Abstract
Background: Diet interventions have shown effectiveness in improving diabetes risk factors; however, little is known about whether the effects of diet intervention are different according to genetic susceptibility.
Objective: We examined interactions between weight-loss diets and the genetic risk score (GRS) for diabetes on 2-y changes in markers of insulin resistance and β cell function in a randomized controlled trial.
Design: Data from the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial were analyzed. A GRS was calculated on the basis of 31 diabetes-associated variants in 744 overweight or obese nondiabetic adults (80% white Americans). We assessed the changes in insulin resistance and β cell function over the 2-y intervention.
Results: Dietary protein significantly interacted with the diabetes GRS on fasting insulin, glycated hemoglobin (HbA1c), the homeostasis model assessment of β cell function (HOMA-B), and the homeostasis model assessment of insulin resistance (HOMA-IR) at 2 y in white Americans (P-interaction = 0.02, 0.04, 0.01, and 0.05, respectively). The lower GRS was associated with a greater decrease in fasting insulin (P = 0.04), HbA1c (P = 0.0001), and HOMA-IR (P = 0.02), and a lesser increase in HOMA-B (P = 0.004) in participants consuming a low-protein diet. Participants with a higher GRS might have a greater reduction in fasting insulin when consuming a high-protein diet (P = 0.03).
Conclusions: Our data suggest that individuals with a lower genetic risk of diabetes may benefit more from consuming a low-protein weight-loss diet in improving insulin resistance and β cell function, whereas a high-protein diet may be more beneficial for white patients with a higher genetic risk. This trial was registered at clinicaltrials.gov as NCT00072995.
Keywords: genetic risk score, weight-loss diets, insulin resistance, β cell function, gene–diet interaction
INTRODUCTION
Type 2 diabetes has been increasing rapidly and is becoming a critical threat to public health in the United States and throughout the world (1). The epidemic of diabetes is attributable mainly to dramatic changes in dietary patterns and lifestyle (2). Diet and lifestyle modifications are considered to be the first option in preventing diabetes (2, 3). A large number of clinical trials have demonstrated that diet and lifestyle interventions prevent or delay the onset of diabetes (3, 4).
Considerable interindividual variation has been noted in response to diet and lifestyle interventions, and previous evidence suggests that such variation may be determined by interactions with genetic factors. For example, genetic variants in the GIPR9 (5), PCSK7 (6), and IRS1 genes (7) associated with diabetes have been found to modify the effect of diet and lifestyle on insulin resistance. However, recent observational studies have found that overall genetic susceptibility to diabetes might interact with diet and lifestyle on diabetes risk factors (8, 9). For example, the NHANES found that macronutrients modified genetic susceptibility to diabetes (8). In addition, the Atherosclerosis Risk in Communities study showed that the high genetic score attenuated the protective effect of physical activity on type 2 diabetes and insulin resistance (9). However, similar relations were not detected in the European Prospective Investigation into Cancer and Nutrition (EPIC) InterAct case-cohort study (http://www.inter-act.eu/) (10). Limited evidence currently is available from clinical trials.
In the present study, we aimed to quantify the combined effects of the genetic risk score (GRS), which was calculated on the basis of 31 variants associated with type 2 diabetes (11, 12), on markers of insulin resistance and β cell function in a 2-y randomized controlled trial. In particular, we tested whether weight-loss diets might modify genetic association with long-term improvement in insulin resistance and β cell function over the intervention.
METHODS
Study participants
The Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial (NCT00072995), conducted in Boston, Massachusetts, and Baton Rouge, Louisiana, in 2004–2007, was a 2-y randomized clinical trial that compared the effects of energy-reduced diets that had different compositions of fat, protein, and carbohydrate with weight change. People with type 2 diabetes controlled with diet, or with hypertension or hyperlipidemia treated with diet or drugs, were eligible to participate. Exclusions were diabetes treated with oral medications or insulin, serious gastrointestinal disease, alcohol or drug abuse, treatment for an eating disorder, unstable or recent onset of cardiovascular disease or other serious illness, the use of medications that affect body weight, and insufficient motivation. A total of 811 overweight and obese subjects were recruited for this trial. The trial was approved by the Human Subjects Committee at the Harvard School of Public Health and Brigham and Women’s Hospital in Boston, the Pennington Biomedical Research Center of the Louisiana State University System in Baton Rouge, Louisiana, and a data and safety monitoring board appointed by the National Heart, Lung, and Blood Institute. All participants gave written informed consent. Detailed information on the study design and methods has been described previously (13).
