Abstract
Fibroblasts are central to wound healing and fibrosis through TGFβ1-triggered differentiation into contractile, α-smooth muscle actin (α-SMA)-positive myofibroblasts. This is mediated by accumulation of a pericellular matrix of hyaluronan (HA) and the HA-dependent co-localization of CD44 with the epidermal growth factor receptor (EGFR). Interactions of HA with hyaladherins, such as inter-α-inhibitor (IαI) and tumor necrosis factor-stimulated gene-6 (TSG-6), are also essential for differentiation. This study investigated the mechanisms involved. TSG-6 and α-SMA had different kinetics of induction by TGFβ1, with TSG-6 peaking before α-SMA. Si CD44 or EGFR inhibition prevented differentiation but had no effect on TSG-6 expression. TSG-6 was essential for differentiation, and mAb A38 (preventing IαI heavy chain (HC) transfer), HA-oligosaccharides, cobalt, or Si bikunin prevented TSG-6 activity, preventing differentiation. A38 also prevented the EGFR/CD44 association. This suggested that TSG-6/IαI HC interaction was necessary for the effect of TSG-6 and that HC stabilization of HA initiated the CD44/EGFR association. The newly described HC5 was shown to be the principal HC expressed, and its cell surface expression was prevented by siRNA inhibition of TSG-6 or bikunin. HC5 was released by hyaluronidase treatment, confirming its association with cell surface HA. Finally, HC5 knockdown by siRNA confirmed its role in myofibroblast differentiation. The current study describes a novel mechanism linking the TSG-6 transfer of the newly described HC5 to the HA-dependent control of cell phenotype. The interaction of HC5 with cell surface HA was essential for TGFβ1-dependent differentiation of fibroblasts to myofibroblasts, highlighting its importance as a novel potential therapeutic target.
Keywords: fibroblast, fibrosis, hyaluronan, myofibroblast, transforming growth factor beta (TGF-B), fibroblast, tumor necrosis factor gene 6
Introduction
The fibroblast is the most abundant cell type in normal connective tissues. It plays a central role in the synthesis, degradation, and remodeling of extracellular matrix both in health and in disease. At sites of tissue damage and in the context of wound healing, activated fibroblasts, termed myofibroblasts, with a contractile phenotype, characterized by the expression of the smooth muscle isoform of α-actin (α-SMA),3 are essential for the synthesis of a collagen-rich scar, providing the force for wound contraction (1). During a healing response, myofibroblasts are a transient cell population (2). Myofibroblasts are also the major effectors of fibrosis; their persistent presence has been established as the best marker of numerous types of progressive organ dysfunction, such as that seen in chronic renal failure of various etiologies (3–7), liver disease (8), and pulmonary fibrosis (9).
The cytokine TGFβ1 is a well recognized mediator of progressive tissue fibrosis, and in vitro and in vivo evidence suggests that it is the primary driving force in fibroblast-myofibroblast phenotypic activation (10, 11). The sequence of events leading up to this transformation is being increasingly characterized at a molecular level in an attempt to understand and eventually control this process.
We have previously demonstrated that TGFβ1-mediated phenotypic activation is dependent on an increase in the synthesis of the extracellular matrix polysaccharide hyaluronan (HA) (12–14). TGFβ1, through stimulation of EGF and subsequent activation of EGFR-associated ERK-MAPK activity, stimulates HA synthesis. This increased HA synthesis is associated with the formation of an organized pericellular HA coat, which subsequently orchestrates assembly of a CD44/EGFR receptor complex on the cell surface. Activation of ERK signaling downstream of this complex subsequently leads to a reorganization of the actin cytoskeleton.
The secreted product of tumor necrosis factor-stimulated gene 6 (TSG-6) is important in the formation and remodeling of HA-rich pericellular coats and matrices (14–16). It is involved in the formation of TSG-6-cross-linked HA networks (17) and may also stabilize the HA matrix either by binding directly as a hyaladherin or through facilitating the covalent transfer of heavy chains (HCs) of inter-α-inhibitor (IαI) onto HA to form an HC·HA complex (18, 19). Thus, in the TGFβ1-triggered generation of the myofibroblast phenotype, we proposed that TSG-6 may play an important part in forming the HA·CD44·EGFR complex, which then signals to induce ERK activation.
The mechanism of HC transfer has been elucidated by Rugg et al. (19). TSG-6 forms an intermediate species with one of the two heavy chains from the IαI complex, which consists of the light chain bikunin and its chondroitin sulfate chain, which is bound to two heavy chains. Classically, three HCs have been described. HC1 and HC2 are known to occur bound to chondroitin sulfate to form IαI, whereas HC3 is thought to bind chondroitin sulfate/bikunin as a single chain forming an assembly known as pre-αI. Previous work (20) has shown that TSG-6 is capable of transferring not only HC2 but also HC1 and HC3 from IαI (or pre-αI) onto HA but that the presence of HC2 appeared to be necessary for this enzymatic action of TSG-6 to take place.
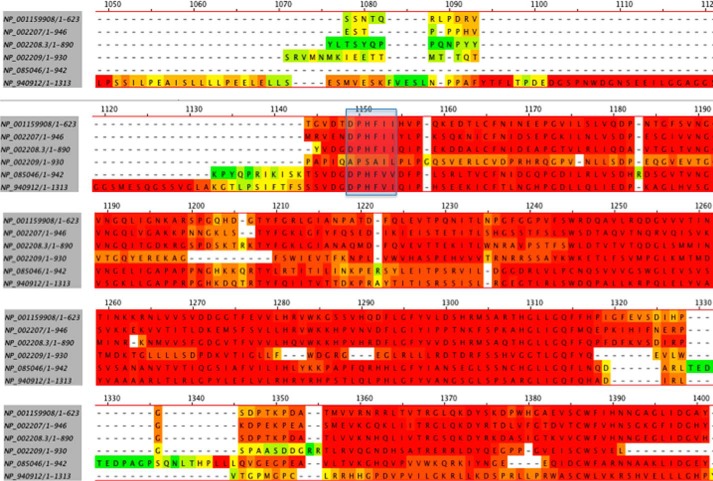
There is a fourth HC (HC4) about which less is known. It does not have the conserved cleavage site of HC1–3 that allows them to link with bikunin. It does, however, bind to and block the polymerization of actin, resulting in inhibition of neutrophil phagocytosis and chemotaxis (21). The more recently described HC5 has not yet been fully characterized; nor has it been shown to complex with chondroitin sulfate to form IαI (or pre-IαI). It does, however, possess the necessary binding sites to enable it to do so (22) (Fig. 1) The existence of HC6 was recently identified in GenBankTM from a report of 2003 (23), but no protein product has yet been described.
FIGURE 1.
The sequences of all six of the IαI heavy chains were obtained from GenBankTM and aligned using MegAlign software. The sequences of the putative binding sites of the heavy chains to chondroitin sulfate to form IαI are highlighted in the blue box. It can be seen that the sequence of HC4 is uniquely distinct from the other heavy chains at this site.
In the current paper, we have examined the role of TSG-6 following TGFβ1 stimulation and highlighted HC transfer activity as being key to form a HA matrix that is anchored by CD44, leading to a CD44·EGFR signaling complex. This activity was specifically dependent on HC5. We demonstrate for the first time that HC5 has a major mechanistic role to play in the phenotypic activation of fibroblasts.
Experimental Procedures
Materials
All reagents were from Sigma-Aldrich (Poole, UK) unless otherwise stated. ERK (MEK) inhibitor PD98059 and EGFR inhibitor AG1478 were purchased from Calbiochem (Darmstadt, Germany). They were prepared as stock solutions of 5 and 3 mg/ml, respectively. Oligo-HA (10 units; i.e. 10 monosaccharides) (purchased from Hyalose, Oklahoma City, OK; HYA-OLIGO10EF-1) and a hexasaccharide control (Iduron UK) were prepared at a concentration of 1 mg/ml and were added to give a final concentration of 50 μg/ml on the cells 2 h before stimulation with TGFβ. Cobalt chloride was stored at a concentration of 500 mm in PBS. All inhibitors were diluted immediately before use and added to the cell cultures at the times given in the legends before the addition of TGFβ1. Anti-IαI heavy chain 5 antibody ab107846 and anti-IαI heavy chain 2 antibody ab118257 together with the loading control anti-GAPDH antibody ab9485 were purchased from Abcam (Cambridge, UK).
