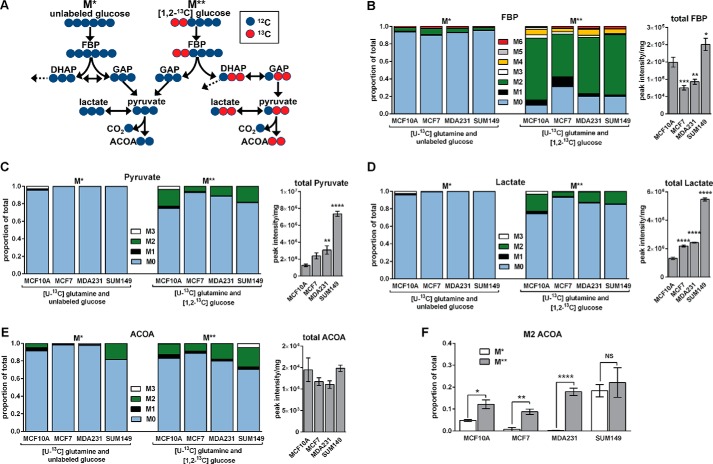
To directly probe steady-state intracellular metabolic fluxes, cells were cultured in one of two forms of 13C-labeled growth media identified as either M* or M** to indicate the presence of one or two labeled substrates, respectively, for 5 h. Medium formulations contained 2.5 mm [U-13C]glutamine and either 10 mm unlabeled glucose (M*) or 10 mm [1,2-13C]glucose (M**). Under the M** condition, 13C enrichment is expected in the M2 isotopomer of fructose 1,6-bisphosphate (FBP) and dihydroxyacetone phosphate (DHAP) because of the presence of [1,2-13C]glucose. The cleavage of FBP (labeled on the first two carbons) into DHAP and glyceraldehyde phosphate (GAP) will result in labeling of the second and third carbon positions of DHAP and no labeling in GAP. If DHAP isomerizes to GAP, then the second and third carbons of GAP become labeled. M2 GAP can then give rise to M2 pyruvate, lactate, and acetyl-CoA via downstream metabolic reactions (Fig. 2A).
FIGURE 2.
Glucose is not a predominant of source of acetyl-CoA in SUM149 cells. Cells were cultured for 5 h in medium containing 2.5 mm [U-13C]glutamine and 10 mm unlabeled glucose (M*) or 2.5 mm [U-13C]glutamine and 10 mm [1,2-13C] glucose (M**). A, glucose-derived M0 acetyl-CoA (ACOA) is possible under either the M* or M** conditions. Under the M** condition, glucose-derived M2 ACOA can be formed when M2 DHAP isomerizes to M2 GAP. Dashed arrows leading from DHAP indicate potential DHAP flux toward triglyceride synthesis. Red circles indicate 13C, and blue circles indicate 12C. B–E, the mass isotopomer distributions of FBP (B), pyruvate (C), lactate (D), and ACOA (E) after M* and M** labeling are presented along with total metabolite pool sizes. For total pool data, the four M* and four M** replicate peak intensities were averaged prior to normalization by total protein. Differences in total pool sizes were assessed by one-way ANOVA and Sidak's multiple comparison tests versus MCF10A. F, M2 ACOA fractional abundances are compared for each cell line. Values are mean ± S.E. (n = 4, except total pool data, which are n = 8). Pairwise differences between M* and M** conditions in M2 ACOA were assessed by a two-tailed Student's t test and the Holm-Sidak method for multiple comparison correction. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; NS, not significant.
Under the in M* condition, all four cell lines exhibited a small proportion of 13C enrichment in FBP and little to no 13C enrichment in either lactate or pyruvate. Under the M** condition, all four cell lines exhibited non-negligible levels of 13C enrichment in FBP, lactate, and pyruvate (Fig. 2, B–D). The difference between the M2 fraction of acetyl-CoA under the M* and M** conditions was statistically significant in MCF10A, MCF7, and MDA231 cells. In contrast, there was no statistical difference between the M2 fractions in SUM149 cells. These data indicate that the majority of M2 acetyl-CoA was derived from glutamine in SUM149 cells (Fig. 2, E and F). Moreover, the total pool sizes of FBP, pyruvate, and lactate were higher in SUM149 compared with the other cell lines, providing further evidence that SUM149 cells are highly glycolytic relative to the other two breast cancer cell lines studied.
Given the high rate of glycolysis, low mitochondrial respiratory capacity, and large glutamine to acetyl-CoA flux of SUM149 cells, we wondered whether there was a defect in mitochondrial pyruvate transport. We therefore performed a separate labeling experiment where cells were exposed to medium containing 10 mm [2,3-13C]pyruvate with unlabeled glucose and glutamine. After 1 h, M2 lactate and M2 acetyl-CoA represented 74.3% ± 2.4% and 40.0% ± 3.4% of the total lactate and acetyl-CoA pools, respectively, in SUM149 cells (Fig. 3, A and B), providing evidence that mitochondrial acetyl-CoA production from pyruvate is functional in SUM149 cells. However, the SUM149 citrate pool was completely unlabeled and very small (Fig. 3C) compared with the other cell lines during this time period. These data indicate that, despite lack of evidence of impaired mitochondrial pyruvate transport in SUM149 cells, pyruvate is not a primary source of citrate production in these cells.
FIGURE 3.
Conversion of pyruvate to acetyl-CoA is functional in SUM149 cells. Cells were cultured for 15 or 60 min in unbuffered DMEM containing 10 mm [2,3-13C]pyruvate, 11 mm unlabeled glucose, and 2.5 mm unlabeled glutamine. The mass isotopomer distributions and total pool sizes of lactate (A), ACOA (B), and citrate (C) citrate are presented. Values are mean ± S.E. (n = 4).
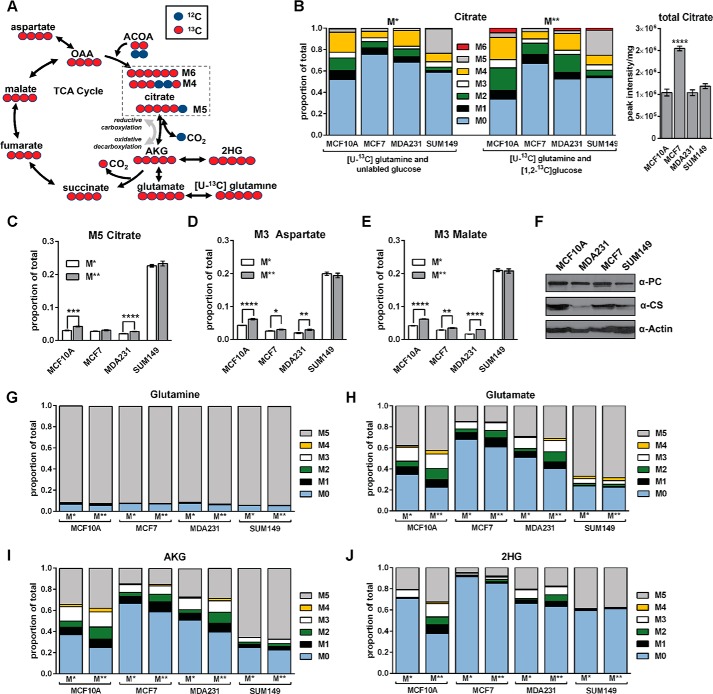
SUM149 Cells Reductively Carboxylate Glutamine-derived α-Ketoglutarate
M4 and M6 citrate are expected to form from the condensation of M4 oxaloacetate (OAA) with M0 or M2 acetyl-CoA, respectively, in the presence of [U-13C]glutamine (Fig. 4A). M5 citrate can form from the reductive carboxylation of M5 AKG with unlabeled CO2 (35, 36). In SUM149 cells, M5 citrate represented 22.6% and 23.3% of the total citrate pool under the M* and M** labeling conditions, respectively. MCF10A produced the next highest M5 citrate abundance (4.2% under the M** condition) after SUM149 (Fig. 4, B and C).
