Abstract
Vanilloids are pain evoking molecules that serve as ligands of the “heat and capsaicin receptor” TRPV1. Binding of either endogenous or exogenous vanilloids evokes channel and subsequent neuronal activation, leading to pain sensation. Despite its pivotal physiological role, the molecular basis of TRPV1 activation and deactivation is not fully understood. The highly conserved tyrosine in position 511 (Tyr511) of the rat TRPV1 (rTRPV1) was the first residue to be identified as a necessary participant in the vanilloid-mediated response. rTRPV1 cryo-EM structures implicated rotation of this residue in the vanilloids bound state. Therefore, we hypothesize that the rTRPV1 Tyr511 residue entraps vanilloids in their binding site, prolonging channel activity. To test our hypothesis, we generated an array of rTRPV1 mutants, containing the whole spectrum of Tyr511 substitutions, and tested their response to both exo- and endovanilloids. Our data show that only substitutions of Tyr511 to aromatic amino acids were able to mimic, albeit partially, the vanilloid-evoked activation pattern of the wt receptor. Although these substitutions reduced the channel sensitivity to vanilloids, a maximal open-channel lifetime could be achieved. Moreover, whereas their current activation rate remains intact, receptors with Tyr511 substitutions exhibited a faster current deactivation. Our findings therefore suggest that the duration of channel activity evoked by vanilloids is regulated by the interaction between Tyr511 and the agonist. To conclude, we suggest that Tyr511-mediated anchoring of vanilloids in their binding pocket is pivotal for TRPV1 activation and subsequent pain sensation.
Keywords: electrophysiology, gating, site-directed mutagenesis, structure-function, transient receptor potential channels (TRP channels)
Introduction
Vanilloids, both endogenous and exogenous, are pain evoking molecules (1, 2). They serve as ligands of the mammalian transient receptor potential vanilloid type 1 (TRPV1)2 protein, a nonselective cation channel also known as the “heat and capsaicin receptor” (3). TRPV1 is mainly expressed on C and Aδ fibers of the somatosensory system, where it plays an essential role in the development of inflammatory hyperalgesia and pain (4–6). Furthermore, this polymodal receptor acts as a molecular sensor for a large array of acute noxious stimuli, of both physical and chemical nature, including, in addition to vanilloids, heat (>42 °C) (7), low pH (pH ≤ 6.5) (8, 9), and peptide toxins (10, 11). Although endovanilloids (such as the endocannabinoids anandamide, N-arachidonoyl dopamine (NADA), and lipoxygenase products of arachidonic acid) have been identified as TRPV1 agonists (12, 13), its most known and studied activator is the exovanilloid capsaicin, the “hot” ingredient in chili peppers (14). However, although elucidating this ligand-receptor interaction will provide a better understanding of the mechanism underlying noxious stimuli detection in the pain pathway, the molecular basis of vanilloids-mediated TRPV1 activation and deactivation is not fully understood.
Since TRPV1 cloning in 1997 (3), several residues that participate in its vanilloid-mediated activation have been identified (15–19). The identification of these residues, mainly through site-directed mutagenesis analyses, laid the basis for the ever increasing body of studies aiming to elucidate various aspects of this important receptor (20–22). These studies have led to the identification of the vanilloid binding site (VBS), a TRPV1 intracellular pocket to which all known vanilloids, endogenous and exogenous, bind and evoke subsequent channel activation (23–28). The solved cryo-EM structure of the rat TRPV1 (rTRPV1) showed that in the presence of vanilloids (capsaicin or resiniferatoxin), residues within the VBS are scattered around the agonist, in proximity to the intracellular domain between the receptor S3 and S4 transmembrane segments (23). Moreover, based on this cryo-EM structure, a recent molecular docking and molecular dynamic studies proposed potential configurations of vanilloids inside the VBS (26–28). However, the VBS mechanism of action and the roles of its different residues in the vanilloid-evoked TRPV1 response are yet to be defined.
The tyrosine at position 511 of the rTRPV1 was the first VBS residue to be identified as a participant in capsaicin and resiniferatoxin-evoked receptor activation (15), as the Y511A substitution alone was sufficient to abolish receptor activation by these exogenous vanilloids (15, 17). The rTRPV1 cryo-EM structures in the presence of exovanilloids clearly points to a close proximity of this highly conserved residue to the agonist (23). Moreover, these structures indicate that Tyr511 assumes two distinct rotamers in apo versus bound states, where its side chain points away either from or into the VBS, respectively (Fig. 1A). Although recent molecular docking and molecular dynamics studies point to an interaction between Tyr511 and the ligand in the bound state (Fig. 1B) (26–28), its position in the apo state suggests a limited role in the initial interaction between vanilloids and their binding site. Therefore, how this residue participates in the vanilloids-evoked TRPV1 activation remains to be determined.
FIGURE 1.
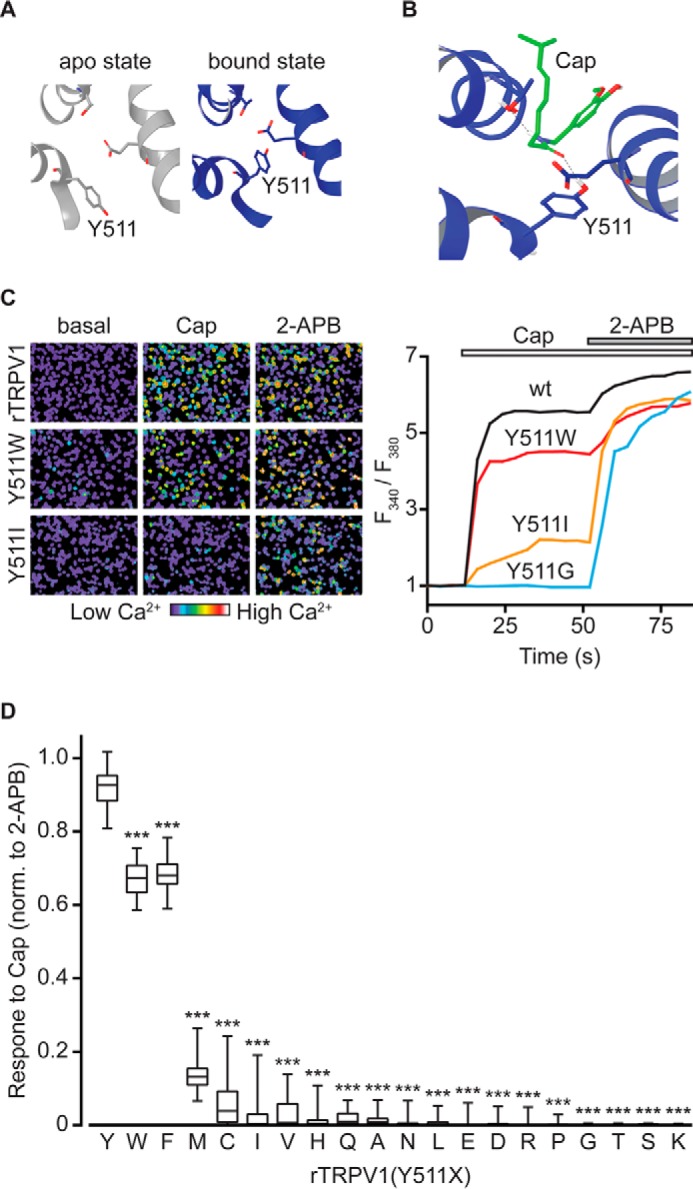
Substitutions in position 511 of rTRPV1 differentially affect capsaicin-evoked response. A, ribbon presentations of top views of rTRPV1 putative VBS in the apo (left; PDB code 3j5p; gray) and capsaicin-bound (right; PDB code 3j5r; blue) states. Tyr511, Thr550, and Glu570 residues are shown as sticks. Note the dramatic rotamer shift of Tyr511. B, a representative configuration of capsaicin-docked VBS (Cap; green). Tyr511, Thr550, and Glu570 residues are shown as sticks. Hydrogen bonds are shown as dashed black lines. C, representative calcium imaging analysis of rTRPV1 response. Left: pseudo-colored images of Fura-2-loaded HEK293T cells expressing the wt rTRPV1 (top panel), and the mutant receptors rTRPV1 (Y511W) (middle panel) and rTRPV1 (Y511I) (bottom panels) before (basal) and after application of capsaicin (2 μm), and after application of 2-APB (0.3 mm). Scale bar indicates levels of intracellular calcium. Right: changes with time of intracellular calcium levels of HEK293T cells expressing the wt (black line), Y511W (red line), Y511I (orange line), and Y511G (blue line) receptors in response to 2 μm capsaicin (empty bar), followed by subsequent application of 0.3 mm 2-APB (gray bar). All graphs represent an average of 50 2-APB responsive cells. D, box and whiskers plot shows capsaicin (2 μm)-evoked calcium response of HEK293T cells expressing rTRPV1 with the indicated Tyr511 substitutions, normalized to the 2-APB (0.3 mm)-evoked response. Boxes represent the mean of 3–4 independent experiments (each n ≥ 50 cells). Statistical significance between normalized responses of wt rTRPV1 and different mutant constructs are indicted as ***, p ≤ 0.001 (ANOVA followed by multiple comparison test).
