Abstract
Atypical protein kinase C (aPKC) isozymes modulate insulin signaling and cell polarity, but how their activity is controlled in cells is not well understood. These enzymes are constitutively phosphorylated, insensitive to second messengers, and have relatively low activity. Here we show that protein scaffolds not only localize but also differentially control the catalytic activity of the aPKC PKCζ, thus promoting activity toward localized substrates and restricting activity toward global substrates. Using cellular substrate readouts and scaffolded activity reporters in live cell imaging, we show that PKCζ has highly localized and differentially controlled activity on the scaffolds p62 and Par6. Both scaffolds tether aPKC in an active conformation as assessed through pharmacological inhibition of basal activity, monitored using a genetically encoded reporter for PKC activity. However, binding to Par6 is of higher affinity and is more effective in locking PKCζ in an active conformation. FRET-based translocation assays reveal that insulin promotes the association of both p62 and aPKC with the insulin-regulated scaffold IRS-1. Using the aPKC substrate MARK2 as another readout for activity, we show that overexpression of IRS-1 reduces the phosphorylation of MARK2 and enhances its plasma membrane localization, indicating sequestration of aPKC by IRS-1 away from MARK2. These results are consistent with scaffolds serving as allosteric activators of aPKCs, tethering them in an active conformation near specific substrates. Thus, signaling of these intrinsically low activity kinases is kept at a minimum in the absence of scaffolding interactions, which position the enzymes for stoichiometric phosphorylation of substrates co-localized on the same protein scaffold.
Keywords: insulin, insulin receptor substrate 1 (IRS-1), p62 (sequestosome 1(SQSTM1)), protein kinase C (PKC), scaffold protein, MARK2, Par6, autoinhibition
Introduction
The coordination of signal transduction by protein scaffolds controls downstream signaling for a multitude of protein kinases (1). Protein scaffolds locally enrich the kinase, positioning it near or sequestering it away from substrates to allow specificity and fidelity in cell signaling. In addition, binding to scaffold proteins can impact the conformation of kinases, tuning their signaling output. The atypical PKC isozymes (aPKC)3 are an example of a kinase whose interaction with a scaffold tethers them in a signaling-competent conformation.
aPKCs make up one of three classes of the PKC family of enzymes. These classes are the canonical diacylglycerol (DAG)-regulated conventional (Ca2+-dependent; α, β, and γ) and novel (Ca2+-independent but also DAG-regulated; δ, ϵ, η, and θ) PKCs and the aPKCs (ζ and ι/λ), which are regulated by neither DAG nor Ca2+ (2–4). aPKCs share the same general architecture of other family members, with a C-terminal kinase domain whose function is controlled by determinants in the N-terminal regulatory moiety. Specifically, all PKCs have an autoinhibitory pseudosubstrate segment immediately preceding a C1 domain that masks the kinase domain to maintain the enzyme in an autoinhibited conformation. In addition, all PKCs are constitutively phosphorylated following their biosynthesis (3, 5). aPKCs differ from the DAG-regulated PKCs in that 1) they are not regulated by second messengers, 2) their catalytic activity is an order of magnitude lower than the DAG-regulated PKCs (5), and 3) they have a protein-protein interacting Phox and Bem1 (PB1) domain at their N terminus. Additionally, they have a type III PDZ ligand at the end of their C terminus (3). For the DAG-regulated PKCs, binding of second messengers results in release of the pseudosubstrate. In the case of aPKCs, binding to protein scaffolds promotes the release of the atypical pseudosubstrate.
Two scaffolds that regulate the activity of aPKCs are p62 (also known as sequestosome 1, SQSTM1) (6) and partitioning-defective protein 6 (Par6), a cell polarity regulator (7). Binding to these scaffolds promotes the open and active conformation of the aPKCs. In the case of p62, an acidic surface unique to its PB1 domain binds the basic pseudosubstrate of aPKCs, tethering it away from the substrate binding cavity to engage aPKC in an active conformation (6). Binding of aPKC to Par6 also displaces the pseudosubstrate to tether aPKC in an open and signaling-competent conformation (7). Par6 is well characterized as a signaling platform to localize aPKC near substrates such as the cell polarity regulators Par3 and Lgl (8–13). As a signaling hub with multiple interacting domains and partners (14, 15), p62 may also function to localize aPKC near its targets, although specific substrates on this platform remain to be identified.
A key function of aPKC is the regulation of insulin-stimulated glucose transport (16–21), yet the mechanism for how it transduces this signal is poorly understood. We have recently shown that PKCζ is not regulated by the insulin-induced PI3-kinase pathway that activates Akt (5), a mechanism previously proposed to regulate aPKC activity (22–24). Rather, scaffold interactions may be the critical regulators of aPKC function in insulin signaling. In this regard, insulin promotes a multitude of phosphorylation-dependent protein interactions, many centralized around the insulin receptor substrate (IRS-1/2) (25). The p62 scaffold has recently been shown to interact with IRS-1 in an insulin-dependent manner (26, 27) and is also known to have functions in metabolism as p62-deficient mice exhibit an insulin-resistant, obese, and diabetic phenotype (28, 29). A metabolic phenotype is also observed in mice lacking microtubule affinity regulating kinase 2 (MARK2), an aPKC substrate (30, 31). These animals are insulin-hypersensitive and resistant to weight gain when placed on a high fat diet (32, 33). MARK2 has a well characterized role in mediating cell polarity, but the mechanism by which it regulates insulin signaling is less understood. How aPKCs control substrate phosphorylation and downstream signaling in response to insulin remains to be elucidated.
Here, we address the role of protein scaffolds in controlling the activity of aPKCs. Specifically, we examine the scaffold-associated activity of full-length PKCζ or constructs lacking the pseudosubstrate, PB1, or the entire N-terminal regulatory moiety using the C kinase activity reporter (CKAR) (34) fused to the PB1 domain of p62 or Par6. Basal activity at each scaffold is assessed by monitoring the drop in activity following addition of a validated aPKC active site inhibitor, PZ09. Our data reveal that aPKC is differentially bound and activated on the two PB1 scaffolds; it binds with higher affinity to Par6, which tethers it in a fully active conformation, compared with binding to p62 which is of lower affinity and which tethers a partially active enzyme. Furthermore, we demonstrate the ability of IRS-1 to regulate the phosphorylation and localization of the aPKC substrate MARK2. Finally, a fluorescent resonance energy transfer (FRET) assay reveals that insulin promotes the association of PKCζ with p62 in addition to p62 and PKCζ with IRS-1. Our data support a model in which scaffolds regulate aPKC signaling by not only providing a signaling scaffold but by differentially tuning activity.
Experimental Procedures
Materials
Staurosporine, forskolin, and Gö6983 were purchased from Calbiochem. PZ09 was a kind gift from Dr. Christopher Hulme and Dr. Sourav Ghosh. Insulin was purchased from Sigma. The following antibodies were purchased from Santa Cruz Biotechnology: anti-p410 PKCζ (sc-12894-R), anti-total PKCζ (sc-216), and anti-MARK2 (sc-98800). Antibodies for p308 Akt (9275), total Akt (9272), PKC serine substrate (2261 lot no. 18), and GAPDH (2118) were purchased from Cell Signaling Technology. The anti-HA (MMS-101P) antibody was from Biolegend. The anti-p595 MARK2 antibody was from Abcam (ab34751). The anti-GFP antibody was from Clontech (catalog no. 632376). The Ser(P)-24 p62 antibody was a kind gift from Dr. George Baillie. HRP-conjugated goat anti-mouse IgG and goat anti-rabbit IgG were from Calbiochem (catalog nos. 401215 and 401315).
Cell Lines and Plasmid Constructs
Human PKCζ and PKCα cDNA were gifts from Dr. Tony Hunter. Human p62 was a gift from Dr. Jorge Moscat. Human MARK2 and human IRS-1 were purchased from Addgene (plasmid no. 66706 and plasmid no. 11025, respectively), whereas human Par6α was a gift from Dr. Sourav Ghosh, originally from Addgene (plasmid no. 15474). The protein CKAR construct was described previously (34, 35). Human PKCζ, PKCα, MARK2, Par6α, IRS-1, and p62 constructs were cloned into the pDONR vector and subsequently recombined with various pDEST vectors constructed in-house to make fusion proteins with HA, mCherry, CKAR, CFP, or YFP tags at the N terminus in pCDNA3 vectors for mammalian cell expression using the Gateway cloning system (Invitrogen). Human PKMζ (residues 183–592) of PKCζ was inserted into pDONR for Gateway recombination. The PB1 domains of both human p62 (residues 3–102) and human Par6α (residues 1–101) were cloned into pDONR and inserted into a Gateway expression vector for expression with N-terminal CFP and CKAR tags. Domain deletion mutations were constructed in PKCζ as follows: ΔPS (deleted pseudosubstrate, residues 113–130) and ΔPB1 (deleted aPKC PB1 domain, residues 25–106). Point and domain mutations were made using QuikChange site-directed mutagenesis (Stratagene). The catalytically inactive spine mutant (V266F (5)) was used for the kinase-dead controls of PKCζ and PKMζ.
Cell Culture and Transfection
Mammalian cells were maintained at 37 °C in 5% CO2. CHO-IR cells were grown in 1× DMEM/F-12 50:50, (Cellgro) supplemented with 5% dialyzed fetal bovine serum (FBS, Atlanta Biologics), 1% penicillin/streptomycin, and 50 μg/ml geneticin (Gibco). COS-7 cells were grown in 1× DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Mammalian cells were transiently transfected using JetPRIME (Polyplus Transfection).