FIGURE 4.
SUM149 cells reductively carboxylate glutamine derived α-ketoglutarate. Cells were cultured for 5 h in medium containing 2.5 mm [U-13C]glutamine and 10 mm unlabeled glucose (M*) or 2.5 mm [U-13C]glutamine and 10 mm [1,2-13C]glucose (M**). A, expected citrate labeling in the TCA cycle. M4 and M6 citrate are formed (clockwise) from M4 OAA and M0 or M2 ACOA, respectively. M5 citrate is formed by reductive carboxylation (counterclockwise) of M5 AKG. Red circles indicate 13C, and blue circles indicate 12C. B, the MIDs of M* and M** citrate and the total citrate pool sizes. For total pool data, the 4 M* and 4 M** replicate peak intensities were averaged prior to normalization by total protein. Differences in total pool sizes were assessed by one-way ANOVA and Sidak's multiple comparison tests versus MCF10A. C–E, the proportion of M5 citrate (C), M3 aspartate (D), and M3 malate (E) under the M* and M** labeling conditions are presented. Pairwise differences between M* and M** conditions in M5 citrate, M3 aspartate, and M3 malate were assessed by a two-tailed Student's t test and the Holm-Sidak method for multiple comparison correction. F, representative Western blots of pyruvate carboxylase (PC) and citrate synthase (CS). G–J, comparison of 13C enrichment in the MIDs of glutamine, glutamate, AKG, and 2-hydroxyglutarate (2HG) indicates that 13C enrichment patterns are nearly identical under the M* and M** conditions in SUM149 cells. Values are mean ± S.E. (n = 4, except total pool data, which are n = 8). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Another possible source of M5 citrate in the presence of [U-13C]glutamine may be mitochondrial condensation of M2 acetyl-CoA and M3 OAA. In this scenario, M3 pyruvate would form from the decarboxylation of M4 malate by malic enzyme. M3 pyruvate would then produce M2 acetyl-CoA and M3 OAA via reactions catalyzed by pyruvate dehydrogenase and pyruvate carboxylase, respectively (35). M3 pyruvate was not observed in SUM149 cells under either the M* or M** growth conditions (Fig. 2C), however, providing strong evidence that SUM149 cells reductively carboxylate glutamine-derived AKG.
Citrate exported from the mitochondria can be cleaved into acetyl-CoA and OAA via cytosolic ATP citrate lyase. Cytosolic acetyl-CoA can then be used to fuel fatty acid synthesis. In SUM149 cells, M5 citrate (Fig. 4C), M3 aspartate (Fig. 4D), and M3 malate (Fig. 4E) were very high relative to the other cell lines under both labeling conditions. The cleavage of glutamine-derived M5 citrate by ATP citrate lyase will produce cytosolic M2 acetyl-CoA and M3 OAA (36). M3 OAA can then be converted to M3 aspartate or M3 malate. In SUM149 cells, the high proportion of M3 malate and M3 aspartate correlated with the high proportion of M5 citrate observed. Together, these data strongly suggest that SUM149 cells rely on reductive carboxylation of glutamine-derived AKG to produce cytosolic acetyl-CoA.
At the molecular level, the lack of 13C enrichment observed in citrate in the presence of [2,3-13C]pyruvate (Fig. 3C) or [1,2-13C]glucose (Fig. 4B) in SUM149 cannot be explained by reduced levels of the mitochondrial enzymes citrate synthase or pyruvate carboxylase. Measured levels of pyruvate carboxylase (which catalyzes the carboxylation of pyruvate to form OAA) were similar across MDA231, MCF7, and SUM149 cells (Fig. 4F). Although levels of citrate synthase (which catalyzes the condensation of acetyl-coA and OAA to form citrate) did appear to be reduced in SUM149 cells compared with MCF10A cells, they were comparable with levels in MDA231 cells, which demonstrated robust flux into the TCA cycle (Figs. 3C and 4B). Thus, citrate synthase levels alone cannot explain the altered TCA cycle phenotype observed in the SUM149 cell line.
The extent of labeling in glutamine (Fig. 4G), which represented ∼95% of the total pool, was uniform across the four cell lines under both the M* and M** labeling conditions. However, differences in the mass isotopomer distributions (MIDs) of glutamate (Fig. 4H), AKG (Fig. 4I), and 2-hydroxyglutarate (Fig. 4J) produced under the M* and M** conditions were observed in MCF10A, MCF7, and MDA231 cells. In contrast, very little difference in the MIDs of these metabolites was observed in SUM149 cells when the two labeling conditions were compared. Thus, the addition of labeled glucose had very little impact on the 13C enrichment of glutamate, AKG, 2-hydroxyglutarate, and citrate in SUM149 cells, which strongly suggests that glutamine is the primary carbon source used in the production of AKG and citrate in these cells.
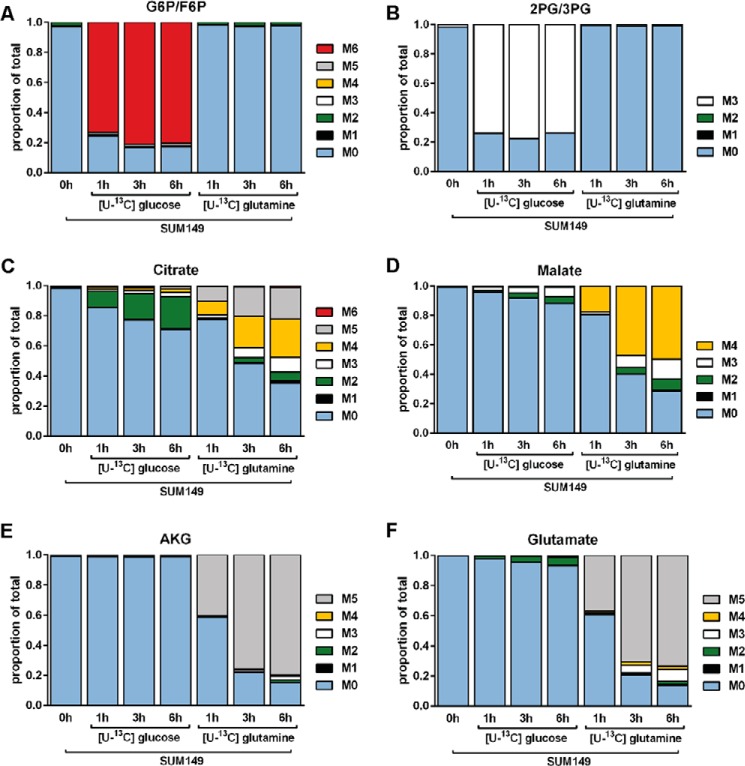
Additionally, SUM149 cells were grown for 72 h in unlabeled DMEM with 8 mm glucose and 2 mm glutamine. Immediately following a PBS wash, cells were changed into fresh DMEM containing either 10 mm [U-13C]glucose and 2.5 mm unlabeled glutamine or 10 mm unlabeled glucose and 2.5 mm [U-13C]glutamine. Cells were harvested after 1, 3, or 6 h. SUM149 cells exposed to [U-13C]glucose exhibited elevated labeling in glycolytic metabolites (Fig. 5, A and B) and little to no labeling in TCA cycle metabolites (Fig. 5, C–F). Conversely, TCA cycle metabolites were highly labeled in cells exposed to [U-13C]glutamine (Fig. 5, C–F), providing further evidence that glutamine is the primary carbon source for TCA cycle metabolism in SUM149 cells.