Taking into account the suggested rotation of the rTRPV1 Tyr511 residue (23) and its interaction with vanilloid in the bound state (26–28), we hypothesize that this residue entraps the vanilloids in the VBS, thus prolonging ligand occupancy in its binding pocket and enabling full duration of channel activation. To test this hypothesis, we generated an array of rTRPV1 mutants, containing the whole spectrum of Tyr511 substitutions, and tested their response to the exovanilloid capsaicin and the endovanilloid NADA by calcium imaging and electrophysiology. Our data showed that only substitutions of Tyr511 to aromatic amino acids were able to mimic, albeit partially, the vanilloid-evoked activation pattern of the wt receptor. Although these substitutions reduced the channel sensitivity to vanilloids, indicated by the rightward shift in capsaicin dose-response, maximal open-channel lifetime was achieved. Moreover, whereas current activation rate remains intact, Tyr511 substitutions lead to a faster current deactivation. Taken together, our data clearly indicate that upon vanilloid binding, Tyr511 rotates and interacts with the ligand to enable a full duration of channel activity. Thus, Tyr511 primarily regulates the deactivation process of vanilloid-mediated TRPV1 response. Furthermore, our findings suggest that substituting this residue to small aliphatic amino acids (such in the widely used Y511A substitution) shorten the duration of ligand occupancy in the VBS, which does not allow adequate channel gating. To conclude, we propose that upon its rotation, Tyr511 binds vanilloids to stabilize ligand-receptor interaction and allow vanilloid-evoked TRPV1 activation and pain sensation.
Experimental Procedures
Structural Representation
To refine the protein structure, capsaicin bound (PDB code 3j5r) (23) and apo (PDB code 3j5p) (29) rTRPV1 reconstructions were downloaded from RCSB Protein Data Bank, and loaded into the Maestro molecular modeling environment (Maestro, version 9.2, Schrödinger, New York). Ionizable residues were set to their normal ionization states at pH 7, and a restrained energy minimization (with a relatively higher convergence threshold of a gradient to ∼0.3 kJ/Å mol) was performed using OPLS-2005 force field.
Molecular Docking Analysis
The capsaicin structure was reconstituted using the Maestro Molecular Modeling Environment 2008 (Maestro, version 9.2, Schrödinger), and energy was minimized using Macromodel minimization panel with a OPLS-2005 force field and GB/SA water model, applying a constant dielectric of 1.0. Polak-Ribiere first derivative, conjugated gradient minimization was employed with maximum of 1,000 iterations and a convergence threshold of a gradient to 0.05 kJ/Å mol. The LigPrep2.0 module of Schrodinger was used to generate possible ionization states at pH 7.0 ± 2.0. Conformers from confgen-ligprep output were then docked in an rTRPV1 VBS using the extra precision (XP) scoring mode of Glide. A grid of center of 10 × 10 × 10 Å on residues 511, 570, and 550 was constructed. Van der Waals radii of ligand and receptor were 0.8 and a partial charge cut-off of 0.15 was used. Ten docked poses were generated to sample a range of possible docking modes. An in situ refinement of the predicted protein-ligand complexes was then performed using Macromodel minimization, using the OPLS-2005 force field and GB/SA water model with a constant dielectric of 1.0. A Polak-Ribiere first derivative, conjugate gradient minimization was employed with a maximum of 5,000 iterations and convergence threshold of a gradient of 0.05 kJ/Å mol. In addition, per-residue free energy contributions (Van der Walls, Columbic, C-C nonbonding, and H bonding) and average distances for the intrinsic docking poses were analyzed.
Site-directed Mutagenesis
Site-directed mutagenesis of the wt rTRPV1 gene cloned into a pCDNA3.1+ plasmid was performed using a Q5® Site-directed Mutagenesis Kit (New England Biolabs) according to the manufacturer's protocol, with primers (Sigma) designed using the NEBaseChanger software (New England Biolabs). Additionally, following digestion with the restriction enzymes HindIII and ApaI (New England Biolabs), wt rTRPV1 and Y511F, Y511M, and Y511W mutants were subcloned into the pCDNA5/FRT/TO plasmid using the T4 DNA ligase (Thermo Scientific), according to the manufacturer's protocol. Following bacterial (TOP 10 Chemically Competent Escherichia coli (Invitrogen)) transformation, all DNA constructs were extracted using a miniprep kit (Purelink®, Invitrogen) according to the manufacturer's protocol, and sequenced (Hy labs, Israel) to verify mutagenesis.
Cell Culturing and Transfection
Cell culture and transfection were carried out as described (11, 25). Briefly, human embryonic kidney 293T (HEK293T) and Flp-in T-REx 293 (Invitrogen) cells were transfected with a total of 1 μg of DNA (100–500 ng of rTRPV1 expressing constructs, 200 ng of enhanced green fluorescent protein in pCDNA3.1+, and an empty pcDNA3.1+ plasmid to reach a total DNA amount of 1000 ng) using Mirus LT1 transfection reagent (Mirus Bio) and Opti-MEM I (reduced serum medium, Invitrogen) 24 h before analysis. Co-transfection with enhanced green fluorescent protein was carried for quick identification of successful transfection (except those used in calcium imaging). Three hours before electrophysiological analysis, transfected Flp-in T-REx 293 cells were treated with doxycycline (0.2–0.7 μg/ml) to induce transgene expression.