Immunoprecipitation and Immunoblotting
Cells transfected with HA-tagged kinases were rinsed in PBS and lysed in Buffer A (100 mm KCl, 50 mm Tris, 3 mm NaCl, 3.5 mm MgCl2, pH 7.3) with freshly added 1% protein-grade Triton X-100 (Calbiochem), 100 μm PMSF, 1 mm DTT, 2 mm benzamidine, 50 μg/ml leupeptin, 1 μm microcystin-LR, and 1 mm sodium orthovanadate. Soluble lysates were incubated with anti-HA antibody (Biolegend) for 1–2 h at 4 °C followed by incubation with protein A/G resin beads (Thermo Scientific) for 1–2 h at 4 °C. Protein-bound beads were washed three times with Buffer A prior to adding 25% sample buffer (62.5 mm Tris, 2% SDS, 10% glycerol, 20 μg/ml bromphenol blue, 2.86 m 2-mercaptoethanol), boiling at 100 °C, and performing SDS-PAGE. For experiments with immunoblotting only, cells were lysed in Buffer B (100 mm NaCl, 50 mm Tris, 10 mm Na4P2O7, 50 mm NaF, 1% Triton X-100, pH 7.2, plus inhibitors), and whole cell lysates were sonicated prior to adding 25% sample buffer, boiling at 100 °C, and performing SDS-PAGE. Gels were transferred to PVDF membrane and blocked with 5% BSA before incubating in antibodies.
Live Cell Fluorescence Imaging
COS-7 cells were plated onto sterilized glass coverslips in 35-mm imaging dishes, co-transfected with the indicated constructs, and imaged in Hanks' balanced salt solution supplemented with 1 mm CaCl2 ∼24 h post-transfection using a ×40 objective. For experiments with insulin stimulation, cells were serum-starved overnight prior to imaging. For experiments with forskolin treatment, cells were treated ∼24 h prior to imaging and transfected the day before. Kinase activity was monitored via intramolecular FRET of the activity reporters (CKAR or CKAR-PB1), and protein translocation was monitored via intermolecular FRET between the YFP-tagged protein and the CFP-tagged target using methods previously described (6, 36).
Statistical Analysis
All statistical analyses were performed using GraphPad Prism software.
Results
PZ09 Inhibits Atypical PKCs but Not Conventional PKCs in Cells
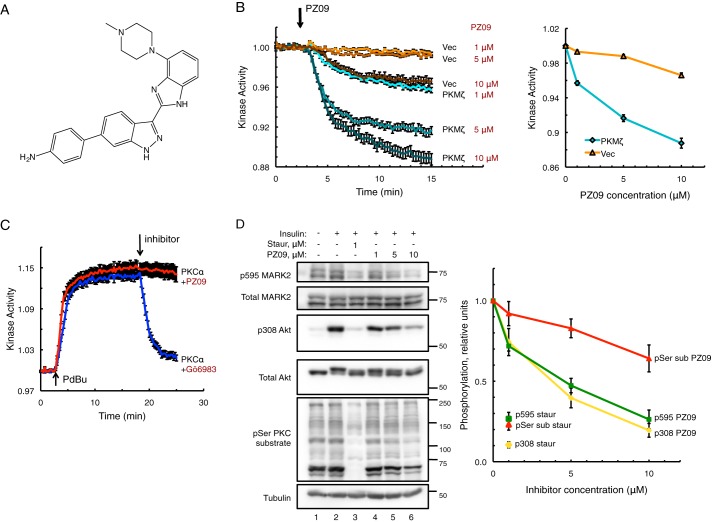
The toolbox of inhibitors that selectively inhibit atypical PKCs is scant compared with the collection of compounds that can effectively inhibit conventional and novel PKCs for pharmacological studies (37). Additionally, certain compounds previously claimed to be inhibitors of aPKC from in vitro studies (ZIP and chelerythrine (38, 39)) have been shown to be ineffective at inhibiting aPKC in cells (40). Given the need for an effective aPKC modulator to investigate its biochemical regulation, and the history within the field of in vitro efficacy not concurring with efficacy in cells, we set out to validate an active site inhibitor of aPKC, PZ09 (Fig. 1A), previously shown to inhibit aPKC in vitro (41) and already used for studies in vivo (42) for selective modulation of aPKC substrate readouts in cells. Using a genetically encoded FRET reporter, the C kinase activity reporter (CKAR) (34) previously validated for measurement of aPKC activity in real time in live cells (5, 6, 40), we examined inhibition of endogenous aPKC or mCherry-tagged constructs of aPKC by PZ09. The drop in activity following inhibitor addition serves as a measure of the constitutive activity of aPKC. Henceforth, this inhibitor-sensitive activity is referred to as “basal activity.” Fig. 1B shows the effect of increasing concentrations of PZ09 on PKMζ, an alternate transcript of PKCζ preferentially expressed in brain tissue that comprises only the kinase domain. PKMζ was effectively inhibited by PZ09, with an IC50 of ∼3 μm. The addition of inhibitor caused a small drop in FRET readout in mCherry-Vector (Vec)-transfected control cells, reflecting inhibition of endogenous aPKCs and background activity on CKAR by other kinases sensitive to PZ09 (Fig. 1B). The larger drop in FRET readout following equal PZ09 treatment for the PKMζ-transfected cells versus the Vec-transfected cells is indicative of higher basal activity for the overexpressed PKMζ on CKAR than for the endogenous kinases on CKAR, as each trace is normalized to its baseline value before inhibitor treatment. As an additional control, a kinase-dead version of mCherry-PKMζ with a catalytically inactive spine mutation V266F (5) was tested on CKAR and displayed the same response as the mCherry-Vec control (data not shown). To examine the selectivity of PZ09 in cells for aPKC versus conventional PKCs, cells expressing CKAR and mCherry-PKCα were treated with 200 nm phorbol dibutyrate to stimulate cPKC activity and then treated with either 5 μm PZ09 or 250 nm Gö6983, a potent inhibitor of cPKCs and novel PKCs (Fig. 1C). Gö6983 reversed stimulated PKCα activity (Fig. 1C, blue trace); in contrast 5 μm PZ09 had no effect on PKCα activity in cells (Fig. 1C, red trace), confirming previous selectivity results performed in vitro (41). Thus, monitoring PZ09-sensitive phosphorylation of CKAR is an effective tool to measure aPKC activity in cells.
FIGURE 1.
PZ09 inhibits atypical PKCs but not conventional PKCs in cells. A, chemical structure of PZ09. B, basal kinase activity measured via changes in CFP over FRET ratio using PKC-specific substrate reporter CKAR and mCherry-tagged PKMζ versus mCherry-Vec control in live COS-7 cells treated with increasing concentrations of aPKC inhibitor PZ09. The trace for each cell imaged was normalized to its t = 0-min baseline value and plotted as means ± S.E. Normalized FRET ratios were combined from three independent experiments (all PKMζ traces and Vec 1 μm and 10 μm traces) or five independent experiments for Vec 5 μm, with each experiment analyzing 6–12 selected cells. Inhibitory response curve is plotted as amplitude of drop from baseline activity at 15-min time point versus PZ09 concentration. C, stimulated kinase activity on CKAR of mCherry-PKCα after treatment with 200 nm phorbol dibutyrate (PDBu) followed by treatment with either cPKC/novel PKC-specific inhibitor Gö6983 (250 nm) or PZ09 (5 μm). The trace for each cell imaged was normalized to its t = 0-min baseline value, and normalized FRET ratios were combined from three independent experiments with analysis of 8–12 selected cells each and plotted as mean ± S.E. D, COS-7 cells were serum-starved overnight and treated with either DMSO, staurosporine (Staur) (1 μm), or increasing concentrations of PZ09 for 30 min prior to stimulation with 100 nm insulin for 10 min before lysis. Immunoblots show endogenous substrate phosphorylation representing aPKC inhibition (p595 MARK2), PDK1 inhibition (insulin-stimulated p308 Akt), and cPKC inhibition (pSer sub, using an antibody for a PKC-specific serine substrate sequence). Quantification of phosphorylated protein substrate over total protein normalized to DMSO + insulin control using n = 5 separate experiments plotted as mean ± S.E. versus inhibitor concentration (PZ09 or staurosporine) is shown. Phosphorylated PKC serine substrate was quantified as the intensity of the total band ensemble detected between 50 and 250 kDa divided by tubulin signal and normalized to DMSO + insulin control.
Higher Concentrations of PZ09 Have Off-target Effects on Endogenous PDK1 in Cells
Although in vitro inhibition studies of PZ09 have demonstrated its selectivity for aPKC over other PKCs (41), PZ09 also inhibited several other non-PKC kinases, including PDK1, PKA, and p70 S6K when used at 10 μm concentration. To examine the effects of increasing PZ09 concentrations on endogenous readouts of aPKC, PDK1, and cPKC, we pre-treated cells with PZ09 followed by stimulation with insulin prior to lysis to achieve optimal readout of PDK1 activity on the agonist-induced site p308 Akt (lane 2 versus lane 1, Fig. 1D). We then blotted for p595 MARK2 (aPKC substrate (30, 31)) and p308 Akt (PDK1 substrate (43)) or phosphorylated PKC serine substrate using an antibody specific for the PKC substrate recognition sequence and quantified each phosphorylation relative to total protein. As a control for general kinase inhibition, we also treated cells with 1 μm staurosporine (Fig. 1D, lane 3), an inhibitor we have previously used to examine aPKC activity in cells (40). Both aPKC and PDK1 were inhibited to the same extent by increasing concentrations of PZ09 up to 10 μm (Fig. 1D, lanes 4–6 compared with lane 2 control, p595, and p308 readouts, respectively), whereas cPKC experienced significantly less inhibition by PZ09 (Ser(P) PKC substrate readout). However, off-target effects of PZ09 on PDK1 and cPKC were considerably less than those of the general kinase inhibitor staurosporine used at 1 μm concentration (quantification, Fig. 1D).