Cell Culture
Primary human lung fibroblasts (AG02262; NIA Aging Cell Respiratory Corriel Institute) were cultured in DMEM and F-12 medium containing 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin supplemented with 10% FCS (Biologic Industries Ltd., Cumbernauld, UK). The cells were maintained at 37 °C in a humidified incubator in an atmosphere of 5% CO2, and fresh growth medium was added to the cells every 3–4 days until confluence. Cells were growth-arrested in serum-free medium for 48 h before use in experiments, and all experiments were performed under serum-free conditions unless otherwise stated. For the induction of differentiation, cells were exposed to a single treatment of TGFβ1 at the concentrations indicated for times up to 72 h as in previous studies (12–14).
RT and Real-time Quantitative PCR (qPCR)
Cells were grown in 35-mm dishes and washed with PBS, pH 7.3, before lysis with TRI Reagent and RNA purification according to the manufacturer's protocol. RNA was precipitated with isopropyl alcohol and washed with 70% ethanol before being dried and resuspended in water. The concentration of RNA was determined by the absorbance at 260 nm. Reverse transcription was performed on 1 μg of RNA, using random primers to initiate cDNA synthesis with High Capacity cDNA reverse transcription kits, according to the manufacturer's protocol (Applied Biosystems, Cheshire, UK). As a negative control, RT was performed with sterile H2O replacing the RNA sample. qPCR was carried out using a 7900HT fast real-time PCR system (Applied Biosystems). cDNA was diluted 4-fold, and 4 μl (equivalent to 0.05 μg of the original RNA) was amplified in a final volume of 20 μl containing 10 μl of Taqman master mix (Applied Biosystems), 5 μl of H2O, and 1 μl of specific primer.
The following primers were purchased from Applied Biosystems: α-SMA primer ACTA2 (catalog no. 4331182 Hs00426835_g1) and TSG-6 primer TNFAIP6 (catalog no. Hs00200180_m1). rRNA was used as the internal control for all samples, and the results are presented as the relative quantification (RQ) calculated by the ΔΔCT method using the equation, RQ = 2−(ΔCT(1) − ΔCT(2)), (where ΔCT(1) is the mean ΔCT calculated for the experimental samples and ΔCT(2) is the mean ΔCT calculated for the control samples (Applied Biosystems, Cheshire, UK).
Heavy chain amplification was carried out by qPCR of the cDNA with specific primers to each IαI heavy chain using SYBR Green (Applied Biosystems).
In selected experiments mRNA copy number was determined by amplifying each heavy chain by conventional PCR and calculating the number of molecules present in the purified amplified product. Serial dilutions from 300,000 to 30 copies were prepared for each heavy chain, and the amplified qPCR signals of the experimental samples were read off from the standard curves produced.
All oligonucleotide primers were purchased from Invitrogen (Paisley, UK). The inter-α-trypsin inhibitor heavy chain (ITIH) qPCR primers used were as follows: ITIH1 (forward), TCCATGGAGAACAACGGACG; ITIH1 (reverse), GGGGGTACTGCAAATCCACA; ITIH2 (forward), GCCATTTCGATGGTGTTCCG; ITIH2 (reverse), TTCTCTGCTGTGCTACCGTG; ITIH3 (forward), CCTTTCGGCTGCTTGGGAAA; ITIH3 (reverse), GACGGCTCTCATGGTGACAA; ITIH4 (forward), AGTCACCAAACCCGATGACC; ITIH4 (reverse), GCTCCAGCTGAGTGGACATT; ITIH5 (forward), ACTGTCGCTGGAGAACTGTG; ITIH5 (reverse), CGGGGTCCTGATTTCATCGT; ITIH6 (forward), CATTGCATGGAAGCCGATGG; ITIH6 (reverse), GGCTCCTGGTTGCTATGGTT.
The FnEDR qPCR primers used were as follows: FnEDR (forward), GCTCAGAATCCAAGCGGAGA; FnEDR (reverse), CCAGTCCTTTAGGGCGATCA.
Inhibition by siRNA
All siRNA was purchased from Life Technologies Ltd. (Paisley, UK): TSG-6 siRNA, catalog no. M16704/s14263; bikunin siRNA, catalog no. 4392420/s1311; CD44 siRNA, catalog no. AM16704/114068; ITIH5 siRNA, catalog no. 4392420/s37361; ITIH2 siRNA, catalog no. 43920/s7604; Smad2 siRNA, catalog no. M16704/107875; Smad3 siRNA, catalog no. M16704/115717; Silencer® negative control No. 2 siRNA, catalog no. AM4613.
Cells to be transfected were deprived of serum for 4 h before the addition of the siRNA. Briefly, Lipofectamine 2000 (Invitrogen) was diluted 1:50 in Opti-MEM (Gibco 31085) and allowed to stand for 5 min. This was then added to a 333 nm dilution of the siRNA prepared immediately before use from a 100 μm stock. The mixture was left at room temperature for 30 min to allow the Lipofectamine·siRNA complexes to form, and then 200 μl of this preparation was added to 800 μl of serum-free antibiotic-free medium on the cells to give a final concentration of 33 nm, and the transfection was allowed to take place overnight. Control cells were transfected with scrambled siRNA. The following day, the appropriate stimuli were added to the cells as described, without removing the siRNA complexes, and the incubation continued for a further 72 h (unless otherwise indicated) before isolating RNA. Typical knockdown ranges using these siRNAs were as follows: Si TSG-6, 51–60%; Si bikunin, 78–89%; Si CD44, 53–67%; Si HC5, 89–98%; HC2, 90–99%.
SDS-PAGE and Western Blotting
Cells were cultured for 72 h under the designated conditions before total cell protein was extracted in 100 μl of lysis buffer as described previously (24). Protein concentration was determined by the Bradford protein assay, and 30 μg of each sample was boiled with reducing buffer for 10 min before being subjected to SDS-PAGE on a 7.5% acrylamide gel. Western blotting with rabbit anti-IαI HC5 primary antibody (Abcam, catalog no. ab136324) was performed by standard methods to detect the presence of HC5. Detection was performed with a specific anti-rabbit HRP-conjugated antibody (Abcam catalog no. 970510) and ECL reagent. Equal loading was confirmed by the use of anti GAPDH antibody (Abcam catalog no. 9485).
Immunocytochemistry
Cells were growth-arrested before incubation with TGFβ1 (10 ng/ml) or serum-free medium alone in the presence or absence of anti-TSG-6 antibody A38 (Santa Cruz Biotechnology catalog no. sc-65886) at 50 μg/ml. After 72 h, cells were fixed with 4% paraformaldehyde at room temperature for 15 min. Fixed cells were stained for EGFR (Abcam GR01, 1:30) (visualized using Alexa Fluor 488) and for CD44 (Ab A020 1:200) (Merck Millipore) (visualized with Alexa Fluor 555). The images were merged to demonstrate the co-localization of CD44 and EGFR.
The unpaired two-tailed Student's t test was used to identify statistical significance. Data were analyzed using the GraphPad Prism version 4.0a (GraphPad Software Inc., La Jolla, CA), and p ≤ 0.05 (*) and p ≤ 0.01 (**) were considered significant.
Results
A Temporal Association between Induction of TSG-6 and Phenotypic Activation of Fibroblasts
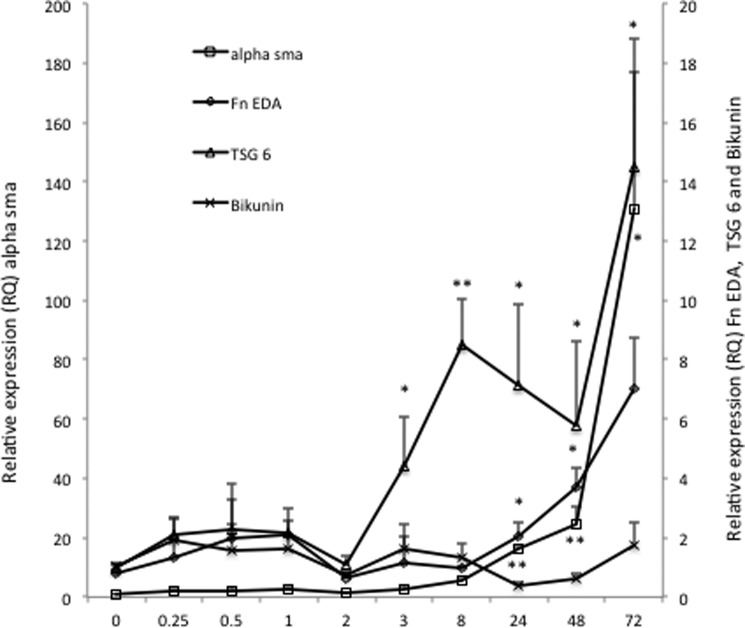
TSG-6 has been shown to be essential for the TGFβ1-dependent phenotypic activation of fibroblasts and renal epithelial cells (14, 16, 25). To determine the temporal relationship between TSG-6 expression and induction of the myofibroblast phenotype, growth-arrested fibroblasts were stimulated with TGFβ1 (10 ng/ml), and mRNA was extracted at times up to 72 h. The addition of TGFβ1 led to a time-dependent induction of TSG-6 that was significant at 3 h, reached a maximum at 8 h, and was sustained to 72 h. In contrast, the induction of the phenotypic markers, α-SMA and the EDA isoform of fibronectin (fnEDA), was slower, becoming significant at 24 h and continuing to increase up to 72 h. There was no significant change in the levels of bikunin over this time course (Fig. 2). These results suggest but do not confirm in themselves a temporal separation between TSG-6 up-regulation and induction of phenotypic activation.