A total of 811 overweight or obese subjects [BMI (in kg/m2) ≥25 and ≤40] aged 30–70 y were randomly assigned to 1 of 4 diets with targeted percentages of energy derived from varying macronutrient composition: 1) 20% fat, 15% protein, and 65% carbohydrate; 2) 20% fat, 25% protein, and 55% carbohydrate; 3) 40% fat, 15% protein, and 45% carbohydrate; and 4) 40% fat, 25% protein, and 35% carbohydrate. Eighty percent of the participants (n = 645) completed the 2-y trial. Among the participants who had genotyping data (n = 744), 594 participants actually completed the 2-y trial. A total of 471 white Americans were included at 24 mo. To assess dietary adherence across the intervention, dietary intake was assessed in a random sample of 50% of the participants by a review of the 5-d diet record at baseline and by 24-h recall during a telephone interview on 3 nonconsecutive days at 6 mo and at 2 y. Total energy, fat, protein, and carbohydrate intake and changes in urinary nitrogen and respiratory quotient (biomarkers of adherence) confirmed that differences in macronutrient intake between the groups were consistent with those recorded in the dietary reports (13).
Measurements
Height was measured at baseline. Body weight and waist circumference were measured, and fasting blood samples, 24-h urine samples, and measurements of resting metabolic rate were obtained in the fasting state at baseline, 6 mo, and 2 y. BMI was calculated as weight in kilograms divided by height in meters squared. Serum glucose, insulin, and urinary nitrogen were measured at the clinical laboratory at the Pennington Biomedical Research Center. Glucose and insulin were measured with the use of an immunoassay with chemiluminescent detection on an Immulite analyzer (Diagnostic Products Corporation). β cell function was estimated by homeostasis model assessment of β cell function (HOMA-B) with the use of the following equation: [20 × fasting insulin (μU/mL)]/{[fasting glucose (mg/dL)/18.01]−3.5}. Insulin resistance was estimated by HOMA-IR as follows: [fasting insulin (μU/mL)] × [fasting glucose (mg/dL)/18.01]/22.5 (14). A 24-h urine sample was collected at baseline, 6 mo, and 2 y for creatinine and urea (as a biomarker of protein intake), and was measured at the Core Laboratory at Pennington.
Genotyping and GRS calculation
DNA was extracted from the buffy coat fraction of blood that was centrifuged at 14,000 × 3 g for 10 min at 5°C with the use of the QIAmp Blood Kit (Qiagen). We selected 31 single-nucleotide polymorphisms (SNPs) previously associated with type 2 diabetes at a genome-wide significance level in white patients (11, 12, 15). We assumed that each SNP in the panel would act independently in an additive manner, and the genetic score was calculated by using a weighted method. Each SNP was weighted by its relative effect size (β coefficient) obtained from the reported genome-wide association study (GWAS) data (11, 12). By using the same method for the previously reported waist-to-hip ratio genetic score (16), we first created a weighted score with the use of the following equation: weighted score = β1 × SNP1 + β2 × SNP2 + … + βn × SNPn, where β is the β coefficient for each individual SNP, and n is number of SNPs. To reflect the number of diabetes risk alleles, we rescaled the weighted score with the use of the following equation: weighted GRS = weighted score × (total number of SNPs/sum of the β coefficients). Most of the SNPs included in the GRS were genotyped (Supplemental Table 1). The SNPs were genotyped successfully in 744 of 811 total participants with the use of the OpenArray SNP Genotyping System (BioTrove). The genotype success rate was 99% in available DNA samples. Replicated quality control samples (10%) were included in every genotyping plate with >99% concordance (17).
Statistical analysis
In the present analysis, we included only those participants who had genotype data (n = 744). The primary endpoints for this study were changes in fasting insulin, insulin resistance, and β cell function over the intervention. General linear models (PROC GLM) for continuous variables and a chi-square test (PROC FREQ) for categorical variables were applied for the comparison according to genotype groups at baseline. We compared the changes in the primary endpoints, biomarkers of adherence and nutrient intake, across genotype groups at 6 mo and 2 y with the use of generalized linear models. To test for interactions, we examined genotype and genotype–diet interactions as independent predictors of changes in diabetes traits, adjusted for age, sex, ethnicity (whole population only), baseline weight, weight change, and the baseline value for the respective outcome trait in the generalized linear models. We assigned the median value of GRS to each tertile of GRS when testing the P-trend in subgroups. Linear mixed models (PROC MIXED) with the use of variance components structure were used to test the genotype effect on the trajectory of changes in diabetes traits in the participants who provide measurements at baseline, 6 mo, and 2 y in each of diet groups over the 2-y intervention by including genotype × time interaction terms. All reported P values were nominal. P < 0.05 was considered to be statistically significant. We used Quanto 1.2.4 (University of Southern California, Los Angeles, California; http://hydra.usc.edu.gxe) to estimate the detectable effect sizes of genotype–diet interactions. In this post hoc analysis, the study had 80% power to detect gene–diet interaction effect sizes of 0.16 units for changes in fasting insulin, and 0.18 units for changes in HOMA-IR at 2 y at a significance level of 0.05. Statistical analyses were performed with SAS version 9.1.