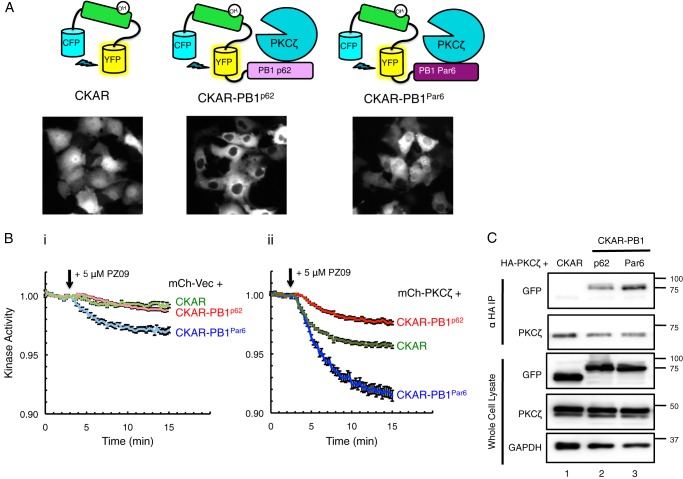
PKCζ Is Constitutively Active on CKAR Substrate Reporter Tethered to Interacting PB1 Domains of Scaffold Proteins p62 and Par6
To investigate the role of scaffolds in regulating the localized activity of PKCζ, we fused CKAR to either the PB1 domain of p62 or the PB1 domain of Par6α (Fig. 2A). We have previously used a fusion construct of CKAR tethered to full-length p62 to measure aPKC activity (6). Because this reporter had a tendency to form large clusters of aggregated p62 within the cells, we fused CKAR to just the PB1 domain of p62 or the PB1 of Par6α (which also forms aggregates when expressed in full-length form (7)) as the PB1 domain of each scaffold is the principal surface for aPKC binding (44, 45). Untethered CKAR was expressed throughout the cell; the CKAR-PB1p62 reporter became excluded from the nucleus, localizing within the cytosol in the same regions as PKCζ, with minimally visible puncta (Fig. 2A). The CKAR-PB1Par6 construct was only partially excluded from the nucleus (Fig. 2A). We next examined the PZ09-sensitive basal activity globally or on the p62 or Par6 scaffolds (Fig. 2B) in cells expressing either mCherry-Vec (representing activity of endogenous aPKC and background kinases on CKAR, Fig. 2B, panel i) or full-length mCherry-PKCζ (representing activity of overexpressed PKCζ on co-expressed CKAR, Fig. 2B, panel ii). Cells chosen for analysis had the same range of expression for the reporter and mCherry. PZ09 caused a small drop in the activity of endogenous global PKC activity (CKAR, Fig. 2, panel i) and activity on the p62 scaffold (CKAR-PB1p62, panel i), and a more significant drop in the activity was measured on the Par6 scaffold (CKAR-PB1Par6, Fig. 2, panel i). Cells overexpressing mCherry-PKCζ displayed much more significant drops in basal activity than for mCherry-Vec on each of the various reporters (compare Fig. 2, panel ii to panel i, same scale on each y axis), indicating aPKC-specific activity. The greatest inhibitor-sensitive activity of mCherry-PKCζ was also observed on the Par6 scaffold (CKAR-PB1Par6, Fig. 2, panel ii). Curiously, lower basal activity was detected on the CKAR-PB1p62 reporter compared with CKAR alone, although there was a pronounced drop in activity compared with its vector-transfected control (Fig. 2, panel ii versus panel i). The pronounced difference in basal activity readout by the two PB1 domain-tagged reporters prompted us to investigate whether the PB1 domain of Par6 was binding more tightly to PKCζ, thus recruiting more of the overexpressed enzyme to phosphorylate CKAR. Co-immunoprecipitation (co-IP) experiments shown in Fig. 2C revealed that PKCζ pulled down significantly more CKAR-PB1Par6 (lane 3) than CKAR-PB1p62 (lane 2) (2.5 ± 0.5-fold, p = 0.0187) as determined through quantification of anti-GFP signal (detecting CKAR) over anti-PKCζ signal from four independent experiments. PKCζ did not pull down the CKAR reporter alone (lane 1), confirming its non-interaction with the untethered substrate and reliance on the PB1 domain for scaffolding. These results reveal that aPKC is basally active at both the Par6 and p62 scaffolds, with significantly more effective binding and thus activity at the Par6 scaffold.
FIGURE 2.
PKCζ is constitutively active on CKAR substrate reporter tethered to interacting PB1 domains of scaffold proteins p62 and Par6. A, diagram of reporters constructed to measure aPKC activity: CKAR, CKAR-PB1p62, and CKAR-PB1Par6, with corresponding images of COS-7 cells transfected with each reporter showing its localization. B, basal kinase activity of mCherry-Vec (mCh-Vec) (panel i) and mCh-PKCζ (panel ii) on each reporter shown in A after treatment with 5 μm PZ09 in live COS-7 cells. The trace for each cell imaged was normalized to its t = 0-min baseline value and plotted as mean ± S.E. Normalized FRET ratios were combined from four independent experiments (CKAR-PB1Par6 traces) or five independent experiments (CKAR and CKAR-PB1p62 traces), with each experiment analyzing 4–12 selected cells. C, HA-PKCζ was co-expressed with CKAR, CKAR-PB1p62, or CKAR-PB1Par6 in COS-7 cells, immunoprecipitated (IP) from soluble lysates using anti-HA antibody, and blotted for co-IP of CKAR tag using anti-GFP antibody; whole cell lysate was loaded at 10% input.
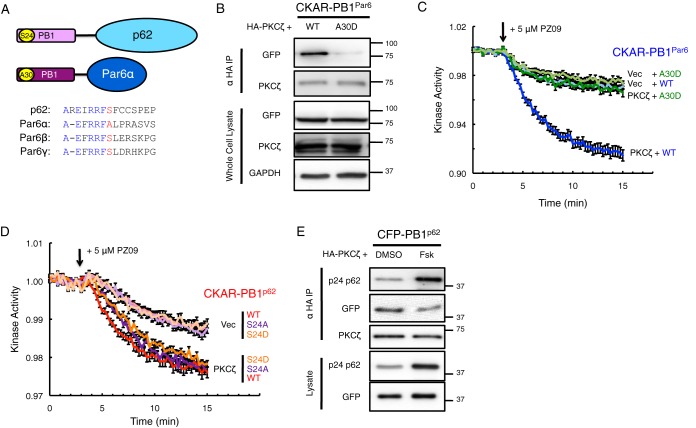
Negative Charge at Ser-24/Ala-30 in the PB1 Domains of p62 and Par6α Impairs Binding of PKCζ and Activity on CKAR-PB1Par6
A protein kinase A (PKA)-mediated phosphorylation site present in the PB1 domain of p62, residue Ser-24, has recently been identified (46). Phosphorylation at this site is proposed to regulate aPKC binding to the PB1 domain of p62; co-IP studies comparing S24A and S24D mutants showed that negative charge at this site dismantled the ability of aPKC to bind the p62 scaffold (46). Intriguingly, the sequence surrounding the Ser-24 site of p62 is present in the PB1 domain of the Par6 isoforms, except that an Ala occupies the position of the Ser for Par6α (residue Ala-30) but not for Par6β and Par6γ (Fig. 3A). To address whether negative charge at this position in the PB1 domain of Par6α would dismantle the capacity to bind PKCζ, we constructed an A30D mutant in our CKAR-PB1Par6 reporter. Indeed, co-IP studies revealed that PKCζ binding to the A30D mutant was dramatically reduced compared with binding to wild-type Par6α PB1 (Fig. 3B; 95 ± 1% reduction, p < 0.0001 from three independent experiments). Subsequently, we compared the activity of full-length mCherry-PKCζ on CKAR-PB1Par6 in which the PB1 domain was wild-type or had the A30D mutation (Fig. 3C). In accordance with the co-IP results, the basal activity of PKCζ was significantly reduced on the A30D mutant compared with the wild-type (WT) version of the Par6α PB1 reporter and comparable with its vector-transfected control. We also examined whether mutating the Ser-24 site on the PB1 domain of p62 affected PKCζ binding and activity. Activity readout by the S24A CKAR-PB1p62 reporter demonstrated no difference in overexpressed or endogenous PKCζ basal activity compared with the wild-type version (Fig. 3D) and did not differ in its ability to bind PKCζ in co-IP (data not shown). The readout of the S24D CKAR-PB1p62 reporter for PKCζ basal activity also did not change from the wild-type version, although this mutation on p62 was previously shown to displace binding of aPKC in co-IP (46). To examine the effect of Ser-24 phosphorylation on PKCζ binding to p62, cells were treated with forskolin (24 h at 50 μm) prior to lysis, previously shown to induce PKA-mediated phosphorylation of Ser-24 (46). Treatment with forskolin increased Ser(P)-24 on CFP-PB1p62 and diminished binding to PKCζ (40 ± 8% reduction, p = 0.005) as demonstrated through co-IP from six independent experiments (Fig. 3E).
FIGURE 3.
Negative charge at Ser-24/Ala-30 in the PB1 domains of p62 and Par6α impairs binding of PKCζ and activity on CKAR-PB1Par6. A, diagram showing consensus within the PB1 domain for the Ser-24 site of p62 versus the Ala-30 site of Par6α and corresponding Ser sites in Par6β and Par6γ. B, HA-PKCζ was co-expressed with either wild-type (WT) or A30D mutation of CKAR-PB1Par6 in COS-7, immunoprecipitated from soluble lysates using anti-HA antibody, and blotted for co-IP of CKAR tag using anti-GFP antibody, and whole cell lysate was loaded at 10% input. C and D, basal kinase activity of mCherry-PKCζ versus mCherry-Vec on either WT or A30D CKAR-PB1Par6 (C) or WT, S24A, or S24D CKAR-PB1p62 (D) after treatment with 5 μm PZ09 in live COS-7 cells. The trace for each cell imaged was normalized to its t = 0-min baseline value and plotted as means ± S.E. Normalized FRET ratios were combined from four independent experiments (all traces in C and CKAR-PB1p62 S24A Vec trace in D), five independent experiments (CKAR-PB1p62 WT traces and CKAR-PB1p62 S24D Vec trace), or six independent experiments (CKAR-PB1p62 S24A and CKAR-PB1p62 S24D PKCζ traces) with each experiment analyzing 6–13 selected cells. E, HA-PKCζ was co-expressed with WT CFP-PB1p62, treated for 24 h prior to lysis with either DMSO or 50 μm forskolin (Fsk) in COS-7, immunoprecipitated from soluble lysates using anti-HA antibody, and blotted for co-IP of CKAR tag using anti-GFP antibody along with a phospho-specific antibody for Ser(P)-24 p62; whole cell lysate was loaded at 10% input.