FIGURE 5.
Glucose is not a significant contributor to TCA cycle flux in SUM149 cells. Cells were cultured in medium containing either 10 mm [U-13C]glucose and 2.5 mm unlabeled glutamine or 10 mm unlabeled glucose and 2.5 mm [U-13C]glutamine for 1, 3, or 6 h as indicated. The mass isotopomer distributions of the combined glucose 6-phosphate and fructose 6-phospate pools (G6P/F6P, A), 2-phosphoglyceric acid/3-phosphoglyceric acid (2PG/3PG, B), citrate (C), malate (D), AKG (E), and glutamate (F) are presented.
Disruption of HIF-1α Increases Oxygen Consumption in SUM149 Cells
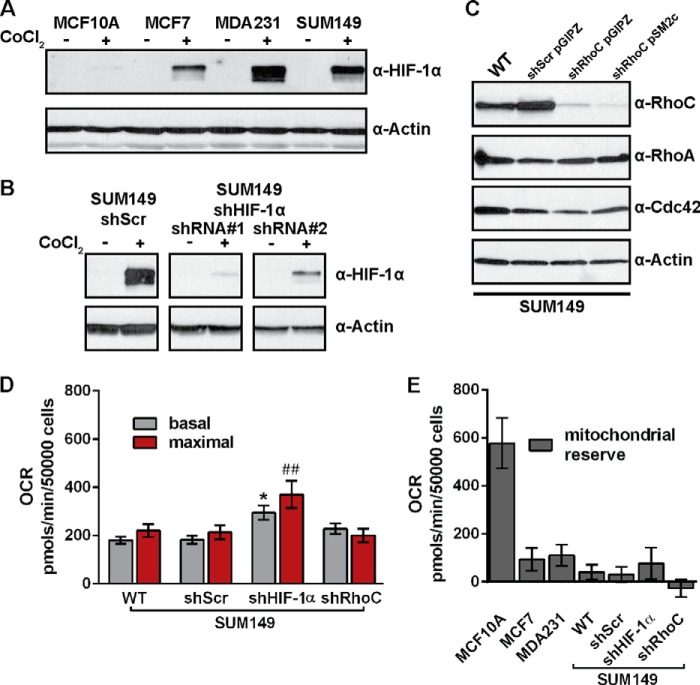
Molecular evidence suggests that IBC cells are highly adapted to hypoxia at the molecular level partly because of VEGF overexpression and elevated hypoxia-inducible factor 1α (HIF-1α) levels (3). Under hypoxic conditions, levels of HIF-1α protein increase rapidly, triggering the up-regulation of a variety of hypoxia-associated genes, including several glycolytic enzymes (37). Knockdown of HIF-1α in SUM149 cells was recently shown to significantly decrease tumor size in mice, demonstrating a potential role for HIF-1α in SUM149 cell proliferation (38). We therefore hypothesized that the metabolic phenotype observed in SUM149 under normal culture conditions may be regulated by HIF-1α stabilization. In contrast to the three cancer cell lines, the non-tumorigenic MCF10A cell line did not exhibit detectable levels of HIF-1α upon treatment with CoCl2, which was used to prevent the proteolytic degradation of HIF-1α (37) (Fig. 6A). Because RhoC is a critical determinant of metastatic potential in breast cancer and is overexpressed in virtually all IBCs (11), we also hypothesized that RhoC regulates the metabolic phenotype observed in SUM149 cells. Thus, stable shRNA cell lines with depletion of HIF-1α (Fig. 6B) or RhoC (Fig. 6C) were produced to test these hypotheses.
FIGURE 6.
Establishment of stable SUM149 shHIF-1α and shRhoC cell lines. A, representative Western blot of endogenous levels of HIF-1α in the MCF10A, MCF7, MDA231, and SUM149 cell lines. Cells were treated with 100 μm CoCl2 for 4 h before protein harvest where indicated. B, representative HIF-1α Western blot of control shRNA (shScr) and HIF-1α shRNA knockdown cell lines. Cells were treated with 100 μm CoCl2 for 4 h before protein harvest where indicated. C, RhoC protein knockdown cell lines were generated using the same shRNA sequence with pGIPZ-derived lentivirus or stable pSM2c vector integration. Representative Western blots against the Rho GTPases RhoC, RhoA, and Cdc42 are shown to demonstrate the specificity of RhoC knockdown in SUM149 WT, control (shScr pGIPZ), and RhoC knockdown stable cell lines (shRhoC pGIPZ and shRhoC pSM2c). D, basal and maximal OCRs were measured via mitochondrial stress tests. E, the mitochondrial reserve respiratory capacity is the difference between basal and maximal OCR. Values are mean ± S.E. Data are from three to six independent experiments. Statistical differences within groups were confirmed by ANOVA and Sidak's multiple comparison tests versus SUM149 WT basal (*, p < 0.05) or maximal (##, p < 0.01).
HIF-1α depletion resulted in higher basal and maximal OCR values compared with the WT cell line (Fig. 6D). In contrast, depletion of RhoC did not alter basal OCR values compared with the WT. However, the SUM149 shRhoC cell line failed to respond to carbonyl cyanide p-trifluoromethoxyphenylhydrazone treatment, an uncoupler of mitochondrial respiration, in any of five independent experiments or in a separate experiment with carbonyl cyanide p-trifluoromethoxyphenylhydrazone titration concentrations that ranged from 300 to 2.9 mm (data not shown). As a consequence, the calculated mitochondrial reserve capacity, which is the difference between basal and maximal OCR, was negative for SUM149 shRhoC cells (Fig. 6E).