Live-cell Calcium Imaging
Transfected HEK293T cells were spotted at poly-d-lysine (0.2 mg/ml)-coated imaging chambers (μ-slide, 8 well, Ibidi, Germany) 3–4 h before being loaded with Fura-2AM (Invitrogen; 2 μm) dissolved in Ringer's salt solution (in mm: 140 NaCl, 2.5 KCl, 1.8 CaCl2, 2 MgSO4, 20 HEPES, and 5 d-glucose, pH 7.4, with NaOH) for 1 h. Cells were then washed twice with Ringer's solution and incubated for 30 min. Stock solutions of 10 mm capsaicin (Tocris Bioscience, UK), 10 mm NADA (Tocris Bioscience), and 100 mm 2-aminoethoxydiphenyl borate (2-APB; Tocris Bioscience) in DMSO were dissolved in Ringer's salt solution to obtain the desired concentrations. Using an inverted microscope (Olympus IX70, Japan), cells were illuminated with a xenon arc lamp, and excitation wavelengths (340/380 nm) were selected by a Lambda DG-4 monochromatic wavelength changer (Sutter Instrument). Fluorescence emission at >480 nm was captured with a front-illuminated interline CCD camera (Exi Blue, QImaging, BC, Canada) and the MetaFluor Fluorescence Imaging Software (Universal Imaging Corp.) that was further used for offline analysis. Background-corrected 340/380 nm ratio images were collected every 4 s.
Electrophysiology
Whole cell and single channel recordings were carried out as described (25). Briefly, HEK293T cells were used for whole cell recordings and Flp-in T-REx 293 cells for single channel recordings, 24 h after transfection. Patch electrodes were fabricated from borosilicate glass using the P1000 Micropipette Puller (Sutter Instrument, CA) and fire polished using the microforge MF-900 (Narishige, Tokyo, Japan). Glass electrodes were fire polished to a resistance of 3–4 megohms for whole cell and 12–15 megohms for outside-out patch. The pipette solution constituted (mm): 130 KCl, 4 NaCl, 2 MgSO4, 0.5 CaCl2, 10 EGTA, and 10 HEPES adjusted to pH 7.2 with KOH (estimated free Ca2+ was 17 nm, using Maxchelator software). Bath solution comprised of (mm): 140 NaCl, 2.3 KCl, 2 MgSO4, 5 HEPES, and 5 MES (pH 7.4) was adjusted with NaOH. Calcium was excluded from external solution to avoid TRPV1 desensitization (30). Following establishment of the whole cell or outside-out configuration, cells were perfused using the ValveBank perfusion system (AutoMate Scientific, CA). Stock solutions of 10 mm capsaicin (Tocris Bioscience) or NADA (Tocris Bioscience) in DMSO were dissolved in bath solution to obtain the desired concentrations. Recordings were performed using Digidata 1440A digitizer and Axopatch 200B amplifier (Molecular Devices, CA). Whole cell currents were recorded at −40 mV, whereas outside-out patches were held at +50 mV. Data were collected and analyzed using pClamp 10.2 software (Molecular Devices). All experiments were carried out at 22–23 °C. Whole cell currents were filtered at 1 kHz using a low-pass Bessel filter and sampled at 2–5 kHz, whereas single channel currents were filtered at 5 kHz and sampled at 50 kHz.
Data Analysis
All statistical data were calculated using SigmaPlot 11 (Systat Software, CA) and Prism 6 (GraphPad Software) software. Student's t test and ANOVA were used to determine statistical significance. To determine the ratio between protons and vanilloid-evoked responses, maximal currents (i.e. peaks) were used for quantification. Dose-responses were calculated using the sigmoidal Hill equation (Eq. 1) as follows,
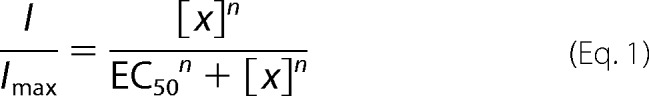 |
when I = measured current; Imax = maximal current at the relevant saturating dose (pre-measured for each construct); x = tested agonist concentration; EC50 = calculated concentration that elicits 50% of maximal current, and n = Hill coefficient. Clampfit 10.2 software (Molecular Devices) was used to calculate rates of whole cell current activation (τon) and washout (τoff), using exponential fitting (using single-exponential function from 10 to 90% of the peak) as previously described (31, 32), and for single-channel analysis. For each construct, 300–1400 events were collected from 4 to 8 separate outside-out patches. To determine channel amplitude, all-point amplitude histograms were generated using data digitally filtered at 1 kHz. To determine channel open probability, traces filtered at 2 kHz were idealized using the half-amplitude threshold crossing method (25, 31, 33).
Results
Determining Capsaicin Sensitivity in rTRPV1 Containing Various Amino Acid Substitutions in Position 511
To determine the role tyrosine 511 plays in the activation mechanism of rTRPV1, we substituted this amino acid with all remaining amino acids by generating an array of mutated rTRPV1 constructs, and transiently expressed them in HEK293T cells (Fig. 1). We determined the sensitivity of rTRPV1 containing the various Tyr511 substitutions to capsaicin (2 μm) by calcium imaging (Fig. 1C). To verify expression, correct folding, and trafficking of the different rTRPV1 mutants, we exposed them to 2-APB (0.3 mm), a non-vanilloid TRPV1 agonist that does not require Tyr511 (34), and analyzed their response by calcium imaging (Fig. 1C). As summarized in Fig. 1D, capsaicin-evoked TRPV1 activation was dramatically impaired in all mutants; whereas substitutions to small or charged amino acids showed the most profound effect, substitutions to the two aromatic amino acids, phenylalanine (F) and tryptophan (W), had only a moderate effect.
We next assessed the activation pattern of the two most capsaicin-sensitive mutants, Y511F and Y511W, as compared with wt rTRPV1 (Fig. 2). We also determined the activation pattern of two of the least capsaicin-sensitive mutated receptors, Y511M and Y511I, which maintained sufficient sensitivity to allow current analysis. Using whole cell recordings (VH = −40 mV), the currents evoked by two capsaicin concentrations, 1 and 30 μm, were determined and normalized to the corresponding proton-evoked currents (pH 5.5) (Fig. 2). These two capsaicin concentrations were chosen because 1 μm capsaicin is a saturating concentration for the wt rTRPV1 (25, 26), whereas 30–100 μm is regarded as the maximal concentrations for this agonist (mainly due to its limited solubility in physiological solutions). To allow comparison between the different constructs, we used protons (pH 5.5) because they activate TRPV1 through a different binding site (the outer pore domain) (35, 36) and via a different activation mechanism (25). Importantly, to avoid cross-contamination between different agonists during sequential applications, protons, rather than 2-APB, were used as they are washed fast and fully. Calcium was omitted from all electrophysiological solutions to avoid calcium-dependent receptor desensitization (30). As shown in Fig. 2A, both 1 and 30 μm capsaicin evoked similar currents in the wt receptor, which was double of the protons-evoked response (IpH 5.5 = 429 pA; I1μm Cap = 882 pA; I30μm Cap = 920 pA), as previously shown (11, 25, 35, 36). However, whereas 30 μm capsaicin evoked a robust response in the Y511W mutated receptor, which was close to double the protons evoked current (IpH 5.5 = 934 pA; I30μm Cap = 1569 pA), the current evoked by the lower capsaicin concentration was half of that evoked by protons (I1μm Cap = 498 pA). This phenomenon was even more profound in receptors harboring a substitution to a non-aromatic amino acid, such as the Y511M substitution. Application of 1 μm capsaicin evoked a small current (I1μm Cap = 19 pA) in the Y511M mutant receptor, and application of 30 μm capsaicin resulted in a current that was half of that evoked by protons (IpH 5.5 = 3044 pA; I30μm Cap = 1794 pA). Of note, this was only obtained when cells with a substantially high receptor expression levels, indicated by high protons current, were chosen for analysis. Fig. 2B summarizes normalized currents of five different rTRPV1 constructs (wt, Y511W, Y511F, Y511M, and Y511I). To exclude that those substitutions affect protons sensitivity, we analyzed the protons dose-response of the wt, Y511W, and Y511F constructs. As shown in Fig. 2C, comparable dose-response curves were obtained. Thus, our calcium imaging and electrophysiological data demonstrate that only substitutions to the two aromatic amino acids (Phe and Trp) can partially preserve TRPV1 response to capsaicin, albeit in high agonist concentration.