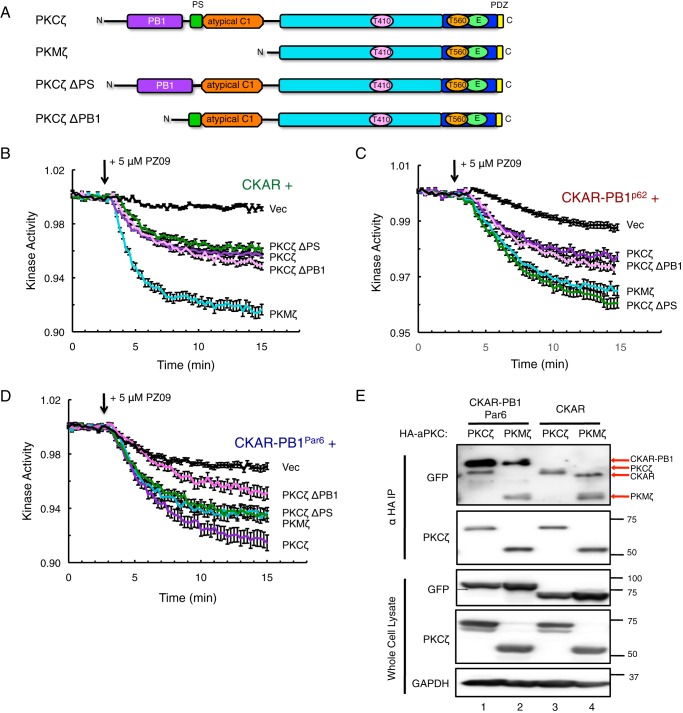
Basal Activity of PKCζ Is Regulated by a Combination of Autoinhibition and Scaffold-regulated Localization or Sequestration
We next investigated the role of the N-terminal regulatory domains, namely the PB1 domain and the pseudosubstrate (PS), in regulating the scaffolded activity of PKCζ on the various CKAR reporters examined in Fig. 2. The alternative transcript PKMζ lacks the N-terminal regulatory domains (PB1, pseudosubstrate, and atypical C1 regions) and consists of the kinase domain, the C-terminal region, and the PDZ ligand (Fig. 4A). In addition to PKMζ, we constructed two deletion mutants, PKCζ ΔPS (deleted pseudosubstrate) and PKCζ ΔPB1 (deleted PB1 domain), to investigate the role of these domains in regulating the activity of PKCζ on scaffolds (Fig. 4A). Co-expressing these constructs first with non-tethered CKAR and measuring basal activity after treatment with PZ09 (Fig. 2B), PKMζ demonstrated the most pronounced basal activity on CKAR. The ΔPS and ΔPB1 constructs displayed equal basal activity to full-length PKCζ, which was significantly increased from the vector-transfected control. Next, we examined the basal activity of the PKCζ constructs on the CKAR-PB1p62 reporter, this time showing equal activity between PKCζ ΔPS and PKMζ that was more pronounced than full-length PKCζ and PKCζ ΔPB1 (Fig. 4C). On CKAR-PB1Par6, PKCζ ΔPS and PKMζ once again displayed equal basal activity but were slightly less active than full-length PKCζ and more active than PKCζ ΔPB1 (Fig. 4D). To compare the binding of PKCζ versus PKMζ on the reporters, we immunoprecipitated HA-PKCζ or HA-PKMζ co-expressed with CKAR-PB1 or CKAR and examined co-IP of the reporters using the αGFP antibody for detection (Fig. 4E). Intriguingly, PKMζ was capable of binding the untethered version of CKAR, although PKCζ did not (Fig. 4E, 4th versus 3rd lane, note identities of bands in αGFP blot of the immunoprecipitate indicated by red arrows as the αGFP antibody also recognized HA-aPKC signal), in addition to binding CKAR-PB1Par6 (2nd lane). This binding interaction was confirmed to be between PKMζ and the CFP/YFP fluorophores present in CKAR, as PKMζ also co-immunoprecipitated CFP alone but not Myc-PB1Par6 (data not shown), thus confirming the expected non-interaction between PKMζ and PB1.
FIGURE 4.
Basal activity of PKCζ is regulated by a combination of autoinhibition and scaffold-regulated localization or sequestration. A, domain schematic of PKCζ, PKMζ, and deletion constructs ΔPS (deleted pseudosubstrate) and ΔPB1 (deleted PB1 domain) used in basal activity assays. B–D, basal kinase activity of mCherry-PKCζ versus mCherry-tagged deletion constructs and mCherry-Vec control on CKAR (B), CKAR-PB1p62 (C), and CKAR-PB1Par6 (D) in COS-7 cells treated with 5 μm PZ09. The trace for each cell imaged was normalized to its t = 0-min baseline value and plotted as means ± S.E. Normalized FRET ratios were combined from three independent experiments (CKAR PKMζ and ΔPB1 traces in B), four independent experiments (CKAR ΔPS trace in B, CKAR-PB1p62 PKMζ and ΔPB1 traces in C, and all traces in D), or five independent experiments (CKAR PKCζ and Vec traces in B and CKAR-PB1p62 PKCζ, ΔPS, and Vec traces in C) with each experiment analyzing 4–12 selected cells. E, HA-PKCζ or HA-PKMζ were co-expressed with CKAR-PB1Par6 or CKAR in COS-7. aPKC was immunoprecipitated from soluble lysates using anti-HA antibody and blotted for co-IP of CKAR using anti-GFP antibody; whole cell lysate loaded at 10% input. The band sizes of CKAR-PB1, HA-PKCζ, CKAR, and HA-PKMζ detected in the overexposed anti-GFP blot are indicated with red arrows, as the αGFP antibody detected overexpressed aPKC as well as the fluorophores.
PKMζ Is More Sensitive to Dephosphorylation at the Activation Loop Thr-410 than PKCζ
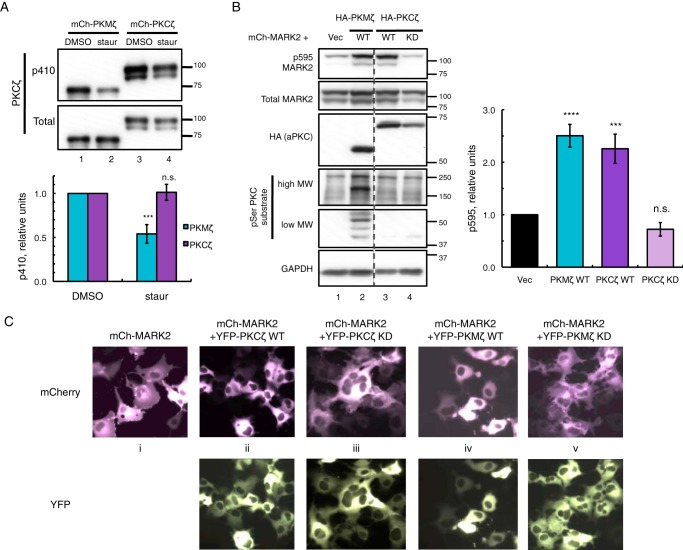
We have previously used the general kinase inhibitor staurosporine to inhibit the basal activity of PKMζ on CKAR (40). The inhibitory action of staurosporine on PKMζ is mostly indirect through its potent active site inhibition of PDK1 thus reducing basal phosphorylation on the PDK1 site of PKMζ, the activation loop Thr-410 (40). Inhibition of PDK1 permits conquest of PKMζ by opposing phosphatases acting basally on Thr-410, a phosphorylation site necessary for aPKC catalytic activity (5, 23). To investigate the role of N-terminally interacting aPKC scaffolds in regulating dephosphorylation of PKCζ at Thr-410, we transfected COS-7 cells with either mCherry-PKMζ or mCherry-PKCζ and treated them with 1 μm staurosporine for 2 h prior to lysis (Fig. 5A). Phosphorylation of Thr-410 was significantly reduced by 2-fold for PKMζ (Fig. 5A, lane 2 versus lane 1 and p410 PKCζ versus total PKCζ blots), whereas staurosporine treatment of PKCζ had no effect on p410 levels (Fig. 5A, lane 4 versus lane 3). Thus, p410 on full-length PKCζ is less sensitive to PDK1 inhibition than PKMζ, indicating a protective mechanism against dephosphorylation provided by the N-terminal regulatory domains of PKCζ.
FIGURE 5.