RhoC Modulates Carbon Flux in SUM149 Cells
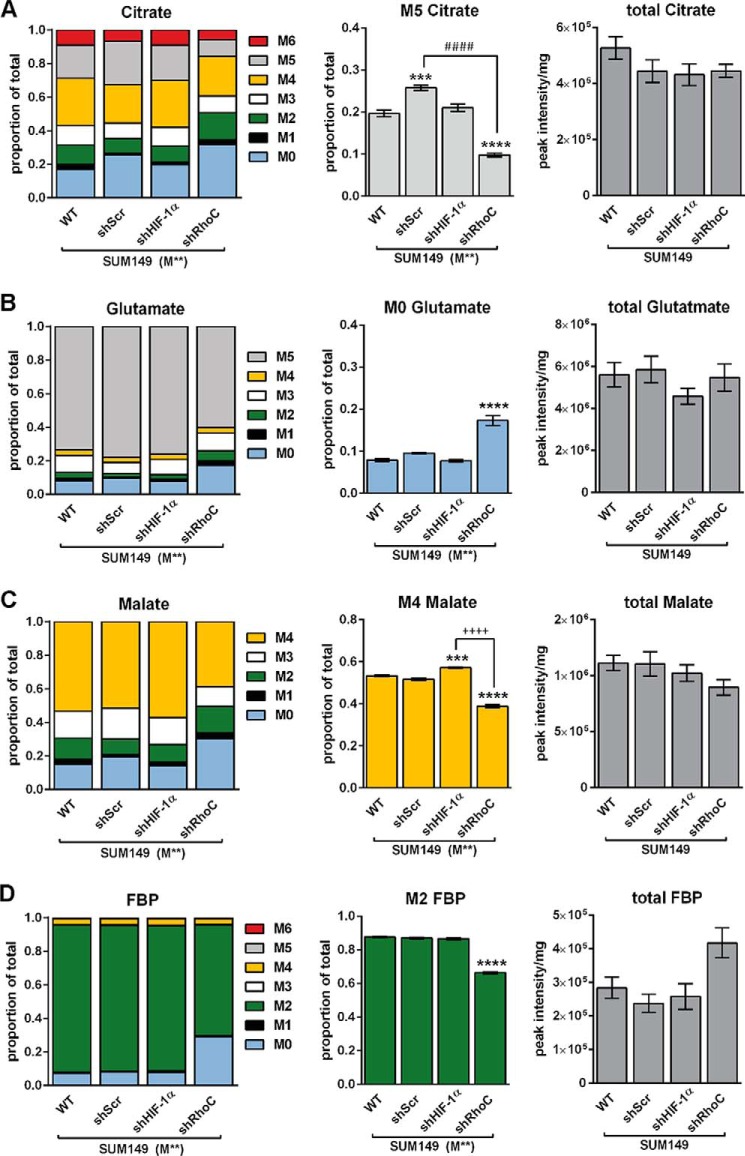
To probe the effects of depletion of HIF-1α or RhoC proteins on metabolic flux, we repeated the 5 h M** isotope labeling experiment with the stable shRNA cell lines. SUM149 shRhoC cells exhibited altered 13C enrichment compared with SUM149 WT in virtually every metabolite measured by LC-MS. In the case of citrate, the M5 isotopomer was significantly reduced in shRhoC cells (Fig. 7A), indicating that RhoC depletion modulated the reductive carboxylation of AKG. Moreover, SUM149 shRhoC cells contained approximately twice as much M0 glutamate as the SUM149 WT (Fig. 7B). Similar labeling alterations were observed in other metabolites, including malate (Fig. 7C) and FBP (Fig. 7D), indicating that RhoC depletion not only altered TCA cycle flux but also glycolytic flux. In contrast, shHIF-1α cells exhibited only small labeling deviations compared with SUM149 WT, suggesting that HIF-1α depletion had little effect on carbon flux.
FIGURE 7.
RhoC modulates metabolic flux in SUM149 cells. SUM149 WT, shScr, shHIF-1α, and shRhoC cells were cultured for 5 h in medium containing 2.5 mm [U-13C]glutamine and 10 mm [1,2 13C]glucose (M**). A–D, citrate MIDs and M5 citrate (A), glutamate MIDs and M0 glutamate (B), malate MIDs and M4 malate (C), and FBP MIDs and M2 FBP (D) are presented along with total metabolite pool sizes. Values are mean ± S.E. (n = 4). For total pool data, the 4 M** replicate peak intensities were averaged prior to normalization by total protein. Differences in total pool sizes were assessed by one-way ANOVA and Sidak's multiple comparison tests versus SUM149 WT. Differences within groups for M0, M2, M4, and M5 labeling were confirmed by ANOVA and Tukey's multiple comparison tests versus SUM149 WT (***, p < 0.001; ****, p < 0.0001), shScr (####, p < 0.0001), and shHIF-1α (++++, p < 0.0001).
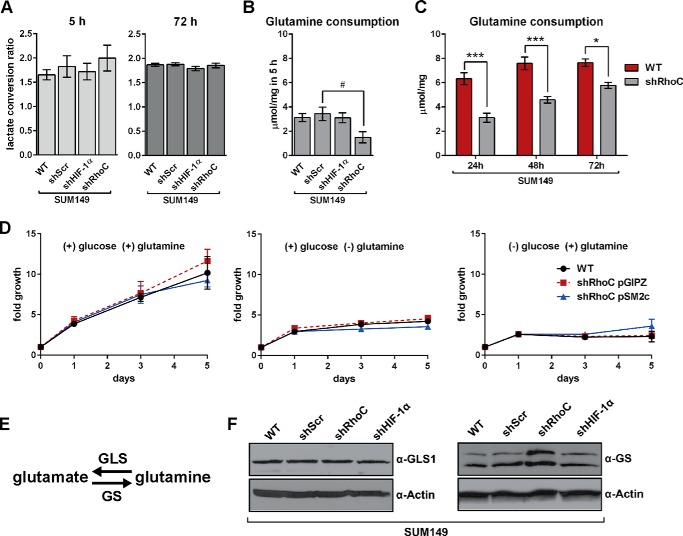
RhoC Modulates Glutamine Uptake in SUM149 Cells
The extracellular flux of glucose, lactate, and glutamine in SUM149 WT, shScr, shHIF-1α, and shRhoC cells was measured as described previously. No statistical differences in glucose uptake or lactate production were observed in the shRNA cell lines compared with the WT. Like SUM149 WT cells (Fig. 1B), shScr, shHIF-1α, and shRhoC cells produced lactate at approximately twice the rate as they consumed glucose (Fig. 8A). There was, however, a marked decrease in glutamine uptake in shRhoC cells (Fig. 8B). Although the difference between WT and shRhoC glutamine uptake after 5 h was not significant at α = 0.05 (p = 0.098), the difference between shScr and shRhoC was significant (p = 0.042). After 72 h, however, the glutamine uptake in the shRhoC cell line was significantly different from WT, shScr, and shHIF-1α cell lines (data not shown). In a separate experiment, where extracellular glutamine was measured at 24, 48, and 72 h (Fig. 8C), glutamine uptake was significantly reduced in the shRhoC cells compared with the WT at all three time points, indicating strong modulation of glutamine uptake in these cells after RhoC depletion. The large increase observed in the M0 glutamate pool in SUM149 shRhoC compared with WT cells (Fig. 7B) is likely a direct result of decreased uptake of labeled glutamine.
FIGURE 8.
RhoC modulates glutamine uptake in SUM149 cells. A, the lactate conversion ratio was calculated by dividing the lactate production by glucose consumption after 5 h (n = 4) and 72 h (n = 3). The extracellular glutamine consumption rate was measured after 5 h (n = 4, B) and after 24, 48, and 72 h (n = 3, C). Values are mean ± S.E. D, SUM149 and shRhoC cell lines were grown for 5 days in medium containing 20 mm glucose (center panel), 4 mm glutamine (right panel), or both (left panel). Cell proliferation was measured at 1, 3, and 5 days. Data are the average of three independent experiments. E, enzymes involved in the reversible conversion of glutamine to glutamate. F, representative Western blots of GLS1 and GS in the SUM149 WT and shRNA cell lines. Statistical differences within groups were confirmed by ANOVA and Tukey's multiple comparison tests (A and B) or Sidak's multiple comparison tests (C) versus SUM149 WT (*, p < 0.05; ***, p < 0.001) and shScr (#, p < 0.05).