FIGURE 2.
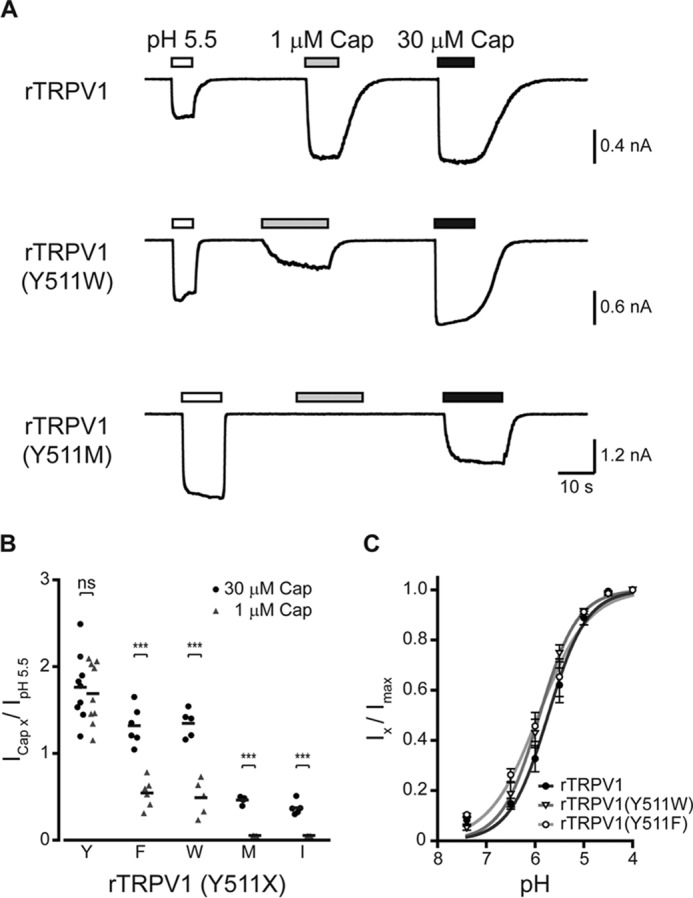
rTRPV1 Tyr511 substitutions exhibit impaired capsaicin sensitivity. A, current traces of whole cell recordings from HEK293T cells expressing the wt rTRPV1 (top panel), and the mutants rTRPV1 containing the Y511W (middle panel), and Y511M (bottom panel) substitutions at a holding potential of −40 mV. Cells were first exposed to pH 5.5 (empty bars; as a reference for channel expression) followed by applications of 1 and 30 μm capsaicin (Cap; gray and black bars, respectively). Bars above the trace indicate the time course of each activator application. B, mean/scatter-dot plot representing the normalized amplitude of whole cell currents in cells expressing rTRPV1 with indicated Tyr511 substitutions, evoked by 1 (gray triangles) or 30 μm (black circles) capsaicin, normalized to the current amplitude of the pH 5.5 response. Statistical significance between responses to different capsaicin concentrations for each construct was determined with paired Student's t test, where *** represents p ≤ 0.001 and ns represents a non-significant difference (n = 5–9 cells). Note a reduction in the capsaicin-evoked response in all tested mutants. Importantly, the recordings from Y511M and Y511I constructs could only be obtained when overexpressed. C, normalized concentration-response relationships to protons of wt and the indicated mutant receptors. Points represent the mean ± S.E. response of 6–7 HEK293T cells and solid lines are fit to the Hill equation: rTRPV1 (full circles, black line; nH = 1.1 ± 0.2; EC50 = pH 5.7 ± 0.3), rTRPV1 (Y511W) (empty triangle, dark gray line; nH = 1.1 ± 0.1; EC50 = pH 5.9 ± 0.1), and rTRPV1 (Y511F) (empty circle, light gray line; nH = 0.9 ± 0.1; EC50 = pH 5.8 ± 0.3). Holding potential of −40 mV.
Substitutions in Position 511 Impair Binding of Capsaicin to rTRPV1
To determine whether the impaired capsaicin sensitivity upon substitutions in position 511 affects rTRPV1 gating and/or binding of the agonist, we compared the dose-responses of the two capsaicin-responsive mutated receptors, Y511F and Y511W, to that of the wt receptor. These mutant receptors were chosen for analysis, as they were the only ones to reach saturated responses in soluble capsaicin concentration (≤50 μm in physiological solutions). Whole cell recordings (VH = −40 mV) of HEK293T cells transiently expressing the wt and mutant receptors were used for this analysis, where cells were exposed to increasing concentrations of capsaicin (to minimize concentration estimation errors arising from its hydrophobicity) until currents were saturated (Fig. 3A). As shown in Fig. 3, the obtained dose-response curves indicate a dramatic shift in the sensitivity of mutant receptors to capsaicin (Table 1). However, clear current saturation was achieved in the mutated receptors and the obtained Hill coefficients were similar to that of the wt rTRPV1 (Fig. 3 and Table 1).
FIGURE 3.
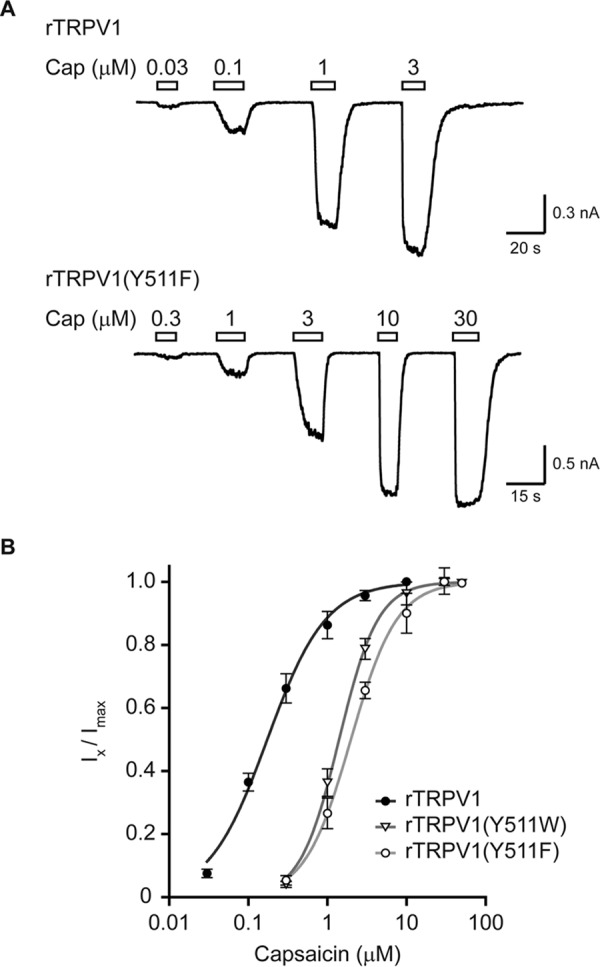
Substitutions of rTRPV1 Tyr511 to aromatic amino acids lead to a parallel rightward shift in capsaicin dose-response. A, current traces of whole cell recordings from HEK293T cells expressing either wt (rTRPV1; top panel), or mutant (rTRPV1 Y511F; bottom panel) receptor at holding potential of −40 mV. Cells were exposed to increasing concentrations of capsaicin (Cap) as indicated. Empty bars above the trace indicate the time course of each concentration application. B, normalized capsaicin concentration-response relationships of wt and the indicated mutated receptors. Points represent the mean (± S.E.) response of 6–9 HEK293T cells and solid lines are fit to the Hill equation: rTRPV1 (full circles, black line; nH = 1.2 ± 0.1; EC50 = 0.17 ± 0.02 μm), rTRPV1 (Y511W) (empty triangle, dark gray line; nH = 1.7 ± 0.1; EC50 = 1.40 ± 0.04 μm), and rTRPV1 (Y511F) (empty circle, light gray line; nH = 1.5 ± 0.1; EC50 = 1.98 ± 0.08 μm). Holding potential of −40 mV.