PKMζ is more sensitive to dephosphorylation than PKCζ and active on global substrates, although both are basally active on MARK2 substrate. A, immunoblots showing activation loop phosphorylation (p410 PKCζ) of mCherry (mCh)-PKMζ versus mCherry-PKCζ expressed in COS-7 cells treated with either DMSO vehicle or 1 μm staurosporine (staur) for 2 h prior to lysis. The p410/total PKCζ ratios were quantified from four (PKMζ) or five (PKCζ) independent experiments, normalized to DMSO controls, and plotted as mean ± S.E. Statistical analysis was performed using two separate t tests comparing staurosporine treatment versus DMSO control for each protein. Significance was notated as ***, p < 0.001, or n.s., not significant. B, immunoblots of COS-7 co-expressing mCherry-MARK2 and either vector, HA-tagged wild-type (WT) PKMζ, or PKCζ or kinase-dead (KD) PKCζ. The p595/total MARK2 ratios were quantified from six (PKCζ KD) or seven (Vec, PKCζ WT, and PKMζ WT) independent experiments, normalized to Vec-transfected control and plotted as means ± S.E. Statistical analysis was performed using ordinary two-way analysis of variance followed by Dunnett's multiple comparison test with Vec as control. Significance notated as **** (p < 0.0001); *** (p < 0.001); or n.s., not significant. C, images of live COS-7 expressing mCherry-MARK2 alone or co-expressed with YFP-tagged WT or KD versions of PKMζ or PKCζ.
PKMζ but Not PKCζ Is Active on Global Substrates although Both Are Basally Active on MARK2 Substrate
PKCζ is known to phosphorylate and interact with microtubule affinity regulating kinase 2 (MARK2) (30, 31), a phosphorylation event at the Thr-595 site of MARK2, which results in the translocation of phosphorylated, inactive MARK2 away from the plasma membrane and into the cytosol (30, 31). However, the activity of aPKC on other non-interacting substrates that contain a PKC phosphorylation site is unclear. Using p595 MARK2 as a read-out for aPKC activity in cells, we compared the catalytic activities of overexpressed PKMζ versus full-length PKCζ on global PKC substrates, using an antibody for PKC serine substrate (Fig. 5B). Expression of PKMζ (Fig. 5B, lane 2) revealed an ensemble of induced serine substrate bands (including a higher molecular weight band previously seen with PKMζ (40)) that were not present in lysates transfected with PKCζ (lane 3), indicating catalytic activity of PKMζ on multiple substrates inaccessible to PKCζ. Both kinase constructs were fully active basally to the same extent on co-expressed mCherry-MARK2 as quantified in Fig. 5B, whereas the kinase-dead (KD) form of PKCζ showed no effect on p595 MARK2 (lane 4). When expressed alone, mCherry-MARK2 is partially localized at the plasma membrane, evident by the ruffled appearance of the cell edges (panel i, Fig. 5C). Both YFP-tagged PKCζ and PKMζ constructs revealed the same ability to induce translocation of MARK2 from the plasma membrane to the cytosol (panels ii and iv, Fig. 5C, smooth appearance of cell edges), a feature that was absent when co-expressed with KD versions of PKCζ and PKMζ (panels iii and v, Fig. 5C).
Scaffold Proteins Differentially Regulate the Phosphorylation and Localization of the aPKC Substrate MARK2
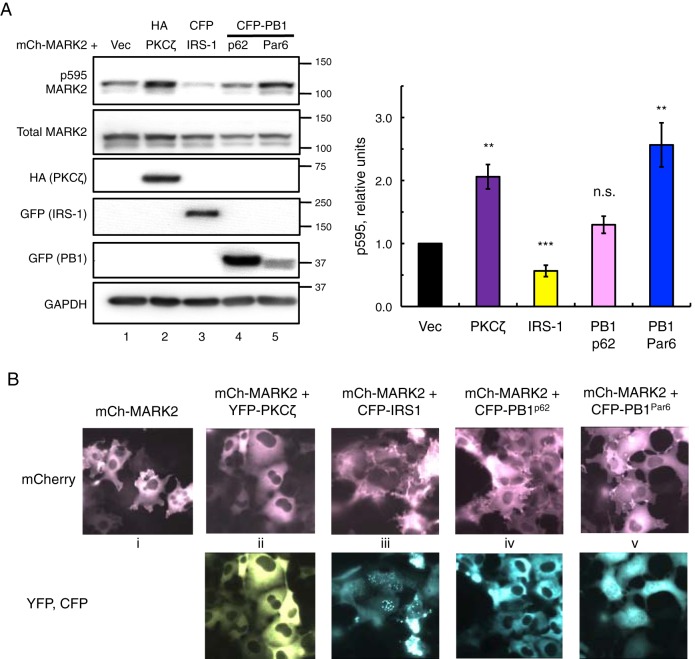
Using the MARK2 co-expression system validated for PKCζ catalytic activity and MARK2 cellular localization from Fig. 5, we set out to examine the effects of PKCζ-regulating scaffolds on MARK2 phosphorylation and localization. In addition to the PB1 domains of p62 and Par6 that directly interact with PKCζ (44, 45), we also examined the effects on MARK2 localization of IRS-1, a large scaffold protein known to interact with PKCζ through its agonist-induced interaction with p62 (26, 27). Intriguingly, co-expression of IRS-1 with MARK2 resulted in a decrease of p595 MARK2 levels (Fig. 6A, compare lanes 1 and 3), contrasting with the increased p595 levels produced with PKCζ co-expression (Fig. 6A, compare lanes 1 and 2). Additionally, co-expression of the PB1 domain of p62 had no effect on p595 MARK2 (Fig. 6A, lane 4), although the PB1 domain of Par6 increased p595 levels (lane 5), similar to PKCζ effects (quantification in Fig. 6A). We also examined the concordant effects on MARK2 cellular localization for each of the scaffolds used in Fig. 6A. In agreement with the phosphorylation results, co-expression of PKCζ resulted in localization of MARK2 into the cytosol (Fig. 6B, panel ii). Expression of the PB1 domain of p62 did not change MARK2 localization (Fig. 6B, panel iv), whereas co-expression of IRS-1 increased the localization of MARK2 at the plasma membrane (Fig. 6B, panel iii). Fully localized plasma membrane staining is evident by the spindle-like appearance of MARK2 in these cells along with the absence of visible nuclei, as decreases in nuclear visibility correlate with increases in plasma membrane localization (47). Curiously, co-expression of the Par6 PB1 domain did not change MARK2 localization relative to the plasma membrane (Fig. 6B, panel v) even though elevated p595 levels were observed. Similar results were also seen when conducted in polarized epithelial MDCK cells (data not shown).
FIGURE 6.
Scaffold proteins differentially regulate the phosphorylation and localization of the aPKC substrate MARK2. A, immunoblots of COS-7 co-expressing mCherry-MARK2 (mCh-MARK2) and either vector, HA-PKCζ, CFP-IRS-1, CFP-PB1p62, or CFP-PB1Par6. The p595/total MARK2 ratios were quantified from five (PB1Par6), six (IRS-1), or seven (Vec, PKCζ, and PB1p62) independent experiments, normalized to Vec-transfected control, and plotted as means ± S.E. Statistical analysis was performed using separate t tests between each co-expression (PKCζ, IRS-1, PB1p62, and PB1Par6) and Vec control. Significance notated as ***, p < 0.001; **, p < 0.01; or n.s., not significant. B, images of live COS-7 expressing mCherry-MARK2 alone or co-expressed with YFP-tagged PKCζ, CFP-IRS-1, CFP-PB1p62, or CFP-PB1Par6.
Insulin Regulates the Localization of Scaffolded PKCζ
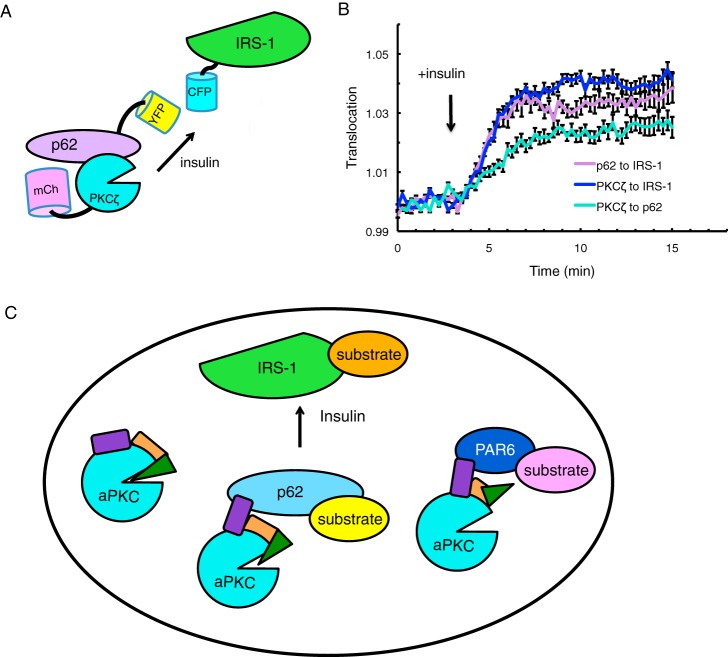
Recent studies have demonstrated through co-IP experiments that insulin promotes the binding of p62 to IRS-1 via an interaction between the PB1 domain of p62 and an insulin-induced YXXM phosphorylation motif on IRS-1 (26). Additionally, insulin stimulates increased binding of PKCζ to p62 (27) and subsequent PKCζ engagement to IRS-1 through p62, also shown through co-IP (48). To investigate these interactions in real time translocation assays, we examined combinations of various YFP, CFP, and mCherry-tagged versions of p62, PKCζ, and IRS-1 in CHO-IR cells serum-starved overnight. The YFP tag was placed on the translocating protein (p62 or PKCζ); the CFP tag was placed on the destination protein (IRS-1 or p62), and the mCherry tag was placed on the third protein to confirm expression (diagram shown in Fig. 7A). Analysis of FRET as a measure of translocation (described previously (6)) revealed that insulin stimulation caused translocation of both YFP-p62 and YFP-PKCζ to CFP-IRS-1, in addition to translocation of YFP-PKCζ to CFP-p62 (Fig. 7B). We note that the increase in FRET upon insulin stimulation was only observed in a sub-fraction of the CHO-IR cells imaged in each experiment; an increase was observed for 26% (n = 94 cells), 40% (n = 50 cells), and 52% (n = 31 cells) of cells imaged for the FRET from PKCζ to IRS-1, PKCζ to p62, and p62-IRS-1, respectively.