RESULTS
Characteristics of the study population according to GRS
Baseline characteristics of participants according to tertiles (low, median, high) of the GRS are presented in Table 1 and Supplemental Table 1. The GRS was calculated on the basis of 31 genetic variants associated with diabetes (Supplemental Table 2). The distribution of the GRS was significantly different by ethnicity and sex. Body weight, BMI, waist circumference, fasting insulin, and HOMA-IR were not related to the GRS at baseline. No associations of GRS with weight loss and change in waist circumference at 6 mo and 2 y were observed, except for a 6-mo change in waist circumference (P = 0.02). Supplemental Figure 1 presents the distribution of GRS and its association with 24-mo changes in fasting insulin, insulin resistance measured by HOMA-IR, and β cell function estimated by HOMA-B. We did not observe significant genetic associations with diabetes traits.
TABLE 1.
Baseline characteristics of white participants in the POUNDS LOST trial1
| Tertile of the genetic risk score |
||||
| T1 (low) (n = 199) | T2 (median) (n = 199) | T3 (high) (n = 198) | P2 | |
| Age, y | 52.1 ± 8.9 | 51.6 ± 9.2 | 52.3 ± 8.9 | 0.70 |
| Female | 120 (60.6) | 106 (73.3) | 112 (56.3) | 0.33 |
| Low-protein diets | 105 (53.0) | 91 (45.7) | 106 (53.3) | 0.23 |
| Height, cm | 168.9 ± 9.0 | 169.6 ± 8.7 | 169.1 ± 8.9 | 0.85 |
| Weight, kg | 92.7 ± 15.8 | 94.4 ± 15.6 | 93.6 ± 16.4 | 0.76 |
| Waist, cm | 103.7 ± 13.3 | 104.9 ± 13.3 | 103.9 ± 13.6 | 0.72 |
| Total energy, kcal/d | 1933 ± 510 | 2066 ± 559 | 1955 ± 558 | 0.87 |
| Carbohydrate, % of energy | 44.8 ± 7.2 | 43.6 ± 8.1 | 44.0 ± 7.7 | 0.06 |
| Fat, % of energy | 36.8 ± 5.5 | 37.2 ± 5.9 | 37.5 ± 6.7 | 0.41 |
| Protein, % of energy | 18.3 ± 3.1 | 18.3 ± 3.6 | 18.1 ± 3.5 | 0.06 |
| Baseline mean respiratory quotient | 0.84 ± 0.04 | 0.84 ± 0.05 | 0.84 ± 0.04 | 0.99 |
| Baseline urinary nitrogen, mg/d | 11.9 ± 4.1 | 12.8 ± 4.4 | 12.7 ± 4.9 | 0.23 |
| Baseline fasting insulin, mIU/L | 11.4 ± 6.9 | 12.9 ± 8.6 | 11.8 ± 7.1 | 0.42 |
| Baseline HOMA-IR | 2.6 ± 1.7 | 3.1 ± 2.3 | 2.8 ± 1.8 | 0.16 |
| Baseline HOMA-B | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.41 |
| Baseline HbA1c, % | 5.3 ± 0.3 | 5.4 ± 0.4 | 5.3 ± 0.4 | 0.44 |
| 6-mo weight change, kg | −7.1 ± 6.2 | −7.6 ± 5.7 | −7.0 ± 5.5 | 0.22 |
| 6-mo waist change, cm | −7.8 ± 6.6 | −7.8 ± 6.1 | −6.9 ± 6.0 | 0.04 |
| 24-mo weight change, kg | −4.4 ± 7.8 | −5.0 ± 7.3 | −4.3 ± 8.2 | 0.31 |
| 24-mo waist change, cm | −6.1 ± 8.0 | −6.7 ± 7.4 | −5.9 ± 8.5 | 0.06 |
Values are means ± SDs or n (%). HbA1c, glycated hemoglobin; HOMA-B, homeostasis model assessment of β cell function; POUNDS LOST, Preventing Overweight Using Novel Dietary Strategies; T, tertile.
P values were calculated by using the chi-square test for categorical variables and F tests after adjusting for sex (except for sex) and age (except for age).
The dietary intake and adherence markers of the white participants are shown in Table 2. We assessed biomarkers to confirm dietary adherence. There were no significant differences in mean values of nutrient intake and biomarkers of adherence at 6 mo and 2 y across the tertiles of GRS (P > 0.05). Total energy, fat, protein, and carbohydrate and changes in urinary nitrogen and respiratory quotient (biomarkers of adherence) confirmed that participants modified their intake of macronutrients in the direction of the goals, although the targets were not fully achieved.