FIGURE 2.
Human lung fibroblasts were growth-arrested and were then stimulated with TGFβ1 (10 ng/ml). The RNA was collected at the times shown, and the RQ of α-SMA, FnEDA, bikunin (Bik), and TSG-6 were determined by RT-qPCR. Data shown are the mean ± S.E. of five independent experiments, each of which generated a mean of three replicates. *, p < 0.05; **, p < 0.005 compared with 0 h.
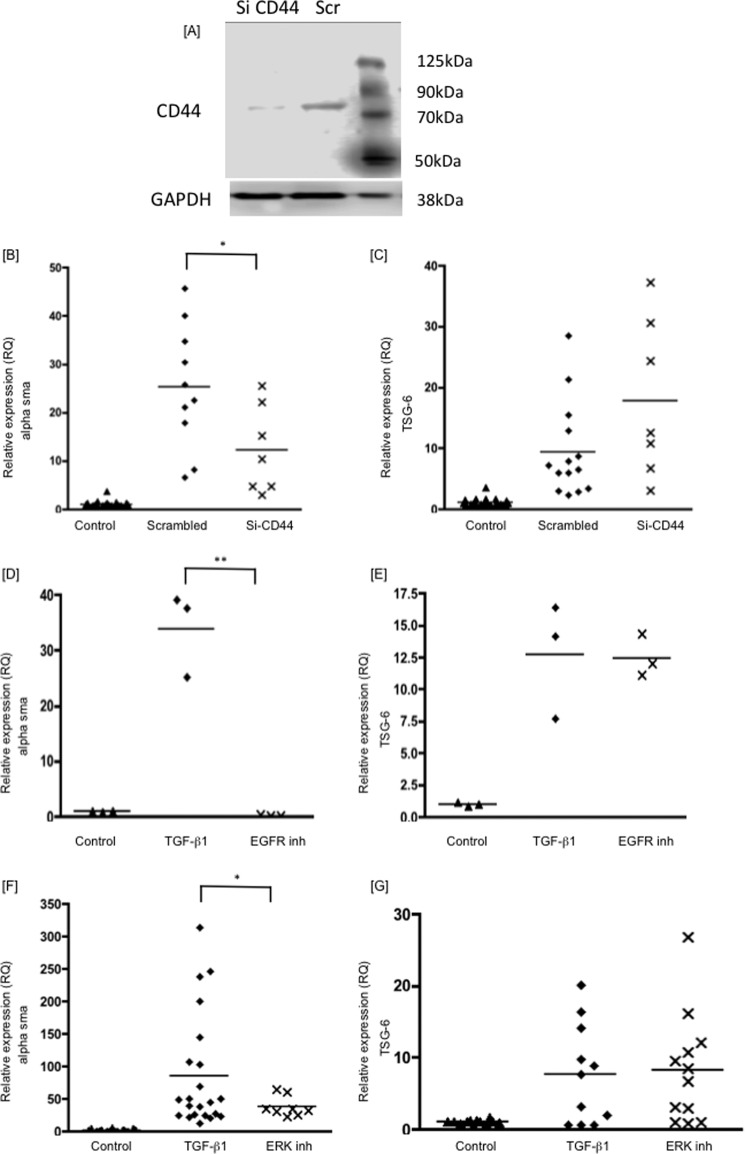
To investigate whether the inductions of TSG-6 and α-SMA were both triggered by a CD44-dependent mechanism, siRNA was used to knockdown CD44 expression. This resulted in >50% inhibition of CD44 protein expression (Fig. 3A), which was associated with a 60% inhibition of α-SMA mRNA (Fig. 3B), as reported previously (14, 23, 26). Knocking down CD44, however, had no inhibitory effect on TSG-6 expression (Fig. 3C). The change of phenotype in response to TGFβ1 is also, however, dependent on EGFR activation (14). Following inhibition of EGFR activity by AG1478, α-SMA induction was inhibited by >99% (Fig. 3D). TSG-6 expression, however, was unaffected (Fig. 3E). Phosphorylation of ERK1 and -2 is downstream from the EGFR-CD44 interaction. To confirm that TSG-6 was not induced as a result of this signaling pathway, ERK activation by MEK was inhibited using PD98059. This inhibited α-SMA induction by ∼60%, but TSG-6 was again unaffected (Fig. 3, F and G). Taken together, these results indicate that TSG-6 was not induced by the same series of activation events as those leading to phenotypic change and, together with our previous work (14, 16, 24), that it may therefore lie upstream of formation of the EGFR·CD44 complex.
FIGURE 3.
Human lung fibroblasts were growth-arrested and were then stimulated with TGFβ1 (10 ng/ml). The RNA was collected after 72 h of treatment, and the RQ values of α-SMA (B, D, and F), and TSG-6 (C, E, and G) were determined by RT-qPCR. The panels show cells treated with Si CD44, lysed, and protein-extracted for CD44 Western blotting (A); Si CD44 (B and C); EFGR inhibitor AG1478 (10 μm) (D and E); and ERK inhibitor PD98059 (10 μm) (F and G). Inhibitors were added 4 h before TGFβ1 stimulation. Data shown are the mean and scatter of values in three independent experiments, each with between two and three replicates, except one experiment with three replicates in D and E. *, p < 0.05; **, p < 0.005 compared with TGFβ1 treatment alone.
Induction of TSG-6 is SMAD-dependent
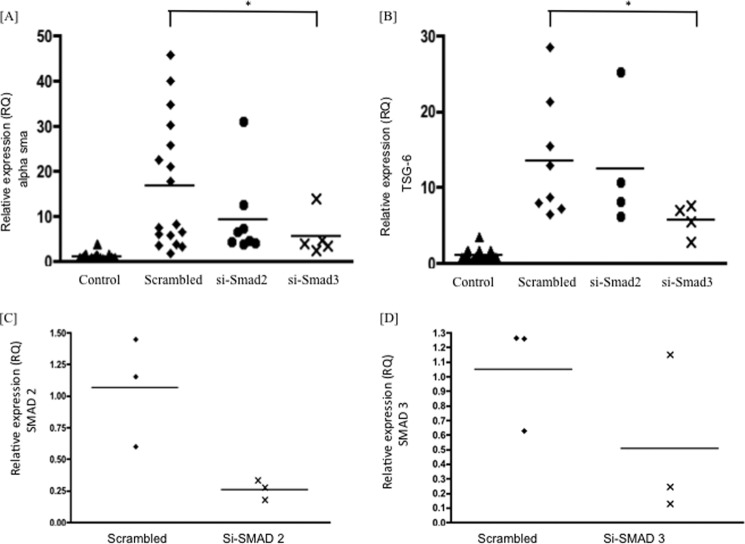
Although induction of HA and HA synthase is dependent on TGFβ1 activation of ERK and subsequently the EGFR, classical TGFβ1 signaling is initiated when the ligand induces assembly of a heteromeric complex of type II and type I receptors. The RII kinase then phosphorylates RI on a conserved glycine-serine-rich domain. This activates the RI kinase, which subsequently recognizes and phosphorylates members of the intracellular receptor-regulated SMADs (SMAD2 and SMAD3). SMAD signaling through the activation of this classical pathway has previously been implicated in phenotypic activation of fibroblasts, through the use of chemical inhibition of the ALK5 receptor (26). The role of SMAD signaling was examined by gene silencing of either SMAD2 or SMAD3 using siRNA. The siRNA to SMAD2 caused between 60 and 70% knockdown of SMAD2 in these cells, with the Si Smad3 giving between 50 and 60% knockdown of SMAD3 (Fig. 4). SMAD gene knockdown inhibited TGFβ1-dependent induction of TSG-6 and phenotypic activation, reflected by an inhibition of TGFβ1 induction of the myofibroblast marker α-SMA (Fig. 4, A and B). These data therefore suggest that both SMAD-dependent and -independent pathways are required for phenotypic activation, with the SMAD pathway alone leading to induction of TSG-6, which is distinct from the signaling pathway leading to HA synthesis and HA-dependent signaling.