To test for glutamine-dependent growth in SUM149 cells, we grew cells for 5 days with glucose and glutamine, with glucose alone, or with glutamine alone (Fig. 8D). We found that SUM149 WT cells required glucose and glutamine for growth, indicating that glutamine is necessary but not sufficient for proliferation in these cells. Depletion of RhoC did not alter this requirement, suggesting that the RhoC-depleted cells remain very reliant on glutamine despite a reduction in uptake flux.
We next probed the effect of RhoC knockdown on enzymes involved in the reversible conversion of glutamine to glutamate (Fig. 8E). RhoC knockdown resulted in a marked increase in glutamine synthetase (GS) levels, but it did not have an effect on glutaminase (GLS1, Fig. 8F). There was also no effect on glutaminase 2 (GLS2) levels because this enzyme was not detected in any SUM149 cells lines (data not shown).
RhoC Regulates N-Acetylaspartate Production in SUM149 Cells
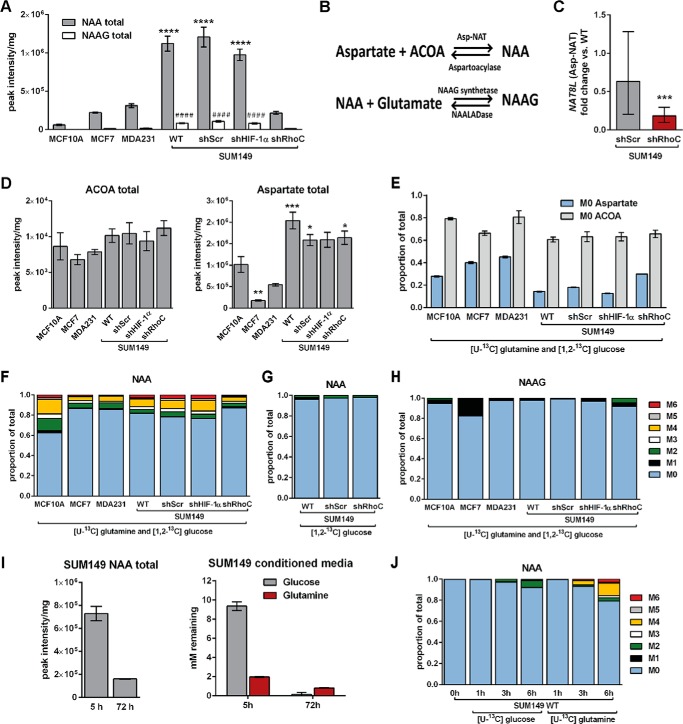
Using an untargeted metabolomics screen performed as described previously (40), NAA levels were found to be very high in SUM149 cells compared with MCF10A and MDA231 cells. Although NAA has long been considered a brain-specific metabolite (18), it was recently linked to ovarian cancer (19–21). NAA was subsequently quantitated via targeted LC-MS, which confirmed that NAA levels are very high in SUM149 WT relative to MCF10A, MCF7, and MDA231 cells. Like SUM149 WT cells, SUM149 shScr and shHIF-1α cells produced very high levels of NAA relative to MCF10A. In contrast, SUM149 shRhoC cells produced low levels of NAA that were not statistically different from MCF10A (Fig. 9A).
FIGURE 9.
RhoC modulates N-acetylaspartate production in SUM149 cells. Cells were cultured for 5 h in 10 mm [1,2-3C]glucose and 2.5 mm [U-13C]glutamine (M**) unless otherwise indicated. A, total pool sizes of NAA and NAAG. B, summary of reactions involved in the metabolism of NAA. C, expression of the Asp-NAT gene NAT8L was analyzed by RT-qPCR. Results were normalized first to two reference cDNAs and then to WT SUM149 cells. Statistical significance was assessed by REST. D and E, the total pool sizes of ACOA and aspartate (D) and the M0 fractions of ACOA and aspartate (E) are summarized. F–H, the MIDs of NAA under the M** condition (F), NAA in 10 mm [1,2-13C]glucose and 2.5 mm unlabeled glutamine for 5 h (G), and NAAG under the M** condition (H) are summarized. I, SUM149 cells were grown for 5 and 72 h in medium containing 10 mm glucose and 2 mm glutamine. The concentration of glucose and glutamine was measured at each time point. Values are mean ± S.E. (n = 4, except SUM149 total NAA in I, where n = 2). J, MIDs of NAA in cells cultured in medium containing 10 mm [U-13C]glucose and 2.5 mm unlabeled glutamine or 10 mm unlabeled glucose and 2.5 mm [U-13C]glutamine for the indicated time points following 72 h depletion of NAA as in I. A and D, differences in total pool sizes were assessed by one-way ANOVA and Sidak's multiple comparison tests versus SUM149 WT (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
NAA is synthesized from aspartate and acetyl-CoA in a condensation reaction catalyzed by aspartate-N-acetyltransferase (Asp-NAT). NAA can also form via hydrolysis of N-acetyl-aspartylglutamate (NAAG) (Fig. 9B). The expression of genes that encode for enzymes involved in NAA metabolism was measured via the RT-qPCR in SUM149 WT, shScr, and shRhoC cell lines. ASPA, the gene that encodes aspartoacylase, which catalyzes cleavage of NAA, was not detected in any of the SUM149 cell lines. RIMKLA and RIMKLB encode enzymes that catalyze the formation of NAAG from NAA and glutamate. RIMKLB expression did not change significantly upon depletion of RhoC, whereas RIMKLA expression was often below the detection limit of the assay.
A significant decrease in NAT8L expression (the gene that encodes Asp-NAT), however, was observed in shRhoC cells (Fig. 9C). Moreover, the levels of acetyl-CoA and aspartate remained relatively fixed in all four SUM149-derived cell lines (Fig. 9, D and E). These data indicate that the decreased NAA levels observed in RhoC-depleted cells may be a result of reduced Asp-NAT levels rather than a consequence of depleted precursor substrates.
In all cell lines, including MCF10A, MCF7, and MDA231, the NAA pool was mostly unlabeled (Fig. 9F). A large unlabeled metabolite pool may arise when the initial pool size at the start of the experiment is very large. A large unlabeled NAA pool was observed in all cell lines, even in those with very small NAA pool sizes (Fig. 9A).
Among the SUM149-derived cell lines, NAA was less labeled in shRhoC cells (Fig. 9F). After 5 h, the percentage of labeled NAA in shRhoC cells was 12.9% ± 1.3% compared with 18.2% ±0.4%, 21.5% ± 0.4%, and 23.5% ± 0.4%, respectively, in SUM149 WT, shScr, and shHIF-1α cells. Additionally, NAA synthesis in SUM149 cells is predominantly derived from glutamine because incubation in [1,2-13C]glucose medium failed to significantly label NAA in WT, shScr, or shRhoC cells (Fig. 9G). The decreased NAA labeling observed in shRhoC cells may be attributed in part to the decrease in aspartate labeling (i.e. an increased M0 mass isotopomer) in shRhoC cells (Fig. 9E). The closely related metabolite NAAG was substantially less labeled than NAA (Fig. 9H) and was present at much lower levels than NAA in all cell lines (Fig. 9A).