FIGURE 7.
Insulin regulates the localization of scaffolded PKCζ. A, schematic showing of FRET-based translocation assay. CFP is tagged to the destination protein (IRS-1 or p62); YFP is tagged to the translocating protein (p62 or PKCζ), and mCherry is tagged to the third protein to confirm cellular expression. B, CHO-IR cells expressing the indicated proteins were serum-starved overnight, and FRET over CFP was measured before and after treatment with 100 nm insulin: 1) YFP-p62 with CFP-IRS-1 and mCherry-PKCζ (p62 to PKCζ); 2) YFP-PKCζ with CFP-IRS-1 and mCherry-p62 (PKCζ to IRS-1); and 3) YFP-PKCζ with CFP-p62 and mCherry-IRS-1 (PKCζ to p62). The trace for each responding cell imaged was normalized to its t = 0-min baseline value and plotted as means ± S.E. Normalized FRET ratios were combined from four independent experiments with each experiment analyzing 4–7 selected cells. C, model showing regulation of the localization and activity of aPKC by protein scaffolds. Non-scaffolded aPKC is effectively autoinhibited by intramolecular interactions that mask the active site of the kinase domain (cyan circle). Binding through the PB1 domain (purple) of scaffolds such as p62 (light blue) or Par6 (dark blue) results in partial or complete removal of the pseudosubstrate (green triangle) from the substrate binding cavity, resulting in low (p62) to maximal (Par6) activity at the scaffold. Agonists such as insulin relocalize these scaffolds, for example recruiting the aPKC:p62 complex to IRS-1 where it can phosphorylate proximal substrates to affect downstream signaling.
Discussion
The lack of regulation by either phosphorylation or second messengers (3, 49, 50)), including the Akt activator, phosphatidylinositol 3,4,5-triphosphate (5, 43, 51)), has confounded the understanding of how aPKCs signal in cells. Here, we show that protein scaffolds relocalize aPKC in response to insulin and that the binding to specific scaffolds differentially tunes the activity of aPKCs. Using FRET-based technologies to study the real time dynamics of aPKC activity and location in live cells, we show that p62 and Par6 tether PKCζ in a partially active or fully active conformation, respectively, to enhance signaling on these PB1 domain scaffolds. Translocation experiments reveal that insulin enhances the interaction of aPKC to p62, in turn recruiting the complex to IRS-1. Additionally, we show that IRS-1 can sequester aPKC away from phosphorylating the membrane-localized substrate MARK2. As part of this study, we validate the use of a suitable, newly available aPKC-specific active site inhibitor, PZ09, for study of aPKC regulation in live cells with PKC-specific substrates, noting that its inhibition of other non-PKC kinases such as PDK1 may cloud its utility for studying aPKC-specific function in vivo. These findings underscore the importance of scaffold interactions in controlling the cellular function of aPKCs and are particularly relevant as the exceptionally low catalytic activity (5 mol of phosphate/min/mol of PKC (5) compared with 200 mol of phosphate/min/mol of PKC for cPKCs (52)) would require localization of substrates in the vicinity of aPKC for effective phosphorylation.
The PB1 domain mediates multiple protein interactions primarily through electrostatic interactions between acidic and basic residues of heterodimerizing PB1 domains (53). p62 and Par6 are the two most characterized PB1 binding partners of aPKC and are thought to regulate separate downstream pathways of aPKC signaling (42). Here, we demonstrate that the basal activity of PKCζ is differentially regulated by these two scaffolds, with maximal signaling when scaffolded to Par6α compared with p62, which tethers a conformation that relieves significant autoinhibition. In addition, we show that the PB1 domain of Par6α binds more tightly to PKCζ than the PB1 domain of p62, indicating that the increased population of PKCζ localized to CKAR-PB1Par6 contributes to the considerably high basal activity on this scaffold. However, the attenuated activity response of PKCζ on bound CKAR-PB1p62 compared with unbound CKAR may also reflect greater PKCζ resistance to active site inhibitors when scaffolded to p62 but not to Par6, a feature known to impede the efficacy of other active site inhibitors on scaffolded PKC (54).
Investigation of the enhanced binding of PKCζ to Par6 compared with p62 led us to identify another key regulatory residue on the PB1 domain; Ser versus Ala at the position corresponding to Ser-24 and Ala-30 on p62 versus Par6, respectively, tunes the affinity for the aPKC PB1 domain. This residue is within a consensus region present only in the PB1 domains of p62 and the Par6 isoforms and not other human PB1 domain proteins (53). Previous work by Christian et al. (46) demonstrated that Ser-24 on p62 is phosphorylated by PKA, a modification that disrupts the interaction of p62 with aPKC. Consistent with phosphorylation at this site controlling protein interactions, we show that introduction of a negative charge at the comparable position on Par6α (A30D mutation) disrupts the binding of PKCζ to this PB1 domain and significantly reduces the basal activity of PKCζ on CKAR-PB1Par6. The Ala residue is only present on the Par6α isoform and is Ser at the corresponding site on Par6β and Par6γ. Although previous work has shown that aPKC binds to all three Par6 isoforms, yeast two-hybrid screens demonstrated the most significant interaction of aPKC to Par6α (also known as Par6C) (45), in agreement with the Ala favoring aPKC binding. Curiously, mutation of Ser-24 to either Asp or Ala on the p62 PB1 domain did not affect PKCζ activity. One possibility is that Asp is a poor phosphomimetic for this site on p62. However, mutation to Asp has been shown to inhibit aPKC binding (46); thus, any drop in activity may be below the detection limit of our cellular activity assays. Importantly, extended forskolin stimulation to promote PKA phosphorylation at this site decreased the binding of PKCζ to PB1 p62. Taken together, these data support a model wherein the unique Ser at position 24 on p62 provides a consensus phosphorylation site that regulates the binding and activity of aPKC.
The deletion of the PB1 domain (ΔPB1), the pseudosubstrate (ΔPS), or both (PKMζ) allows dissection of how scaffold interactions relieve the autoinhibition of aPKC to tune its the level of localized basal activity. For the non-scaffolded CKAR, PKMζ demonstrates the highest basal activity of all the constructs as it is neither autoinhibited (no PS) nor sequestered on an endogenous scaffold (no PB1), and therefore unrestricted to phosphorylate a globally expressed substrate such as CKAR. Supporting the model of PKMζ's unrestricted access to CKAR, co-IP results reveal that PKMζ interacts with CKAR whereas PKCζ does not, suggesting sequestration of the full-length kinase on a scaffold that restricts its interaction with CKAR. Similarly, analysis with an antibody targeted to specific PKC phosphorylation sites reveals that PKMζ exerts highly enhanced phosphorylation of global endogenous PKC substrates compared with PKCζ. However, PKCζ still has the ability to phosphorylate interactive specific substrates such as MARK2 (30) with equal capacity as PKMζ and to regulate its localization from the plasma membrane into the cytosol. Deletion of either the PS or the PB1 domain did not significantly affect the basal activity of PKCζ on non-scaffolded CKAR. In contrast, activity at the protein scaffolds was sensitive to deletion of the PB1 domain and, depending on the scaffold, the pseudosubstrate. On CKAR-PB1p62, deletion of the pseudosubstrate significantly enhanced basal activity, revealing some autoinhibition by the pseudosubstrate when PKCζ is bound to p62. This is consistent with our previous study showing that full-length PKCζ displays ∼25% of its maximal unrestrained activity on p62 as assessed with a p62-scaffolded CKAR (6). Deletion of the pseudosubstrate had no significant effect on the activity on the Par6 scaffold, revealing that the majority of the PKCζ bound to Par6 is in the open conformation, as reported previously (7), with the pseudosubstrate tethered away from the substrate-binding cavity. Activity on both scaffolds was sensitive to deletion of the PB1 domain, consistent with release of the enzymes from the scaffolds. These results reveal that interaction with regulatory determinants autoinhibit PKCζ and that binding to protein scaffolds differentially relieves these inhibitory constraints.
Further supporting autoinhibitory constraints of PKCζ, we show that PKMζ is more sensitive to dephosphorylation at its activation loop phosphorylation site, Thr-410 (5, 23), compared with PKCζ (5, 23). Thus, similar to conventional PKCs (55), intramolecular autoinhibitory interactions with regulatory domains mask this phosphorylation site not only in vitro (5) but also in cells. Additionally, binding to scaffolds may sequester aPKCs from phosphatases, although localization of phosphatases to protein scaffolds also serves as a mechanism to control signaling output of kinases (56).
Finally, we show that scaffold interactions can regulate the sequestration or localization of PKCζ to restrict or enhance its phosphorylation of physiological substrates. This phenomenon is analogous to the well known differential targeting of the protein phosphatase 1 catalytic subunit (PP1c) by its diverse arsenal of regulatory subunits to a variety of subcellular locations, including glycogen, spliceosome, and cytoskeleton where activity toward specific localized substrates is enhanced (57–63). In our studies, overexpression of IRS-1 inhibited the aPKC-dependent phosphorylation of MARK2, thus preventing the release of this substrate from the plasma membrane. In contrast, overexpression of the PB1 domain of Par6 enhanced the phosphorylation on the aPKC-regulated site of MARK2, suggesting that this scaffold may localize aPKC near MARK2. Note that this Par6-induced MARK2 phosphorylation did not displace the substrate from the plasma membrane, likely because shuttling phosphorylated MARK2 away from the apical surface of polarized cells depends on a second step, binding to the polarity regulator Par5 (also known as 14-3-3) (31, 64). Overexpression of Par6 may indeed localize more endogenous aPKC to phosphorylate MARK2 yet interfere with the Par5 ability to bind MARK2.