TABLE 1.
Whole cell and single channel properties of wt rTRPV1 and indicated Tyr511 substitutions (Y511X)
| rTRPV1 (Y511X) | Whole cell |
Single channel |
||||
|---|---|---|---|---|---|---|
| EC50 | Hill coefficient | τon | τoff | Open probability | Conductance | |
| μm | n | s | Po | pS | ||
| Y | 0.17 ± 0.02 | 1.2 ± 0.1 | 0.28 ± 0.07 | 7.60 ± 0.94 | 0.93 ± 0.01 (1 μm) | 93 ± 3 |
| 0.97 ± 0.01 (30 μm) | ||||||
| F | 1.98 ± 0.08 | 1.5 ± 0.1 | 0.21 ± 0.05 | 2.46 ± 0.09 | 0.37 ± 0.06 (1 μm) | 88 ± 2 |
| 0.96 ± 0.01 (30 μm) | ||||||
| W | 1.40 ± 0.04 | 1.7 ± 0.1 | 0.21 ± 0.04 | 3.29 ± 0.28 | 0.34 ± 0.05 (1 μm) | 95 ± 3 |
| 0.94 ± 0.01 (30 μm) | ||||||
| M | NDa | ND | ND | ND | NRb (1 μm) | 86 ± 5 |
| 0.03 ± 0.01 (30 μm) | ||||||
a ND, not determined.
b NR, no response.
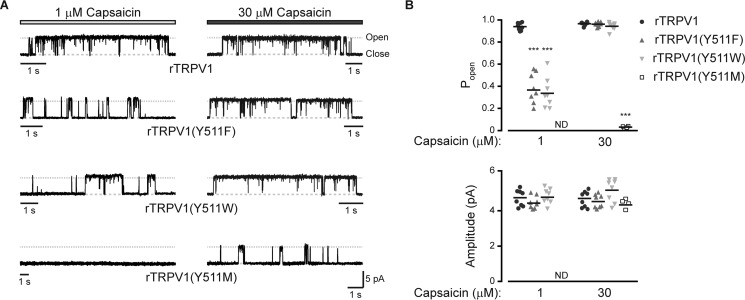
To test for potential impairment of vanilloids-evoked channel gating in the mutated TRPV1, we analyzed the open probability and conductance of the capsaicin-evoked response (Fig. 4). To this end, we explored the single channel properties of the wt receptor, the highest capsaicin-sensitive mutants containing the Y511F or Y511W substitutions, and a relatively capsaicin-insensitive mutant containing the Y511M substitution (Figs. 1 and 2). Constructs were expressed using the Flp-in T-REx 293 system for controlled protein expression, because it allows low and time-dependent channel expression in levels suitable for single-channel recordings (11, 25, 37). Using the outside-out patch configuration of the patch clamp technique from Flp-in T-REx 293 cells transiently expressing the different constructs, we analyzed the conductance and open probability of capsaicin-evoked currents. We initially exposed the patch to 1 μm capsaicin, which was followed by exposure to 30 μm capsaicin (Fig. 4A). As shown in Fig. 4, the saturating capsaicin concentration for the wt receptor (1 μm) resulted in low open probability of the Y511F and Y511W receptors (Table 1). In contrast, the maximal capsaicin concentration (30 μm) resulted in a similar open probability in wt, Y511F, and Y511W receptors (Table 1). No activity was detected for the Y511M receptor in response to 1 μm capsaicin, whereas low open probability was observed in response to 30 μm capsaicin (Fig. 4 and Table 1). Notably, no changes in the conductance of the channel were obtained in the different mutant receptors (Fig. 4B and Table 1). Thus, our data indicate that substitutions in position 511 of rTRPV1 only minimally affected channel gating.
FIGURE 4.
A maximal open-channel lifetime is achieved when an aromatic residue occupies position 511. A, representative current traces from outside-out patches of capsaicin-exposed Flp-in T-REx HEK293 cells transiently expressing the indicated constructs. Upward (outward) currents indicate channel opening (gray dash line). Shown are representative channel activities upon exposing the patches to rTRPV1-saturating (1 μm; light gray bar; left panel) and maximal (30 μm; dark gray; right panel) capsaicin concentrations. Holding potential at +50 mV were sampled at 50 kHz and filtered at 1 kHz for display. B, mean/scatter-dot plot (n = 4–8 patches) representing the open probability (top panel, Popen) and amplitude (bottom panel) of the indicated rTRPV1 single-channel currents activated by 1 or 30 μm capsaicin. Statistical significance between the wt and mutants rTRPV1 in each capsaicin concentration are indicted as ***, p ≤ 0.001 (ANOVA followed by multiple comparison test). ND, not determined due to lack of activity.
Substitutions at rTRPV1 Position 511 Increase the Rate of Current Washout
Our results indicate that only aromatic substitutions maintain relatively high sensitivity to capsaicin (Figs. 1–3). Taken together with the suggested rotation of the rTRPV1 Tyr511 residue upon vanilloid binding and the unaffected channel gating (23) (Figs. 1–4), we hypothesize that the observed impaired binding of capsaicin to mutated rTRPV1 resulted from impaired entrapping of the agonist in the VBS. To test whether the substitutions in this residue affect the duration of agonist binding, we measured the rates of activation and deactivation (at 30 μm capsaicin) of the wt receptor and those containing Y511F and Y511W substitutions. As shown in Figs. 3 and 4, the activation of these mutants was saturated at maximal capsaicin concentration and their open probability was similar to that of the wt receptor at the relevant maximal concentration. Using the whole cell configuration, we recorded capsaicin-evoked currents from HEK293T cells transiently expressing the different constructs (VH = −40 mV). To verify similar expression levels, cells were initially exposed to protons (pH 5.5; 5 s) and only those with a protons-evoked current of 0.8–1 nA (corresponded to maximal capsaicin-evoked current of 1.8–2 nA) were further analyzed. As shown in Fig. 5, wt and mutated receptors exhibited similar activation rates when exposed to capsaicin (Table 1). Nevertheless, washout rates were dramatically increased in mutated receptors as compared with wt (Table 1). These results indicate that the Y511F and Y511W substitutions did not interfere with the initial binding of capsaicin to TRPV1; rather, they shorten the duration of agonist occupancy by increasing its dissociation rate from the VBS.
FIGURE 5.