FIGURE 4.
Confluent fibroblasts were growth-arrested in serum-free medium for 48 h and then treated with either scrambled siRNA or siRNA specific for SMAD2 or SMAD3, as detailed under “Experimental Procedures.” The cells were then incubated with TGFβ1 at 10 ng/ml in serum-free medium for 72 h. Control cells remained in serum-free conditions, without TGFβ1. The cells were lysed, RNA-extracted, and subjected to RT-qPCR for α-SMA (A) or TSG-6 (B), carried out as described under “Experimental Procedures.” Results are expressed as mean and scatter of values in two independent experiments, each with two or three replicates. *, p < 0.05, compared with scrambled. Scrambled in A and B was independently analyzed more frequently. C and D, inhibition of SMAD2 and SMAD3 by Si SMAD2 or Si SMAD3, respectively, in three independent experiments.
The Mechanism of TSG-6 Action in Phenotypic Activation of Fibroblasts
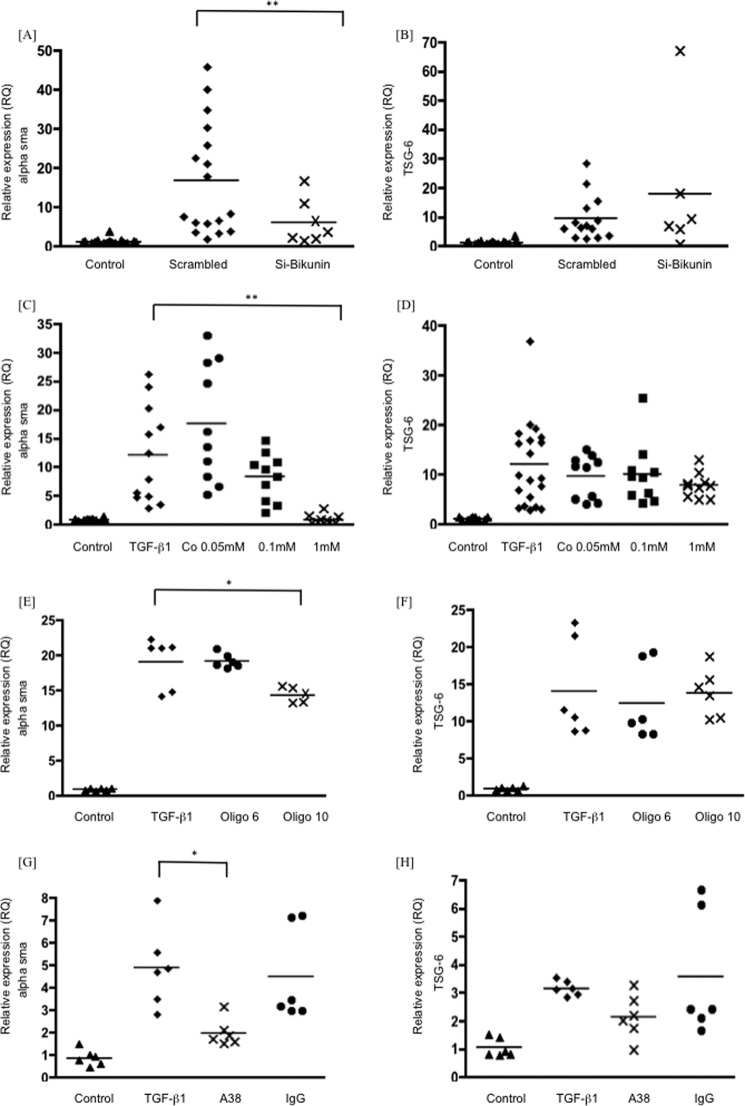
TSG-6 binds directly to HA through its link module. In addition, by a separate mechanism, TSG-6 also supports the covalent transfer of HCs of the IαI family members to HA, both of which roles are likely to stabilize HA matrices. The IαI family exists as a distinct assembly of one bikunin chain with one or more unique HCs designated HC1, HC2, HC3, HC5, and HC6 (HC4 does not bind bikunin). Covalent transfer of heavy chains to TSG-6 requires the bikunin component of IαI and is irreversibly inhibited by cobalt (19). Gene silencing of bikunin significantly inhibited TGFβ1-dependent phenotypic activation of fibroblasts, as did the presence of 1 mm cobalt (Fig. 5, A and C). Neither intervention influenced TGFβ1-dependent induction of TSG-6 (Fig. 5, B and D). TSG-6 function has been previously shown to be inhibited by the rat anti-human TSG-6 monoclonal antibody A38, which blocks HC binding to TSG-6 (27) and inhibits the formation of TSG-6·HC complexes in vitro (28). Similarly, HA decasaccharides (oligo-HA) irreversibly compete with HC binding to HA, preventing HA assembly, whereas hexasaccharides are ineffective. Stimulation of fibroblasts with TGFβ1 in the presence of either oligo-HA, to prevent HA binding by HC (Fig. 5, E and F), or of the A38 antibody, at a concentration previously demonstrated to inhibit TSG-6 activity in vitro (16) (Fig. 5, G and H), inhibited phenotypic activation without affecting TGFβ1 induction of TSG-6. These data suggest that the function of TSG-6 that is critical for phenotypic activation is covalent transfer of heavy chains of IαI to HA, which subsequently forms an organized and stable HA matrix.
FIGURE 5.
Human lung fibroblasts were growth-arrested and were then stimulated with TGFβ1 (10 ng/ml). The RNA was collected after 72 h of treatment, and the RQ values of α-SMA (A, C, E, and G) and TSG-6 (B, D, F, and H) were determined by RT-qPCR. The panels show cells treated for 4 h before TGFβ1 stimulation with Si bikunin (A and B), cobalt chloride (0.05–1 mm) (C and D), oligo-HA (at 50 μg/ml) (E and F), anti-TGS-6 antibody A38 (G and H), and non-immune IgG control (50 μg/ml). Data shown are the mean and scatter of values in three independent experiments, each with more than two replicates. *, p < 0.05; **, p < 0.005 compared with scrambled (A and B) or TGFβ1-stimulated cells (C–H). Oligo 6 and Oligo 10, hyaluronan oligosaccharides (6 and 10 monosaccharide units, respectively).
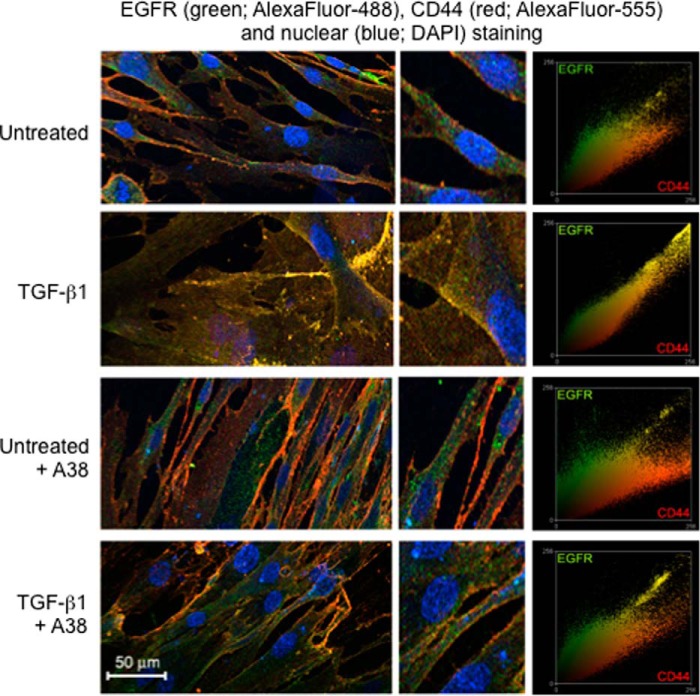
To confirm that TSG-6-dependent covalent heavy chain transfer onto HA was required to drive receptor co-localization, fibroblasts were stimulated with TGFβ1 in the presence of the A38 antibody. After 72 h, cells were dual-labeled for CD44 (red) and EGFR (green). The results demonstrate an increased association of CD44 and EGFR (yellow) following TGFβ1 stimulation, which was inhibited by the presence of the anti-TSG-6 A38 antibody (Fig. 6). These results are therefore consistent with TSG-6-mediated heavy chain stabilization of the HA matrix driving CD44/EGFR co-localization. The phenotypic change triggered by TGFβ1 is therefore dependent on the induction of TSG-6, which, through catalytic transfer of IαI HCs, orchestrates HA-dependent CD44-EGFR interactions and triggers downstream ERK signaling pathways.