How insulin controls aPKC function has been difficult to reconcile with the inability of this agonist to alter the phosphorylation state or activity of aPKC (5). Live cell imaging studies reveal that insulin promotes the association of PKCζ to both p62 and PKCζ, with a half-time on the order of 5 min under the conditions of our assays. This time frame agrees with aPKC-dependent changes in functions such as glucose transport, observed within 5–15 min of insulin stimulation (17, 18, 21). The insulin-dependent association of aPKC with these scaffolds supports previous co-IP results (26, 27). IRS-1 is a large docking hub for downstream insulin signaling events to occur (25) and possesses Ser/Thr phosphorylation sites previously shown to be regulated by aPKC (65–68). Taken together, these results suggest that the major effect of insulin on controlling aPKC function is by re-localizing the kinase to cellular signaling hubs such as IRS-1, poising it next to relevant downstream substrates.
In summary, our data support a model in which aPKC is regulated by binding to specific protein scaffolds that differentially control its activity. When unscaffolded, aPKC has low basal activity because of efficient autoinhibition by its regulatory domains, such as the pseudosubstrate (Fig. 7C, green triangle) masking the kinase domain (cyan circle). Binding to Par6 tethers the kinase in an open conformation, with the pseudosubstrate removed from the substrate-binding cavity in the kinase domain to allow maximal activity. Lower affinity binding to p62 results in less effective tethering of the pseudosubstrate, so that the scaffolded enzyme has ∼25% maximal activity (6). This sequestration on protein scaffolds can either enhance phosphorylation of co-localized substrates or suppress phosphorylation of other substrates (e.g. the inhibition of MARK2 phosphorylation upon sequestration of aPKC by IRS-1 overexpression). The particularly low catalytic activity of aPKCs ensures that signaling is kept at a minimum in the absence of regulated scaffold interactions, which poise the enzyme for stoichiometric phosphorylation of substrates recruited to the same signaling platform.
Author Contributions
I. S. T. and A. C. N. conceived the experiments and wrote the manuscript. I. S. T. performed and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Maya Kunkel and Charles King for helpful discussions and Lisa Tsai for generation of p62 constructs.
This work was supported in part by National Institutes of Health Grants P01DK054441 (to A. C. N.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- aPKC
- atypical protein kinase C
- cPKC
- conventional PKC
- CKAR
- C kinase activity reporter
- DAG
- diacylglycerol
- CFP
- cyan fluorescent protein
- Vec
- vector
- co-IP
- co-immunoprecipitation
- PS
- pseudosubstrate
- ΔPS
- deleted PS
- KD
- kinase-dead.
References
- 1. Langeberg L. K., and Scott J. D. (2015) Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol. 16, 232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Newton A. C. (2001) Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 101, 2353–2364 [DOI] [PubMed] [Google Scholar]
- 3. Newton A. C. (2010) Protein kinase C: poised to signal. Am. J. Physiol. Endocrinol. Metab. 298, E395–E402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parker P. J., and Murray-Rust J. (2004) PKC at a glance. J. Cell Sci. 117, 131–132 [DOI] [PubMed] [Google Scholar]
- 5. Tobias I. S., Kaulich M., Kim P. K., Simon N., Jacinto E., Dowdy S. F., King C. C., and Newton A. C. (2016) Protein kinase Cζ exhibits constitutive phosphorylation and phosphatidylinositol-3,4,5-triphosphate-independent regulation. Biochem. J. 473, 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai L. C., Xie L., Dore K., Xie L., Del Rio J. C., King C. C., Martinez-Ariza G., Hulme C., Malinow R., Bourne P. E., and Newton A. C. (2015) ζ inhibitory peptide disrupts electrostatic interactions that maintain atypical protein kinase C in its active conformation on the scaffold p62. J. Biol. Chem. 290, 21845–21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graybill C., Wee B., Atwood S. X., and Prehoda K. E. (2012) Partitioning-defective protein 6 (Par-6) activates atypical protein kinase C (aPKC) by pseudosubstrate displacement. J. Biol. Chem. 287, 21003–21011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin D., Edwards A. S., Fawcett J. P., Mbamalu G., Scott J. D., and Pawson T. (2000) A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2, 540–547 [DOI] [PubMed] [Google Scholar]
- 9. Hirose T., Izumi Y., Nagashima Y., Tamai-Nagai Y., Kurihara H., Sakai T., Suzuki Y., Yamanaka T., Suzuki A., Mizuno K., and Ohno S. (2002) Involvement of ASIP/PAR-3 in the promotion of epithelial tight junction formation. J. Cell Sci. 115, 2485–2495 [DOI] [PubMed] [Google Scholar]
- 10. Nagai-Tamai Y., Mizuno K., Hirose T., Suzuki A., and Ohno S. (2002) Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells 7, 1161–1171 [DOI] [PubMed] [Google Scholar]
- 11. Yamanaka T., Horikoshi Y., Sugiyama Y., Ishiyama C., Suzuki A., Hirose T., Iwamatsu A., Shinohara A., and Ohno S. (2003) Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr. Biol. 13, 734–743 [DOI] [PubMed] [Google Scholar]
- 12. Betschinger J., Mechtler K., and Knoblich J. A. (2003) The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422, 326–330 [DOI] [PubMed] [Google Scholar]
- 13. Plant P. J., Fawcett J. P., Lin D. C., Holdorf A. D., Binns K., Kulkarni S., and Pawson T. (2003) A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5, 301–308 [DOI] [PubMed] [Google Scholar]
- 14. Moscat J., and Diaz-Meco M. T. (2009) p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Komatsu M., Kageyama S., and Ichimura Y. (2012) p62/SQSTM1/A170: physiology and pathology. Pharmacol. Res. 66, 457–462 [DOI] [PubMed] [Google Scholar]
- 16. Bandyopadhyay G., Standaert M. L., Zhao L., Yu B., Avignon A., Galloway L., Karnam P., Moscat J., and Farese R. V. (1997) Activation of protein kinase C (α, β, and ζ) by insulin in 3T3/L1 cells. Transfection studies suggest a role for PKC-ζ in glucose transport. J. Biol. Chem. 272, 2551–2558 [DOI] [PubMed] [Google Scholar]
- 17. Kotani K., Ogawa W., Matsumoto M., Kitamura T., Sakaue H., Hino Y., Miyake K., Sano W., Akimoto K., Ohno S., and Kasuga M. (1998) Requirement of atypical protein kinase clambda for insulin stimulation of glucose uptake but not for Akt activation in 3T3-L1 adipocytes. Mol. Cell. Biol. 18, 6971–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bandyopadhyay G., Kanoh Y., Sajan M. P., Standaert M. L., and Farese R. V. (2000) Effects of adenoviral gene transfer of wild-type, constitutively active, and kinase-defective protein kinase C-λ on insulin-stimulated glucose transport in L6 myotubes. Endocrinology 141, 4120–4127 [DOI] [PubMed] [Google Scholar]
- 19. Bandyopadhyay G., Sajan M. P., Kanoh Y., Standaert M. L., Quon M. J., Lea-Currie R., Sen A., and Farese R. V. (2002) PKC-ζ mediates insulin effects on glucose transport in cultured preadipocyte-derived human adipocytes. J. Clin. Endocrinol. Metab. 87, 716–723 [DOI] [PubMed] [Google Scholar]
- 20. Sajan M. P., Rivas J., Li P., Standaert M. L., and Farese R. V. (2006) Repletion of atypical protein kinase C following RNA interference-mediated depletion restores insulin-stimulated glucose transport. J. Biol. Chem. 281, 17466–17473 [DOI] [PubMed] [Google Scholar]
- 21. Farese R. V., Sajan M. P., Yang H., Li P., Mastorides S., Gower W. R. Jr., Nimal S., Choi C. S., Kim S., Shulman G. I., Kahn C. R., Braun U., and Leitges M. (2007) Muscle-specific knockout of PKC-λ impairs glucose transport and induces metabolic and diabetic syndromes. J. Clin. Invest. 117, 2289–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bandyopadhyay G., Standaert M. L., Sajan M. P., Karnitz L. M., Cong L., Quon M. J., and Farese R. V. (1999) Dependence of insulin-stimulated glucose transporter 4 translocation on 3-phosphoinositide-dependent protein kinase-1 and its target threonine-410 in the activation loop of protein kinase C-ζ. Mol. Endocrinol. 13, 1766–1772 [DOI] [PubMed] [Google Scholar]
- 23. Standaert M. L., Bandyopadhyay G., Kanoh Y., Sajan M. P., and Farese R. V. (2001) Insulin and PIP3 activate PKC-ζ by mechanisms that are both dependent and independent of phosphorylation of activation loop (T410) and autophosphorylation (T560) sites. Biochemistry 40, 249–255 [DOI] [PubMed] [Google Scholar]
- 24. Hirai T., and Chida K. (2003) Protein kinase Cζ (PKCζ): activation mechanisms and cellular functions. J. Biochem. 133, 1–7 [DOI] [PubMed] [Google Scholar]
- 25. Copps K. D., and White M. F. (2012) Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 55, 2565–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geetha T., Zheng C., Vishwaprakash N., Broderick T. L., and Babu J. R. (2012) Sequestosome 1/p62, a scaffolding protein, is a newly identified partner of IRS-1 protein. J. Biol. Chem. 287, 29672–29678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xi G., Shen X., Rosen C. J., and Clemmons D. R. (2016) IRS-1 functions as a molecular scaffold to coordinate IGF-I/IGFBP-2 signaling during osteoblast differentiation. J. Bone Miner. Res. 10.1002/jbmr.2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodriguez A., Durán A., Selloum M., Champy M. F., Diez-Guerra F. J., Flores J. M., Serrano M., Auwerx J., Diaz-Meco M. T., and Moscat J. (2006) Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 3, 211–222 [DOI] [PubMed] [Google Scholar]