Importantly, we observed that the NAA pool in SUM149 WT cells was depleted after 72 h, concurrent with glucose and glutamine reduction in the growth medium (Fig. 9I). These data suggest that when extracellular nutrients are depleted, these cells may break down NAA to facilitate continued survival. NAA-depleted SUM149 WT cells were next incubated in fresh medium containing either [U-13C]glucose or [U-13C]glutamine for 1, 3, or 6 h (Fig. 9J). Under these conditions, NAA synthesis was again observed to be primarily glutamine-derived in SUM149 WT cells.
Discussion
The molecular characteristics of IBC cells are highly suggestive of a phenotype constitutively adapted to hypoxia. IBC-derived SUM149 cells are resistant to protein synthesis inhibition under hypoxia, IBC tumor emboli have higher intratumoral microvessel density than other tumors, and IBC cells overexpress VEGF and IL-8 growth factors that strongly promote angiogenesis (3, 41). These and other molecular adaptations may be the primary mechanism by which IBC emboli invade the poorly oxygenated lymphatic system and promote neolymphangiogenesis under hypoxia (3, 42). Using SUM149 cells, the primary in vitro model for the study of IBC, we have provided the first in-depth investigation into the underlying metabolic alterations associated with these adaptations.
We found that, under normal culture conditions (∼20% O2), SUM149 cells convert nearly all consumed glucose to lactate, exhibit very low mitochondrial respiratory capacity, and perform reductive carboxylation of glutamine-derived α-ketoglutarate to form citrate. Citrate is used to maintain anaplerotic reactions in the TCA cycle as well as to fuel fatty acid synthesis upon cleavage by ATP citrate lyase. Under sufficient oxygen, normal cells generate citrate from glucose-derived acetyl-CoA following condensation with OAA. However, our data indicate that glucose-derived acetyl-CoA is not a significant source of citrate in SUM149 cells (Figs. 2, 3, and 5).
Under hypoxic conditions, glutamine can become a primary source of citrate via reductive metabolism in cancer cells (36), and several studies have demonstrated the importance of glutamine-dependent reductive carboxylation of AKG in aggressive cancers (36, 43–45). In this type of metabolism, the conversion of glutamine-derived AKG to isocitrate and CO2 is catalyzed by isocitrate dehydrogenase (IDH) running in the “reverse” direction to form isocitrate that isomerizes to citrate, which is then used for fatty acid synthesis. A variety of mutations in isoforms of IDH have been identified in cancer cells (36, 43, 44), underscoring the importance of this reaction to cancer metabolism. We found no mutations in the IDH1, IDH2, or IDH3α genes sequenced from cDNA templates purified from the cell lines studied in this work, however. Thus, SUM149 cells reductively carboxylate AKG under normal culture conditions (∼20% O2) in the presence of wild-type IDH.
Because SUM149 cells exhibit elevated levels of HIF-1α (Fig. 6A) (38), we initially hypothesized that HIF-1α stabilization was a driver of the observed metabolic phenotype. However, although a moderate increase in basal and maximal oxygen consumption rates in HIF-1α-depleted SUM149 cells was observed (Fig. 6D), HIF-1α depletion had very little effect on carbon flux and no observable effect on reductive carboxylation of AKG to citrate (Fig. 7, A–D). Our data instead strongly point to RhoC as a potent regulator of carbon metabolism and glutamine uptake in SUM149 cells. Given the decrease in glutamine uptake (Fig. 8, B and C) and increase in M0 glutamate observed after RhoC depletion (Fig. 7B), it is possible that RhoC directly or indirectly regulates glutamine transport. Activation of Rho GTPases by addition of the Rho-family GEF Dbl was recently shown to regulate the activity of GLS (10), however. Thus, another potential mechanism for RhoC's modulation of glutamine flux is via direct regulation of glutaminase. We did not observe a modulation in GLS protein levels when RhoC was depleted, however. In contrast, an appreciable increase in GS (the enzyme that operates in the reverse direction) was apparent (Fig. 8F) after RhoC depletion, suggesting RhoC may regulate GS rather than GLS in SUM149 cells.
This work also revealed for the first time that RhoC plays an integral role in the regulation of NAA metabolism in SUM149 cells. NAA, which is the second most abundant free amino acid in mammalian nervous systems, produces very high signals in MRI in vivo (18). Abnormalities in the MRI peak of NAA are associated with a number of neuronal disorders (46) and gliomas (47). Although generally considered brain-specific, NAA was found in an ovarian mucinous cystadenoma sample (19) and in the cystic fluid of serous ovarian tumors (21). Moreover, in a large-scale metabolomics analysis of 100 ovarian tumor biopsy samples, NAA and NAAG levels were found to increase with ovarian cancer aggressiveness (20). Recent work has further implicated elevated NAA and NAT8L levels as markers of decreased survival in ovarian cancer (48).
In our study, ∼20% of the total NAA pool was labeled in SUM149 WT cells, and almost no label was detected in NAAG after 5 h of growth in [U-13C]glutamine and [1,2-13C]glucose (Fig. 9, F and H). The most likely explanation for the large unlabeled pools of NAA and NAAG is a high proportion of NAA stored in microsomes or other subcellular compartments (49) that do not directly participate in the carbon-based metabolism probed over the course of the 5-h labeling experiment. Nevertheless, the very large NAA pool in SUM149 cells suggests that NAA plays an active metabolic role in these cells. The large reductive carboxylation flux in these cells may generate an excess of acetyl-CoA and acetate that is stored as NAA until needed for fatty acid synthesis. Indeed, when we grew SUM149 cells for 72 h, we found that the NAA pool was depleted along with glucose and glutamine levels (Fig. 9I). Although NAA has been proposed as a potential marker for neuronal loss and mitochondrial dysfunction (50), its precise function is still debated. Nonetheless, there is evidence that NAA is used as an acetyl donor to myelin lipid synthesis in neuronal tissue (18, 49). Our data, combined with other recent work, point toward NAA functioning as an oncometabolite (20, 48, 51) that may promote tumor progression by providing a potential reservoir to accommodate varying nutritional needs during the diverse steps of the metastatic process.
The plasticity of SUM149 cells renders them highly adaptive to new environments. NAA storage may help promote survival when resources are scarce. In contrast, the RhoC-depleted SUM149 cell line, with its lower glutamine uptake and decreased reductive carboxylation flux, would be expected to have far less free acetate available for fatty acid synthesis and thus would lack elevated levels of NAA. If the model of NAA as a storage metabolite for acetate is correct, then RhoC-depleted SUM149 cells likely funnel their glutamine-derived acetyl-CoA directly to fatty acid synthesis rather than to storage as NAA.
IBC is an extremely lethal cancer that rapidly metastasizes. The 5-year survival rate after diagnosis with metastatic breast cancer is only 24% compared with more than 80% for other stages of breast cancer (39). Continued research into the increasingly complex role RhoC plays in driving the molecular and metabolic adaptations that promote metastasis as well as the functional role of NAA in tumor cells has the potential to yield novel clinical interventions for highly lethal malignancies such as IBC.
Author Contributions
M. L. W., J. A. Y., C. R. E., L. D. V. W., Z. F. W., S. B., L. B., C. F., S. A., and M. J. M. performed the research. M. L. W., S. D. M., C. F. B., and S. S. conceived the study. M. L. W., J. A. Y., C. R. E., L. D. V. W., S. D. M., C. F. B., M. J. M., and L. S. S. designed the experiments. M. L. W. and J. A. Y. wrote the manuscript. All authors interpreted the data and approved the final version of the manuscript.