FIGURE 6.
Human lung fibroblasts were growth-arrested and were then incubated with either TGFβ1 (10 ng/ml) or serum-free medium alone in the presence (A38) or absence of anti-TSG-6 antibody A38 at 50 μg/ml. Cells were stained for EGFR (antibody GR01, 1:30), visualized using Alexa Fluor 488, and CD44 (antibody A020 1:200) (Merck Millipore), visualized with Alexa Fluor 555. The images were merged to demonstrate the co-localization of CD44 and EGFR (yellow). Nuclei were stained with DAPI, and the central images show a ×4 magnification of sections of the images. Co-localization scatter plots were performed to confirm co-localization (depicted as yellow areas). Data shown are representative of three independent experiments.
The Heavy Chain 5 of Inter-α-inhibitor Is Highly Expressed by Fibroblasts and Is Linked to Myofibroblast Formation
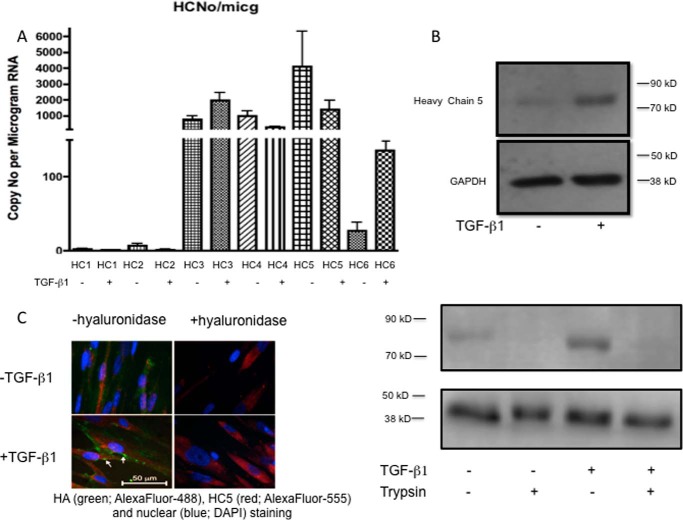
To investigate the role of each of the heavy chains of IαI in phenotypic activation, qPCR was performed for each of the chains HC1–HC6. All of the chains were expressed, and repeated analysis to obtain absolute values for the copy numbers of each transcript showed that HC5 was consistently very highly expressed compared with minimal expression of either HC1 or HC2 (Fig. 7A). HC1 and HC2 proteins were not detected on the cells either by immunocytochemistry or Western blotting of cell lysates (not shown). In contrast, HC5 protein expression was examined by Western blot of cell lysates, and, although low in unstimulated cells, it was strongly induced by TGFβ1 treatment (Fig. 7B). Following treatment of the fibroblasts with a low concentration of trypsin before lysis, HC5 became undetectable by Western blotting, suggesting that the majority of the HC5 detected in the original lysates was extracellular and associated with the cell surface. Cell surface expression of HC5 was confirmed by immunocytochemistry in both untreated and TGFβ1-treated cells (Fig. 7C) and was reduced by exposure of the cells to Streptomyces hyaluronidase.
FIGURE 7.
A, the absolute copy number of each IαI heavy chain was determined by reading standard curves prepared by adding known numbers of molecules to the qPCR. Heavy chain 5 was shown to be the predominant isoform. Results are mean ± S.E. of four independent experiments. B, human lung fibroblasts were growth-arrested and were then cultured in the presence or absence of TGFβ1 (10 ng/ml) for 72 h. The cells were lysed with radioimmune precipitation assay buffer, and 30 μg of protein was separated on 7.5% polyacrylamide gels. GAPDH antibody was used as a loading control. Blots shown are representative of four experiments. Top, Western blot showing that the amount of IαI heavy chain 5 present was markedly increased following stimulation with TGFβ1 Bottom, following TGFβ1 incubation, the cells were treated with 10 μg/ml trypsin in PBS for 10 min at room temperature as shown previously to remove any cell surface/CD44-associated HA (43). C, cells were incubated with or without TGFβ1 (10 ng/ml) for 72 h. The cells were then either left untreated or exposed to Streptomyces hyaluronidase (0.4 units in 400 μl of PBS for 2 h). All cells were then fixed with 4% paraformaldehyde in PBS for 20 min and stained with anti-HC5 antibody followed by FITC-conjugated anti-rabbit IgG to demonstrate the presence of heavy chain 5 on the cell surface. Data shown are representative of three independent experiments.
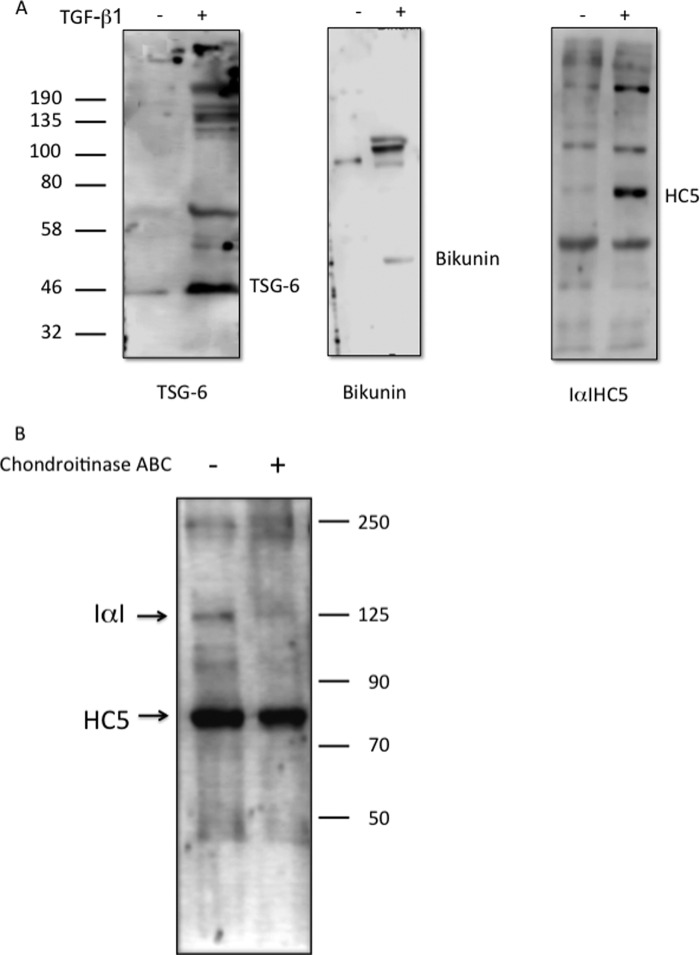
The data generated above all suggest the importance of interactions between TSG-6 and HC5 for controlling the change in phenotype of the fibroblasts. To investigate whether these proteins could be found in higher molecular weight complexes, supernatants from the cells incubated in serum-free medium or treated with TGFβ1 were probed for TSG-6, HC5, and bikunin. TGFβ1 induced the expression of all three proteins in the cell supernatants (Fig. 8A), and they were all also present as components of higher molecular weight complexes. Treatment of supernatants with chondroitinase ABC resulted in a reduction of HC5-containing complexes (molecular mass approximately 100–130 kDa) without affecting the non-complexed HC5 band, confirming that HC5 is present in IαI complexes with bikunin.
FIGURE 8.
Human lung fibroblasts were growth-arrested and were then incubated in the presence (+) or absence (−) of TGFβ1 (10 ng/ml) for 72 h. A, the culture supernatant was collected and concentrated using a Micron centrifugal filter (Millipore) with a 3-kDa cut-off. The equivalent of 1 ml of supernatant was then loaded into each well of a 7.5% polyacrylamide gel. Western blotting was carried out to demonstrate the presence of the native proteins and higher molecular weight complexes of TSG-6, bikunin, and heavy chain 5. B, TGFβ1-treated cells were lysed and split into two identical samples of 30 μg. One half was treated with chondroitinase ABC (0.166 units/ml; Sigma-Aldrich), and both samples were incubated at 37 °C overnight. The samples were then separated on a 7.5% polyacrylamide gel, and Western blotting was carried out using anti-HC5 to demonstrate the degradation of the higher molecular weight complexes containing heavy chain 5. All blots in A and B are representative of three independent experiments.