- 29. Okada K., Yanagawa T., Warabi E., Yamastu K., Uwayama J., Takeda K., Utsunomiya H., Yoshida H., Shoda J., and Ishii T. (2009) The α-glucosidase inhibitor acarbose prevents obesity and simple steatosis in sequestosome 1/A170/p62 deficient mice. Hepatol. Res. 39, 490–500 [DOI] [PubMed] [Google Scholar]
- 30. Hurov J. B., Watkins J. L., and Piwnica-Worms H. (2004) Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 14, 736–741 [DOI] [PubMed] [Google Scholar]
- 31. Suzuki A., Hirata M., Kamimura K., Maniwa R., Yamanaka T., Mizuno K., Kishikawa M., Hirose H., Amano Y., Izumi N., Miwa Y., and Ohno S. (2004) aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 14, 1425–1435 [DOI] [PubMed] [Google Scholar]
- 32. Hurov J., and Piwnica-Worms H. (2007) The Par-1/MARK family of protein kinases: from polarity to metabolism. Cell Cycle 6, 1966–1969 [DOI] [PubMed] [Google Scholar]
- 33. Hurov J. B., Huang M., White L. S., Lennerz J., Choi C. S., Cho Y. R., Kim H. J., Prior J. L., Piwnica-Worms D., Cantley L. C., Kim J. K., Shulman G. I., and Piwnica-Worms H. (2007) Loss of the Par-1b/MARK2 polarity kinase leads to increased metabolic rate, decreased adiposity, and insulin hypersensitivity in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 5680–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Violin J. D., Zhang J., Tsien R. Y., and Newton A. C. (2003) A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 161, 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kunkel M. T., Ni Q., Tsien R. Y., Zhang J., and Newton A. C. (2005) Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J. Biol. Chem. 280, 5581–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu-Zhang A. X., Murphy A. N., Bachman M., and Newton A. C. (2012) Isozyme-specific interaction of protein kinase Cδ with mitochondria dissected using live cell fluorescence imaging. J. Biol. Chem. 287, 37891–37906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu-Zhang A. X., and Newton A. C. (2013) Protein kinase C pharmacology: refining the toolbox. Biochem. J. 452, 195–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ling D. S., Benardo L. S., Serrano P. A., Blace N., Kelly M. T., Crary J. F., and Sacktor T. C. (2002) Protein kinase Mζ is necessary and sufficient for LTP maintenance. Nat. Neurosci. 5, 295–296 [DOI] [PubMed] [Google Scholar]
- 39. Herbert J. M., Augereau J. M., Gleye J., and Maffrand J. P. (1990) Chelerythrine is a potent and specific inhibitor of protein kinase C. Biochem. Biophys. Res. Commun. 172, 993–999 [DOI] [PubMed] [Google Scholar]
- 40. Wu-Zhang A. X., Schramm C. L., Nabavi S., Malinow R., and Newton A. C. (2012) Cellular pharmacology of protein kinase Mζ (PKMζ) contrasts with its in vitro profile: implications for PKMζ as a mediator of memory. J. Biol. Chem. 287, 12879–12885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trujillo J. I., Kiefer J. R., Huang W., Thorarensen A., Xing L., Caspers N. L., Day J. E., Mathis K. J., Kretzmer K. K., Reitz B. A., Weinberg R. A., Stegeman R. A., Wrightstone A., Christine L., Compton R., and Li X. (2009) 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: design and synthesis of a potent and isoform selective PKC-ζ inhibitor. Bioorg. Med. Chem. Lett. 19, 908–911 [DOI] [PubMed] [Google Scholar]
- 42. Kusne Y., Carrera-Silva E. A., Perry A. S., Rushing E. J., Mandell E. K., Dietrich J. D., Errasti A. E., Gibbs D., Berens M. E., Loftus J. C., Hulme C., Yang W., Lu Z., Aldape K., Sanai N., et al. (2014) Targeting aPKC disables oncogenic signaling by both the EGFR and the proinflammatory cytokine TNFα in glioblastoma. Sci. Signal. 7, ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., and Cohen P. (1997) Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 44. Puls A., Schmidt S., Grawe F., and Stabel S. (1997) Interaction of protein kinase Cζ with ZIP, a novel protein kinase C-binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 6191–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Joberty G., Petersen C., Gao L., and Macara I. G. (2000) The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2, 531–539 [DOI] [PubMed] [Google Scholar]
- 46. Christian F., Krause E., Houslay M. D., and Baillie G. S. (2014) PKA phosphorylation of p62/SQSTM1 regulates PB1 domain interaction partner binding. Biochim. Biophys. Acta 1843, 2765–2774 [DOI] [PubMed] [Google Scholar]
- 47. Antal C. E., Violin J. D., Kunkel M. T., Skovsø S., and Newton A. C. (2014) Intramolecular conformational changes optimize protein kinase C signaling. Chem. Biol. 21, 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xi G., Shen X., Wai C., Vilas C. K., and Clemmons D. R. (2015) Hyperglycemia stimulates p62/PKCζ interaction, which mediates NF-κB activation, increased Nox4 expression, and inflammatory cytokine activation in vascular smooth muscle. FASEB J. 29, 4772–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kazanietz M. G., Bustelo X. R., Barbacid M., Kolch W., Mischak H., Wong G., Pettit G. R., Bruns J. D., and Blumberg P. M. (1994) Zinc finger domains and phorbol ester pharmacophore. Analysis of binding to mutated form of protein kinase Cζ and the vav and c-raf proto-oncogene products. J. Biol. Chem. 269, 11590–11594 [PubMed] [Google Scholar]
- 50. Pu Y., Peach M. L., Garfield S. H., Wincovitch S., Marquez V. E., and Blumberg P. M. (2006) Effects on ligand interaction and membrane translocation of the positively charged arginine residues situated along the C1 domain binding cleft in the atypical protein kinase C isoforms. J. Biol. Chem. 281, 33773–33788 [DOI] [PubMed] [Google Scholar]
- 51. Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., and Hemmings B. A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 52. Johnson J. E., Edwards A. S., and Newton A. C. (1997) A putative phosphatidylserine binding motif is not involved in the lipid regulation of protein kinase C. J. Biol. Chem. 272, 30787–30792 [DOI] [PubMed] [Google Scholar]
- 53. Sumimoto H., Kamakura S., and Ito T. (2007) Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Science's STKE 2007, re6. [DOI] [PubMed] [Google Scholar]
- 54. Hoshi N., Langeberg L. K., Gould C. M., Newton A. C., and Scott J. D. (2010) Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol. Cell 37, 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dutil E. M., Keranen L. M., DePaoli-Roach A. A., and Newton A. C. (1994) In vivo regulation of protein kinase C by trans-phosphorylation followed by autophosphorylation. J. Biol. Chem. 269, 29359–29362 [PubMed] [Google Scholar]
- 56. Scott J. D., and Newton A. C. (2012) Shedding light on local kinase activation. BMC Biology 10, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cohen P. T. (2002) Protein phosphatase 1–targeted in many directions. J. Cell Sci. 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 58. Hubbard M. J., and Cohen P. (1993) On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem. Sci. 18, 172–177 [DOI] [PubMed] [Google Scholar]
- 59. Armstrong C. G., Browne G. J., Cohen P., and Cohen P. T. (1997) PPP1R6, a novel member of the family of glycogen-targetting subunits of protein phosphatase 1. FEBS Lett. 418, 210–214 [DOI] [PubMed] [Google Scholar]
- 60. Wu J., Kleiner U., and Brautigan D. L. (1996) Protein phosphatase type-1 and glycogen bind to a domain in the skeletal muscle regulatory subunit containing conserved hydrophobic sequence motif. Biochemistry 35, 13858–13864 [DOI] [PubMed] [Google Scholar]
- 61. Hirano K., Erdödi F., Patton J. G., and Hartshorne D. J. (1996) Interaction of protein phosphatase type 1 with a splicing factor. FEBS Lett. 389, 191–194 [DOI] [PubMed] [Google Scholar]
- 62. Allen P. B., Kwon Y. G., Nairn A. C., and Greengard P. (1998) Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. J. Biol. Chem. 273, 4089–4095 [DOI] [PubMed] [Google Scholar]
- 63. Schillace R. V., and Scott J. D. (1999) Association of the type 1 protein phosphatase PP1 with the A-kinase anchoring protein AKAP220. Curr. Biol. 9, 321–324 [DOI] [PubMed] [Google Scholar]
- 64. Macara I. G. (2004) Parsing the polarity code. Nat. Rev. Mol. Cell Biol. 5, 220–231 [DOI] [PubMed] [Google Scholar]
- 65. Ravichandran L. V., Esposito D. L., Chen J., and Quon M. J. (2001) Protein kinase C-ζ phosphorylates insulin receptor substrate-1 and impairs its ability to activate phosphatidylinositol 3-kinase in response to insulin. J. Biol. Chem. 276, 3543–3549 [DOI] [PubMed] [Google Scholar]
- 66. Sommerfeld M. R., Metzger S., Stosik M., Tennagels N., and Eckel J. (2004) In vitro phosphorylation of insulin receptor substrate 1 by protein kinase C-ζ: functional analysis and identification of novel phosphorylation sites. Biochemistry 43, 5888–5901 [DOI] [PubMed] [Google Scholar]
- 67. Moeschel K., Beck A., Weigert C., Lammers R., Kalbacher H., Voelter W., Schleicher E. D., Häring H. U., and Lehmann R. (2004) Protein kinase C-ζ-induced phosphorylation of Ser318 in insulin receptor substrate-1 (IRS-1) attenuates the interaction with the insulin receptor and the tyrosine phosphorylation of IRS-1. J. Biol. Chem. 279, 25157–25163 [DOI] [PubMed] [Google Scholar]
- 68. Lee S., Lynn E. G., Kim J. A., and Quon M. J. (2008) Protein kinase C-ζ phosphorylates insulin receptor substrate-1, -3, and -4 but not -2: isoform specific determinants of specificity in insulin signaling. Endocrinology 149, 2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]