Acknowledgments
This work utilized core services of the Michigan Regional Comprehensive Metabolomics Resource Core, supported by National Institutes of Health Grant U24 DK097153. We thank Firas Midani for help with natural abundance correction algorithms and Mariana Rodriguez Ortiz for help with figure preparation.
This work was supported in part by the Metavivor Foundation (to S. D. M., J. A. Y., and M. L. W.); the Avon Foundation (to S. D. M., J. A. Y., and L. D. V. W.); the Breast Cancer Research Foundation (to S. D. M., J. A. Y., and L. D. V. W.); the James S. McDonnell Foundation (to M. L. W.); the Liz and Eric Lefkofsky Innovative Research Fund (to S. D. M. and M. L. W.); National Institutes of Health Grants T32 CA140044 (to M. L. W.) and 5T32 GM008353 (to L. D. V. W.); NIDDK, National Institutes of Health Grant K25 DK092558 (to C. R. E.); National Institutes of Health Grant RO1 DK46409 (to L. S. S.); and University of Michigan Cancer Center Support Grant P30 CA046592. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- IBC
- inflammatory breast cancer
- TCA
- tricarboxylic acid
- AKG
- α-ketoglutarate
- NAA
- N-acetylaspartate
- GS
- glutamine synthetase
- OCR
- oxygen consumption rate
- RT-qPCR
- quantitative RT-PCR
- PKAR
- pyruvate kinase M2 activity reporter
- KCN
- potassium cyanide
- FBP
- fructose 1,6-bisphosphate
- DHAP
- dihydroxyacetone phosphate
- GAP
- glyceraldehyde phosphate
- OAA
- oxaloacetate
- MID
- mass isotopomer distribution
- HIF
- hypoxia-inducible factor
- NAT
- N-acetyltransferase
- NAAG
- N-acetyl-aspartylglutamate
- IDH
- isocitrate dehydrogenase
- GLS
- glutaminase
- ANOVA
- analysis of variance
- ACOA
- acetyl-CoA.
References
- 1. Wecsler J. S., Tereffe W., Pedersen R. C., Sieffert M. R., Mack W. J., Cui H., Russell C. A., Woods R. R., Viscusi R. K., Sener S. F., and Lang J. E. (2015) Lymph node status in inflammatory breast cancer. Breast Cancer Res. Treat. 151, 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woodward W. A., and Cristofanilli M. (2009) Inflammatory breast cancer. Semin. Radiat. Oncol. 19, 256–265 [DOI] [PubMed] [Google Scholar]
- 3. Silvera D., and Schneider R. J. (2009) Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle 8, 3091–3096 [DOI] [PubMed] [Google Scholar]
- 4. Vander Heiden M. G. (2011) Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 10, 671–684 [DOI] [PubMed] [Google Scholar]
- 5. Gillies R. J., Robey I., and Gatenby R. A. (2008) Causes and consequences of increased glucose metabolism of cancers. J. Nucl. Med. 49, 24S–42S [DOI] [PubMed] [Google Scholar]
- 6. Ferreira L. M. (2010) Cancer metabolism: the Warburg effect today. Exp. Mol. Pathol. 89, 372–380 [DOI] [PubMed] [Google Scholar]
- 7. Vaupel P., and Mayer A. (2012) Availability, not respiratory capacity governs oxygen consumption of solid tumors. Int. J. Biochem. Cell Biol. 44, 1477–1481 [DOI] [PubMed] [Google Scholar]
- 8. Eagle H. (1955) Nutrition needs of mammalian cells in tissue culture. Science 122, 501–514 [DOI] [PubMed] [Google Scholar]
- 9. Sauer L. A., and Dauchy R. T. (1983) Ketone body, glucose, lactic acid, and amino acid utilization by tumors in vivo in fasted rats. Cancer Res. 43, 3497–3503 [PubMed] [Google Scholar]
- 10. Wang J. B., Erickson J. W., Fuji R., Ramachandran S., Gao P., Dinavahi R., Wilson K. F., Ambrosio A. L., Dias S. M., Dang C. V., and Cerione R. A. (2010) Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18, 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Golen K. L., Davies S., Wu Z. F., Wang Y., Bucana C. D., Root H., Chandrasekharappa S., Strawderman M., Ethier S. P., and Merajver S. D. (1999) A novel putative low-affinity insulin-like growth factor-binding protein, LIBC (lost in inflammatory breast cancer), and RhoC GTPase correlate with the inflammatory breast cancer phenotype. Clin. Cancer Res. 5, 2511–2519 [PubMed] [Google Scholar]
- 12. van Golen K. L., Wu Z. F., Qiao X. T., Bao L. W., and Merajver S. D. (2000) RhoC GTPase, a novel transforming oncogene for human mammary epithelial cells that partially recapitulates the inflammatory breast cancer phenotype. Cancer Res. 60, 5832–5838 [PubMed] [Google Scholar]
- 13. Kleer C. G., Griffith K. A., Sabel M. S., Gallagher G., van Golen K. L., Wu Z. F., and Merajver S. D. (2005) RhoC-GTPase is a novel tissue biomarker associated with biologically aggressive carcinomas of the breast. Breast Cancer Res. Treat. 93, 101–110 [DOI] [PubMed] [Google Scholar]
- 14. Clark E. A., Golub T. R., Lander E. S., and Hynes R. O. (2000) Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406, 532–535 [DOI] [PubMed] [Google Scholar]
- 15. Griner E. M., Dancik G. M., Costello J. C., Owens C., Guin S., Edwards M. G., Brautigan D. L., and Theodorescu D. (2015) RhoC is an unexpected target of RhoGDI2 in prevention of lung colonization of bladder cancer. Mol. Cancer Res. 13, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ikoma T., Takahashi T., Nagano S., Li Y. M., Ohno Y., Ando K., Fujiwara T., Fujiwara H., and Kosai K. (2004) A definitive role of RhoC in metastasis of orthotopic lung cancer in mice. Clin. Cancer Res. 10, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 17. Suwa H., Ohshio G., Imamura T., Watanabe G., Arii S., Imamura M., Narumiya S., Hiai H., and Fukumoto M. (1998) Overexpression of the rhoC gene correlates with progression of ductal adenocarcinoma of the pancreas. Br. J. Cancer 77, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mehta V., and Namboodiri M. A. (1995) N-acetylaspartate as an acetyl source in the nervous system. Brain Res. Mol. Brain Res. 31, 151–157 [DOI] [PubMed] [Google Scholar]
- 19. Hascalik S., Celik O., Sarac K., Alkan A., and Mizrak B. (2006) Clinical significance of N-acetyl-l-aspartate resonance in ovarian mucinous cystadenoma. Int. J. Gynecol. Cancer 16, 423–426 [DOI] [PubMed] [Google Scholar]
- 20. Fong M. Y., McDunn J., and Kakar S. S. (2011) Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS ONE 6, e19963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kolwijck E., Wevers R. A., Engelke U. F., Woudenberg J., Bulten J., Blom H. J., and Massuger L. F. (2010) Ovarian cyst fluid of serous ovarian tumors contains large quantities of the brain amino acid N-acetylaspartate. PLoS ONE 5, e10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosenthal D. T., Zhang J., Bao L., Zhu L., Wu Z., Toy K., Kleer C. G., and Merajver S. D. (2012) RhoC impacts the metastatic potential and abundance of breast cancer stem cells. PLoS ONE 7, e40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lorenz M. A., Burant C. F., and Kennedy R. T. (2011) Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal. Chem. 83, 3406–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Noguchi Y., Young J. D., Aleman J. O., Hansen M. E., Kelleher J. K., and Stephanopoulos G. (2009) Effect of anaplerotic fluxes and amino acid availability on hepatic lipoapoptosis. J. Biol. Chem. 284, 33425–33436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Young J. D. (2014) INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics 30, 1333–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fernandez C. A., Des Rosiers C., Previs S. F., David F., and Brunengraber H. (1996) Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31, 255–262 [DOI] [PubMed] [Google Scholar]