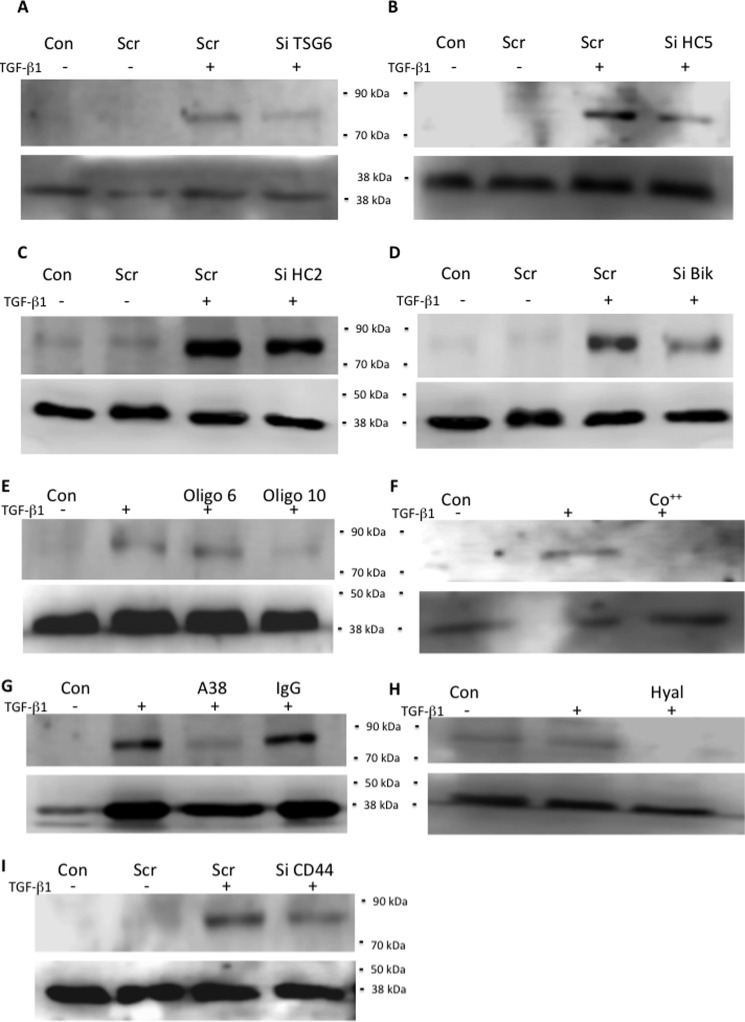
To confirm the importance of TSG-6 in making HC5 an integral part of the pericellular HA matrix, cells were treated with Si TSG-6 and then lysed, and Western blotting was performed for HC5 (Fig. 9A). There was a marked reduction in HC5 levels similar to the effect of direct knockdown of HC5 (Fig. 9B), when compared with treatment with scrambled control or Si HC2 (Fig. 9C). Treatment with Si bikunin also markedly reduced HC5 levels, suggesting that HC5 must be part of an IαI or pre-αI complex to be functional (Fig. 9D). To confirm that active TSG-6 was needed for HC5 incorporation into the pericellular matrix, cells were treated with oligo-HA (Fig. 9E), cobalt (Fig. 9F), or mAb A38 (Fig. 9G). All reduced HC5 protein levels markedly. To examine the mechanisms holding HC5 at the cell surface, cells were treated with hyaluronidase (Fig. 9H) or with Si CD44 (Fig. 9I). Both treatments reduced HC5 levels detected by Western blot, demonstrating that an association with HA and the presence of this HA binding receptor were essential for anchoring HC5 at the cell surface.
FIGURE 9.
Human lung fibroblasts were growth-arrested and were then stimulated with TGFβ1 (10 ng/ml) or placed in serum-free medium only (Con) for 72 h. The cells were then lysed (150 μl of radioimmune precipitation assay buffer/well of a 6-well plate), and 30 μg was separated on a 7.5% polyacrylamide gel. They were then subjected to Western blotting for HC5 with GAPDH used as a loading control. Blots shown are representative of four independent experiments. Upper panels show HC5, and lower panels show GAPDH. A–D, cells were cultured in the absence (−) or presence (+) of TGFβ1 (10 ng/ml) plus either scrambled siRNA (scr) or target-specific siRNA: Si TSG-6 (A), Si HC5 (B), Si HC2 (C), and Si bikunin (D). E–I, cells were cultured in the absence (Con) or presence of TGFβ1 (10 ng/ml) together with either oligosaccharides of HA (E), cobalt chloride (1 mm) (F), or mAb A38 (50 μg/ml) (G), compared with non-immune IgG or Streptomyces hyaluronidase (0.4 units in 400 μl of PBS for 2 h) (H). Si CD44 was also used as above to specifically knock down CD44 compared with scrambled control (I) as above (A–D). Oligo 6 and Oligo 10, hyaluronan oligosaccharides (6 and 10 monosaccharide units, respectively).
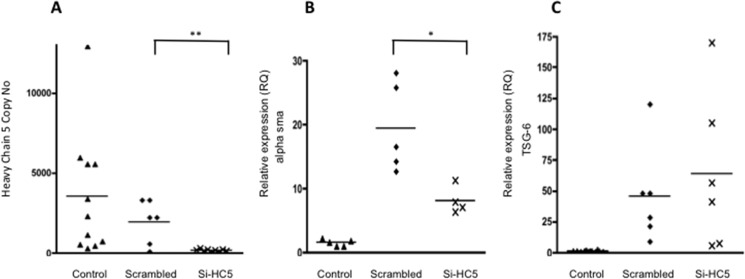
HC5 knockdown was used to investigate the functional significance of the HC5 associations at the cell surface described above. Knockdown of HC5 was >95% (Fig. 10A) and resulted in a reduction in TGFβ1-simulated α-SMA expression of >50% (Fig. 10B). There was no effect of knockdown on TSG-6 expression (Fig. 10C), confirming the independence of the HA-dependent differentiation pathway and the TSG-6 induction pathway.
FIGURE 10.
Human lung fibroblasts were growth-arrested and were then stimulated with TGFβ1 (10 ng/ml) in the presence or absence of Si HC5. The RNA was collected after 72 h of incubation, and the effect of Si HC5 on copy number of heavy chain 5 (A) and the RQ of α-SMA (B) or TSG-6 (C) was determined by RT-qPCR. Data shown are mean and scatter of values from three independent experiments. *, p < 0.05; **, p < 0.005 for Si HC5 compared with scrambled.
Discussion
Hyaluronan is a ubiquitous connective tissue glycosaminoglycan that, in vivo, is present as a high molecular mass component of the extracellular matrix. In addition to its role in providing cellular support, it is known that under normal circumstances, HA regulates cell-cell adhesion, migration, proliferation, differentiation, and the movement of interstitial fluid and macromolecules (reviewed in Ref. 29). As a result, it is likely to be an important contributor to and a regulator of tissue remodeling. Our primary interest is understanding the pathogenesis of progressive fibrosis, particularly in the context of renal disease. In a study of patients with biopsy-proven diabetic nephropathy, we have previously demonstrated an association between the deposition of hyaluronan and the degree of renal fibrosis, which is a known predictor of clinical outcome (30). This led us to examine the role of HA in regulation of fibroblast phenotype.
We have demonstrated that the phenotypic effects of TGFβ1 in fibroblasts are orchestrated by its stimulation of HA synthesis and accumulation in a pericellular coat (12–14, 26). The formation of this coat and the resulting change in cell phenotype is shown in this study to be dependent on the induction and activity of the hyaladherin, TSG-6. Inhibition of TSG-6 expression or its HC transfer ability strongly inhibited the phenotypic change of fibroblasts to myofibroblasts. Many cell types assemble HA in an organized pericellular coat in vitro, in which the HA is anchored to the cell surface by CD44 (16, 31–35). Our recent studies in proximal tubular epithelial cells have investigated the roles of the IαI family of hyaladherin proteins, together with TSG-6 and the HA-binding proteoglycans bikunin and versican, in the macromolecular assembly of HA by PTC (16, 25, 35, 36). The results demonstrated that the TSG-6-mediated formation of IαI HC·HA complexes was critical for the formation of a pericellular HA matrix.