TABLE 2.
Nutrient intake and biomarkers of adherence according to tertiles of genetic risk score at 6 mo and 24 mo in white Americans in the POUNDS LOST trial1
| At 6 mo |
At 24 mo |
|||||
| T1 (low) | T2 (median) | T3 (high) | T1 (low) | T2 (median) | T3 (high) | |
| Genetic risk score | 35.69 ± 2.70 | 40.07 ± 1.20 | 45.49 ± 4.53 | 35.47 ± 2.67 | 40.12 ± 1.27 | 46.12 ± 4.76 |
| Dietary intake per day2 | ||||||
| Energy, kcal | 1679 ± 471 | 1651 ± 591 | 1513 ± 457 | 1602 ± 536 | 1516 ± 423 | 1441 ± 486 |
| Carbohydrate, % | 49.8 ± 10.2 | 50.4 ± 10.2 | 52.2 ± 10.7 | 48.6 ± 10.6 | 47.0 ± 8.7 | 52.3 ± 11.3 |
| Fat, % | 30.7 ± 8.5 | 30.8 ± 8.1 | 28.4 ± 8.0 | 29.9 ± 8.5 | 33.2 ± 7.6 | 28.6 ± 8.9 |
| Protein, % | 19.8 ± 4.8 | 20.3 ± 4.2 | 20.0 ± 4.6 | 21.1 ± 5.2 | 20.3 ± 4.3 | 19.3 ± 4.2 |
| Biomarkers of adherence3 | ||||||
| Urinary nitrogen, g | 11.1 ± 3.9 | 12.2 ± 4.9 | 11.3 ± 4.6 | 12.1 ± 3.9 | 11.8 ± 4.8 | 11.8 ± 4.8 |
| Respiratory quotient | 0.80 ± 0.01 | 0.81 ± 0.01 | 0.81 ± 0.01 | 0.81 ± 0.01 | 0.80 ± 0.01 | 0.81 ± 0.01 |
Values are means ± SDs. All P-trends across tertiles were >0.5. The tertiles for the genetic risk score were defined in white Americans. General linear models (PROC GLM) were applied for the comparison according to groups. POUNDS LOST, Preventing Overweight Using Novel Dietary Strategies; T, tertile.
Sample sizes at 6 mo: T1, n = 91; T2, n = 99; and T3, n = 96. Sample sizes at 24 mo: T1, n = 49; T2, n = 46; and T3, n = 51.
Sample sizes at 6 mo: T1, n = 199; T2, n = 199; and T3, n = 198. Sample sizes at 24 mo: T1, n = 175; T2, n = 166; and T3, n = 130.
Gene–diet interactions on improvement in insulin resistance and β cell function
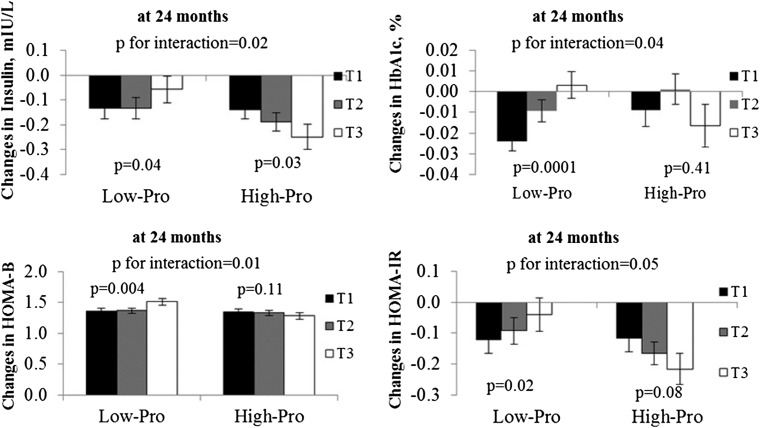
In the POUNDS LOST trial, the high-carbohydrate diets are the same as the low-fat diets and vice versa. In the present analysis, participants from the diet groups were combined in order to compare low-carbohydrate diets (35% and 45% carbohydrate diet groups combined) and high-carbohydrate diets (55% and 65% carbohydrate diet groups combined), and to compare low-protein diet (15% protein) and high-protein diets (25% protein). We tested the interactions between the GRS and the intervention diets (2-factor comparisons: low-protein compared with high-protein, and high-fat compared with low-fat) on changes in markers of insulin resistance and β cell function over 24 mo. Dietary protein significantly modified the genetic effects of diabetes on fasting insulin, glycated hemoglobin (HbA1c), HOMA-B, and HOMA-IR (P-interaction = 0.02, 0.04, 0.01, and 0.05, respectively) at 2 y in white Americans, after adjustment for age, sex, weight change, and baseline perspective phenotype (Figure 1). Further adjustment for baseline BMI and dietary fat in the model yielded similar results. In the adjusted model, the lower GRS was associated with a greater decrease in fasting insulin (P-trend = 0.04), HbA1c (P-trend = 0.0001), and HOMA-IR (P-trend = 0.02), and a lesser increase in HOMA-B (P-trend = 0.004) between participants consuming the low-protein diet. We observed the opposite genetic effects on changes in fasting insulin (P-trend =0.03) in the high-protein diet group. Participants with a higher GRS might have a greater reduction in fasting insulin when choosing a high-protein diet. A similar interaction pattern was found for HbA1c, but not for other measures, in all participants (Supplemental Table 3). We did not find a significant interaction for other weight-loss diets in either white Americans alone or the study population as a whole.