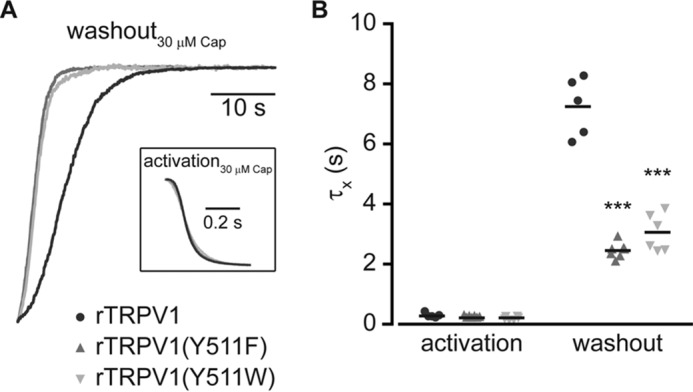
Substitutions of the Tyr511 increase the current washout rate. A, superimposed normalized whole cell washout currents of indicated constructs (wt rTRPV1, black line; rTRPV1 (Y511F), dark gray line; rTRPV1 (Y511W), light gray line) exposed to 30 μm capsaicin (Cap). Inset, superimposed normalized whole cell activation currents of the same constructs. Holding potential was −40 mV. B, mean/scatter-dot plot (n = 5–7 cells) representing the rate (τ) of activation (left) and washout (right) of the whole cell currents from the indicated constructs evoked by 30 μm capsaicin. Rates were determined by exponential fitting of the data. Statistical significances between the wt and mutants rTRPV1 are indicted as ***, p ≤ 0.001 (ANOVA followed by multiple comparison test).
Substitutions in rTRPV1 Tyr511 Similarly Affect Exo- and Endovanilloids Evoked Response
To assess whether the role of the Tyr511 residue is unique to capsaicin or also applicable to other vanilloids, we extended our analysis to the endovanilloid NADA. To this end, we determined NADA-evoked response of the rTRPV1 Tyr511 substitutions Trp, Phe, Met, Ile, and Gly (Fig. 6). Throughout our analysis, the maximum concentration of NADA was used (i.e. 2 μm) to allow VBS saturation (38). Calcium imaging analysis revealed similar reduction in receptor sensitivity to NADA upon substitutions of the Tyr511 residue, as obtained for capsaicin (compare Figs. 1D and 6A). In addition, the ratio between the currents evoked by NADA and protons in receptors containing aromatic substitutions was significantly smaller than the wt, similar to the ratios in maximal capsaicin concentration (compare Figs. 2B and 6B). Furthermore, similarly to capsaicin, a dramatic increase in the NADA washout rate was observed for the aromatic substitutions (compare Fig. 5 to 6, C and D). Notably, the washout rates of NADA for the wt receptor and the analyzed substitutions were slower than the capsaicin washout rates (for wt rTRPV1, capsaicin: τoff = 7.60 ± 0.94 s, NADA: τoff = 22.10 ± 4.20 s). This is likely due to the longer aliphatic chain of NADA as compared with capsaicin, resulting in its higher hydrophobicity, which retains it in proximity to the receptor. In summary, our findings indicate that residue Tyr511 of rTRPV1 is required for entrapping the agonist to its binding site, and suggest this tyrosine forms favorable interactions with the ligand to allow full duration of channel activation.
FIGURE 6.
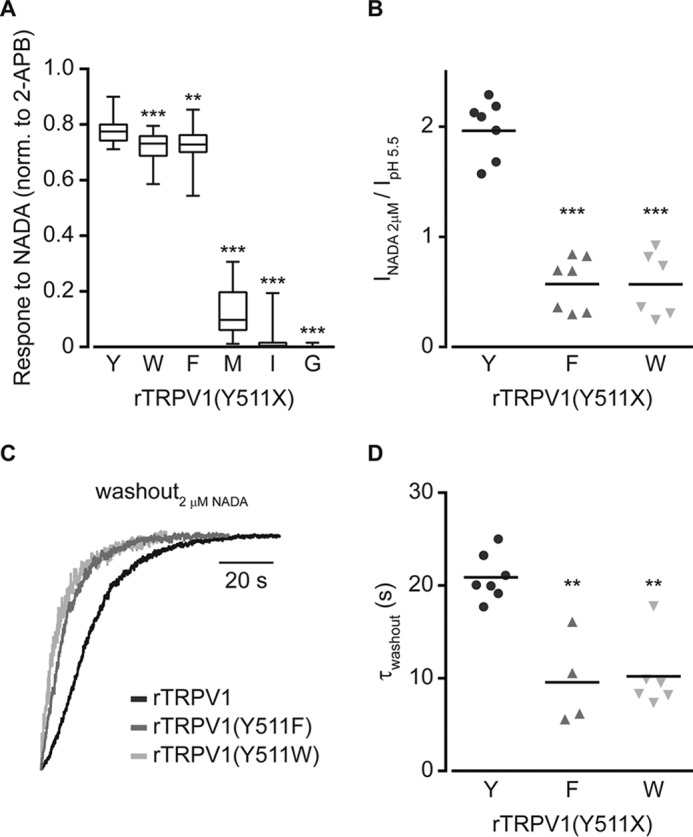
Sensitivity and washout rate of the endovanilloid NADA are affected by Tyr511 substitutions. A, box and whiskers plot shows NADA (2 μm)-evoked calcium response of HEK293T cells expressing rTRPV1 with or without the indicated Tyr511 substitutions, normalized to the response to 2-APB (0.3 mm). Boxes are mean of 3 independent experiments (each n ≥ 50 cells). Statistical significance between normalized responses of wt and mutated rTRPV1 are indicted as **, p ≤ 0.01 and ***, p ≤ 0.001 (ANOVA followed by a multiple comparison test). B, mean/scatter-dot plot (n = 6–7 cells) representing normalized amplitudes of the whole cell currents in cells expressing rTRPV1 with or without the indicated Tyr511 substitutions, evoked by 2 μm NADA, normalized to the current amplitude evoked by pH 5.5. Statistical significance between normalized responses of wt rTRPV1 and different mutant constructs is indicted as ***, p ≤ 0.001 (ANOVA followed by multiple comparison test). C, superimposed normalized whole cell washout currents of the indicated constructs (wt rTRPV1, black line; rTRPV1 (Y511F), dark gray line; rTRPV1 (Y511W), light gray line) exposed to 2 μm NADA. Holding potential at −40 mV. D, mean/scatter-dot plot (n = 4–7 cells) representing the washout rate (τ) of whole cell currents evoked by 2 μm NADA. Rates were determined by exponential fitting of the data. Statistical significances between the wt and mutants rTRPV1 are indicted as **, p ≤ 0.01 (ANOVA followed by multiple comparison test).
Discussion
Endogenous and exogenous vanilloids activate sensory nerve terminals by opening TRPV1 channels, eliciting receptor generator potentials, initiating action potentials, and leading to pain sensation (2, 6, 21). Although efforts to elucidate the molecular basis of these sensory processes have identified several amino acids in the TRPV1 protein that participate in vanilloids binding and subsequent channel gating (15, 17, 19), their mechanism(s) of action remained largely unknown. Here, we analyzed the role of a conserved tyrosine, Tyr511 in the rTRPV1 sequence, in the vanilloid-evoked response. We show that substitutions to other aromatic residues only partly mimic tyrosine activity, implying that this amino acid directly interacts with the agonist. Furthermore, we provide evidence that rTRPV1 Tyr511 secures the agonist in its binding pocket, allowing full duration of the vanilloid-evoked response.
Tyr511 of rTRPV1 was the first amino acid to be identified as part of the VBS (15). Although the Y511A substitution was extensively used to study physiological, structural, and biophysical processes related to the VBS, the exact role of this residue in the activation mechanism of TRPV1 by vanilloids remained largely unknown. To outline the specific role of Tyr511 in the vanilloid-evoked TRPV1 response, we initially substituted this residue with all of the different amino acids. Our findings clearly indicate that only substitution to the aromatic residues, phenylalanine and tryptophan, can mimic, albeit partly, the vanilloid-evoked response of the wt receptor (Figs. 1, 2, and 6). Although receptors containing substitutions to other aliphatic residues such as isoleucine, methionine, and cysteine were also activated by capsaicin and NADA, they did so to a much lower extent than substitutions to aromatic residues, whereas no activation was obtained upon substitutions to small aliphatic, charged or polar amino acids (Figs. 1, 2, and 6).