The data included in this paper suggest that it is the HC·HA catalytic activity of TSG-6 that is the key for the phenotypic activation of fibroblasts. Both TSG-6·HC formation and the subsequent HC transfer onto HA are metal ion-dependent. Previous studies have demonstrated the reaction to be dependent on either Mg2+ or Mn2+ and to be inhibited by Co2+ (19). Inhibition of TGFβ1-dependent phenotypic activation without any effect on the induction of TSG-6 itself is therefore consistent with an HC transfer function of TSG-6 orchestrating matrix assembly and phenotypic activation. This is further supported by the inhibition of phenotypic activation by the stimulation of fibroblasts with TGFβ1 in the presence of the rat anti-human monoclonal antibody A38. This antibody has previously been demonstrated to block HA binding to TSG-6 (27) and inhibit the formation of TSG-6·HC complexes in vitro (28). The transfer of HC requires an intact IαI protein, as evidenced by female mice lacking the bikunin gene, which express the HCs but are unable to assemble neither IαI nor pre-αI and which do not form cumulus·oocyte HC·HA complexes (37, 38). Our data using bikunin RNA silencing to inhibit the phenotypic activation of fibroblasts are therefore again compatible with the HC transfer catalytic activity of TSG-6 being a key component of acquisition of the myofibroblast phenotype.
TSG-6-mediated organization of the HA matrix has been reported previously to alter the interaction of HA with its principle cell surface receptor CD44. For example, in solution, TSG-6·HA complexes enhance or induce binding of HA to cell surface CD44 (39). It is well established that an organized pericellular HA coat is anchored to cell surface CD44 (34). We have previously demonstrated that phenotypic activation of fibroblasts requires HA-driven formation of CD44·EGFR complexes, and this, together with the data in this paper, suggests that TSG-6-dependent HC-HA interactions lead to the formation of a pericellular HA matrix, which facilitates CD44 relocalization within the cell membrane. This receptor complex then activates a downstream signaling cascade that is responsible for reorganization of the actin cytoskeleton (24).
Although this study demonstrates the importance of TSG-6-dependent HC-HA interactions, rather than the well described HC1–HC3 involvement seen in epithelial cells, the essential and necessary HC for triggering HA transfer and the phenotypic change in fibroblasts was HC5. Its importance was further emphasized by the fact that in cell supernatants, HC5 was only found in high molecular weight complexes and not as a monomer. In addition, whereas the mRNA levels of HC5 decrease following TGFβ1 treatment, the cell-associated protein levels increase, suggesting that the most important regulation of HC5 is not transcriptional but post-transcriptional through its incorporation into the HA pericellular matrix of the cell.
HC5 plays a role in tumor suppression with dysregulated or reduced expression of HC5 directly linked to tumor development (22, 40, 41) and its overexpression leading to suppression of cell migration and colony spreading (42). A recent study also found that HC5 was the major heavy chain expressed in skin, predominantly due to its production by dermal fibroblasts (43). The authors conclude that HC5 may play an important role in inflammation through its interaction with HA. To our knowledge, this report of Huth et al. (43) is the first description of a role for HC5 outside the area of tumorigenesis and metastasis and adds to the important role for HC5 described in our current study.
In summary, much has been previously published on the anti-inflammatory role of the hyaladherin TSG-6. The data in this paper provide evidence that it is also involved in driving a profibrotic response through its catalytic transfer of HC5 to HA and that the mechanisms regulating TSG-6 and HA synthesis during this profibrotic, TGFβ1-dependent process are distinct.
Author Contributions
J. M. performed the majority of the experimental work with experimental and technical advice from S. M., T. B., E. W., and A. M. A. O. P. and R. S. jointly supervised the work. R. S. and J. M. wrote the manuscript with contributions from A. O. P.
This work was supported by Kidney Wales Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
- α-SMA
- α-smooth-muscle actin
- EGFR
- epidermal growth factor receptor
- IαI
- inter-α-inhibitor
- TSG-6
- tumor necrosis factor-stimulated gene-6
- HC
- IαI heavy chain
- HA
- hyaluronan
- qPCR
- quantitative PCR
- FnEDR
- EDA isoform of fibronectin
- oligo-HA
- hyaluronan oligosaccharide
- ITIH
- inter-α-trypsin inhibitor heavy chain
- RQ
- relative quantification.
References
- 1. Gabbiani G. (2003) The myofibroblast in wound healing and fibrocontractive diseases. J. Pathol. 200, 500–503 [DOI] [PubMed] [Google Scholar]
- 2. Tomasek J. J., Gabbiani G., Hinz B., Chaponnier C., and Brown R. A. (2002) Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 3, 349–363 [DOI] [PubMed] [Google Scholar]
- 3. Eddy A. A. (2005) Progression in chronic kidney disease. Adv. Chronic Kidney Dis. 12, 353–365 [DOI] [PubMed] [Google Scholar]
- 4. Essawy M., Soylemezoglu O., Muchaneta-Kubara E. C., Shortland J., Brown C. B., and el Nahas A. M. (1997) Myofibroblasts and the progression of diabetic nephropathy. Nephrol. Dial. Transplant. 12, 43–50 [DOI] [PubMed] [Google Scholar]
- 5. Goumenos D., Tsomi K., Iatrou C., Oldroyd S., Sungur A., Papaioannides D., Moustakas G., Ziroyannis P., Mountokalakis T., and El Nahas A. (1998) Myofibroblasts and the progression of crescentic glomerulonephritis. Nephrol. Dial. Transplant. 13, 1652–1661 [DOI] [PubMed] [Google Scholar]
- 6. Goumenos D. S., Brown C. B., Shortland J., and el Nahas A. M. (1994) Myofibroblasts, predictors of progression of mesangial IgA nephropathy? Nephrol. Dial. Transplant. 9, 1418–1425 [PubMed] [Google Scholar]
- 7. Roberts I. S., Burrows C., Shanks J. H., Venning M., and McWilliam L. J. (1997) Interstitial myofibroblasts: predictors of progression in membranous nephropathy. J. Clin. Pathol. 50, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desmoulière A., Darby I. A., and Gabbiani G. (2003) Normal and pathologic soft tissue remodeling: the role of the myofibroblast, with special emphasis on liver and kidney fibrosis. Lab. Invest. 83, 1689–1707 [DOI] [PubMed] [Google Scholar]
- 9. Kuhn C., and McDonald J. A. (1991) The role of the myofibroblast in idiopathic pulmonary fibrosis. Am. J. Pathol. 138, 1257–1265 [PMC free article] [PubMed] [Google Scholar]
- 10. Evans R. A., Tian Y. C., Steadman R., and Phillips A. O. (2003) TGF-β1-mediated fibroblast-myofibroblast terminal differentiation: the role of Smad proteins. Exp. Cell Res. 282, 90–100 [DOI] [PubMed] [Google Scholar]
- 11. Vaughan M. B., Howard E. W., and Tomasek J. J. (2000) Transforming growth factor β1 promotes the morphological and functional differentiation of the myofibroblast. Exp. Cell Res. 257, 180–189 [DOI] [PubMed] [Google Scholar]
- 12. Meran S., Thomas D., Stephens P., Martin J., Bowen T., Phillips A., and Steadman R. (2007) Involvement of hyaluronan in regulation of fibroblast phenotype. J. Biol. Chem. 282, 25687–25697 [DOI] [PubMed] [Google Scholar]
- 13. Simpson R. M., Meran S., Thomas D., Stephens P., Bowen T., Steadman R., and Phillips A. (2009) Age-related changes in peri-cellular hyaluronan organisation leads to impaired dermal fibroblast to myofibroblast differentiation. Am. J. Pathol. 175, 1915–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simpson R. M. L., Wells A., Thomas D., Stephens P., Steadman R., and Phillips A. (2010) Ageing fibroblasts resist phenotypic maturation due to impaired hyaluronan-dependent CD44/EGF receptor signaling. Am. J. Pathol. 176, 1215–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fülöp C., Szántó S., Mukhopadhyay D., Bárdos T., Kamath R. V., Rugg M. S., Day A. J., Salustri A., Hascall V. C., Glant T. T., and Mikecz K. (2003) Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development 130, 2253–2261 [DOI] [PubMed] [Google Scholar]