FIGURE 1.
Effects of the genetic risk score for diabetes and weight-loss diets on changes in fasting insulin and HOMA-IR during a 2-y intervention in white Americans. Values are means ± SEs. High-Pro group sample sizes: T1, n = 87; T2, n = 84; and T3, n = 63. Low-Pro group sample sizes: T1, n = 88; T2, n = 82; and T3, n = 67. The tertiles for the genetic risk score were defined in white Americans. To test for interactions, we examined genotype and genotype–diet interactions as independent predictors of changes in diabetes traits, adjusted for age, sex, ethnicity (in the whole population analysis), baseline weight, weight change, and the baseline value for the respective outcome trait in the generalized linear models. P-interaction refers to the interaction between protein diets and genetic risk score. P-trends across the tertile groups were tested in Low-Pro and High-Pro groups after adjustment for age, sex, weight change, and baseline values for respective phenotypes. Fasting insulin, HOMA-IR, HOMA-B, and HbA1c were log-transformed before analysis. HbA1c, glycated hemoglobin; High-Pro, high protein; HOMA-B, homeostasis model assessment of β cell function; Low-Pro, low protein; T, tertile.
Trajectory of changes in fasting insulin and insulin resistance and β cell function
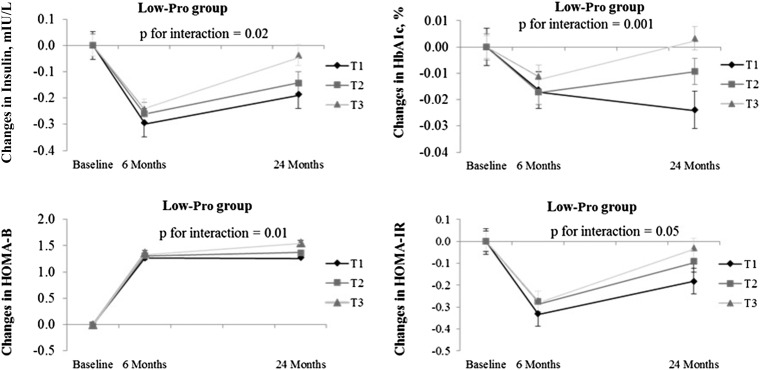
We used linear mixed models to assess the genetic effect on insulin resistance and β cell function by time effect over the 2-y intervention in low- and high-protein diet groups. Participants with a lower genetic risk of diabetes had a greater improvement in insulin resistance than did those with a higher genetic risk across the 2-y intervention in the low-protein diet group. Among white participants, those with a higher genetic risk may benefit more in improving their β cell function than may those with a lower genetic risk when choosing a low-protein diet. However, we did not find any significant differences in insulin resistance and β cell function across tertiles of GRS in the high-protein diet group (Figure 2).
FIGURE 2.
Effects of the genetic risk score for diabetes on the trajectory of changes in fasting insulin and HOMA-IR in white Americans over 2 y in the Low-Pro group. Values are means ± SEs after adjustment for age, sex, weight change, and baseline values for respective phenotypes, n = 471 white Americans. Low-Pro group sample sizes—6 mo: T1, n = 100; T2, n = 99; and T3, n = 98; 24 mo: T1, n = 88; T2, n = 82; and T3, n = 67. P values were tested for the interaction between genotype and intervention time. Linear mixed models (PROC MIXED) were used to test the genotype effect on the trajectory of changes in diabetes traits. HbA1c, glycated hemoglobin; HOMA-B, homeostasis model assessment of β cell function; Low-Pro, low protein; T, tertile.