A possible caveat of our analysis is the limited solubility of vanilloids, which precludes analysis in response to high concentrations. Therefore, it is possible that mutated receptors that did not respond to 30 μm capsaicin in our analysis maintained low sensitivity to this agonist and are capable to respond to higher concentrations. However, all of the known VBS-associated activators or inhibitors are highly hydrophobic and the membrane proximity of this site indicates that more hydrophilic agents might not reach this site (2). Nevertheless, our findings demonstrate that a maximal vanilloid-evoked TRPV1 activation could only be achieved if an aromatic residue occupies position 511.
What is the role of Tyr511 in the activation mechanism of TRPV1 by vanilloids? An elegant, simplified model was proposed by Del Castillo and Katz (39) (recently summarized by William Zagotta (40)) (Equation 2), which separates ligand binding from activation gating of ligand-gated ion channels,
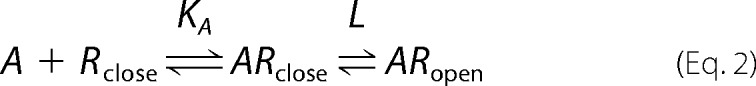 |
where A is the agonist, R is the receptor, KA is the association equilibrium constant for binding, and L is the equilibrium constant of the bound channel. The parallel shift in the apparent EC50 of receptors containing the Y511F or Y511W substitutions (with no change in the Hill coefficient; Table 1 and Fig. 3), together with their unaffected open probability (at maximal capsaicin concentration; Fig. 4), clearly indicate the binding (KA) was most affected in these TRPV1 mutants (“K phenotype”) (41). Furthermore, we found a dramatic shift of ∼2–3-fold in the deactivation rate, but no shift in the activation rate under saturation conditions of both capsaicin and NADA (Figs. 5 and 6). Thus, our results point to a pivotal role of rTRPV1 Tyr511 in ligand binding, mainly in stabilizing the ligand-receptor complex, with no apparent role in receptor gating.
Our data suggest that Tyr511 entraps the agonist to its binding pocket. Thus, agonist binding leads to movement of this residue, as suggested by the recently published rTRPV1 structure and molecular docking and dynamic analyses (23, 26–28), resulting in favorable interactions and stabilization of the agonist in its pocket. Surprisingly, substitutions of tyrosine in this position with either phenylalanine or tryptophan caused a comparable shift in the apparent EC50 (Fig. 3) and current washout (Figs. 5 and 6). This may reflect a role of the tyrosine hydroxyl group in anchoring the agonist. Indeed, the formation of a hydrogen bond between the Tyr511 hydroxyl group and capsaicin amide oxygen was suggested by a recently reported molecular dynamics simulation (27, 28) (Fig. 1B). Our docking analysis suggests that the aromatic interaction between the phenyl ring of capsaicin and rTRPV1 Tyr511 allows a significant gain in free energy (−6 kcal/mol, with an average distance of ∼2 Å), which was further synergized by the hydrogen bond (by −1 kcal/mol). However, whereas the docking score of the Y511F substitution (−4.1 kcal/mol) was only marginally lower as compared with the wt receptor (−4.8 kcal/mol), its free energy gain was significantly lower (−2 kcal/mol, with an average distance of ∼2.5 Å). These observations suggest that whereas aryl-aryl interactions with aromatic rings favor vanilloid binding, the hydrogen bond between the agonist and the tyrosine residue further strengthen this interaction.
Thus, in combination with the structure analysis and molecular dynamic simulation (23, 27, 28), our mutagenesis analysis points to a two-step mechanism of Tyr511-depdendent TRPV1 activation. First, agonist binding induces conformational changes, resulting in an aryl-aryl interaction between the ligand and Tyr511 and enabling channel activation (as evident by the similar activation pattern of the aromatic substitutions, Figs. 4 and 5). Ligand-receptor interactions involving aromatic rings, as we propose for vanilloids and Tyr511 of TRPV1, are key processes in biological recognition (42). Second, the formation of hydrogen bonds between Tyr511 and the agonist, which prolongs agonist occupancy in the VBS (as suggested by the increased deactivation rate upon substitution to other aromatic acids, Figs. 5 and 6). In summary, we suggest that TRPV1 Tyr511 is required for anchoring the agonist to the VBS through both aryl-aryl interaction and hydrogen bond formation, which increase the duration of the ligand-receptor complex, enabling adequate channel gating.
Author Contributions
A. P. conceived, coordinated the study, and wrote the paper. R. K. designed, performed, and analyzed the experiments in Figs. 2–5. A. H. designed, performed, and analyzed the experiments in Fig. 6. A. B. and N. Z. designed, performed, and analyzed the experiments in Fig. 1. H. M. provided technical assistance and contributed to the electrophysiological analysis. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Amiram Goldblum for help with the structural analysis and members of the Priel laboratory for helpful discussion and comments. A. P. is affiliated with the Brettler Center for Research in Molecular Pharmacology and Therapeutics and the David R. Bloom Center for Pharmacy, School of Pharmacy, The Hebrew University of Jerusalem.
This work was supported in part by the Israel Science Foundation Grants 1721/12 and 1368/12 (to A. P.), Marie-Curie Career Integration Grant FP7-CIG-321899 (to A. P.), a Jerusalem Brain Committee (JBC) Postdoctoral Fellowship (to R. K.), the Kaete Klausner fellowship (to A. H), and the Fellowship Program for Outstanding Post-Doctoral Researchers of the Planning and Budget Commission of Israel (to A. B.). The authors declare that they have no conflicts of interest with the contents of this article.
- rTRPV1
- rat transient receptor potential vanilloid 1
- 2-APB
- 2-aminoethoxydiphenyl borate
- Cap
- capsaicin
- NADA
- N-arachidonoyl dopamine
- VBS
- vanilloid binding site
- PDB
- Protein Data Bank
- ANOVA
- analysis of variance.