- 16. Selbi W., Day A. J., Rugg M. S., Fülöp C., de la Motte C. A., Bowen T., Hascall V. C., and Phillips A. O. (2006) Over-expression of hyaluronan synthase 2 alters hyaluronan distribution and function in proximal tubular epithelial cells. J. Am. Soc. Nephrol. 17, 1553–1567 [DOI] [PubMed] [Google Scholar]
- 17. Baranova N. S., Nilebäck E., Haller F. M., Briggs D. C., Svedhem S., Day A. J., and Richter R. P. (2011) The inflammation-associated protein TSG-6 cross-links hyaluronan via hyaluronan-induced TSG-6 oligomers. J. Biol. Chem. 286, 25675–25686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blom A. M., Mörgelin M., Oyen M., Jarvet J., and Fries E. (1999) Structural characterization of inter-α-inhibitor. J. Biol. Chem. 274, 298–304 [DOI] [PubMed] [Google Scholar]
- 19. Rugg M. S., Willis A. C., Mukhopadhyay D., Hascall V. C., Fries E., Fülöp C., Milner C. M., and Day A. J. (2005) Characterization of complexes formed between TSG-6 and inter-α-inhibitor that act as intermediates in the covalent transfer of heavy chains on to hyaluronan. J. Biol. Chem. 280, 25674–25686 [DOI] [PubMed] [Google Scholar]
- 20. Sanggaard K. W., Sonne-Schmidt C. S., Krogager T. P., Lorentzen K. A., Wisniewski H.-G., Thøgersen I. B., and Enghild J. J. (2008) The transfer of heavy chains from bikunin proteins to hyaluronan requires both TSG-6 and HC2. J. Biol. Chem. 283, 18530–18537 [DOI] [PubMed] [Google Scholar]
- 21. Choi-Miura N. H., Takahashi K., Yoda M., Saito K., Hori M., Ozaki H., Mazda T., and Tomita M. (2000) The novel acute phase protein, IHRP, inhibits actin polymerization and phagocytosis of polymorphonuclear cells. Inflamm Res. 49, 305–310 [DOI] [PubMed] [Google Scholar]
- 22. Himmelfarb M., Klopocki E., Grube S., Staub E., Klaman I., Hinzmann B., Kristiansen G., Rosenthal A., Dürst M., and Dahl E. (2004) ITIH5, a novel member of the inter-α-trypsin inhibitor heavy chain family is downregulated in breast cancer. Cancer Lett. 204, 69–77 [DOI] [PubMed] [Google Scholar]
- 23. Clark H. F., Gurney A. L., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Currell B., Deuel B., Dowd P., Eaton D., Foster J., et al. (2003) The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 13, 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Midgley A. C., Rogers M., Hallett M. B., Clayton A., Bowen T., Phillips A. O., and Steadman R. (2013) Transforming growth factor-β1 (TGF-β1)-stimulated fibroblast to myofibroblast differentiation is mediated by hyaluronan (HA)-facilitated epidermal growth factor receptor (EGFR) and CD44 co-localization in lipid rafts. J. Biol. Chem. 288, 14824–14838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bommaya G., Meran S., Krupa A., Phillips A. O., and Steadman R. (2011) Tumour necrosis factor-stimulated gene (TSG)-6 controls epithelial-mesenchymal transition of proximal tubular epithelial cells. Int J Biochem. Cell Biol. 43, 1739–1746 [DOI] [PubMed] [Google Scholar]
- 26. Webber J., Meran S., Steadman R., and Phillips A. (2009) Hyaluronan orchestrates of TGFβ1-dependent maintenance of myofibroblast phenotype. J. Biol. Chem. 284, 9083–9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lesley J., English N. M., Gál I., Mikecz K., Day A. J., and Hyman R. (2002) Hyaluronan binding properties of a CD44 chimera containing the link module of TSG-6. J. Biol. Chem. 277, 26600–26608 [DOI] [PubMed] [Google Scholar]
- 28. Ochsner S. A., Day A. J., Rugg M. S., Breyer R. M., Gomer R. H., and Richards J. S. (2003) Disrupted function of tumor necrosis factor-α-stimulated gene 6 blocks cumulus cell-oocyte complex expansion. Endocrinology 144, 4376–4384 [DOI] [PubMed] [Google Scholar]
- 29. Jiang D., Liang J., and Noble P. W. (2011) Hyaluronan as an immune regulator in human diseases. Physiol. Rev. 91, 221–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lewis A., Steadman R., Manley P., Craig K., de la Motte C., Hascall V., and Phillips A. O. (2008) Diabetic nephropathy, inflammation, hyaluronan and interstitial fibrosis. Histol. Histopathol. 23, 731–739 [DOI] [PubMed] [Google Scholar]
- 31. Banerji S., Wright A. J., Noble M., Mahoney D. J., Campbell I. D., Day A. J., and Jackson D. G. (2007) Structures of the Cd44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 14, 234–239 [DOI] [PubMed] [Google Scholar]
- 32. Day A. J., and de la Motte C. A. (2005) Hyaluronan cross-linking: a protective mechanism in inflammation? Trends Immunol. 26, 637–643 [DOI] [PubMed] [Google Scholar]
- 33. Evanko S. P., Tammi M. I., Tammi R. H., and Wight T. N. (2007) Hyaluronan-dependent pericellular matrix. Adv. Drug Deliv. Rev. 59, 1351–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Knudson W., Aguiar D. J., Hua Q., and Knudson C. B. (1996) CD-44 anchored hyaluronan-rich pericellular matrices: an ultrastructural and biochemical analysis. Exp. Cell Res. 228, 216–228 [DOI] [PubMed] [Google Scholar]
- 35. Selbi W., de la Motte C. A., Hascall V. C., Day A. J., Bowen T., and Phillips A. O. (2006) Characterization of hyaluronan cable structure and function in renal proximal tubular epithelial cells. Kidney Int. 70, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 36. Selbi W., de la Motte C., Hascall V., and Phillips A. (2004) BMP-7 modulates hyaluronan-mediated proximal tubular cell-monocyte interaction. J. Am. Soc. Nephrol. 15, 1199–1211 [DOI] [PubMed] [Google Scholar]
- 37. Sato H., Kajikawa S., Kuroda S., Horisawa Y., Nakamura N., Kaga N., Kakinuma C., Kato K., Morishita H., Niwa H., and Miyazaki J. (2001) Impaired fertility in female mice lacking urinary trypsin inhibitor. Biochem. Biophys. Res. Commun. 281, 1154–1160 [DOI] [PubMed] [Google Scholar]
- 38. Zhuo L., Yoneda M., Zhao M., Yingsung W., Yoshida N., Kitagawa Y., Kawamura K., Suzuki T., and Kimata K. (2001) Defect in SHAP-hyaluronan complex causes severe female infertility: a study by inactivation of the bikunin gene in mice. J. Biol. Chem. 276, 7693–7696 [DOI] [PubMed] [Google Scholar]
- 39. Lesley J., Gál I., Mahoney D. J., Cordell M. R., Rugg M. S., Hyman R., Day A. J., and Mikecz K. (2004) TSG-6 modulates the interaction between hyaluronan and cell surface CD44. J. Biol. Chem. 279, 25745–25754 [DOI] [PubMed] [Google Scholar]
- 40. Hamm A., Veeck J., Bektas N., Wild P. J., Hartmann A., Heindrichs U., Kristiansen G., Werbowetski-Ogilvie T., Del Maestro R., Knuechel R., and Dahl E. (2008) Frequent expression loss of inter-α-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a systematic expression analysis. BMC Cancer 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Veeck J., Chorovicer M., Naami A., Breuer E., Zafrakas M., Bektas N., Dürst M., Kristiansen G., Wild P. J., Hartmann A., Knuechel R., and Dahl E. (2008) The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node-negative breast cancer and its aberrant expression is caused by promoter hypermethylation. Oncogene 27, 865–876 [DOI] [PubMed] [Google Scholar]
- 42. Rose M., Gaisa N. T., Antony P., Fiedler D., Heidenreich A., Otto W., Denzinger S., Bertz S., Hartmann A., Karl A., Knüchel R., and Dahl E. (2014) Epigenetic inactivation of ITIH5 promotes bladder cancer progression and predicts early relapse of pT1 high-grade urothelial tumours. Carcinogenesis 35, 727–736 [DOI] [PubMed] [Google Scholar]
- 43. Huth S., Heise R., Vetter-Kauczok C. S., Skazik C., Marquardt Y., Czaja K., Knüchel R., Merk H. F., Dahl E., and Baron J. M. (2015) Inter-α-inhibitor heavy chain 5 (ITIH5) is overexpressed in inflammatory skin diseases and affects epidermal morphology in constitutive knockout mice and murine 3D skin models. Exp. Dermatol. 24, 663–668 [DOI] [PubMed] [Google Scholar]