DISCUSSION
In a 2-y randomized weight loss intervention trial, dietary protein intake significantly modified genetic association with improvement in insulin resistance and β cell function in white patients. Our findings showed that individuals with a low genetic risk of diabetes might benefit more from consuming a low-protein weight-loss diet through improved insulin resistance, although with less improvement in β cell function.
To our knowledge, most previous studies investigating gene–environment interactions in relation to diabetes have focused extensively on a single locus (5–7, 18, 19). However, findings from the GWAS showed that single variants only had modest effects on diabetes (12). Recent observational studies calculated the genetic score based on diabetes loci and their interaction with diet and lifestyle in relation to risk of diabetes (8–10). In the present analysis, we took advantage of a 2-y clinical trial to investigate genetic susceptibility to insulin resistance. We found that dietary protein intake significantly modified genetic association with insulin resistance, suggesting that individuals with a low genetic risk of diabetes might improve insulin resistance more by consuming a low-protein weight-loss diet. Our results are supported by studies that demonstrated that weight-loss diets modulate the genetic effects of gastric inhibitory polypeptide receptor (GIPR) (5) and proprotein convertase subtilisin/kexin type 7 (PCSK7) (6), with improved insulin resistance. In contrast to insulin resistance, we found that participants with a low genetic risk of diabetes experienced less improvement in β cell function when eating a lower protein (15%) diet. The different modification by weight-loss diets on genetic associations may reflect the distinct biological functions of impaired β cell function and insulin sensitivity in the pathogenesis of type 2 diabetes. Our results can be explained by previous evidence showing that most GWAS-identified loci (20) exert their effect on the risk of type 2 diabetes through impaired β cell function, although a few may be involved in a reduction in insulin sensitivity (20, 21).
Even though the biological basis underlying the observed interaction between the diabetes-related genotypes and protein intake remains unclear at this point, several lines of evidence suggest that such interactions are plausible. For example, the Diet, Obesity, and Genes trial reported that transcription factor AP-2 β (TFAP2B) rs987237, which is associated with obesity, interacted with dietary protein and carbohydrate to modify weight maintenance after weight loss in individuals with obesity (22). Our previous results showed that insulin resistance was modulated by the genotype–macronutrient interaction during the weight regain phase (6). Animal studies have demonstrated that protein intake might regulate the expression of several diabetes-associated genes. A study in rats suggested that maternal protein restriction during pregnancy affected cyclin-dependent kinase inhibitor 2A (CDKN2A) gene expression (23). A group of these loci is associated with impaired B cell function [Wolfram syndrome 1 (WFS1), ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 (CENTD2), solute carrier family 30, member 8 (SLC30A8), cyclin-dependent kinase 5 regulatory subunit-associated protein 1-like (CDKAL1), insulin-like growth factor 2 MRNA binding protein 2 (IGF2BP2), CDKN2A, cyclin-dependent kinase inhibitor 2B (CDKN2B), Notch homolog 2 (NOTCH2), thyroid adenoma associated (THADA), potassium channel, voltage gated KQT-like subfamily Q, member 1 (KCNQ1), melatonin receptor 1B (MTNR1B), glucokinase regulator (GCKR), glucokinase (GCK), and Prospero homeobox protein 1 (PROX1)], whereas several other loci are related to impaired insulin sensitivity [peroxisome proliferator–activated receptor γ (PPARG), insulin receptor substrate 1 (IRS1), insulin-like growth factor 1 (IGF1), ADAM metallopeptidase with thrombospondin type 1 motif, 9 (ADAMTS9), and Krüppel-like factor 14 (KLF14)]. In addition, compelling evidence from observational studies and clinical trials showed that protein intake regulated insulin secretion and glucose metabolism (24–27). Therefore, the biological pathways linking protein intake and genotype to diabetes largely are overlapped, and interactions between them might occur on these pathways. The observed gene–diet interaction on the improvement of insulin resistance and β cell function might reflect the cumulative effects of multiple genetic variants, rather than a single variant. In addition, participants had difficulty achieving the goals of their assigned group for macronutrient intake, and the differences in protein intake and in urinary nitrogen between the low- and high-protein groups at 24 mo were marginally significant. However, the difference in protein intake was significant during the majority of the intervention course, which is the driving force of the observed interaction. If such a difference remained significant at 2 y, the interaction might be stronger. The precise underlying mechanisms explaining the interaction between the weight-loss diets and a GRS in relation to the improvement in insulin resistance and β cell function need to be clarified further in future studies, especially through functional experiments.