References
- 1. Caterina M. J., and Julius D. (2001) The vanilloid receptor: a molecular gateway to the pain pathway. Annu. Rev. Neurosci. 24, 487–517 [DOI] [PubMed] [Google Scholar]
- 2. Van Der Stelt M., and Di Marzo V. (2004) Endovanilloids: putative endogenous ligands of transient receptor potential vanilloid 1 channels. Eur. J. Biochem. 271, 1827–1834 [DOI] [PubMed] [Google Scholar]
- 3. Caterina M. J., Schumacher M. A., Tominaga M., Rosen T. A., Levine J. D., and Julius D. (1997) The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824 [DOI] [PubMed] [Google Scholar]
- 4. Basbaum A. I., Bautista D. M., Scherrer G., and Julius D. (2009) Cellular and molecular mechanisms of pain. Cell 139, 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dubin A. E., and Patapoutian A. (2010) Nociceptors: the sensors of the pain pathway. J. Clin. Invest. 120, 3760–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Julius D. (2013) TRP channels and pain. Annu. Rev. Cell Dev. Biol. 29, 355–384 [DOI] [PubMed] [Google Scholar]
- 7. Caterina M. J. (2007) Transient receptor potential ion channels as participants in thermosensation and thermoregulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R64–76 [DOI] [PubMed] [Google Scholar]
- 8. Tominaga M., Caterina M. J., Malmberg A. B., Rosen T. A., Gilbert H., Skinner K., Raumann B. E., Basbaum A. I., and Julius D. (1998) The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543 [DOI] [PubMed] [Google Scholar]
- 9. Ryu S., Liu B., and Qin F. (2003) Low pH potentiates both capsaicin binding and channel gating of VR1 receptors. J. Gen. Physiol. 122, 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siemens J., Zhou S., Piskorowski R., Nikai T., Lumpkin E. A., Basbaum A. I., King D., and Julius D. (2006) Spider toxins activate the capsaicin receptor to produce inflammatory pain. Nature 444, 208–212 [DOI] [PubMed] [Google Scholar]
- 11. Bohlen C. J., Priel A., Zhou S., King D., Siemens J., and Julius D. (2010) A bivalent tarantula toxin activates the capsaicin receptor, TRPV1, by targeting the outer pore domain. Cell 141, 834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zygmunt P. M., Petersson J., Andersson D. A., Chuang H., Sørgård M., Di Marzo V., Julius D., Högestätt E. D., Marzo V. Di, and Ho E. D. (1999) Vanilloid receptors on sensory nerves mediate the vasodilator action of anandamide. Nature 400, 452–457 [DOI] [PubMed] [Google Scholar]
- 13. Piomelli D., and Sasso O. (2014) Peripheral gating of pain signals by endogenous lipid mediators. Nat. Neurosci. 17, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Neill J., Brock C., Olesen A. E., Andresen T., Nilsson M., and Dickenson A. H. (2012) Unravelling the mystery of capsaicin: a tool to understand and treat pain. Pharmacol. Rev. 64, 939–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jordt S. E., and Julius D. (2002) Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell 108, 421–430 [DOI] [PubMed] [Google Scholar]
- 16. Chou M. Z., Mtui T., Gao Y.-D., Kohler M., and Middleton R. E. (2004) Resiniferatoxin binds to the capsaicin receptor (TRPV1) near the extracellular side of the S4 transmembrane domain. Biochemistry 43, 2501–2511 [DOI] [PubMed] [Google Scholar]
- 17. Gavva N. R., Klionsky L., Qu Y., Shi L., Tamir R., Edenson S., Zhang T. J., Viswanadhan V. N., Toth A., Pearce L. V., Vanderah T. W., Porreca F., Blumberg P. M., Lile J., Sun Y., Wild K., Louis J.-C., and Treanor J. J. (2004) Molecular determinants of vanilloid sensitivity in TRPV1. J. Biol. Chem. 279, 20283–20295 [DOI] [PubMed] [Google Scholar]
- 18. Boukalova S., Marsakova L., Teisinger J., and Vlachova V. (2010) Conserved residues within the putative S4-S5 region serve distinct functions among thermosensitive vanilloid transient receptor potential (TRPV) channels. J. Biol. Chem. 285, 41455–41462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Winter Z., Buhala A., Ötvös F., Jósvay K., Vizler C., Dombi G., Szakonyi G., and Oláh Z. (2013) Functionally important amino acid residues in the transient receptor potential vanilloid 1 (TRPV1) ion channel: an overview of the current mutational data. Mol. Pain 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Szallasi A., Cortright D. N., Blum C. A., and Eid S. R. (2007) The vanilloid receptor TRPV1: 10 years from channel cloning to antagonist proof-of-concept. Nat. Rev. Drug Discov. 6, 357–372 [DOI] [PubMed] [Google Scholar]
- 21. Vriens J., Appendino G., and Nilius B. (2009) Pharmacology of vanilloid transient receptor potential cation channels. Mol. Pharmacol. 75, 1262–1279 [DOI] [PubMed] [Google Scholar]
- 22. Bevan S., Quallo T., and Andersson D. A. (2014) TRPV1. Handb. Exp. Pharmacol. 222, 207–245 [DOI] [PubMed] [Google Scholar]
- 23. Cao E., Liao M., Cheng Y., and Julius D. (2013) TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanson S. M., Newstead S., Swartz K. J., and Sansom M. S. (2015) Capsaicin interaction with TRPV1 channels in a lipid bilayer: molecular dynamics simulation. Biophys. J. 108, 1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hazan A., Kumar R., Matzner H., and Priel A. (2015) The pain receptor TRPV1 displays agonist-dependent activation stoichiometry. Sci. Rep. 5, 12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang F., Xiao X., Cheng W., Yang W., Yu P., Song Z., Yarov-Yarovoy V., and Zheng J. (2015) Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol. 11, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Darré L., and Domene C. (2015) Binding of capsaicin to the TRPV1 ion channel. Mol. Pharm. 12, 4454–4465 [DOI] [PubMed] [Google Scholar]
- 28. Elokely K., Velisetty P., Delemotte L., Palovcak E., Klein M. L., Rohacs T., and Carnevale V. (2016) Understanding TRPV1 activation by ligands: Insights from the binding modes of capsaicin and resiniferatoxin. Proc. Natl. Acad. Sci. U.S.A. 113, E137–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liao M., Cao E., Julius D., and Cheng Y. (2013) Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Touska F., Marsakova L., Teisinger J., and Vlachova V. (2011) A “cute” desensitization of TRPV1. Curr. Pharm. Biotechnol. 12, 122–129 [DOI] [PubMed] [Google Scholar]
- 31. Priel A., and Silberberg S. D. (2004) Mechanism of ivermectin facilitation of human P2X4 receptor channels. J. Gen. Physiol. 123, 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Priel A., Kolleker A., Ayalon G., Gillor M., Osten P., and Stern-Bach Y. (2005) Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J. Neurosci. 25, 2682–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Studer M., and McNaughton P. A. (2010) Modulation of single-channel properties of TRPV1 by phosphorylation. J. Physiol. 588, 3743–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colton C. K., and Zhu M. X. (2007) 2-Aminoethoxydiphenyl borate as a common activator of TRPV1, TRPV2, and TRPV3 channels. Handb. Exp. Pharmacol. 179, 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jordt S. E., Tominaga M., and Julius D. (2000) Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc. Natl. Acad. Sci. U.S.A. 97, 8134–8139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ryu S., Liu B., Yao J., Fu Q., and Qin F. (2007) Uncoupling proton activation of vanilloid receptor TRPV1. J. Neurosci. 27, 12797–12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trapani J. G., and Korn S. J. (2003) Control of ion channel expression for patch clamp recordings using an inducible expression system in mammalian cell lines. BMC Neurosci. 4, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Premkumar L. S., Qi Z.-H., Van Buren J., and Raisinghani M. (2004) Enhancement of potency and efficacy of NADA by PKC-mediated phosphorylation of vanilloid receptor. J. Neurophysiol. 91, 1442–1449 [DOI] [PubMed] [Google Scholar]
- 39. Del Castillo J., and Katz B. (1957) Interaction at end-plate receptors between different choline derivatives. Proc. R. Soc. Lond. B Biol. Sci. 146, 369–381 [DOI] [PubMed] [Google Scholar]
- 40. Zagotta W. N. (2015) Ligand-dependent gating mechanism. in Handbook of Ion Channels (Zheng J., and Trudeau M. C., eds) pp. 41–52, CRC Press, Boca Raton, FL [Google Scholar]
- 41. Galzi J. L., Edelstein S. J., and Changeux J. (1996) The multiple phenotypes of allosteric receptor mutants. Proc. Natl. Acad. Sci. U.S.A. 93, 1853–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meyer E. A., Castellano R. K., and Diederich F. (2003) Interaction with aromatic rings in chemical and biological recognition. Angew. Chem. Int. Ed. Engl. 42, 1210–1250 [DOI] [PubMed] [Google Scholar]