- 27. Chen Y. C., Allen S. G., Ingram P. N., Buckanovich R., Merajver S. D., and Yoon E. (2015) Single-cell migration chip for chemotaxis-based microfluidic selection of heterogeneous cell populations. Sci. Rep. 5, 9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merrins M. J., Van Dyke A. R., Mapp A. K., Rizzo M. A., and Satin L. S. (2013) Direct measurements of oscillatory glycolysis in pancreatic islet β-cells using novel fluorescence resonance energy transfer (FRET) biosensors for pyruvate kinase M2 activity. J. Biol. Chem. 288, 33312–33322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rocheleau J. V., Head W. S., and Piston D. W. (2004) Quantitative NAD(P)H/flavoprotein autofluorescence imaging reveals metabolic mechanisms of pancreatic islet pyruvate response. J. Biol. Chem. 279, 31780–31787 [DOI] [PubMed] [Google Scholar]
- 30. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., and Cantley L. C. (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 [DOI] [PubMed] [Google Scholar]
- 31. Hitosugi T., Kang S., Vander Heiden M. G., Chung T. W., Elf S., Lythgoe K., Dong S., Lonial S., Wang X., Chen G. Z., Xie J., Gu T. L., Polakiewicz R. D., Roesel J. L., Boggon T. J., et al. (2009) Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2, ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mazurek S., Boschek C. B., Hugo F., and Eigenbrodt E. (2005) Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin. Cancer Biol. 15, 300–308 [DOI] [PubMed] [Google Scholar]
- 33. Gao X., Wang H., Yang J. J., Liu X., and Liu Z. R. (2012) Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 45, 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mazurek S. (2011) Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int. J. Biochem. Cell Biol. 43, 969–980 [DOI] [PubMed] [Google Scholar]
- 35. Poore B., Siegel N., Park J., Hwang B., Afif I., and Le A. (2015) In Glutamine in Clinical Nutrition (Rajendram R., Preedy V. R., and Patel V. B., eds.) pp 77–85, Springer, New York [Google Scholar]
- 36. Wise D. R., Ward P. S., Shay J. E., Cross J. R., Gruber J. J., Sachdeva U. M., Platt J. M., DeMatteo R. G., Simon M. C., and Thompson C. B. (2011) Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 108, 19611–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yuan Y., Hilliard G., Ferguson T., and Millhorn D. E. (2003) Cobalt inhibits the interaction between hypoxia-inducible factor-α and von Hippel-Lindau protein by direct binding to hypoxia-inducible factor-α. J. Biol. Chem. 278, 15911–15916 [DOI] [PubMed] [Google Scholar]
- 38. Chu K., Boley K. M., Moraes R., Barsky S. H., and Robertson F. M. (2013) The paradox of E-cadherin: role in response to hypoxia in the tumor microenvironment and regulation of energy metabolism. Oncotarget 4, 446–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Howlader N., Noone A. M., Krapcho M., Garshell J., and Miller D., A. S. F., Kosary C. L., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D. R., Chen H. S., Feuer E. J., Cronin K. A. (eds). (2015) SEER Cancer Statistics Review, 1975–2012. National Cancer Institute, Bethesda, MD [Google Scholar]
- 40. Evans C. R., Karnovsky A., Kovach M. A., Standiford T. J., Burant C. F., and Stringer K. A. (2014) Untargeted LC-MS metabolomics of bronchoalveolar lavage fluid differentiates acute respiratory distress syndrome from health. J. Proteome Res. 13, 640–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Golen K. L., Wu Z. F., Qiao X. T., Bao L., and Merajver S. D. (2000) RhoC GTPase overexpression modulates induction of angiogenic factors in breast cells. Neoplasia 2, 418–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Levine P. H., Portera C. C., Hoffman H. J., Yang S. X., Takikita M., Duong Q. N., Hewitt S. M., and Swain S. M. (2012) Evaluation of lymphangiogenic factors, vascular endothelial growth factor D and E-cadherin in distinguishing inflammatory from locally advanced breast cancer. Clin. Breast Cancer 12, 232–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Metallo C. M., Gameiro P. A., Bell E. L., Mattaini K. R., Yang J., Hiller K., Jewell C. M., Johnson Z. R., Irvine D. J., Guarente L., Kelleher J. K., Vander Heiden M. G., Iliopoulos O., and Stephanopoulos G. (2012) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mullen A. R., Wheaton W. W., Jin E. S., Chen P. H., Sullivan L. B., Cheng T., Yang Y., Linehan W. M., Chandel N. S., and DeBerardinis R. J. (2012) Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holleran A. L., Briscoe D. A., Fiskum G., and Kelleher J. K. (1995) Glutamine metabolism in AS-30D hepatoma cells: evidence for its conversion into lipids via reductive carboxylation. Mol. Cell Biochem. 152, 95–101 [DOI] [PubMed] [Google Scholar]
- 46. Yalcinkaya C., Benbir G., Salomons G. S., Karaarslan E., Rolland M. O., Jakobs C., and van der Knaap M. S. (2005) Atypical MRI findings in Canavan disease: a patient with a mild course. Neuropediatrics 36, 336–339 [DOI] [PubMed] [Google Scholar]
- 47. Tong Z., Yamaki T., Harada K., and Houkin K. (2004) In vivo quantification of the metabolites in normal brain and brain tumors by proton MR spectroscopy using water as an internal standard. Magn. Reson. Imaging 22, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 48. Zand B., Previs R. A., Zacharias N. M., Rupaimoole R., Mitamura T., Nagaraja A. S., Guindani M., Dalton H. J., Yang L., Baddour J., Achreja A., Hu W., Pecot C. V., Ivan C., Wu S. Y., et al. (2016) Role of increased N-acetylaspartate levels in cancer. J. Natl. Cancer Inst. 108, djv426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Satrústegui J., Pardo B., and Del Arco A. (2007) Mitochondrial transporters as novel targets for intracellular calcium signaling. Physiol. Rev. 87, 29–67 [DOI] [PubMed] [Google Scholar]
- 50. Clark J. B. (1998) N-acetyl aspartate: a marker for neuronal loss or mitochondrial dysfunction. Dev. Neurosci. 20, 271–276 [DOI] [PubMed] [Google Scholar]
- 51. Lou T. F., Sethuraman D., Dospoy P., Srivastva P., Kim H. S., Kim J., Ma X., Chen P. H., Huffman K. E., Frink R. E., Larsen J. E., Lewis C., Um S. W., Kim D. H., Ahn J. M., et al. (2016) Cancer-specific production of N-acetylaspartate via NAT8L overexpression in non-small cell lung cancer and its potential as a circulating biomarker. Cancer Prev. Res. (Phila.) 9, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]