To the best of our knowledge, this is the first study to investigate the interaction between overall genetic susceptibility to diabetes and weight-loss diets on long-term changes in markers of insulin resistance and β cell function in a randomized clinical trial. Our findings provide new insights into the role of genetic susceptibility in determining insulin resistance and β cell function. However, several limitations need to be considered. Despite the intensive behavioral counseling in this trial, participants experienced difficulty achieving the goals of their assigned group for macronutrient intake, which might have introduced misclassification, as previously discussed (13). Furthermore, a large number of loci for diabetes (28) and related biomarkers (29) were identified through the GWAS over the past several years; however, the weighted GRS calculated in the present study did not include all of these diabetes loci. This is a limitation of the current study. In addition, although we conducted analysis in both white and whole-population groups, we acknowledge that replication in diverse populations is required to verify our findings. Finally, we did not apply stringent control for the limited number of multiple testings. Therefore, this may lead to potential overstatement of our findings.
In conclusion, our data suggest that individuals with a lower GRS for diabetes may benefit from consuming a low-protein weight-loss diet through an improvement in insulin resistance, although with less improvement in β cell function. In contrast, a high-protein diet may be more beneficial for improving insulin resistance in white patients with a higher GRS. These findings, which, to our knowledge, are novel, provide supportive evidence for the notion of personalized nutrition in preventing diabetes.
Acknowledgments
The authors’ responsibilities were as follows—TH, YZ, TW, and LQ: contributed to the study concept and design, acquisition of data, analysis and interpretation of the data, drafting of the manuscript, and critical revision of the manuscript; SHL, GAB, and FMS: contributed to the study concept and design and critical revision of the manuscript; TH: contributed to the statistical analysis; GAB and FMS: involved in the collection and analysis of data and funding of the initial project; FMS and LQ: contributed to administration, material support, and study supervision; LQ: had full access to all the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis; and all authors: read and approved the final manuscript. None of the authors reported a conflict of interest related to the study.
Footnotes
Abbreviations used: ADAMTS9, ADAM metallopeptidase with thrombospondin type 1 motif, 9; CDKAL1, cyclin-dependent kinase 5 regulatory subunit-associated protein 1-like; CDKN2A, cyclin-dependent kinase inhibitor 2A; CDKN2B, cyclin-dependent kinase inhibitor 2B; CENTD2, ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1; GCK, glucokinase; GCKR, glucokinase regulator; GIPR, gastric inhibitory polypeptide receptor; GRS, genetic risk score; GWAS, genome-wide association study; HbA1c, glycated hemoglobin; HOMA-B, homeostasis model assessment of β cell function; IGF1, insulin-like growth factor 1; IGF2BP2, insulin-like growth factor 2 MRNA binding protein 2; IRS1, insulin receptor substrate 1; KCNQ1, potassium channel, voltage gated KQT-like subfamily Q, member 1; KLF14, Krüppel-like factor 14; MTNR1B, melatonin receptor 1B; NOTCH2, Notch homolog 2; PCSK7, proprotein convertase subtilisin/kexin type 7; POUNDS LOST, Preventing Overweight Using Novel Dietary Strategies; PPARG, peroxisome proliferator–activated receptor γ PROX1, Prospero homeobox protein 1; SLC30A8, solute carrier family 30, member 8; SNP, single-nucleotide polymorphism; TFAP2B, transcription factor AP-2 β THADA, thyroid adenoma associated; WFS1, Wolfram syndrome 1.
REFERENCES
- 1.International Diabetes Federation IDF diabetes atlas (6th edn.) [Internet]. 2013 [cited 2014 Jan 30]. Available from: http//www.idf.org/diabetesatlas.
- 2.Ley SH, Hamdy O, Mohan V, Hu FB. Prevention and management of type 2 diabetes: dietary components and nutritional strategies. Lancet 2014;383:1999–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramachandran A, Snehalatha C, Mary S, Mukesh B, Bhaskar AD, Vijay V. The Indian Diabetes Prevention Programme shows that lifestyle modification and metformin prevent type 2 diabetes in Asian Indian subjects with impaired glucose tolerance (IDPP-1). Diabetologia 2006;49:289–97. [DOI] [PubMed] [Google Scholar]
- 5.Qi Q, Bray GA, Hu FB, Sacks FM, Qi L. Weight-loss diets modify glucose-dependent insulinotropic polypeptide receptor rs2287019 genotype effects on changes in body weight, fasting glucose, and insulin resistance: the Preventing Overweight Using Novel Dietary Strategies trial. Am J Clin Nutr 2012;95:506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang T, Huang J, Qi Q, Li Y, Bray GA, Rood J, Sacks FM, Qi L. PCSK 7 Genotype Modifies Effect of a Weight-Loss Diet on 2-Year Changes of Insulin Resistance: The POUNDS LOST Trial. Diabetes Care 2015;38:439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng JS, Arnett DK, Parnell LD, Smith CE, Li D, Borecki IB, Tucker KL, Ordovas JM, Lai CQ. Modulation by dietary fat and carbohydrate of IRS1 association with type 2 diabetes traits in two populations of different ancestries. Diabetes Care 2013;36:2621–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villegas R, Goodloe RJ, McClellan BE Jr, Boston J, Crawford DC. Gene-carbohydrate and gene-fiber interactions and type 2 diabetes in diverse populations from the National Health and Nutrition Examination Surveys (NHANES) as part of the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) study. BMC Genet 2014;15:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klimentidis YC, Chen Z, Arora A, Hsu CH. Association of physical activity with lower type 2 diabetes incidence is weaker among individuals at high genetic risk. Diabetologia 2014;57:2530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langenberg C, Sharp SJ, Franks PW, Scott RA, Deloukas P, Forouhi NG, Froguel P, Groop LC, Hansen T, Palla L, et al. Gene-lifestyle interaction and type 2 diabetes: the EPIC interact case-cohort study. PLoS Med 2014;11:e1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med 2010;363:2339–50. [DOI] [PubMed] [Google Scholar]
- 12.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sacks FM, Bray GA, Carey VJ, Smith SR, Ryan DH, Anton SD, McManus K, Champagne CM, Bishop LM, Laranjo N, et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 2009;360:859–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–9. [DOI] [PubMed] [Google Scholar]
- 15.Qi Q, Meigs JB, Rexrode KM, Hu FB, Qi L. Diabetes genetic predisposition score and cardiovascular complications among patients with type 2 diabetes. Diabetes Care 2013;36:737–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang T, Qi Q, Zheng Y, Ley SH, Manson JE, Hu FB, Qi L. Genetic predisposition to central obesity and risk of type 2 diabetes: two independent cohort studies. Diabetes Care 2015;38:1306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Q, Bray GA, Smith SR, Hu FB, Sacks FM, Qi L. Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial: the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial. Circulation 2011;124:563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dashti HS, Smith CE, Lee YC, Parnell LD, Lai CQ, Arnett DK, Ordovas JM, Garaulet M. CRY1 circadian gene variant interacts with carbohydrate intake for insulin resistance in two independent populations: Mediterranean and North American. Chronobiol Int 2014;31:660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sotos-Prieto M, Luben R, Khaw KT, Wareham NJ, Forouhi NG. The association between Mediterranean Diet Score and glucokinase regulatory protein gene variation on the markers of cardiometabolic risk: an analysis in the European Prospective Investigation into Cancer (EPIC)-Norfolk study. Br J Nutr 2014;112:122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat genet 2012;44:981–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Florez JC. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: where are the insulin resistance genes? Diabetologia 2008;51:1100–10. [DOI] [PubMed] [Google Scholar]
- 22.Stocks T, Angquist L, Hager J, Charon C, Holst C, Martinez JA, Saris WH, Astrup A, Sorensen TI, Larsen LH. TFAP2B -dietary protein and glycemic index interactions and weight maintenance after weight loss in the DiOGenes trial. Hum Hered 2013;75:213–9. [DOI] [PubMed] [Google Scholar]
- 23.Zheng S, Pan YX. Histone modifications, not DNA methylation, cause transcriptional repression of p16 (CDKN2A) in the mammary glands of offspring of protein-restricted rats. J Nutr Biochem. 2011;22:567–73. [DOI] [PubMed] [Google Scholar]
- 24.Linn T, Geyer R, Prassek S, Laube H. Effect of dietary protein intake on insulin secretion and glucose metabolism in insulin-dependent diabetes mellitus. J Clin Endocrinol Metab 1996;81:3938–43. [DOI] [PubMed] [Google Scholar]
- 25.Linn T, Santosa B, Gronemeyer D, Aygen S, Scholz N, Busch M, Bretzel RG. Effect of long-term dietary protein intake on glucose metabolism in humans. Diabetologia 2000;43:1257–65. [DOI] [PubMed] [Google Scholar]
- 26.Azadbakht L, Atabak S, Esmaillzadeh A. Soy protein intake, cardiorenal indices, and C-reactive protein in type 2 diabetes with nephropathy: a longitudinal randomized clinical trial. Diabetes Care 2008;31:648–54. [DOI] [PubMed] [Google Scholar]
- 27.Larsen RN, Mann NJ, Maclean E, Shaw JE. The effect of high-protein, low-carbohydrate diets in the treatment of type 2 diabetes: a 12 month randomised controlled trial. Diabetologia 2011;54:731–40. [DOI] [PubMed] [Google Scholar]
- 28.Mahajan A, Go MJ, Zhang WH, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat genet 2014;46:234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan JA, Magi R, Strawbridge RJ, Rehnberg E, Gustafsson S, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat genet 2012;44:991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]